Abstract
We describe a new Drosophila gene, mini spindles (msps) identified in a cytological screen for mitotic mutant. Mutation in msps disrupts the structural integrity of the mitotic spindle, resulting in the formation of one or more small additional spindles in diploid cells. Nucleation of microtubules from centrosomes, metaphase alignment of chromosomes, or the focusing of spindle poles appears much less affected. The msps gene encodes a 227-kD protein with high similarity to the vertebrate microtubule-associated proteins (MAPs), human TOGp and Xenopus XMAP215, and with limited similarity to the Dis1 and STU2 proteins from fission yeast and budding yeast. Consistent with their sequence similarity, Msps protein also associates with microtubules in vitro. In the embryonic division cycles, Msps protein localizes to centrosomal regions at all mitotic stages, and spreads over the spindles during metaphase and anaphase. The absence of centrosomal staining in interphase of the cellularized embryos suggests that the interactions between Msps protein and microtubules or centrosomes may be regulated during the cell cycle.
Keywords: spindle, microtubule, mitosis, cytoskeleton, Drosophila
The mitotic spindle is essential for the accurate segregation of chromosomes in eukaryotes. At the onset of mitosis cytoplasmic microtubules are disassembled and a bipolar spindle is assembled in their place. The bipolarity of the spindle ensures the equal separation of replicated DNAs into two daughter cells. The multiple microtubule motors associated with the spindle have been shown to be required for different aspects of assembly and function of the bipolar spindle (Walczak et al. 1998).
It is also recognized that spatial and temporal regulation of microtubule dynamics is essential for the structure and function of the spindle (Hyman and Karsenti 1996). Microtubule dynamics change dramatically upon entry into mitosis (Saxton et al. 1984), and continue to be modulated within the spindle during progression through mitosis (Zhai et al. 1995). Centrosomes and kinetochores act as microtubule organizing centres to capture microtubules and modify their dynamics (Mitchison and Kirschner 1984; Hyman and Mitchison 1990). In addition, there is evidence that motor proteins or a protein interacting with tubulin dimers contributes to the regulation of microtubule dynamics (Belmont et al., 1996; Walczak et al. 1996).
Microtubule-associated proteins (MAPs)1 have long been known to be potential regulators of microtubule dynamics but their involvement in the formation of the mitotic spindle is less appreciated. Classical MAPs, such as MAP1, MAP2, and tau, were identified as proteins copurifying with microtubules from mammalian brains and were shown to promote microtubule assembly in vitro (see Hirokawa 1994, and Mandelkow and Mandelkow 1995, for review). Although direct evidence is missing, it is thought that they are important for axon structure and function. Several MAPs have been purified from Xenopus egg extracts and shown to modulate microtubule dynamics in vitro (Gard and Kirschner 1987; Shiina et al. 1992; Andersen et al. 1994; Andersen and Karsenti, 1997).
In contrast to these in vitro studies, much less is known about the function of MAPs in vivo. Genetic approaches should complement these in vitro studies but have been limited mainly to yeasts. Such work has identified genes encoding MAPs, some of which are conserved in higher eukaryotes (Berlin et al. 1990; Beinhauer et al. 1997; Wang and Huffaker 1997). As the structures of the mitotic apparati of the yeasts differ from those of higher eukaryotes, it remains to be seen whether the conclusions reached from work with yeasts can be directly applied to higher eukaryotes.
Drosophila is an ideal organism to study the function and regulation of MAPs in vivo. It offers sophisticated classical and molecular genetics and its mitotic apparatus, centrosomes, kinetochores and spindle, are very similar to that of mammalian cells (e.g., Moritz et al. 1995). In addition, the function of MAPs can be studied in the context of development in a living organism. Kellogg et al. 1989 purified a number of MAPs from Drosophila embryos and determined their subcellular localizations. Some of them were cloned and shown to be associated with centrosomes (Whitfield et al. 1988; Kellogg and Alberts 1992), but their function in vivo remains unknown as no mutants have been identified. Few genetic studies on Drosophila MAPs have been reported. One analysis has indicated that a 205-kD MAP associated with the spindle and interphase microtubules is not essential for development (Pereira et al. 1992). Molecular cloning of the asp (abnormal spindle) gene, required for mitotic and meiotic function (Ripoll et al. 1985), enabled its gene product to be identified as a MAP that localizes to the polar regions of the spindle (Saunders et al. 1997). Subsequently, it was shown to associate with centrosomes and, together with the γ-tubulin ring complex, can restore microtubule nucleation activity to salt stripped centrosome preparations in vitro (Moritz et al. 1998; Avides et al., 1999).
Here we report a novel Drosophila mutation, mini spindles (msps), which disrupts the integrity of the mitotic spindle and show that it encodes a MAP that is conserved among eukaryotes. We show that Msps protein localizes on mitotic spindles and to centrosomal regions.
Materials and Methods
Fly Maintenance and P Element Remobilization
All stocks were grown at 258C in cornmeal media. Standard techniques of fly manipulation (Ashburner 1989) were used. w 1118 or Oregon R was used as wild-type. Details of balancer chromosomes and mutations are described in Lindsley and Zimm 1992. A reversion test of msps P was carried out as described in Ohkura et al. 1997. yw;msps P /TM6C females were crossed with l(3)/TM3, P(Δ2-3) males to obtain jumpstarter males, yw;msps P /TM3, Sb P(Δ2-3). Individual males were crossed back to yw;msps P /TM6C females. From 21 crosses, 18 gave viable revertants without balancer chromosomes marked with Sb. Out of 26 viable revertants examined from 17 independent crosses, 22 have lost the w + marker derived from the P-lacW (Bier et al. 1989) at 89B in msps P, indicating the P-lacW is responsible for the mutation. To isolate new alleles of the msps mutant, remobilization of the P-lacW was performed as above. Each independent w − revertant chromosome was tested over the msps P mutation.
Cytological Analysis
Squashed preparations of central nervous systems from third instar larvae were examined by orcein staining as previously described (Gonzalez and Glover 1993). Whole mount preparations of larval central nervous systems were prepared for immunostaining according to Gonzalez and Glover 1993 with the following modifications. Central nervous systems were dissected in 0.7% NaCl with 5 mM EGTA (pH 8) and fixed in 10% formaldehyde in 0.7% NaCl followed by two washes in 0.7% NaCl. This method gave an identical spindle structure in wild-type or the mspsP mutant to that obtained by other methods, such as by incubation in taxol followed by 3.7% formaldehyde fixation, or by dissection and fixing in 10% formaldehyde. Immunostaining of embryos was carried out using methanol or 37% formaldehyde as a fixative according to Gonzalez and Glover 1993 with omission of taxol. Although both fixation methods gave identical staining, methanol fixation consistently gave stronger and clearer Msps staining and was routinely used.
TAT1 (Woods et al. 1989; a kind gift from Professor K. Gull, Manchester, UK), YL1/2 or YOL1/34 (Kilmartin et al. 1982; SeraLab or Serotec) were used for α-tubulin staining. Rb188 (Whitfield et al. 1988) or Bx63 (Frasch et al. 1986) were used for CP190 staining. Monoclonal and polyclonal antibodies were used at 1/10–1/100 and 1/500–1/1,000, respectively. FITC-, Cy3-, Cy5- (Jackson Lab), or Alexa488-conjugated (Molecular Probes) secondary antibody was used at 1/500–1/2,000. Absence of cross-species reaction was confirmed by ourselves. DNA was counterstained with 0.2 μg/μl of DAPI or 1 μg/μl of propidium iodide with 100 μg/μl of RNase. Microtubules were artificially induced in embryos by incubation with PBS containing 10 μM of paclitaxel (ICN) and heptane for 5 min before fixation. To depolymerize microtubules, embryos were incubated with 30 μg/ml of colchicine (Sigma Chemical Co.) for 10 min after permeabilization by octane according to Gonzalez and Glover 1993. Weak tubulin staining remained around centrosomes even after longer incubation, treatment with nocodazole, 1 mM calcium, or cold temperature, alone or in combination.
Samples were examined and images were collected using an Optiphot (Nikon) microscope with a confocal scan head (MRC 1024; Bio-Rad Laboratories), or an Axioskop or Axioplan2 (Zeiss) attached with CCD camera (Princeton or Hamamatsu). Figures were prepared using Photoshop (Adobe).
Molecular Analysis of msps
Standard DNA manipulation techniques (Sambrook et al. 1989) were followed.
A genomic fragment flanking the P-lacW was isolated by plasmid rescue from msps P /TM6C adult flies after digestion of the genomic DNA with EcoRI. This was then used to isolate two overlapping cosmids (104B04, 129C05) from a wild-type library (Siden-Kiamos, 1990). In situ hybridization to wild-type polytene chromosomes was carried out according to Saunders et al. 1989 to confirm that the plasmid-rescued fragment and the cosmids were derived from chromosomal region 89B.
The 12-kb SpeI fragment and the 4-kb EcoRI fragment around the P-lacW insertion site was subcloned and sequenced. One of four transcription units in the region was shown to correspond to the msps gene through rescue of msps P mutation by the transgenic wild-type genomic fragment. The 5′ end of the transcript was determined using 5'/3'RACE kit (Boehringer). EST cDNA clones made by the Berkeley Drosophila Genome Project were obtained through Genome Systems or Research Genetics.
DNA sequences were determined using the ABI dye termination kit and automatic sequencer. Database searches were carried out using BLAST (Altschul et al. 1990) and prediction of coiled-coil structure was carried out using the method by Lupas et al. 1991.
P Element–mediated Transformation and Rescue of msps Mutant
The SpeI–NotI 12-kb fragment from cosmid 104B04 was subcloned into pW8 (Klemenz et al. 1987). The resulting plasmid (pHN267) was used for germline transformation of w 1118 flies by coinjection with Δ2-3 plasmid (pπ25.1) into embryos. Two independent insertions on the 2nd chromosome, were used to test for rescue of the msps P mutation. P[w +, HN267]/+; msps P/TM6B males (or msps P /TM6B sibling males as a control) were crossed with msps P /TM6C females. Both rescue mitotic defects in central nervous systems and fully restored growth of imaginal discs in homozygous third instar larvae of msps P, although one of them supported development only until the pharate stage, suggesting the insertion site may affect expression of the gene. A control plasmid (pHN276 containing a SpeI-EcoRI 3.3-kb fragment) that lacks most of the msps gene was tested for rescue of the msps P mutation by the same procedure. Two independent insertions tested failed to restore viability, disc growth or normal mitosis. Presence of the transgenic construct in each larvae examined for mitotic defects was positively identified by PCR.
Protein and Immunological Techniques
Standard protein and immunological techniques (Sambrook et al. 1989; Harlow and Lane 1988) were followed. For immunoblotting, peroxidase-conjugated secondary antibodies (Jackson Lab) were used and detected using the ECL kit (Amersham). Total protein samples from Drosophila tissues were prepared by homogenization in SDS sample buffer. Preheating the samples at 1008C helps to prevent protein degradation. Embryos were dechorionated by bleach before preparation of samples. pHN264 containing the BamHI-XhoI 1.5-kb fragment of cDNA in pET-23a (Pharmacia) was used to express amino acids 1,349–1,784 of Msps protein in E. coli. The polypeptide was initially purified in inclusion bodies then further purified by elution from an SDS gel. A rabbit was immunized with the antigen every 4 wk and antisera were taken a week after immunization by Scottish Antibody Production Unit. The antisera collected on 24.9.98 was used after affinity purification throughout the experiments described in this paper. The antibody was affinity-purified from antisera by incubation with the antigen on nitrocellulose strips followed by low pH elution according to Smith and Fisher 1984.
Crude antiserum and affinity-purified antibody both gave similar staining predominantly on mitotic spindles and centrosomal regions in embryos. Affinity purification of the antibodies using Msps protein resulted in reduced staining of cytoplasm without affecting the intensity or pattern of staining of the mitotic spindles or the centrosomal region. Preimmune serum taken from the same rabbit did not stain mitotic spindles or the centrosomal region even at a high concentration of the serum. Exclusion of the primary antibody against Msps eliminates the staining even when it was costained with other primary and secondary antibodies, indicating that the secondary antibody alone, cross reaction of secondary antibodies or leaking through between the channels is not responsible for the staining.
Microtubule Sedimentation Assay
Microtubule preparation was carried out according to Barton et al. 1995 and Saunders et al. 1997 with some modification. After dechorionation with bleach, 0–6-h-old embryos were homogenized in PEM buffer containing a cocktail of protease inhibitors and 1 mM dithiothreitol. The homogenate was incubated at 4°C for 30 min and spun at 140,000 g at 4°C for 30 min. A final concentration of 20 μM of taxol (paclitaxel; ICN) and 1 mM GTP was added before incubation at room temperature to polymerize microtubules. The microtubules and associated proteins were pelleted by spinning at 80,000 g for 30 min through a 30% sucrose cushion in the buffer. The supernatant and pellet fractions were analyzed by SDS–polyacrylamide gel electrophoresis.
Results
Isolation and Basic Characterization of the mini spindles (msps) Mutant
The mini spindles (msps) mutant was identified through a cytological screen for mutants with mitotic defects in the larval central nervous systems from ∼1,000 lethal or semi-lethal lines in a collection of third chromosome P-insertion mutants (Deák et al., 1997). As this mutant is induced by a P-lacW (Bier et al. 1989) insertion, we call this allele msps P. msps P homozygotes die around the larval/pupal transition. Mutant third instar larvae are superficially normal in size and behavior (they were capable of feeding and crawling) although they grow more slowly than wild-type. They die before or after pupation and no development beyond early pupal stage is observed. Dissection of late third instar larvae from msps P homozygotes revealed that imaginal discs are missing or very small and the size of the central nervous systems is also reduced. Polytenised tissues, such as the salivary glands and fat bodies, do not appear to be affected. These observations suggest that msps gene activity is essential for mitotic cycles but is required at significantly lower levels or not at all for endoreduplication cycles.
Chromosome Segregation Is Disrupted in Mitosis of msps Mutants
To examine mitosis in the msps P mutant, we dissected central nervous systems from late third instar larvae and stained chromosomes of squashed preparations with aceto-orcein. In most mitotic cells, the degree of chromosome condensation is considerably higher than that seen in wild-type (Fig. 1 b), and sister chromatids are attached together at heterochromatic regions. A high level of chromosome condensation is typical when the cells are blocked in mitosis as chromosome condensation continues during the arrest. Consistent with this, higher frequencies of mitotic cells are detected in central nervous systems from the msps P mutant (Fig. 1 e), where an average of the mitotic index is roughly twice that of wild-type. The most striking feature observed in the msps P mutant is the very low frequency of anaphases (Fig. 1 f). The frequency of anaphases among mitotic cells is only 3% in comparison to the 23.5% seen in wild-type. In addition, polyploid cells were found but the frequency is low (2%) in msps P mutants. The high degree of chromosome condensation, high mitotic index, and low frequencies of anaphases indicate that cells are blocked in mitosis before sister chromatid separation.
Figure 1.
Chromosome segregation is disrupted in the msps P mutant. Aceto-orcein was used to stain chromosomes in cells of squashed preparations of the central nervous system from late third instar larvae of wild-type (a and c) and msps P homozygotes (b and d). The msps P mutant shows overcondensation of mitotic chromosomes (b). A quarter of anaphases show V-shaped alignment of chromosomes (d). (e and f) Open bars and solid lines represent mean and standard deviation calculated from the quantitation of mitotic figures in five individuals of wild-type or the msps P mutant. This corresponds to roughly five to ten thousand cells from each individual. (e) Mitotic index is calculated as number of mitotic cells per microscope field using a 100× objective (a field area of 0.05 mm2 containing typically 200–400 cells). (f) % Anaphases is calculated as 100 × (number of anaphases)/(number of all mitotic cells). Bar, 10 μm.
Among the rare anaphases we observed V-shaped configurations in which chromosomes appeared to be distributed to three poles in a significant proportion (25%) of cells (Fig. 1 d). These anaphase cells appear to have a diploid complement of chromosomes that have undergone separation of sister chromatids. Although one set of sister chromatids moves to one pole, the other set of sister chromatids appear to be divided in their movement to two distinct poles. All chromatids appear to move synchronously and none of the chromosomes are left behind.
Integrity of the Mitotic Spindle Is Disrupted in msps Mutant
To examine mitotic spindle structure, we fixed whole central nervous systems dissected from late third instar larvae for immunostaining to visualize microtubules, chromosomes, and the centrosomal antigen, CP190 (Whitfield et al. 1988; Kellog and Alberts, 1992; Whitfield et al. 1995; Oegema et al. 1995).
We found that only 28% of mitotic cells formed an apparently normal bipolar spindle (Fig. 2 a) while the rest showed abnormal structures (Fig. 2 g). The most common abnormalities (36% of total mitotic cells) are cells containing more than one bipolar spindle forms (Fig. 2, b–d). As shown in Fig. 2 b, most chromosomes are aligned at the metaphase plate and associated with a bipolar spindle, but a few of them have become separated from other chromosomes and associate with an additional smaller bipolar spindle. The two bipolar spindles typically share one of the poles. The major bipolar spindle usually has CP190 staining at both poles, whereas the unshared pole of the other spindle typically has no CP190 staining. The poles of both bipolar spindles are focused. In some cases, chromosomes appeared still to be aligned on a common metaphase plate even though the spindle had bifurcated and the metaphase plate appears to be kinked (Fig. 2 c). In other cases, a large bipolar spindle is no longer observed, but instead there are multiple small bipolar spindles associated with individual chromosomes (Fig. 2 d).
Figure 2.

Structural integrity of mitotic spindle is disrupted in the msps P mutant. Whole mount preparations of larval central nervous system were prepared from msps P and stained to reveal tubulin (green), DNA (red), and a centrosomal antigen, CP190 (blue). The focal planes shown display the overall spindle structure and CP190 staining for each mitotic figure. (a) Category I. Apparently normal bipolar spindle with CP190 at both poles. (b) Category II. One bipolar spindle and one smaller bipolar spindle (arrowhead). CP190 is found at both poles of larger spindle and is missing from one pole of the smaller one. (c) Category II. Two bipolar spindles with one shared pole. CP190 is found at both poles of one spindle, while it is absent from the unshared pole of the other spindle (arrowhead). (d) Category II. At least three chromosomes are associated with each of small bipolar spindles. One spindle has CP190 at both poles, another has the protein at one pole, and the other shows no CP190 at the poles. (e) Category IV. One bipolar spindle and a monopolar spindle. The bipolar spindle has CP190 at both poles. (f) Category III. Short disorganized microtubule bundles associated with a chromosome mass. No discrete CP190 staining is seen. (g) Frequencies of each category of spindle structure observed. Bar, 5 μm.
Multipolar spindles are often observed in polyploid cells which can be caused by either failure of mitosis or cytokinesis. However, the multiple bipolar spindle phenotype seen in the msps P mutant is unlikely to be a consequence of the cell becoming polyploidy because orcein staining of squashed samples from the same strain indicates that polyploid cells are rare (2%).
12% of mitotic cells have one bipolar spindle and one monopolar spindle (Fig. 2 e), but a monopolar spindle is rare (3%). A significant proportion (16%) of mitotic cells have mitotic spindles that have disintegrated so that their exact structure cannot be determined. They contain many short but thick microtubule bundles associated with the chromosome mass (Fig. 2 f). Often no discrete CP190 staining was observed. These could represent different classes of structural abnormality, or alternatively such cells may belong to the class with small bipolar spindles associated with the mass of chromosomes.
Molecular Cloning of msps Gene
To gain insight of the molecular nature of the msps mutation, we intended to clone the wild-type msps gene. First we mapped the position of the P-lacW to 89B by in situ hybridization to polytene chromosomes. To confirm that the P-lacW insertion at 89B is responsible for the mutation we remobilized the P-lacW in msps P and obtained viable revertants at a high frequency (see Methods and Materials). Reversion of the lethality correlates with loss of the w + marker derived from the P-lacW insertion at 89B. The revertants have no mitotic defects or disc abnormalities. These results indicate that the P-lacW at 89B is responsible for the lethality and the mitotic defects seen in msps P mutants.
A fragment of genomic DNA flanking the P-lacW was isolated by plasmid rescue and used as a probe to isolate overlapping cosmid clones from a genomic library (Siden-Kiamos, 1990). Two cosmid clones covering over 30 kb around the P-lacW insertion were identified. We have sequenced a total of 14 kb around the P-lacW insertion site and identified four transcription units in the region (Fig. 3 a). To determine which transcription unit is responsible for the msps phenotype, we undertook rescue of the msps phenotype by wild-type genomic fragments. A 12-kb wild-type genomic fragment containing two of the transcription units was introduced into the germline by P element–mediated transformation. This construct HN267 was able to restore the viability and fertility of the msps P mutant. In contrast HN276, in which one of the transcription units is truncated, was not capable of rescuing the viability and fertility of the msps P mutant. We then examined whether HN267 can rescue the mitotic defects of msps P and found the frequency of anaphases to be restored from 4 to 24% comparable to wild-type levels (Fig. 3 b) and all other mitotic defects rescued. In contrast, the control, HN276, failed to rescue the mitotic defects of msps P.
Figure 3.
Molecular cloning and identification of the msps gene. (a) Genomic region at 89B around the P-lacW insertion site in the msps P mutant. The arrows represent four transcription units in this region. Introns are represented by kinked lines. A question mark on the far left indicates uncertainty of the location of the second exon of this transcript that lies beyond the sequenced region. The first exon of this particular transcript does not contain an open reading frame. The lower lines represent the genomic regions used for transgenic constructs (HN267, HN276). HN267 rescued both lethality and mitotic defects of msps P mutation, while HN276 rescued neither, indicating that the shaded transcript corresponds to msps gene. (b) Rescue of mitotic defects in msps P by transgenic constructs. Transgenes were tested for rescue of mitotic defects in msps P. Squashed preparation of central nervous systems was prepared for orcein staining from wild-type, msps P homozygotes, msps P homozygotes carrying the HN267 transgenic construct and msps P homozygotes carrying HN276. Means and standard deviations of the frequencies of anaphases are represented by open bars and solid lines, respectively.
These results indicate that one of the transcription units (shaded in Fig. 3 a) corresponds to the msps gene. RACE (rapid amplification of cDNA end) analysis indicates that msps gene has a 0.9-kb intron in the 5′ nontranslated region and that the P-lacW in msps P is inserted in the intron. It is likely that the P-lacW insertion interferes with the transcription or splicing in the msps P mutant, and therefore disrupts the production of msps protein (see below).
msps Encodes a Microtubule-associated Protein of the dis1-TOG family
The sequence of the msps gene predicts it to encode a protein of 2,050 amino acids, with an estimated molecular mass of 227 kD and an isoelectric point of 8.4. Database searches revealed that the entire region of the Msps protein has striking similarity to the human TOG protein (Charrasse et al. 1995, Charrasse et al. 1998; Fig. 4 a). Further analysis identified a family of proteins which share small regions of similarity. This family, which here we call the dis1-TOG family, consists of proteins encoded by Schizosaccharomyces pombe dis1 (Ohkura et al. 1988; Nabeshima et al. 1995), Saccharomyces cerevisiae STU2 (Wang and Huffaker, 1998), Caenorhabditis elegans zyg-9 (Wood et al. 1980; Mathews et al., 1998), Xenopus laevis XMAP215 (Gard and Kirschner 1987; Charrasse et al. 1998; the entire sequence not published), and human ch-TOG, all of which have been reported to have microtubule binding activity. The common signature among all members of the family is limited to four separate motifs within a repeated sequence unit (Fig. 4b and Fig. c). The repeats are each ∼200 amino acids long and tandemly arranged in the amino terminus with ∼100 residue spacers. Interestingly the two yeast proteins and the C. elegans ZYG-9 have only two repeats, while those from Drosophila and vertebrates have four.
Figure 4.

The Msps protein belongs to the dis1-TOG family. (a) Sequence comparison between Msps and human TOGp. Only identical residues are marked. Five putative cdc2 phosphorylation sites (S/TPXK/R) were identified in the COOH-terminal portion of the Msps protein (amino acids 1,564, 1,568, 1,803, 1,859, and 2,029). (b) Domain structure of members of the dis1-TOG family. From the top, human TOGp, D. melanogaster Msps, C. elegans ZYG-9, S. pombe Dis1, and S. cerevisiae STU2. Shaded boxes represent repeats (see c) common to all the dis1-TOG family. The COOH-terminal portions of the human and Drosophila proteins can be divided into two domains, one of which is also conserved in C. elegans protein, and the other found in Drosophila and vertebrates. (c) Amino acid sequences of the NH2-terminal repeats from the different proteins are aligned. Numbers in parentheses indicate the length of intervening amino acid sequences. The sequence of msps gene is available from GenBank/EMBL/DDBJ under accession number AJ249115.
The similarity between human TOGp and Drosophila Msps extends along nearly the entire COOH-terminal half, while there is no similarity beyond the repeats between the yeast proteins and the higher eukaryotic proteins (Fig. 4 b). The C. elegans ZYG-9 protein shares similarity with only half of the COOH-terminal region of the higher eukaryotic proteins. Although there are no sequence similarities in the COOH-terminal half at the primary sequence level between Dis1 and STU2, the entire region is predicted to have extensive coiled-coil structure. In contrast the Msps and TOG proteins do not have an extensive predicted coiled-coil structure.
These sequence characteristics suggest that Drosophila Msps and vertebrate proteins form a distinct higher eukaryotic subfamily of the dis1-TOG family.
Msps Protein during Drosophila Development
To characterize the msps gene product, we raised a polyclonal antibody against Msps protein (Materials and Methods). The affinity-purified antibody recognizes mainly one polypeptide of ∼220-kD in immnoblots. To confirm that the antibody specifically recognizes Msps protein, protein samples were prepared from wild-type and msps P homozygous late third instar larvae (Fig. 5 a, left). The 220-kD band and other minor bands are greatly reduced in the msps P homozygote. It can be argued, however, that as the sizes of imaginal discs are reduced in the mutant, any proteins highly expressed in discs can be reduced. To eliminate this possibility we used a semi-lethal allele, msps MJ15 which we created by P-element remobilization. Adult males from this allele are morphologically normal except for weak rough eyes and a few missing bristles, and have normal testis with no spermatogenesis defects. The quantity of 220-kD protein and smaller proteins recognized by the antibody is significantly reduced (Fig. 5 a, right). These minor smaller proteins sometimes recognized by this antibody are likely to be degradation products as their quantity is greatly reduced in msps mutants and is variable in one preparation to another. These results indicate that the affinity-purified antibody specifically recognizes Msps protein.
Figure 5.
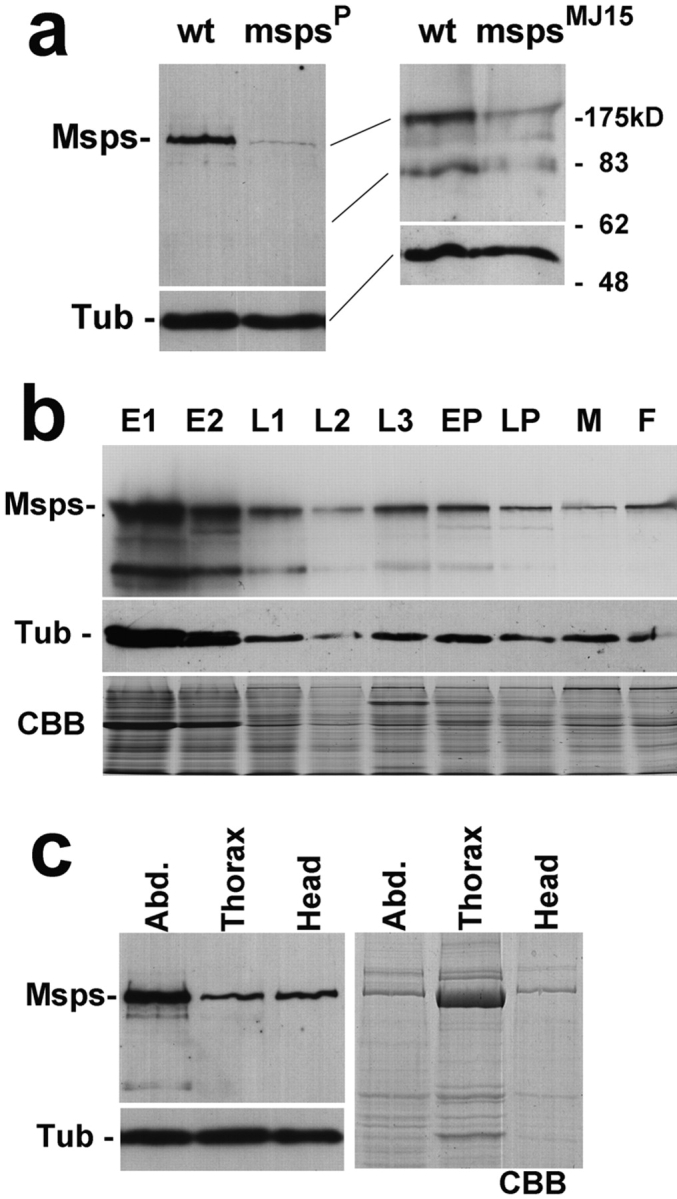
Expression of Msps protein. (a) Specific immunoidentification of Msps protein. (Left) Total protein samples were prepared from late third instar larvae of wild-type and msps P, immunoblotted and probed with the Msps antibody. (Right) Protein samples were prepared from adult males of wild-type and msps MJ15. The levels of 220-kD protein and putative degradation fragments were greatly reduced in the msps mutants. The amounts and profiles of total proteins were identical between wild-type and msps mutants judged by Coomassie blue staining. An α-tubulin antibody was used to give a loading and blotting control. (b) Levels of Msps protein during development. Protein samples were prepared from successive developmental stages of wild-type. E1, 0–4-h embryos; E2, 4–20-h embryos; L1, 1st instar larvae; L2, 2nd instar larvae; L3, 3rd instar larvae; EP, early pupae; LP, late pupae/pharate adult; M, adult males; F, adult females. In the upper panel, the immunoblot is probed with the Msps antibody. Msps protein is most abundant in embryos but a significant amount is also found in other developmental stages. In the middle panel, the immunoblot is probed with α-tubulin antibody. In the bottom panel, Coomassie blue staining shows that each lane has a comparable amount of protein except L2 which is underloaded. (c) Msps protein in adults. Adult females were dissected into three parts, abdomen, thorax, and head. One-tenth of each part from individual flies was loaded in each lane. In the top left panel, the immunoblot was probed with Msps antibody. In the lower left panel, the immunoblot was probed with an α-tubulin antibody. The right panel shows Coomassie blue staining.
To examine the expression of Msps protein during wild-type development, protein samples were prepared from various stages of Drosophila development, and comparable amount of total protein was loaded for immunoblotting (Fig. 5 b). As we expected, a large amount of Msps protein is found in embryos that undertake rapid cycles of nuclear and then cell divisions. However, a significant amount of Msps protein was also detected throughout larval, pupal, and adult stages, when cell division is limited to certain tissues. Next, protein samples were prepared separately from head, thorax, and abdomen of adult females for immunoblotting (Fig. 5 c). A high level of Msps protein was detected in the abdomen, probably reflecting high expression in gonads. It is surprising to see that a significant level of Msps protein is detected in both the thorax and heads, especially considering that the amount of protein from the head is relatively small and that there are supposed to be no cell divisions in the central nervous system at this stage. These results raise the possibility that Msps protein may have some function in non dividing cells. Further analysis will be required to test this possibility.
Msps Protein Binds to Microtubules In Vitro
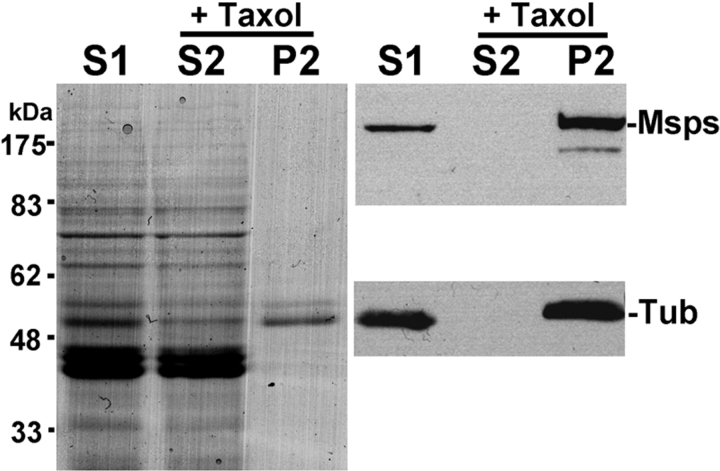
As the predicted sequence of the Msps protein showed high similarity to microtubule-associated proteins from other organisms, we wished to determine whether it was capable of binding to microtubules. We therefore prepared a soluble protein extract (S1 in Fig. 6) from embryos under conditions that depolymerize microtubules. Taxol (paclitaxel) and GTP were then added to repolymerize microtubules, and after 30 min incubation at room temperature, the microtubule fraction (P2) and the soluble fraction (S2) were separated by centrifugation. These protein fractions were run on an SDS–polyacrylamide gel and visualized by Coomassie blue staining. The protein profiles of soluble fractions before and after taxol addition are almost identical, indicating the specific effect of taxol. The microtubule fraction consisted predominantly of proteins of ∼55 kD corresponding to tubulins, together with a number of minor proteins that can be seen by silver staining (data not shown). Immunoblotting indicates that both α-tubulin and Msps are found exclusively in the microtubule fraction (P2), confirming that Msps has microtubule binding activity.
Figure 6.
Msps protein binds to microtubules in vitro. Crude protein extract from 0–8-h-old embryos was incubated on ice to depolymerize microtubules. The high speed supernatant (S1) was incubated with taxol (paclitaxel) and GTP at 20°C to repolymerize the microtubules. Microtubules and associated proteins (P2) were separated from soluble proteins (S2) by centrifugation. (Left) Coomassie blue staining. (Top right) Immunoblot probed by Msps antibody. (Bottom right) Immunoblot probed by α-tubulin antibody. After taxol treatment, Msps protein of unaltered size is exclusively detected on the microtubule fraction.
Msps Protein Localizes on the Mitotic Spindle
To examine whether Msps protein is a constituent of the mitotic spindle, we used the antibody against Msps to localize Msps protein with respect to microtubules in wild-type embryos.
We first examined mitotic cycles 11–13 which take place at the cortex of syncytial embryos (Fig. 7). In prophase, when chromosomes start condensing, we observed microtubules radiating from two discrete regions around the nucleus. That these discrete regions correspond to centrosomes was confirmed using an antibody against a centrosomal antigen, CP190 (data not shown). Affinity-purified antibody against Msps protein revealed that Msps protein localizes predominantly around the centrosomes at this stage.
Figure 7.

Msps protein localizes to mitotic spindles and the centrosomal regions. Wild-type syncytial embryos were immunostained with α-tubulin and Msps antibodies and stained with DAPI to reveal DNA. In merged images, DNA, tubulin, and Msps staining are represented as blue, green, and red, respectively. Bar, 10 μm.
In metaphase, mitotic chromosomes are fully condensed and aligned on the metaphase plate, and the bipolar spindle has formed between the centrosomes. Msps protein appears to spread over the mitotic spindles although it is still concentrated in the polar regions or in the vicinity of the centrosomes. This concentration at the centrosomal regions is more evident in merged images, where the polar region appears orange rather than yellowish green seen along the spindles (Fig. 7).
During anaphase when sister chromatids are separated and move to each pole, Msps staining is still observed on the mitotic spindles and the centrosomal region.
In telophase the chromosomes become decondensed, the nuclear membrane reformed, and the mitotic spindle disassembles except for the midbody at the equator. In syncytial cycles, the centrosomes duplicate and start separating at telophase. Msps protein still appears to be tightly localized in the vicinity of the centrosomes, while a lower level of staining is observed at the midbody.
During interphase, chromatin is decondensed, except for the heterochromatin which lies in the apical region of the nuclei. Thin microtubules emanate from two centrosomes that have already separated at this time. Intense staining of Msps protein is observed in the vicinity of these centrosomes. Interphase embryos without discrete Msps staining at the centrosomal region were not observed, suggesting Msps protein localizes to the centrosomal region throughout interphase in these cycles.
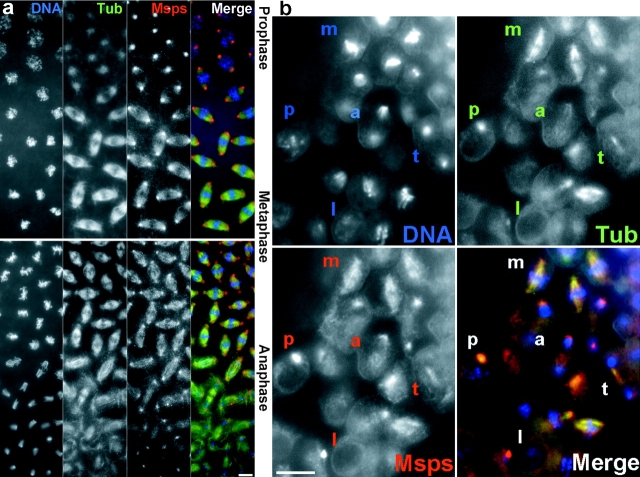
In syncytial embryos, there is often a gradient of mitotic progression as mitotic waves start from the two ends of the embryos. This allowed us to follow the detailed changes in Msps distribution between the different mitotic stages in neighboring nuclei (Fig. 8 a). From prophase to metaphase, Msps staining is strong at the centrosomal regions throughout, gradually spreading out along the mitotic spindles. There is little change in Msps staining at the metaphase/anaphase transition. In particular, the midzone between separating chromatids is stained throughout. This suggests that Msps protein is predominantly localized on pole to pole microtubules. At late anaphase to telophase, Msps staining at the mitotic spindles gets weaker but staining at the centrosomal region stays strong and becomes discrete.
Figure 8.
Msps protein changes its localization during mitotic progression in syncytial and cellularized embryos. Wild-type embryos were immunostained with α-tubulin and Msps antibodies and stained with DAPI to reveal DNA. In the merged images, DNA, tubulin, and Msps staining are represented as blue, green, and red, respectively. (a) Two syncytial embryos are shown to represent progression through mitosis from prophase to late anaphase. (b) Mitotic domains from cellularized embryos at cycle 14. p, prophase; m, metaphase; a, anaphase; t, telophase; I, interphase. Bars, 10 μm.
The cell cycles in syncytial embryos are unique in several respects. The cell cycle is the shortest among any known eukaryotic cells, there are no gap phases in interphase, no DNA replication checkpoints, and cytokinesis does not take place after nuclear division. To test whether the observed localization of Msps is unique to the syncytial cycles, we also examined its localization in cellularized embryos. Cellularization takes place after completion of the 13th mitosis, after which the length of interphase dramatically increases from 10 min to more than an hour. In cellularized embryos, mitotic activity is seen in domains of incompletely synchronized cell divisions.
During mitosis, the localization of Msps in cellularized embryos is basically the same as that observed in syncytial embryos (Fig. 8 b). In prophase (p) the anti-Msps antibody strongly stains the centrosomal region. This persists through metaphase (m) and anaphase (a) when Msps spreads along the mitotic spindles. At telophase (t), it is accumulated on centrosomal regions and weakly on the midbody. The significant difference between syncytial and cellularized embryos is observed in interphase (I). No accumulation of Msps staining at the centrosomal regions is observed in cellularized embryos, contrasting with syncytial embryos in which Msps protein is associated with centrosomal regions throughout interphase. This may simply be a reflection of short interphase in syncytial embryos.
Msps Localization upon Treatment with Drugs Affecting Microtubule Stability
To determine whether the localization of Msps protein is dependent on microtubules, we looked at the effects of microtubule inhibitors. When we treated syncytial embryos with colchicine, microtubules appeared depolymerized but weak tubulin staining remained around the centrosomes (Fig. 9 a). This is the case even when microtubules were depolymerized by other methods (Materials and Methods). In all cases a significant amount of Msps protein was detected around the centrosomes. This result indicates that long microtubules are not essential for the centrosomal localization of at least some of Msps protein. It is possible, however, that short microtubules or tubulin may be required for Msps localization on centrosomes.
Figure 9.
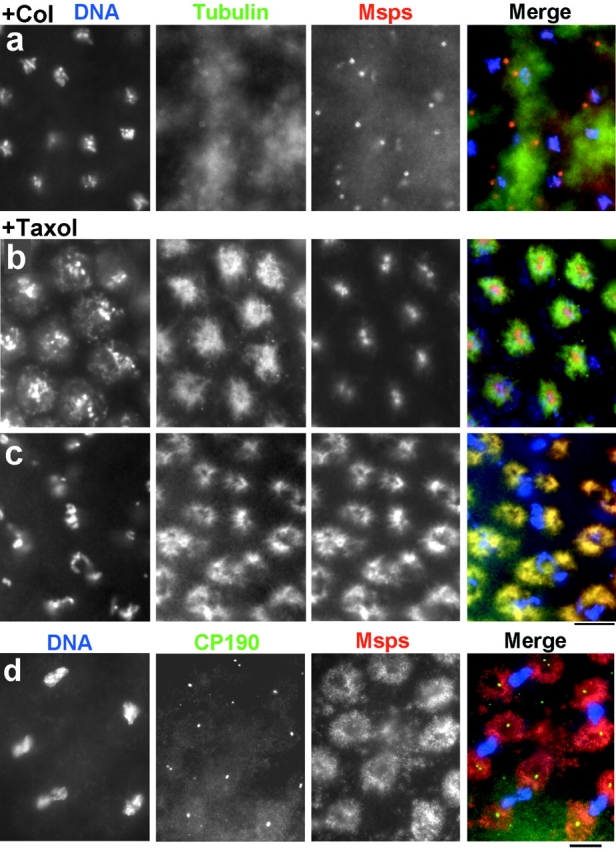
Localization of Msps protein in colchicine or taxol treated embryos. Syncytial embryos were incubated with colchicine (a) or taxol (b–d). (a–c) DNA (left, blue in merge), tubulin (middle, green), and Msps (right, red) were visualized by immunostaining. Overlap between tubulin and Msps staining resulted in yellow color. (d) DNA (left, blue), CP190 (middle, green), and Msps (right, red) were visualized. (a) Microtubules were depolymerized except weak tubulin staining of centrosomes. Msps protein remains on centrosomes. (b) After taxol treatment, Msps protein remains in the centrosomal region during interphase. (c and d) In mitotic cells, Msps protein follows taxol induced microtubules, while CP190 stays on centrosomes. Bars, 10 μm.
The dynamic changes of Msps protein localization in the embryonic cell cycles may reflect cyclical regulation of this process. Alternatively, it may simply be a reflection of changes in concentration, distribution, and dynamics of microtubules. In an attempt to address this question, we artificially induced the formation of microtubules in both mitotic and interphase syncytial embryos using the microtubule stabilizing drug, taxol (paclitaxel).
In interphase embryos, taxol treatment induces extra microtubules from one of two discrete regions around the nuclei, which are likely to be centrosomes (Fig. 9 b). Msps protein remained tightly associated with centrosomal regions in interphase, and there was little staining along the microtubules. In mitotic embryos, extra microtubules appear to be polymerized around the centrosomal regions (Fig. 9 c). In contrast to interphase embryos, Msps protein follows the distribution of microtubules faithfully, as the tubulin and Msps staining overlaps. Thus Msps protein shows a higher affinity for taxol-induced microtubules during mitosis, while during interphase it shows a higher affinity for centrosomes.
We then followed the distribution of CP190 in relation to Msps protein. In untreated embryos, CP190 localizes on centrosomes during mitosis (not shown). CP190 also has microtubule binding activity, so in addition to a strong accumulation on centrosomes a small proportion of the protein localizes on the mitotic spindle during metaphase (Whitfield et al. 1988). After incubation with taxol, CP190 was still tightly associated with centrosomes during mitosis (Fig. 9 d), confirming that centrosomes are not disrupted by taxol treatment. In contrast only weak Msps staining was observed in the vicinity of centrosomes. Although both Msps protein and CP190 are capable of binding to microtubules and localize on or around centrosomes, both proteins behave in different ways when microtubules are induced by taxol during mitosis. Msps protein appears to have a higher affinity to taxol induced microtubules than centrosomes while CP190 has a higher affinity for centrosomes.
Discussion
We have identified a new Drosophila gene, msps, which is essential for spindle formation and function. It encodes a protein belonging to a family of MAPs that can bind to microtubules and associates with the mitotic spindle and centrosomal regions in a cell cycle–dependent manner.
Msps Encodes a Microtubule-associated Protein
Molecular cloning revealed that the msps gene encodes a protein which belongs to the dis1-TOG family. This family of proteins is very divergent at the primary sequence level. Conserved residues are limited to four motifs that form part of repeating regions in the NH2-terminal portion of the molecule. These repeats with their limited conservation could be a structural or functional module in the sense of the tetratrico peptide repeats (TPR), which are found in proteins with various functions (Goebel and Yanagida, 1991). In support of this idea, it was shown that the repeats are dispensable for in vivo function of S. pombe Dis1 (Nakaseko et al. 1996). This raises the question as to whether these repeats do indeed have a common function and whether the dis1-TOG family shares a conserved function in vivo.
The vertebrate and fly proteins share features that are distinct from lower eukaryotic proteins. They have four repeats in the NH2-terminal region, while the yeast and C. elegans have only two repeats. The Msps protein is highly homologous to human TOGp along its entire length. Their COOH-terminal regions are conserved and share some homology with the C. elegans protein. In contrast, the COOH-terminal regions of the yeast protein has a coiled-coil structure bearing no sequence homology. Drosophila genetics should provide a unique opportunity to study in vivo function of higher eukaryotic members of this MAP family.
Msps Protein Localizes to Mitotic Spindle and Centrosomal Region
Considering the divergence of protein sequence of the family members and structure of the mitotic apparatus among eukaryotes, localization of the dis1-TOG proteins is surprisingly similar among dis1-TOG family from yeasts to human (Wang and Huffaker 1997; Charrasse et al. 1998; Mathews et al., 1998; Nabeshima et al. 1998). They concentrate in the vicinity of the centrosome or SPB during the early stages of mitosis, transiently spread along the whole length of the mitotic spindles during mid-mitosis, before localizing back to the vicinity of the centrosome/SPB region in late mitosis.
The cell cycle–dependent interaction between Msps and microtubules or centrosomes could occur as a result of posttranslational modification of Msps protein. Although we have no evidence for such modification of Msps, cell cycle–dependent phosphorylation of the XMAP215 and Dis1 proteins has been observed (Gard and Kirschner 1987; Nabeshima et al. 1995). In the case of Dis1, cdc2 kinase appears responsible for this phosphorylation. However, the effect of phosphorylation on the interaction of these proteins with microtubules and in vivo function remains to be determined.
As we expected, Msps protein is abundant in tissues that contain many dividing cells. However, we also found a significant amount of Msps protein in nonproliferating tissues, such as the adult head. This is also seen in vertebrates. Both human ch-TOG and Xenopus XMAP215 are highly expressed in adult brains (Charrasse et al. 1998; Gard and Kirschner 1987). As these proteins can regulate microtubule dynamics, it is an attractive possibility that these proteins may also function in post mitotic cells, a question that could in future be addressed in vivo in Drosophila.
What Is the Function of Msps Protein?
The msps mutation affects only a limited aspect of spindle formation. It does not appear to have a strong impact on microtubule nucleation, bipolarity of the spindle, focusing of the poles, or chromosome alignment. Rather the mutant appears defective in holding the mitotic spindle together.
As the Msps protein is localized to centrosomal regions, it is possible that it is involved in the nucleation of microtubules around centrosomes. If centrosomal microtubule nucleation were defective, the effects of chromosomes on stabilizing microtubules would become dominant, resulting in the mini spindles phenotype. Such chromosome driven bipolar spindle formation has been demonstrated in centrosome-free systems. Beads coated with DNA are capable of organizing a bipolar spindle in Xenopus egg extracts and single meiotic chromosomes expelled from the spindle in various mutants can organize a bipolar mini spindle during Drosophila female meiosis (Hatsumi and Endow 1992; McKim and Hawley 1995; Heald et al. 1996). Moreover, mini spindle formation is triggered when chromosomes are detached by micromanipulation from the Drosophila male meiotic spindle which contains centrosomes (Church et al., 1986). However, this model is not consistent with the phenotypes seen in either Drosophila γ-tubulin mutants or asp mutants. The γ-tubulin complex and the Asp protein appear essential for the integrity of microtubule nucleation activity of centrosomes (Moritz et al. 1995; Zheng et al. 1995; Avides and Glover 1999). The Drosophila γ-tubulin mutant shows a variety of defects but no mini spindles phenotype has been reported (Sunkel et al. 1995). Similarly the poles of asp mutant spindles are highly disorganized but the spindles are largely intact and bipolar.
Alternatively, it may be possible that Msps protein is required for anchoring spindle microtubules to centrosomes. It is known that many microtubules are not directly attached to centrosomes (Mastronarde et al. 1993), and partial loss of Msps protein may cause a set of spindle microtubules to detach from centrosomes. However, neither this nor the previous model explain the conserved localization of Msps protein along the spindle microtubules during mid-mitosis. We have also found that Msps protein localizes on the female meiotic spindle at metaphase I (Cullen, C.F., and H. Ohkura, unpublished data). As the female meiotic spindle at metaphase I lacks centrosomes and its formation is driven by chromosomes (Matthies et al. 1996), this observation supports the possibility that Msps protein has functions that are independent of centrosomes.
The msps phenotype may be best explained by a failure in the microtubule bundling that holds the mitotic spindle together. Microtubule bundling combined with minus end motors has been proposed as a mechanism to focus the spindle at the polar region. The dynein–dynactin complex in association with NuMA (Merdes et al. 1996), Ncd (Hatsumi and Endow 1992; Matthies et al. 1996), and CTK2 (Heald et al. 1996) are proposed to fulfil such roles. The msps phenotype may suggest that focusing of the polar region requires two steps. In one step, the microtubules that emanate from each chromosome are bundled together, whereas in the second these microtubule bundles are held together. Msps protein may be required mainly for the second step. Although human TOGp and Xenopus XMAP215 have high sequence similarity to Msps protein, no such in vitro activity has been reported for either of the purified proteins. However, it is possible that interaction with other proteins is required for this activity.
The most simple model is that Msps protein is required for formation of long microtubules during mitosis. When cells fail to make long microtubules, the mitotic spindle cannot hold all of its chromosomes and it collapses to form small spindles. This model is supported by the in vitro activity of purified XMAP215 and human TOGp. Purified XMAP215 dramatically increases elongation and shortening velocity and decreases the frequency of the rescue at the plus ends of microtubules while effects on the minus end are much less dramatic. In total, it promotes plus end assembly and turnover, resulting in a population of extremely long but highly dynamic microtubules (Vasquez et al. 1994). In contrast, it was reported that TOGp increases the elongation rate of both ends equally and appears to inhibit catastrophes. As a result, TOGp promotes microtubule assembly (Charrasse et al. 1998). Although it is not clear whether these differences reflect experimental approaches, it is evident that both proteins can promote microtubule assembly.
Learning from Lower Eukaryotes
Apart from a limited sequence similarity, dis1-TOG family members in eukaryotes share an in vitro microtubule binding activity, localization to the spindle and SPB or centrosomes, and function in mitosis. Do lower eukaryotic proteins have the same function in mitosis as Msps protein, and do the studies on lower eukaryotic proteins give clues to the function of Msps?
C. elegans ZYG-9 shows intermediate organization between the higher eukaryotic and yeast proteins at the primary sequence level. Crucially, C. elegans has similar mitotic apparatus to the higher eukaryotes. zyg-9 mutant embryos exhibit disorganized spindles and numerous cytoplasmic clusters of short microtubules during meiosis. Subsequently, pronuclear migration and the migration and rotation of the centrosome-nuclear complex fails (Kemphues et al. 1986). Zyg-9 gene activity is less important for the second or subsequent mitosis and dispensable after gastrulation (Mains et al. 1990). Based on these observations, it was proposed that zyg-9 is required for the formation of long microtubules during the first division. This is supported by the observation that nocodazole, which destabilizes microtubules, mimics the zyg-9 phenotype (Albertson 1984; Kemphues et al. 1986).
In contrast to the zyg-9 gene, msps gene activity appears to be universally required in mitotic cells throughout the development. We observed mitotic defects in the central nervous system of third instar larvae in the msps mutant and severe growth defects of imaginal discs. In addition, mutants with semi-lethal alleles of msps show defects indicative of the failure of cell division of sensory mother cells or histoblast cells. Moreover, female homozygous for those alleles laid eggs that showed mitotic defects during the embryonic divisions (Cullen, C.F., and H. Ohkura, unpublished data). Although the developmental requirement for msps and Zyg-9 differs, an attractive model that reflects the cellular phenotypes of both mutants is that both gene products promote the formation of long microtubules during mitosis.
S. cerevisiae STU2 was originally identified as a dominant chromosomal suppressor of tub2-423, a cold sensitive allele of the β-tubulin gene (Wang and Huffaker 1997). Although these dominant mutations on their own do not affect growth, the disruption of the gene is lethal. The cytological phenotype of this disruptant has not been studied, so the in vivo function of STU2 has not been established. As STU2p was shown to bind microtubules laterally and localizes on the SPB in a microtubule-independent manner, it was proposed that STU2p tethers the microtubules to the SPB while allowing exchange of tubulin subunits at their minus ends.
In contrast to the msps mutant, S. pombe dis1 mutation does not affect the integrity of the mitotic spindle, at least at the light microscope level (Ohkura et al. 1988; Nabeshima et al. 1995), but sister chromatids fail to separate. Real time analysis showed it to be defective in the oscillation of centromeres during metaphase and in restraining spindle elongation (Nabeshima et al. 1998). It was proposed that Dis1 may be required for interactions of microtubules with the kinetochore and SPB, consistent with a microtubule anchoring model. This model is also consistent with observation that NH2-terminally truncated Dis1 protein localizes only on SPB but not on the mitotic spindle and can complement the cold sensitivity of dis1 deletion mutants (Nakaseko et al. 1996). It will be interesting to see whether this is also true for the higher eukaryotic proteins. Studies of the relationship between domain structure, localization, and in vivo function in higher eukaryotic protein is an exciting future possibility in Drosophila. It should be noted that the dis1 + gene is not an essential gene. The dis1 deletion mutant is viable but shows cold sensitive lethality (Nabeshima et al. 1995). This may suggest a role in promoting the assembly of microtubules that are unstable at low temperatures.
Studies of the dis1-TOG family members indicate that they share many common features as well as some apparent differences. At this moment, it is not clear whether they execute exactly the same function. Further studies are needed to understand their exact molecular function and the regulation of localization with respect to protein structure. The availability of Drosophila mutants and the feasibility of genetic manipulation will be invaluable for studying the higher eukaryotic forms in vivo.
Acknowledgments
We thank Dr. Karen May for critical reading of the manuscript, Dr. Robert Saunders for helping with cosmid screening, and Professor Keith Gull for an antibody.
This work was supported by the Wellcome Trust, the Cancer Research Campaign, and the Medical Research Council. H. Ohkura receives a Wellcome Senior Research Fellowship for Basic Biomedical Sciences.
Footnotes
1.used in this paper: asp, abnormal spindle; MAP, microtubule-associated protein; msps, mini spindles
References
- Albertson D.G. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen S.S., Ashford A.J., Tournebize R., Gavet O., Sobel A., Hyman A.A., Karsenti E. Mitotic chromatin regulates phosphorylation of stathmin/Op18. Nature. 1997;389:640–643. doi: 10.1038/39382. [DOI] [PubMed] [Google Scholar]
- Andersen S.S., Buendia B., Dominguez J.E., Sawyer A., Karsenti E. Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laevis eggs and dividing cells. J. Cell Biol. 1994;127:1289–1299. doi: 10.1083/jcb.127.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M. Drosophila 1989. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: pp. 1–1331 [Google Scholar]
- Avides M.C., Glover D.M. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science. 1999;12:1733–1735. doi: 10.1126/science.283.5408.1733. [DOI] [PubMed] [Google Scholar]
- Barton N.R., Pereira A.J., Goldstein L.S. Motor activity and mitotic spindle localization of the Drosophila kinesin-like protein KLP61F. Mol. Biol. Cell. 1995;6:1563–1574. doi: 10.1091/mbc.6.11.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinhauer J.D., Hagan I.M., Hegemann J.H., Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont L.D., Mitchison T.J. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–631. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- Berlin V., Styles C.A., Fink G.R. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J. Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E., Vaessin H., Shepherd S., Lee K., McCall K., Barbel S., Ackerman L., Carretto R., Uemura T., Grell E. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Mazel M., Taviaux S., Berta P., Chow T., Larroque C. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur. J. Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Schroeder M., Gauthier-Rouviere C., Ango F., Cassimeris L., Gard D.L., Larroque C. The TOGp protein is a new human microtubule-associated protein homologous to the Xenopus XMAP215. J. Cell Sci. 1998;111:1371–1383. doi: 10.1242/jcs.111.10.1371. [DOI] [PubMed] [Google Scholar]
- Church K., Nicklas R.B., Lin H.P. Micromanipulated bivalents can trigger mini-spindle formation in Drosophila melanogaster spermatocyte cytoplasm. J. Cell Biol. 1996;103:2765–2773. doi: 10.1083/jcb.103.6.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak P., Omar M.M., Saunders R.D., Pal M., Komonyi O., Szidonya J., Maroy P., Zhang Y., Ashburner M., Benos P. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogastercorrelation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics. 1997;147:1697–1722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Glover D.M., Saumweber H. Nuclear antigens follow different pathways into daughter nuclei during mitosis in early Drosophila embryos. J. Cell Sci. 1986;82:155–172. doi: 10.1242/jcs.82.1.155. [DOI] [PubMed] [Google Scholar]
- Gard D.L., Kirschner M.W. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M., Yanagida M. The TPR snap helixa novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- Gonzalez C., Glover D.M. Techniques for studying mitosis in Drosophila . In: Fantes P., Brooks R., editors. The Cell CycleA Practical Approach. IRL Press; Oxford: 1993. pp. 163–168. [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Hatsumi M., Endow S.A. The Drosophila ncd microtubule motor protein is spindle-associated in meiotic and mitotic cells. J. Cell Sci. 1992;103:1013–1020. doi: 10.1242/jcs.103.4.1013. [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Microtubule organization and dynamics dependent on microtubule-associated proteins. Curr. Opin. Cell Biol. 1994;6:74–81. doi: 10.1016/0955-0674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Mitchison T.J. Modulation of microtubule stability by kinetochores in vitro. J Cell Biol. 1990;110:1607–1616. doi: 10.1083/jcb.110.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Karsenti E. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 1996;84:401–410. doi: 10.1016/s0092-8674(00)81285-4. [DOI] [PubMed] [Google Scholar]
- Kellogg D.R., Field C.M., Alberts B.M. Identification of microtubule-associated proteins in the centrosome, spindle, and kinetochore of the early Drosophila embryo. J. Cell Biol. 1989;109:2977–2991. doi: 10.1083/jcb.109.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Alberts B.M. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol. Biol. Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K.J., Wolf N., Wood W.B., Hirsh D. Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans . Dev. Biol. 1986;113:449–460. doi: 10.1016/0012-1606(86)90180-6. [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V., Wright B., Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R., Weber U., Gehring W.J. The white gene as a marker in a new P-element vector for gene transfer in Drosophila . Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley D.L., Zimm G.G. The genome of Drosophila melanogaster. Academic Press; New York: 1992. [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mains P.E., Sulston I.A., Wood W.B. Dominant maternal-effect mutations causing embryonic lethality in Caenorhabditis elegans . Genetics. 1990;125:351–369. doi: 10.1093/genetics/125.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E., Mandelkow E. Microtubules and microtubule associated proteins. Curr. Opin. Cell Biol. 1995;7:72–81. doi: 10.1016/0955-0674(95)80047-6. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N., McDonald K.L., Ding R., McIntosh J.R. Interpolar spindle microtubules in PTK cells. J. Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L.R., Carter P., Thierry-Mieg D., Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H.J., McDonald H.B., Goldstein L.S., Theurkauf W.E. Anastral meiotic spindle morphogenesisrole of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Hawley R.S. Chromosomal control of meiotic cell division. Science. 1995;270:1595–1601. doi: 10.1126/science.270.5242.1595. [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M.B., Sedat J.W., Alberts B., Agard D.A. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M., Zheng Y., Alberts B.M., Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka H., Takeuchi M., Kinoshita K., Nakaseko Y., Yanagida M. p93Dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A.F., Murray A., Chikashige Y., Yamashita Y.M., Hiraoka Y., Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeastDis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Nabeshima K., Kinoshita K., Yanagida M. Dissection of fission yeast microtubule associating protein p93dis1regions implicated in regulated localization and microtubule interaction. Genes Cells. 1996;1:633–644. doi: 10.1046/j.1365-2443.1996.00253.x. [DOI] [PubMed] [Google Scholar]
- Oegema K., Whitfield W.G., Alberts B. The cell cycle-dependent localization of the CP190 centrosomal protein is determined by the coordinate action of two separable domains. J. Cell Biol. 1995;131:1261–1273. doi: 10.1083/jcb.131.5.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Adachi Y., Kinoshita N., Niwa O., Toda T., Yanagida M. Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:1465–1473. doi: 10.1002/j.1460-2075.1988.tb02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Torok T., Tick G., Hoheisel J., Kiss I., Glover D.M. Mutation of a gene for a Drosophila kinesin-like protein, Klp38B, leads to failure of cytokinesis. J. Cell Sci. 1997;110:945–954. doi: 10.1242/jcs.110.8.945. [DOI] [PubMed] [Google Scholar]
- Pereira A., Doshen J., Tanaka E., Goldstein L.S. Genetic analysis of a Drosophila microtubule-associated protein. J. Cell Biol. 1992;116:377–383. doi: 10.1083/jcb.116.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll P., Pimpinelli S., Valdivia M.M., Avila J. A cell division mutant of Drosophila with a functionally abnormal spindle. Cell. 1985;41:907–912. doi: 10.1016/s0092-8674(85)80071-4. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular CloningA Laboratory Manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Saunders R.D.C., Avides M.C., Howard T., Gonzalez C., Glover D.M. The Drosophila gene abnormal spindle encodes a novel microtubule- associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 1997;137:881–890. doi: 10.1083/jcb.137.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R.D.C., Glover D.M., Ashburner M., Siden-Kiamos I., Louis C., Monastirioti M., Savakis C., Kafatos F. PCR amplification of DNA microdissected from a single polytene chromosome banda comparison with conventional microcloning. Nucleic Acids Res. 1989;17:9027–9037. doi: 10.1093/nar/17.22.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton W.M., Stemple D.L., Leslie R.J., Salmon E.D., Zavortink M., McIntosh J.R. Tubulin dynamics in cultured mammalian cells. J. Cell Biol. 1984;99:2175–2186. doi: 10.1083/jcb.99.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N., Moriguchi T., Ohta K., Gotoh Y., Nishida E. Regulation of a major microtubule-associated protein by MPF and MAP kinase. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:3977–3984. doi: 10.1002/j.1460-2075.1992.tb05491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siden-Kiamos I., Saunders R.D.C., Spanos L., Majerus T., Treanear J., Savakis C., Louis C., Glover D.M., Ashburner M., Kafatos F.C. Towards a physical map of the Drosophila melanogaster genomemapping of cosmid clones within defined genomic divisions. Nucleic Acids Res. 1990;18:6261–6270. doi: 10.1093/nar/18.21.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.E., Fisher P.A. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryosapplication of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J. Cell Biol. 1984;99:20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkel C.E., Gomes R., Sampaio P., Perdigao J., Gonzalez C. Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:28–36. doi: 10.1002/j.1460-2075.1995.tb06972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R.J., Gard D.L., Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J. Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Mitchison T.J., Desai A. XKCM1a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wang P.J., Huffaker T.C. Stu2pA microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield W.G., Millar S.E., Saumweber H., Frasch M., Glover D.M. Cloning of a gene encoding an antigen associated with the centrosome in Drosophila . J. Cell Sci. 1988;89:467–480. doi: 10.1242/jcs.89.4.467. [DOI] [PubMed] [Google Scholar]
- Whitfield W.G., Chaplin M.A., Oegema K., Parry H., Glover D.M. The 190 kDa centrosome-associated protein of Drosophila melanogaster contains four zinc finger motifs and binds to specific sites on polytene chromosomes. J. Cell Sci. 1995;108:3377–3387. doi: 10.1242/jcs.108.11.3377. [DOI] [PubMed] [Google Scholar]
- Wood W.B., Hecht R., Carr S., Vanderslice R., Wolf N., Hirsh D. Parental effects and phenotypic characterization of mutations that affect early development in Caenorhabditis elegans . Dev. Biol. 1980;74:446–469. doi: 10.1016/0012-1606(80)90445-5. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin T., Sasse R., MacRae T.H., Baines A.J., Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Kronebusch P.J., Borisy G.G. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J. Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]