Abstract
Human cells express two kinases that are related to the yeast mitotic checkpoint kinase BUB1. hBUB1 and hBUBR1 bind to kinetochores where they are postulated to be components of the mitotic checkpoint that monitors kinetochore activities to determine if chromosomes have achieved alignment at the spindle equator (Jablonski, S.A., G.K.T. Chan, C.A. Cooke, W.C. Earnshaw, and T.J. Yen. 1998. Chromosoma. 107:386–396). In support of this, hBUB1 and the homologous mouse BUB1 have been shown to be important for the mitotic checkpoint (Cahill, D.P., C. Lengauer, J. Yu, G.J. Riggins, J.K. Willson, S.D. Markowitz, K.W. Kinzler, and B. Vogelstein. 1998. Nature. 392:300–303; Taylor, S.S., and F. McKeon. 1997. Cell. 89:727–735). We now demonstrate that hBUBR1 is also an essential component of the mitotic checkpoint. hBUBR1 is required by cells that are exposed to microtubule inhibitors to arrest in mitosis. Additionally, hBUBR1 is essential for normal mitotic progression as it prevents cells from prematurely entering anaphase. We establish that one of hBUBR1's checkpoint functions is to monitor kinetochore activities that depend on the kinetochore motor CENP-E. hBUBR1 is expressed throughout the cell cycle, but its kinase activity is detected after cells have entered mitosis. hBUBR1 kinase activity was rapidly stimulated when the spindle was disrupted in mitotic cells. Finally, hBUBR1 was associated with the cyclosome/anaphase-promoting complex (APC) in mitotically arrested cells but not in interphase cells. The combined data indicate that hBUBR1 can potentially provide two checkpoint functions by monitoring CENP-E–dependent activities at the kinetochore and regulating cyclosome/APC activity.
Keywords: kinetochore, hBUBR1, mitotic checkpoint •anaphase-promoting complex, CENP-E
Accurate chromosome segregation relies on the interactions between the kinetochore and microtubules of the spindle. The process by which kinetochores capture a microtubule is stochastic in nature, a chance encounter, which is prone to error (Nicklas 1997). Thus, it is critical that cells are able to delay exit from mitosis until all chromosomes have aligned at the spindle equator. The mitotic checkpoint is able to determine whether chromosomes have aligned by monitoring the kinetochore, a structure that mediates attachment of the chromosome to microtubules and is directly responsible for chromosome motility (Inoue and Salmon 1995; Yen and Schaar 1996; Rieder and Salmon 1998). The checkpoint is sensitive to the presence of even a single unattached kinetochore, as this is sufficient to delay cells from exiting mitosis (Rieder et al. 1995). The precise biochemical mechanism by which an unattached kinetochore blocks entry into anaphase remains undefined, but it appears to be sensitive to kinetochore microtubule attachments (Rieder et al. 1994) and to the level of tension that is exerted on the kinetochore (Li and Nicklas 1995).
Molecular insights into the mitotic checkpoint have come from the identification of seven genes in budding yeast that are essential for the mitotic checkpoint. In the presence of spindle or kinetochore defects, the MPS1 kinase (Weiss and Winey 1996) and the BUB1/BUB3 kinase complex (Hoyt et al. 1991; Roberts et al. 1994), along with MAD1, MAD2, MAD3, and BUB2, are required to block cells in mitosis (Li and Murray 1991; Hardwick and Murray 1995). The mitotic checkpoint appears to be highly conserved throughout evolution, as homologues of many of the yeast checkpoint proteins have also been identified in Drosophila and vertebrates. Analysis of MAD1 (Chen et al. 1998; Jin et al. 1998), MAD2 (Chen et al. 1996; Li and Benezra 1996), BUB1, BUBR1, and BUB3 (Taylor and McKeon 1997; Basu et al. 1998; Cahill et al. 1998; Chan et al. 1998; Jablonski et al. 1998; Taylor et al. 1998) in higher eukaryotes show that these proteins accumulate at kinetochores where they are postulated to monitor kinetochore functions and participate in generating the wait anaphase signal. Consistent with this possibility, unattached kinetochores exhibited a higher level of some of these checkpoint proteins than kinetochores that were aligned at the spindle equator. Functional studies have shown that MAD1 and MAD2 are essential components of the mitotic checkpoint in vertebrate cells and in cycling Xenopus egg extracts (Chen et al. 1996; Li and Benezra 1996; Waters et al. 1998). Similarly, mouse BUB1 (mBUB1) has also been shown to be essential for the mitotic checkpoint (Taylor and McKeon 1997).
The target of the mitotic checkpoint in both yeast and vertebrates is the cyclosome/anaphase-promoting complex (APC),1 a multisubunit E3 ubiquitin-ligase that specifies the proteolytic destruction of specific proteins to initiate the onset of anaphase (Sudakin et al. 1995; King et al. 1996; Hershko and Ciechanover 1998). MAD2 was found to interact with the cyclosome/APC in mitotically arrested cells and inhibit its ubiquitination activity in vitro and in vivo (Li et al. 1997; Chen et al. 1998; Fang et al. 1998; Gorbsky et al. 1998). Genetic and biochemical studies have shown that the association between MAD2 and the cyclosome/APC is mediated by p55CDC/cdc20 (Fang et al. 1998; Hwang et al. 1998; Kallio et al. 1998; Kim et al. 1998), an evolutionarily conserved protein that is essential for the metaphase–anaphase transition (Dawson et al. 1995; Visintin et al. 1997; Kallio, 1998). The mechanism by which unaligned chromosomes specify the inhibition of the cyclosome/APC by MAD2 is unclear, but a tentative model suggests that unattached kinetochores serve to convert MAD2 into an inhibitor of the cyclosome/APC (Chen et al. 1998; Gorbsky et al. 1998). This possibility is partly supported by the finding that recombinant human MAD2 can form a homotetramer and this complex is more efficient at inhibiting the cyclosome/APC than monomeric forms of MAD2 (Fang et al. 1998).
In spite of the significant advances in our understanding of MAD2 function, the picture remains incomplete due to the lack of understanding of the functions of the other checkpoint proteins. The situation in mammalian cells may be even more complex than in budding yeast as the function and structure of mammalian kinetochores is vastly more complicated and may require a more elaborate checkpoint monitoring system. This possibility is consistent with the recent finding that mammalian cells express two BUB1-related kinases that appear to have evolved from a common ancestral BUB1 kinase. hBUB1 (the homologue of mBUB1) and hBUBR1 are human BUB1-related kinases that were found to be mutated in 2 out of 20 colorectal carcinomas that exhibited a chromosome instability phenotype (Cahill et al. 1998). The mutations identified in hBUB1 were confirmed to interfere with the mitotic checkpoint as the mutant protein disrupted the activity of the wild-type hBUB1 in a dominant negative fashion (Cahill et al. 1998). Although colorectal carcinomas that were heterozygous for hBUBR1 mutations were also identified in the study (Cahill et al. 1998), the role of hBUBR1 in the mitotic checkpoint was not tested.
hBUBR1 was also independently isolated based on its similarities with a portion of the yeast checkpoint protein MAD3 (Taylor et al. 1998). The significance of this similarity is unknown but it is noteworthy that other members of the BUB1 kinase family also share the same MAD3 homology domain (Roberts et al. 1994; Taylor and McKeon 1997; Chan et al. 1998). Clues to hBUBR1 function came when it was found to associate with the kinetochore motor CENP-E (Chan et al. 1998). Although this interaction was initially identified in a yeast two-hybrid screen for proteins that associate with CENP-E, a stable complex of hBUBR1 and CENP-E was detected in Hela cells. CENP-E is a kinetochore-associated kinesin motor protein whose function is in microtubule attachment to kinetochores and in the alignment of chromosomes to the metaphase plate (Schaar et al. 1997; Wood et al. 1997). The function of CENP-E is likely to be monitored by the mitotic checkpoint as cells invariably were arrested in mitosis when their kinetochores lacked CENP-E functions (Schaar et al. 1997). Taken together, we postulated that the CENP-E–hBUBR1 complex might be integral parts of a mechanosensor that link kinetochore motility with checkpoint control (Chan et al. 1998).
In this paper, we establish the importance of the hBUBR1 kinase in the mitotic checkpoint and provide mechanistic insights into its checkpoint functions. We show that one function of hBUBR1 is to monitor kinetochore activities that require CENP-E. We also demonstrate that hBUBR1 is a mitotic-specific kinase that associates with a large fraction of the cyclosome/APC during mitosis. We propose a working model that integrates these two activities to account for how local defects at the kinetochore are detected, relayed, and amplified to a global level to block premature entry into anaphase. The similarities shared between hBUBR1 and the MAD2–p55CDC complex suggests that they can function as an integrated complex or act along parallel pathways in the mitotic checkpoint.
Materials and Methods
Cell Culture
Hela cells were grown in DME supplemented with 10% FBS. Hela cells adapted for growth in suspension were cultured in SME supplemented with 10% FBS and 1% pluronic acid. K562 suspension cultures were grown in RPMI 1640 supplemented with 10% FBS. All cells were grown in a humidified 37°C chamber at 5% CO2. Centrifugal elutriation was performed using a Beckman centrifuge (J6-MC) with the JE5.0 rotor as described (Gately et al. 1998). Samples were taken from each fraction and analyzed for cell cycle distribution by FACS. Hela cells arrested in mitosis were obtained by growing them in the presence of nocodazole (0.06 μg/ml) for 12–16 h. Both normal and nocodazole-blocked mitotic cells were harvested from the culture dishes by mechanical shakeoff.
Construction and Expression of Mutant hBUBR1
Full-length gfp-hBUBR1 expression construct was described previously (Chan et al. 1998). The mutant gfp:hBUBR1Δkinase construct was made by deleting nine amino acids (amino acids 795–803, KVSSQPVPW) from the most conserved kinase subdomain II (Hunter 1987; Hanks and Quinn 1991). Deletion was accomplished by PCR-based mutagenesis. Transient transfections were performed by lipofection using LT2 (PanVera) as described (Chan et al. 1998).
Microinjection
Hela cells were plated onto gridded coverslips and were synchronized at the G1/S boundary by a double thymidine block. Before injection, the media was replaced with fresh Hepes buffered DME plus 10% FBS. Cells were injected 1–2 h after they were released from the G1/S boundary. Microinjection was performed with a semi-automatic injector (model 5412; Eppendorf Scientific, Inc.) and the Eppendorf automated micromanipulator (model 5402). For each coverslip, 200–300 cells were injected with each antibody mix over a course of ∼30 min. Multiple coverslips were injected to accommodate time course experiments where samples had to be taken at different times. The affinity-purified hBUBR1 and CENP-E antibodies used for the injection experiments have been previously described (Schaar et al. 1997; Chan et al. 1998). Nonimmune IgG was isolated from sera, which was originally used to affinity purify hBUBR1 antibodies, and verified by immunofluorescence and Western blots to no longer contain detectable levels of hBUBR1 antibodies. Antibodies were kept in Ca++- and Mg++-free PBS and were filtered through a 0.22-μm Millipore microfiltration cup before use. To visualize the cells that had been injected with antibodies, cells were fixed for 7 min in 3.5% paraformaldehyde in PBS and permeabilized in 0.2% Triton X-100 for 3–5 min before proceeding with antibody incubations. To minimize loss of loosely attached mitotic or apoptotic cells, the coverslips were centrifuged at 200 g for 3 min in a clinical centrifuge (GPKR; Beckman). For time-lapse microscopy, Hela cells were plated onto 35-mm glass bottom microwell dishes (MatTek Corp.), and cells were monitored with a 60× PlanApo objective mounted on a Nikon Eclipse TE300 inverted microscope. Cells were kept at 37°C with a 35-mm tissue culture dish stage warmer regulated by a temperature controller (TC-102; Medical Systems Corp.). Time-lapse images were captured using a Sensys CCD camera (Photometric) that was controlled by Image-Pro Plus software (Phase 3 Imaging Systems).
Immunofluorescence Microscopy
Cells on coverslips were fixed, permeabilized, and stained as described (Chan et al. 1998). Injected rabbit antibodies were detected with Cy5-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories). Endogenous hBUBR1 was visualized with a rat anti–hBUBR1 antibody that was generated and affinity-purified the same way as the previously described rabbit hBUBR1 antibodies (Chan et al. 1998; Jablonski et al. 1998). Anticentromere autoimmune serum was a gift of K. Sullivan (Scripps Research Institute, La Jolla, CA). Microtubules were stained with anti–α-tubulin monoclonal antibody (Sigma Chemical Co.). The primary antibodies were visualized using an appropriate secondary antibody that was coupled to Cy2 or Texas red (Jackson ImmunoResearch Laboratories). DNA was stained with 0.1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI). Stained cells were examined with either a 40 or 100× PlanNeofluor objective mounted on a Nikon Microphot SA that was equipped with epifluorescence optics. Images were captured with a TEC-1 CCD camera (Dage-MTI) that was controlled with IP LabSpectrum v2.0.1 (Scanalytics Inc.) and contrast enhanced with Adobe Photoshop 5.0 (Adobe Systems Inc.).
Immunoprecipitation and Western Blots
Cells were harvested, washed, and lysed in NP-40 lysis buffer (1% NP-40, 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM DTT) supplemented with protease and phosphatase inhibitors as described previously (Gately et al. 1998). Cell lysates were centrifuged at 10,000 g at 4°C for 15 min. The protein concentrations of the lysates were determined (BCA protein assay; Pierce Chemical Co.) and adjusted to the same protein concentration. Immunoprecipitation was performed with 300 or 500 μg of lysate and the final concentration of hBUBR1 antibody was ∼2 μg/ml. For FPLC separation, S100 was loaded onto a Superose 6 column (Amersham Pharmacia Biotech Inc.), and proteins were eluted in NP-40 lysis buffer with protease and phosphatase inhibitors. Fractions containing cyclosome/APC were combined, concentrated with a Centricon 100 (Amicon Inc.), and incubated with hBUBR1 antibodies. To solubilize hBUBR1 that is associated with the kinetochore, chromosomes were boiled in 2% SDS for 2 min, diluted 10-fold with NP-40 lysis buffer, clarified, and processed for immunoprecipitation. Antibody was incubated with lysates for 3 h at 4°C, and then 30 μl of 50% slurry of protein A–Sepharose (Repligen) was added and incubation proceeded for another 30 min. The beads were recovered by low speed centrifugation, washed five times in 0.5 ml of ice-cold lysis buffer before SDS gel sample buffer was added. In some cases, hBUBR1 immunoprecipitates were incubated with 400 units of λ protein phosphatase (New England Biolabs Inc.) for 30 min at 30°C in buffer supplied by the manufacturer. Samples from the immunoprecipitates or whole cell lysates were separated through 4–12% gradient SDS-PAGE, transferred onto Immobilon P (Millipore), and then processed for Western blots.
Antibodies to cyclosome/APC subunits hscdc16 and hscdc27 were gifts of Dr. P. Hieter (University of British Columbia, Vancouver, British Columbia, Canada). APC7 antibodies were a gift from Dr. J.M. Peters (IMP, Vienna, Austria). Rabbit hBUB3 antibodies were generated against GST:hBUB3 fusion protein and affinity-purified as described (Chan et al. 1998). Primary antibodies were detected with alkaline phosphatase–conjugated anti-rabbit secondary antibodies used at 1:30,000 (Sigma Chemical Co.), and then processed for CDP-Star chemiluminescence detection (Tropix).
Kinase Assays
hBUBR1 was immunoprecipitated from lysates containing identical amounts of protein as described above. After washing the pelleted beads five times in NP-40 lysis buffer, the beads were washed three times in kinase buffer (50 mM Tris-Cl, pH 7.4, 10 mM MgCl2, 10 mM β glycerophosphate). Beads were incubated in kinase buffer that contained 50 μM ATP and 20 μCi γ-[32P]ATP at 25°C for 15 min. Cold ATP was added to a final concentration of 0.5 mM and the reaction was allowed to proceed for another 15 min before SDS sample buffer was added. The immunoprecipitation–kinase samples were boiled and separated through a 4–15% gradient gel, fixed, dried, and then subjected to autoradiography. Quantitation was done using a Bio-Imaging analyzer (Fujix BAS1000; Fuji PhotoFilm Co., Ltd.).
Results
hBUBR1 Is Required for the Mitotic Checkpoint
Two complementary approaches were used to examine if hBUBR1 function is required for the mitotic checkpoint in Hela cells. Our first strategy to inhibit hBUBR1 activity was to microinject hBUBR1 antibodies into cells and test the response of the injected cells to the microtubule inhibitor nocodazole. The hBUBR1 antibodies used for the injection experiments have been shown not to cross-react with hBUB1 as determined by Western blots and immunoprecipitations (Chan et al. 1998; Jablonski et al. 1998). Hela cells synchronized at the G1/S boundary were microinjected with hBUBR1 or nonimmune antibodies shortly after they were released from the G1/S boundary. Approximately 2 h before the synchronized cells were expected to enter mitosis, nocodazole was added to the medium and the injected cells were sampled at various times during the ensuing 8 h (Fig. 1a and Fig. b). Mitotic cells were scored by the presence of condensed chromosomes which aggregated near the center of the cell when the spindle is abolished by nocodazole (Fig. 1 A, rows 1 and 2, left). Approximately 12 h after release from the G1/S boundary, most cells have entered mitosis. At this time, there was no significant difference between the mitotic indices of the cells that were injected with hBUBR1 or the nonimmune antibodies. Thus, injection of antibodies did not interfere with mitotic entry. Nonimmune antibodies were diffusely distributed throughout the mitotic cell (Fig. 1 A, row 1, middle) and did not interfere with the localization of hBUBR1 at kinetochores of the unaligned chromosomes (Fig. 1 A, row 1, right). In contrast, the injected hBUBR1 antibodies were concentrated at the kinetochores of the unaligned chromosomes (Fig. 1 A, row 2, middle). The presence of hBUBR1 at kinetochores of these cells was independently confirmed by staining with hBUBR1 antibodies (Fig. 1 A, row 2, right).
Figure 1.
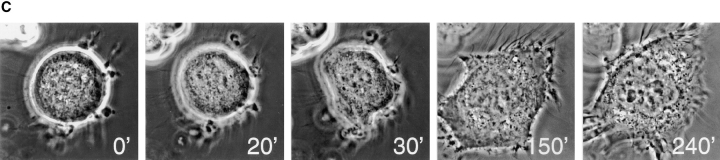
hBUBR1 is essential for cells to arrest in mitosis in the presence of nocodazole. (A) Immunofluorescence staining of Hela cells injected with antibodies and exposed to nocodazole. (row 1 and 2) 12 h after release from the G1/S boundary, nonimmune– and hBUBR1 antibody–injected cells have entered mitosis in the presence of nocodazole with unaligned chromosomes (left). Injected antibodies were visualized with Cy5 anti-rabbit secondary antibodies (middle) and endogenous hBUBR1 was visualized with a rat anti–hBUBR1 antibody and Cy2 anti-rat secondary antibodies (right). (row 3) Cells injected with hBUBR1 (middle) exited mitosis in the presence of nocodazole and formed aberrantly shaped nuclei (left) that contained paired centromeres as determined by ACA and Cy2 anti-human secondary antibody staining (right, arrowheads in the inset). Chromosomes and nuclei were visualized by DAPI staining (left). Images in rows 1 and 2 were captured with a 100× objective, whereas row 3 was visualized with a 40× objective. (B) Histogram comparing the fates of Hela cells injected with nonimmune or hBUBR1 antibodies after exposure to nocodazole. Synchronized Hela cells were injected within 2 h after their release from the G1/S boundary, nocodazole was added 8 h after release from the G1/S boundary and sampled 4, 6, and 8 h later. Hela cells normally enter mitosis between 10–12 h after release from the G1/S boundary. The histogram compares the mitotic (open bars), multi-nucleated (striped bars), and apoptotic (dotted bars) indices of cells injected with hBUBR1 and nonimmune antibodies at 16 h after release from the G1/S boundary. Approximately 80–120 injected cells were counted for each experiment. The plotted values are the means plus SD from at least three experiments. (C) A single cell microinjected with hBUBR1 antibodies and incubated in the presence of nocodazole was followed by time-lapse videomicroscopy shortly after it entered mitosis (time 0). Frames were selected from a 4.5-h long time-lapse experiment. Actual time in minutes is labeled in the bottom right corners of each frame. Bars, 10 μm.
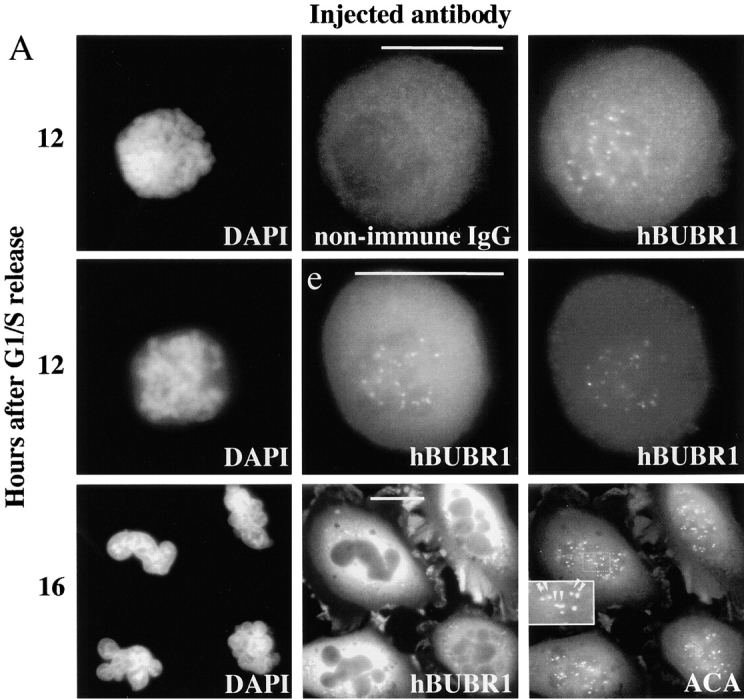
When cells were examined at later times after nocodazole treatment, the mitotic index of cells injected with nonimmune IgG remained high, whereas the mitotic index of cells injected with hBUBR1 antibodies was about threefold lower (Fig. 1 B). The decrease in the mitotic index of cells that were injected with the hBUBR1 antibodies was accompanied by a fivefold rise in the number of interphase cells with aberrantly shaped nuclei relative to the negative control (Fig. 1 A, row 3, and B). The flattened morphology as well as the presence of nuclei indicated that these cells exited mitosis (Fig. 1 A, row 3). The presence of paired centromeres, as revealed by staining with an anticentromere autoimmune (ACA) serum, suggested that either the chromosomes did not separate or had separated but re-replicated (Fig. 1 A, row 3, right, inset).
To directly confirm that inhibition of hBUBR1 abrogated the mitotic checkpoint and caused cells to exit mitosis, time-lapse videomicroscopy was used to follow the fate of nocodazole-treated cells that were microinjected with hBUBR1 antibodies. Fig. 1 C shows that within 30 min after the cell entered mitosis, the cell began to flatten out. 2 h later, the presence of a nucleus demonstrated that it had clearly exited mitosis (Fig. 1 C, 150'). At this level of resolution, we could not determine how rapidly cells injected with hBUBR1 antibodies exited mitosis. Uninjected cells on the same coverslip as well as cells injected with nonimmune antibodies remained blocked in mitosis for up to 16 h (data not shown).
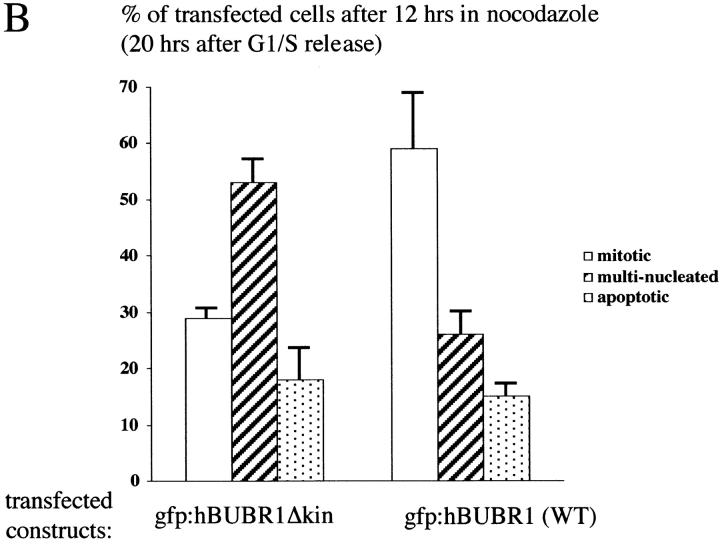
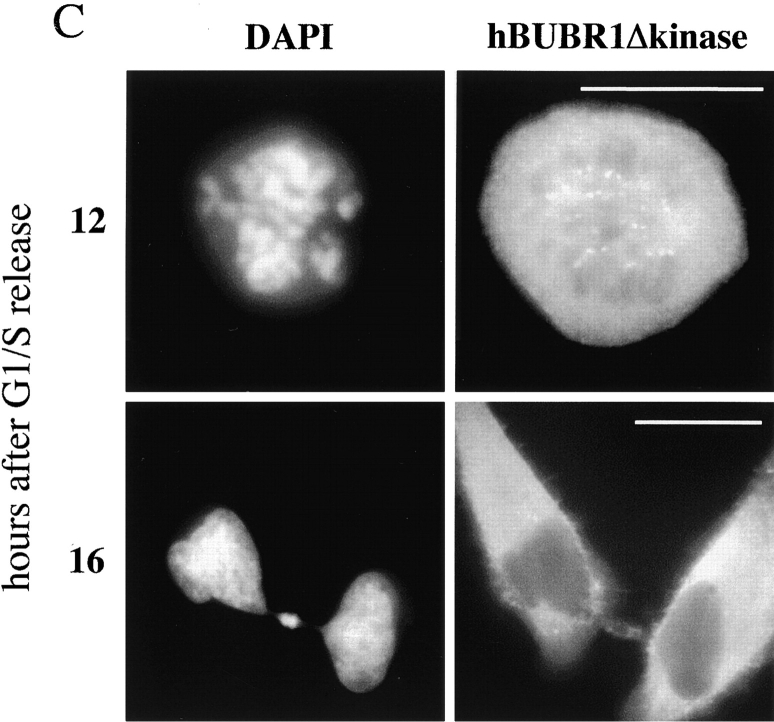
To obtain independent verification of the microinjection results, we attempted to disrupt endogenous hBUBR1 function by targeting an hBUBR1 mutant to kinetochores. We created a mutant by deleting nine amino acids (residues 795–803) from subdomain II of the conserved catalytic core of the hBUBR1 kinase. Western blots confirmed that both mutant and wild-type hBUBR1 were expressed at equivalent levels (data not shown). Both mutant and wild-type gfp:hBUBR1 were transfected into synchronized Hela cells shortly after they were released from the G1/S boundary (Fig. 2). Nocodazole was added ∼2 h before the cells were expected to enter mitosis. By 12 h after release from the G1/S boundary, mitotic cells expressing both mutant and wild-type gfp-hBUBR1 at their kinetochores were identified (Fig. 2 A, rows 1 and 2). After overnight incubation in the presence of nocodazole, the mitotic index of cells expressing the mutant gfp:hBUBR1 was ∼2 times lower than for wild-type gfp:hBUBR1–transfected cells (Fig. 2 B). In concordance with the microinjection results, many of the cells expressing the mutant hBUBR1 exited mitosis and formed aberrant shaped nuclei (Fig. 2 A, row 3, and B). The accumulation of the gfp:hBUBR1Δkinase in the cytoplasm of these aberrant interphase cells is consistent with the distribution of endogenous hBUBR1 in interphase cells as previously reported (Chan et al. 1998). The microinjection and transfection experiments demonstrate that hBUBR1 kinase is likely to be an essential component of the mitotic checkpoint.
Figure 2.
Overexpression of a hBUBR1Δkinase prevents cells from arresting in mitosis in the presence of nocodazole. (A; rows 1 and 2) 12 h after release from the G1/S boundary, cells expressing gfp:hBUBR1 (WT) and the gfp:hBUBR1Δkinase mutant enter mitosis in the presence of nocodazole. Both the gfp:hBUBR1 (WT) and the gfp:hBUBR1Δkinase mutant were concentrated at kinetochores (right). (row 3) After overnight incubation in nocodazole, cells that expressed the gfp:hBUBR1Δkinase mutant had exited mitosis and formed aberrantly shaped nuclei (left). Gfp:fusion constructs were directly visualized by autofluorescence. Chromosomes and nuclei were visualized with DAPI. (B) Fates of Hela cells that expressed wild-type gfp:hBUBR1 or gfp:hBUBR1Δkinase and exposed to nocodazole. Synchronized Hela cells were transfected ∼2 h after release from the G1/S boundary with wild-type gfp:hBUBR1 or gfp:hBUBR1Δkinase; after 5 h, the transfection mix was removed and nocodazole was added. The mitotic (open bars), multinucleated (striped bars), and apoptotic (dotted bars) indices of wild-type gfp:hBUBR1 and gfp:hBUBR1Δkinase after overnight incubation (∼20 h after release from the G1/S boundary) in nocodazole was compared. Approximately 120–150 transfected cells were counted and scored for each experiment. The plotted values are the means plus SD from at least three experiments. Bars, 10 μm.

hBUBR1 Is Essential for a Normal Mitosis
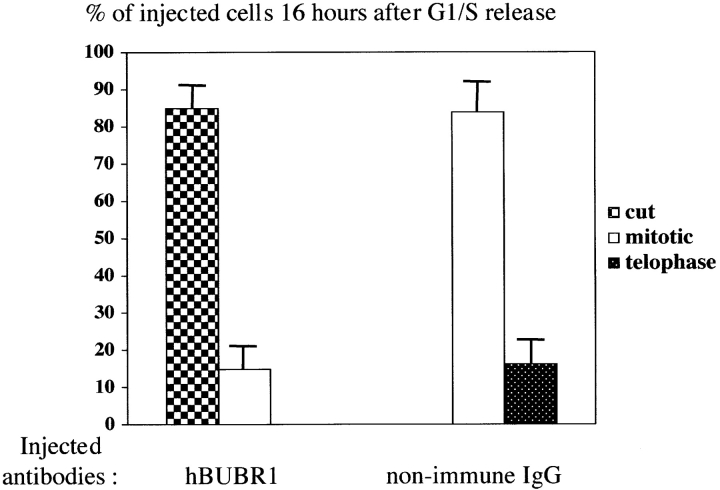
We have previously shown that hBUBR1 can form a complex with the kinetochore motor CENP-E (Chan et al. 1998). If hBUBR1 can monitor kinetochore microtubule interactions mediated by CENP-E as part of its normal checkpoint function, disruption of hBUBR1 might cause cells to exit mitosis prematurely. Alternatively, hBUBR1 might regulate CENP-E functions that are important for chromosome alignment. In this case, loss of hBUBR1 should interfere with CENP-E functions at the kinetochore and block cells in mitosis with unaligned chromosomes (Schaar et al. 1997). To test the importance of hBUBR1 during normal mitosis, the fates of synchronized cells that were either injected with hBUBR1 antibodies or transfected with the gfp:hBUBR1Δkinase were determined. Approximately 12 h after release from the G1/S boundary, cells that were injected with nonimmune IgG were found in prometaphase and metaphase with the majority being at metaphase (Fig. 3 A, row 1). Immunofluorescence staining revealed that the metaphase cells exhibited detectable levels of hBUBR1 at their kinetochores despite the presence of nonimmune antibodies throughout the cell (Fig. 3 A, row 1, middle and right). For cells injected with the hBUBR1 antibody, most of the mitotic cells were in prometaphase. The injected hBUBR1 antibodies were found throughout the cell including kinetochores (Fig. 3 A, row 2, middle). The presence of hBUBR1 antibodies at the kinetochores did not interfere with CENP-E localization (Fig. 3 A, row 2, right). Although a few metaphase cells injected with hBUBR1 were seen, we consistently identified anaphase cells that contained lagging chromosomes (Fig. 3 A, row 3, left). These cells appeared to be in late anaphase as they contained a mature cleavage furrow that was concentrated with CENP-E (Fig. 3 A, row 3, right). The presence of lagging chromosomes suggested that these cells exited mitosis in the presence of unaligned chromosomes and gave rise to the high frequency of aberrantly divided cells that accumulated at later time points (Fig. 3 A, row 4, and B). The nuclei of the cells injected with hBUBR1 antibodies were abnormally shaped and DNA was invariably found in between the two divided cells (Fig. 3 A, row 4, left). The DNA in between the cells was derived from chromosomes because they contained centromeres as determined by ACA staining (Fig. 3 A, row 5, right, inset). However, we cannot distinguish whether the foci of ACA staining represent separated centromeres or fragmented centromeres. This cut phenotype was specific for hBUBR1 antibodies as this was not observed in uninjected cells or cells injected with nonimmune antibodies. In fact, hBUBR1 antibody injection did not produce any normal telophase cells (Fig. 3 B).
Figure 3.
hBUBR1 is essential for cells to proceed normally through mitosis. (A) Synchronized Hela cells released from the G1/S boundary were injected with nonimmune or hBUBR1 antibodies and sampled at 12 or 16 h later. (rows 1–3) 12 h after release from the G1/S boundary, injected cells entered mitosis. (row 1) Metaphase cell injected with nonimmune IgG (middle) stained for endogenous hBUBR1 (right). (rows 2 and 3) Prometaphase and anaphase cells injected with hBUBR1 antibodies (middle) and stained for CENP-E (right). (row 4) 16 h after release from the G1/S boundary, hBUBR1 injected cells (middle) divided with lagging chromosomes trapped in the cleavage furrow. Centromeres (arrowheads) stained with ACA and visualized with Texas red anti-human secondary antibodies. Insets in rows 3 and 4 are longer exposures and enlargements of boxed areas. Injected antibodies were visualized with Cy5 anti-rabbit secondary antibody (middle). Endogenous hBUBR1 was stained with rat anti–hBUBR1 and detected with Cy2 anti-rat (row 1, right). CENP-E was stained with mouse monoclonal anti-CENP-E (mAb177) and detected with Cy2 anti-mouse secondary antibodies (rows 2 and 3, right). Chromosomes and nuclei were stained with DAPI (left). (B) 16 h after release from the G1/S boundary, nonimmune IgG– and hBUBR1 antibody–injected cells were scored for mitotic (open bar), normal telophase (dotted bar) or aberrantly divided cut (checkered bar) based on a combination of DAPI staining and phase-contrast images. Histogram compares relative percentages of each of the three phenotypes. The plotted values are the means plus SD from at least three experiments. (C) Cells expressing gfp:hBUBR1Δkinase exit mitosis with prematurely. Synchronized Hela cells transfected with gfp:hBUBR1Δkinase enter mitosis (top row) 12 h after release from the G1/S boundary. By 16 h, most cells expressing the kinase mutant produced cells that divided with lagging chromosomes trapped in the cleavage furrow (bottom). Bars, 10 μm.
Similar results were obtained when synchronized Hela cells were transfected with the mutant hBUBR1Δkinase. In transfected cells that had progressed into mitosis, the mutant hBUBR1 was concentrated at kinetochores (Fig. 3 C, top right). At later times, there was a large increase in aberrantly divided cells with the cut phenotype (Fig. 3 C, bottom left). Both mock and wild-type gfp:hBUBR1–transfected cells divided normally and did not exhibit the cut phenotype (data not shown). The combined data suggest that disruption of hBUBR1 function caused the cells to exit mitosis in the presence of unaligned chromosomes.
hBUBR1 Monitors CENP-E Kinetochore Functions
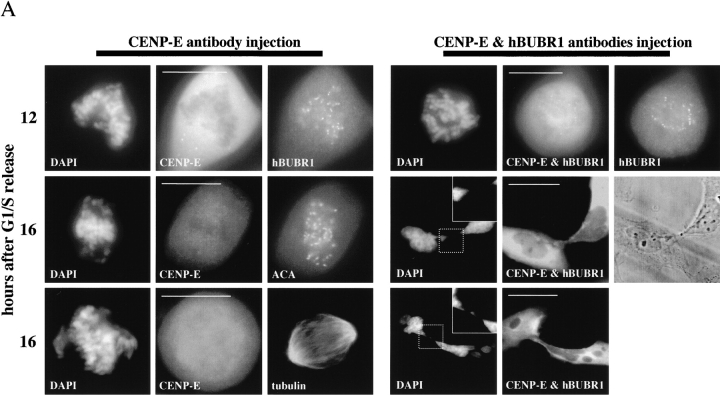
We have previously shown that disruption of CENP-E functions at kinetochores prevented chromosome alignment and arrested cells in mitosis for >12 h (Schaar et al. 1997). To test whether hBUBR1 is required for this checkpoint, we coinjected hBUBR1 and CENP-E antibodies into synchronized Hela cells, and then compared the fates of these cells to those that were injected with CENP-E antibodies alone. Injection of CENP-E antibodies blocked the assembly of CENP-E onto kinetochores (Fig. 1 A, left and middle) but did not interfere with kinetochore localization of hBUBR1 (Fig. 4 A, left gallery, row 1) or the distribution of other centromere components that were recognized by ACA (Fig. 4 A, left gallery, row 2). As shown previously, the mitotic block induced by injection of CENP-E antibodies is the direct result of disruption of kinetochore function and not bipolar spindle formation (Fig. 4 A, left gallery, row 3).
Figure 4.
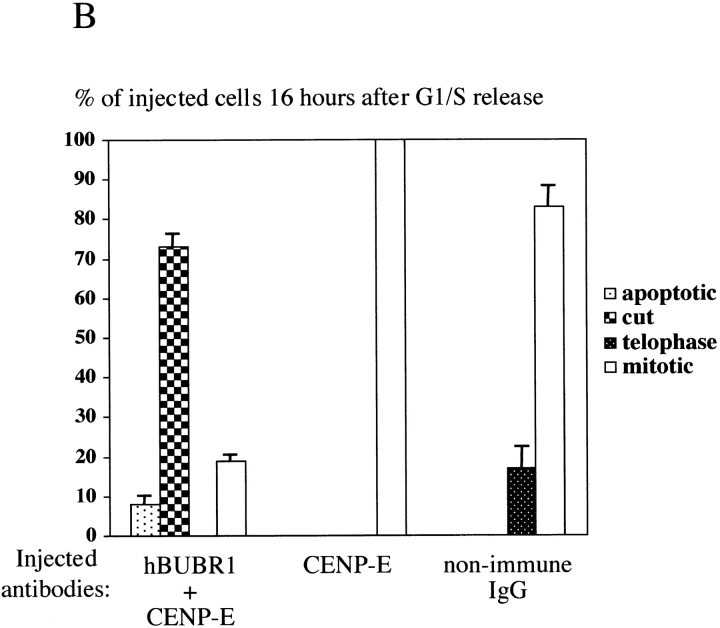
hBUBR1 is required for mitotic arrest resulting from loss of CENP-E functions at kinetochores. Synchronized Hela cells were injected with rabbit anti–CENP-E and hBUBR1 antibodies and sampled at 12 or 16 h after release from the G1/S boundary. (A) Immunofluorescence staining of injected cells at these time points. (left gallery) Cells injected with CENP-E antibodies arrest in mitosis with unaligned chromosomes. Cells were stained for the injected CENP-E antibodies (middle), localization of endogenous hBUBR1, ACA, and tubulin (rows 1–3, respectively, right). (right gallery) Cells coinjected with CENP-E and hBUBR1 antibodies (middle) enter mitosis with hBUBR1 at kinetochores (right, row 1). At the 16-h time points (rows 2 and 3), the cells coinjected with CENP-E and hBUBR1 antibodies exit mitosis with unaligned chromosomes to produce divided cells with lagging chromosomes (inset in left) and multiple nuclei that reform around unsegregated or incompletely separated chromosomes. Phase-contrast image show cells were tethered at the stembody (row 2, right). Injected antibodies were stained with Cy5 anti-rabbit secondary antibodies (middle columns of both galleries). Endogenous hBUBR1 was stained with anti-rat hBUBR1 and visualized with Cy2 anti-rat secondary antibody. ACA was visualized with Cy2 anti-human secondary antibodies. Microtubules were detected with antitubulin mAbs and Cy2 anti-mouse secondary antibodies. Chromosomes and nuclei were stained with DAPI (left column). (B) Comparison of the fates of cells injected with CENP-E antibodies, hBUBR1+CENP-E antibodies, and nonimmune IgG. After cells were released from the G1/S boundary for 16 h, the percentage of cells injected with the various antibodies that were in mitosis (open bars), normal telophase (white dotted bar), cut (checkered bar), and apoptosis (black dotted bar) were quantitated and compared. The plotted values are the means plus SDs from at least three experiments. Bars, 10 μm.
Cells coinjected with CENP-E and hBUBR1 antibodies entered mitosis. Immunofluorescence staining confirmed that hBUBR1 remained at kinetochores (Fig. 4 A, right gallery, row 1, right) of the injected cells. 16 h after release from the G1/S boundary, we counted cells that were either in mitosis, telophase, aberrantly divided cut, or apoptosis. Cells injected with just CENP-E antibodies were arrested in mitosis with unaligned chromosomes (Fig. 4 B). No cells were found to be apoptotic, in telophase, or cut. Examination of the cells that were coinjected with hBUBR1 and CENP-E antibodies showed an ∼5-fold reduction in the number of mitotic cells relative to cells injected with CENP-E alone (Fig. 4 B). The reduction of mitotic cells was accompanied by a large increase in the number of aberrantly divided cells that exhibited the cut phenotype (Fig. 4 A, right gallery, rows 2 and 3) along with a slight increase in apoptotic cells. Normal telophase cells that were coinjected with the hBUBR1 and CENP-E antibodies were not found. These data showed that hBUBR1 is an essential component of the mitotic checkpoint that is sensitive to kinetochore functions that were specified by CENP-E.
Characterization of hBUBR1 Protein Expression and Kinase Activity
Having established that hBUBR1 is an essential component of the mitotic checkpoint, we wanted to investigate its mechanism of action by characterizing its expression and kinase activity as a function of the cell cycle. hBUBR1 was immunoprecipitated from K562 erythroleukemic cells and Hela cells that were separated into different phases of the cell cycles by centrifugal elutriation. Portions of the immunocomplexes were used to determine steady-state levels of protein and kinase activity. We assayed the immunoprecipitated hBUBR1 for autokinase activity as a recent study showed that a transfected hBUBR1 exhibited autokinase activity (Taylor et al. 1998). However, hBUBR1 isolated from K562 and Hela (data not shown) exhibited no detectable autokinase activity (Fig. 5 A, top) or kinase activities towards exogenous substrates (data not shown) even though it was expressed throughout the cell cycle of both cell types (Fig. 5 A, middle and bottom). hBUBR1 levels remained fairly constant throughout the cell cycle of K562 cells, whereas hBUBR1 levels in Hela cells fluctuated with the cell cycle. hBUBR1 level was lowest in G1 and steadily increased as cells progressed towards mitosis.
Figure 5.

hBUBR1 expression, phosphorylation, and kinase activity during the cell cycle. (A) K562 and Hela cells were separated according to their position in the cell cycle by centrifugal elutriation. hBUBR1 was immunoprecipitated from equal amounts of protein from each fraction, and then tested for autokinase activity (top) or probed with hBUBR1 antibodies to compare its cell cycle expression profile (middle and bottom). (B) Comparison of autokinase activity (top) and levels of hBUBR1 (middle) and hBUB3 found in hBUBR1 immunoprecipitates from asynchronous interphase cells (A) or cells blocked in mitosis by nocodazole (N). (C) hBUBR1 is hyperphosphorylated in mitosis. hBUBR1 immunoprecipitated from mitotically blocked Hela cells were incubated in the presence (lane 1) or absence (lane 2) of lambda protein phosphatase (λPP). (lanes 3–8) Comparison of hyperphosphorylation status of hBUBR1 in normal mitotic Hela cells (lane 3, M), nocodazole-blocked mitotic cells (lane 4, N), or cells released from the nocodazole block for 1, 2, 3, and 4 h (lanes 5–8, respectively). Cells were determined to have reentered G1 by phase-contrast microscopy.
If hBUBR1 is a mitosis-specific kinase, its activity would have escaped detection because the vast majority of the cells that were elutriated into the late fractions were in G2 and only a very minor fraction was in mitosis. Therefore, we compared the hBUBR1 kinase activity between interphase and mitotically arrested Hela cells. hBUBR1 autokinase activity remained undetectable in interphase cells, but was highly active in mitotically arrested cells (Fig. 5 B). The same profile of hBUBR1 kinase activity was obtained with exogenous substrates such as myelin basic protein and a carboxyl-terminal fragment of CENP-E (data not shown). The increase in hBUBR1 kinase activity in mitotic cells was not due to increased expression of the protein or its association with the hBUB3 subunit relative to interphase cells (Fig. 5 B, bottom). However, hBUBR1 mobility was reduced in mitotically blocked cells relative to the interphase cells (Fig. 5 B, middle right). The upshift was due to phosphorylation as the electrophoretic mobility of hBUBR1 was no longer retarded after phosphatase treatment (Fig. 5 C, compare lanes 1 and 2). In vivo labeling experiments showed that hBUBR1 was also phosphorylated during interphase but became hyperphosphorylated in mitosis (data not shown). To further test if the hyperphosphorylation of hBUBR1 was related to the mitotic checkpoint, we compared hBUBR1 in mitotically blocked cells to cells that were released from the block and had exited mitosis (Fig. 5 C, right). The majority of hBUBR1 was hyperphosphorylated in mitotically blocked cells relative to normal mitotic cells. 1 h after their release from the mitotic block, hBUBR1 was no longer hyperphosphorylated (Fig. 5 C, compare lanes 4 and 5).
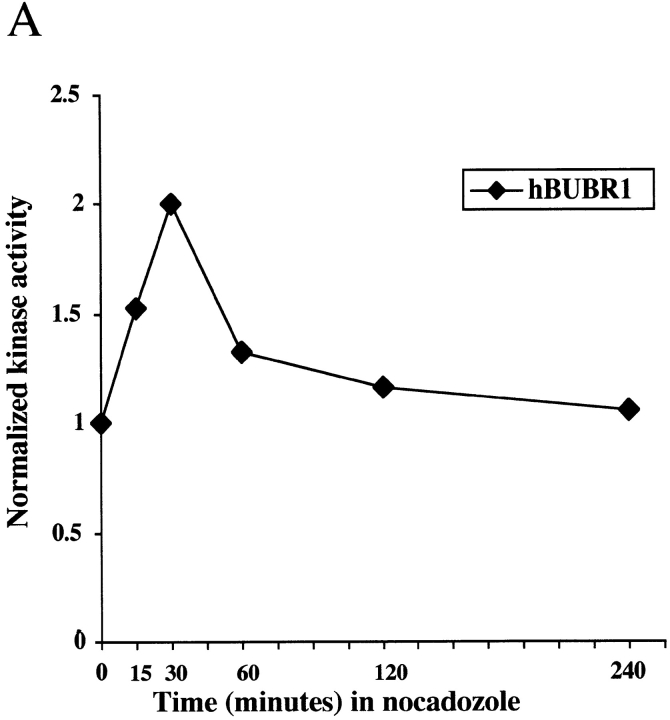
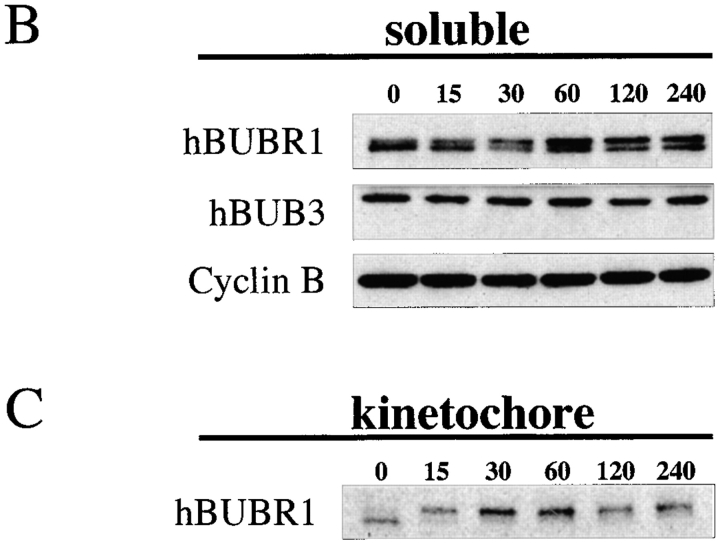
In the previous experiments, hBUBR1 kinase activity was determined in cells that had already been blocked in mitosis for many hours. As a checkpoint kinase, hBUBR1 should respond to spindle defects even after cells have reached metaphase. To test this, we isolated mitotic Hela cells by mechanical shakeoff, and then exposed them to nocodazole for various times. Microscopic analysis revealed that >90% of the mechanically detached cells were in metaphase with some prometaphase and very few (<1%) anaphase cells. hBUBR1 was immunoprecipitated from cell lysates that were harvested at different times of incubation with nocodazole, and then assayed for kinase activities and protein expression (Fig. 6 A). Comparison of hBUBR1 kinase activity between metaphase cells and those that were exposed to nocodazole showed that kinase activity was rapidly stimulated within 15 min of nocodazole treatment (Fig. 6 A). At this time, the spindle was absent and chromosomes were unaligned (data not shown). hBUBR1 kinase activity reached a peak at 30 min after nocodazole treatment and gradually declined. By 4 h, hBUBR1 kinase activity had decayed to a level that was slightly above that of normal metaphase cells. During the course of the experiment, cells were confirmed to be blocked in mitosis by phase-contrast microscopy and expression levels of cyclin B (Fig. 6 B, bottom). Western blots of the hBUBR1 immunoprecipitates revealed that the levels of hBUBR1 and hBUB3 remained fairly constant over the course of the experiment (Fig. 6 B). The increase of kinase activity was not due to changes in levels of hBUBR1 or its subunit, hBUB3. However, we detected an increase in the level of hyperphosphorylated hBUBR1 (as determined by retarded migration) between metaphase cells and those that were incubated in nocodazole (Fig. 6 B, compare top bands of 0 min and 120 min). The increase in the level of hyperphosphorylated hBUBR1 did not correlate with its peak kinase activities (Fig. 6a and Fig. B, compare 30 min and 120 min time points).
Figure 6.
hBUBR1 kinase in metaphase Hela cells are stimulated by disruption of the spindle. Mitotic shakeoff cells that were nearly uniformly at metaphase were exposed to nocodazole for the times indicated and hBUBR1 complexes were immunoprecipitated from cell lysates and probed for autokinase activity. (A) hBUBR1 autokinase activity from each time point was quantitated with a PhosphorImager and normalized to metaphase cells (time 0). This is a valid comparison as equal amounts of hBUBR1 were used for the kinase assay. (B) Western blots of hBUBR1 immunoprecipitates probed with hBUBR1 (top) and hBUB3 (middle). Lysates obtained from the same experiment were separately probed for cyclin B expression (bottom). (C) Western blot of the insoluble chromosome fractions obtained from the experiment described above was probed for hBUBR1.
The hBUBR1 kinase activities described above were derived from the soluble pool of proteins and excluded those that were associated with kinetochores. As the kinetochore is a critical component of the checkpoint, we probed the insoluble fractions that were enriched in chromosomes for hBUBR1 to see how this population responded to spindle disruption (Fig. 6 C). Western blots revealed that hBUBR1 levels increased slightly after 30 and 60 min in nocodazole, but returned to the initial level after 2 and 4 h of nocodazole treatment. The most dramatic change was that hBUBR1 was quantitatively hyperphosphorylated within 15 min of spindle disruption and this high level of phosphorylation was maintained throughout the block.
hBUBR1 Associates with the Cyclosome/APC
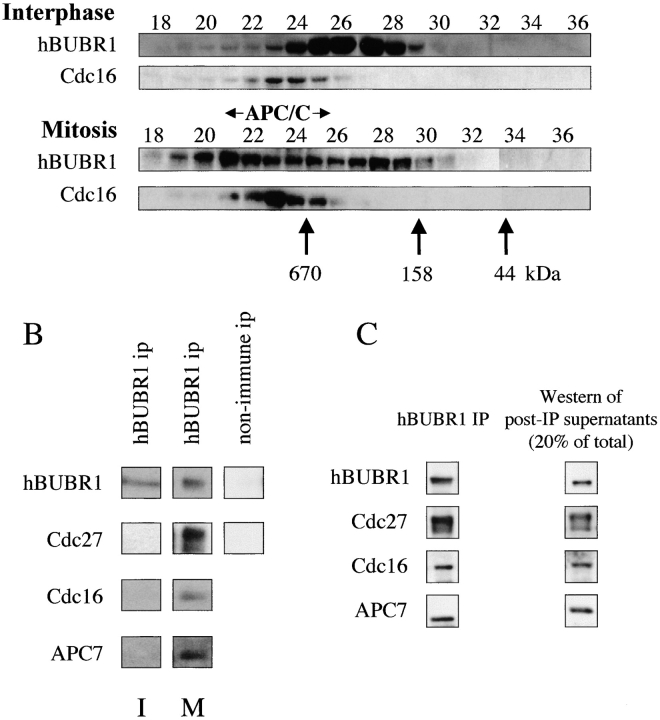
Next, we examined the native size of hBUBR1 between interphase and mitotically blocked cells. Gel filtration chromatography revealed that hBUBR1 exists primarily in an ∼500-kD complex in interphase cells (Fig. 7 A, top). In mitotically arrested cells, hBUBR1 was associated with the ∼500-kD complex, but was also found in a larger complex that overlapped with the cyclosome/APC (Fig. 7 A, bottom). The possibility that hBUBR1 was associated with the cyclosome/APC was tested by immunoprecipitating hBUBR1 from the fractions containing the cyclosome/APC and probing for the presence of the APC subunits hscdc27, hscdc16 (Tugendreich et al. 1995), and APC7 (Yu et al. 1998). The results showed that all three of the APC subunits, coimmunoprecipitated with hBUBR1 in the mitotically blocked lysates (Fig. 7 B, middle). Although some hBUBR1 was detected in the cyclosome/APC containing fractions in interphase cells (Fig. 7 A, top), this was not associated with the cyclosome/APC (Fig. 7 B, left), and most likely resulted from smearing of the 500-kD complex in the gel filtration column. This finding along with the absence of cyclosome/APC subunits in immunoprecipitates from nonimmune IgG demonstrated that the cyclosome/APC specifically associates with hBUBR1 only in mitosis.
Figure 7.
hBUBR1 associates with the cyclosome/APC during mitosis. (A) Lysates prepared from Hela cells enriched in G1 by centrifugal elutriation (interphase) or blocked in mitosis with nocodazole were separated on a Superose 6 column by FPLC. Individual fractions were probed with hBUBR1 antibodies (top) and hscdc16 antibodies (bottom). Fractions 21–25 contain the cyclosome/APC as determined by size markers and presence of cyclosome/APC subunits, hscdc16 (bottom), hscdc27, and APC7 (not shown). The column was calibrated with three different size markers as indicated by the arrows. (B) Fractions 21–25 from the interphase and mitotic gel filtration columns that contained peak levels of cyclosome/APC were pooled and immunoprecipitated with hBUBR1 antibodies (left and middle columns) or nonimmune antibodies (right column, mitotic samples only). Immunoprecipitates were probed with antibodies to hBUBR1 and three different cyclosome/APC subunits. (C) hBUBR1 immunoprecipitates from nocodazole-blocked mitotic Hela lysates were probed for hBUBR1, and the cyclosome/APC subunits hscdc27, hscdc16, and APC7. 20% of the remaining supernatant (after removal of the IP) was also probed for the presence of these proteins by Western blots. Samples were probed at the same time and exposed to film for identical times. Exposures were chosen that had not saturated the film. Relative band intensities were estimated. For example, the Western blots indicated that there was approximately twice the amount of hBUBR1 in the IP than in the supernatant. Taking into consideration that the hBUBR1 signal in the supernatant represented only one fifth of the input, the ratio of hBUBR1 between IP and supernatant was ∼2:5. From this ratio, we calculated that 28% of the total pool of hBUBR1 was found in the IP. The broad cdc27 bands in the mitotic samples has been shown to be hyperphosphorylation (Yu et al. 1998).
We estimated the proportion of the cellular pool of the cyclosome/APC that was associated with hBUBR1 in mitotically arrested cells by comparing the relative levels of various APC subunits in hBUBR1 immunoprecipitates and the remaining supernatant. When ∼30% of the total pool of hBUBR1 was immunoprecipitated from mitotic lysates, roughly 16–20% of the total pool of hscdc16, hscdc27, and APC7 was also found in the precipitate. Assuming that the remaining pool of hBUBR1 that was not immunoprecipitated can also associate with the APC, we can extrapolate that ∼50% of the total pool of APC can be associated with hBUBR1. Since the gel filtration data show that ∼50% of the total pool of hBUBR1 comigrates with the APC, we estimate that about 25% of the total pool of APC is associated with hBUBR1 in mitosis.
Discussion
hBUBR1 Is Essential for the Mitotic Checkpoint
The results from this study provide details about the function and biochemical properties of hBUBR1 kinase. We demonstrate that hBUBR1 is essential for the mitotic checkpoint as disruption of hBUBR1 function prevented cells from arresting in mitosis in the presence of nocodazole. Instead, the cells exited mitosis without dividing and formed highly aberrant nuclei that contained unsegregated chromosomes. The behavior of hBUBR1-defective cells is very similar to that exhibited by cells with defective MAD1, MAD2, or BUB1 functions. Depletion of MAD2 and MAD1 from Xenopus egg extracts resulted in the inability to block mitosis in the presence of nocodazole (Chen et al. 1996, Chen et al. 1998). Introduction of anti-MAD2 antibodies into cells abrogated their ability to arrest in mitosis in the presence of nocodazole (Li and Benezra 1996) or taxol (Waters et al. 1998). Overexpression of a mutant form of mBUB1 (Taylor and McKeon 1997) and hBUB1 (Cahill et al. 1998) prevented cells from arresting in mitosis in the presence of nocodazole.
Our studies also showed that hBUBR1 provided an essential function during normal mitosis by preventing cells from prematurely exiting mitosis. It was unlikely that disrupting hBUBR1 functions directly interfered with chromosome alignment since we were able to identify metaphase cells that were injected with hBUBR1 antibody or expressed the hBUBR1 kinase–defective mutant. As the frequency of such metaphase cells was rare, the most likely fate of many of the prometaphase cells that we saw was premature exit from mitosis. This interpretation is supported by the identification of aberrant anaphase cells with lagging chromosomes. The lagging chromosomes most likely were chromosomes that had not achieved alignment at the metaphase plate when cells decided to exit mitosis. This defect would explain the high frequency of abnormally divided cells that contained chromatin trapped by the cleavage furrow. At present, we have not directly determined precisely how rapidly cells progress through mitosis when hBUBR1 function is inhibited. However, studies have shown that disruption of mBUB1 appeared to accelerate cells through mitosis by ∼25 min (Taylor and McKeon, 1998). Likewise, microinjection of MAD2 antibodies into prophase human keratinocyte cells caused them to precociously exit mitosis before chromosomes were properly aligned (Gorbsky et al. 1998). High resolution videomicroscopy showed that disruption of MAD2 in human keratinocyte cells accelerated mitotic progression by 6.5 min. Despite the similarity in the outcomes of all these studies, it remains to be seen how MAD2, BUB1, and BUBR1 relate to each other at a mechanistic level. As all of these proteins are localized at the kinetochore, they may act in parallel by monitoring different activities of the kinetochore or work as an integrated complex to generate the wait anaphase signal from the kinetochore. These two possibilities are supported by the fact that the two BUB1-related kinases do not share overlapping functions as neither one is able to maintain the mitotic checkpoint alone.
hBUBR1 Is Sensitive to CENP-E Functions at Kinetochores
Our data show that one checkpoint function of hBUBR1 is to monitor kinetochore functions that are specified by CENP-E. When kinetochores are depleted of CENP-E, cells arrest in mitosis with unaligned chromosomes but a normal spindle. Our data suggest that hBUBR1 activity at the kinetochore is important for generating the wait anaphase signal. This possibility is supported by the finding that hBUBR1 was still present on kinetochores that lacked CENP-E. Furthermore, microinjection of hBUBR1 antibodies abrogated the mitotic arrest and caused cells to exit mitosis. If hBUBR1 was contributing to the wait anaphase signal from the CENP-E–depleted kinetochores, it must be able to do so when it is not in direct contact with CENP-E. Under normal circumstances, we speculate that interactions between CENP-E and microtubules can directly regulate hBUBR1 activity by altering its interactions with CENP-E.
Although such a feedback mechanism can link kinetochore attachments to the checkpoint, the threshold level of CENP-E activity that is required to activate or inactivate hBUBR1 activity remains to be determined. We have shown that when kinetochore–microtubule attachments of metaphase cells are dissolved by nocodazole, the most dramatic change was the quantitative hyperphosphorylation of the kinetochore-bound hBUBR1. Perhaps the phosphorylation state of hBUBR1 at the kinetochore is related to the number of kinetochore microtubules that are attached to CENP-E and the level of tension that results from these interactions. At metaphase, the levels of both CENP-E and hBUBR1 have been shown to be reduced (Chan et al. 1998; Jablonski et al. 1998) along with other checkpoint proteins. Under these circumstances, there may be insufficient hBUBR1 kinase activity that is generated from the kinetochore to sustain the checkpoint components that lie downstream of the kinetochore. Studies have shown that it takes ∼30 min from the time the last chromosome becomes aligned at the metaphase plate until the onset of anaphase (Rieder et al. 1995). This may be the time that is required for the checkpoint signal to fall and to allow cyclosome/APC activity to reach a critical level that is necessary to drive cells into anaphase. Before this critical level of cyclosome/APC activity is reached, the checkpoint can be reactivated by unattached kinetochores to once again inhibit cyclosome/APC. Although we proposed that hBUBR1 kinase activity is sensitive to the mechanical activities of the kinetochore, activation of its kinase may require additional biochemical changes such as mitosis-specific phosphorylations. As kinetochores are now known to also contain cdc2 (Rattner et al. 1990), MAP kinase (Shapiro et al. 1998; Zecevic et al. 1998), and hBUB1 (mBUB1) kinases (Taylor and McKeon 1997; Jablonski et al. 1998), any one of these may be important for regulating hBUBR1 kinase at the kinetochore.
hBUBR1 Is Associated with the Cyclosome/APC in Mitosis
We infer from the interaction between hBUBR1 and cyclosome/APC that hBUBR1 might provide global checkpoint functions by directly inhibiting cyclosome/APC. This is supported by the finding that partially purified hBUBR1 from Hela cells inhibited ubiquitination activity of the APC in vitro (Sudakin, V., and T. Yen, unpublished results). By comparing the amount of APC that coimmunoprecipitated with hBUBR1 to that in the remaining supernatant, we estimated that 25% of the total pool of APC was bound to hBUBR1. As MAD2–p55cdc was also found to be associated with only a fraction of the total pool of APC, it is possible that the hBUBR1–APC complex represents a different subpopulation of the APC. As hBUBR1 kinase activity is stimulated when the spindle is disrupted, hBUBR1 may directly phosphorylate and inhibit cyclosome/APC activity or phosphorylate specific substrates so that they are no longer recognized by the cyclosome/APC.
As proposed for MAD2–p55cdc (Gorbsky et al. 1998), the interactions between hBUBR1 and the cyclosome/APC should be inherently unstable so that cells can proceed into anaphase when the checkpoint is extinguished. As hBUBR1 appears to form a fairly stable complex with the cyclosome/APC in cells arrested in mitosis, its kinase activity may be labile. This is supported by the observation that the in vitro hBUBR1 kinase activity had decayed when it was isolated from cells whose spindle had been disrupted by nocodazole for an hour as compared with the mitotic cells that were examined 30 min earlier. As the phosphorylation status of the soluble pool of hBUBR1 did not strongly correlate with its kinase activity, additional factors may influence hBUBR1 kinase activity. Despite the decay in the soluble pool of hBUBR1 kinase activity, sufficient kinase activity was probably maintained in the mitotically arrested cells by the continued presence of unattached kinetochores.
Proposed Mechanism of Action of hBUBR1
The cumulative results obtained for hBUBR1 suggest that it can provide checkpoint functions at the local and global levels. In the simplest model, unattached kinetochores contain active hBUBR1 kinase that can rapidly activate the soluble pool of hBUBR1 through autophosphorylation. This then leads to inhibition of the cyclosome/APC and exit from mitosis. This model of hBUBR1 function shares features proposed for MAD2–p55cdc and MAD2/MAD1 (Chen et al. 1998; Gorbsky et al. 1998; Kallio et al. 1998). The overall view is that a soluble pool of checkpoint proteins is used to block cyclosome/APC activity, but the inhibitory status of these checkpoint proteins must be maintained through interactions with the unattached kinetochore. We propose that hBUBR1 can accomplish this through an autokinase loop that may also include MAD2–p55cdc complex. As MAD2 is not known to be hyperphosphorylated in mitotically blocked cells, hBUBR1 might phosphorylate one of its associated subunits such as p55CDC (Weinstein 1997) or MAD1 (Chen et al. 1998; Jin et al. 1998). Biochemical analysis of yeast MAD1 showed that it is hyperphosphorylated in mitotically arrested cells and that this is dependent on the MPS1 and BUB1 kinases (Hardwick and Murray 1995). Although vertebrate MAD1 does not undergo phosphorylation-dependent shifts in its migration in SDS-PAGE (Chen et al. 1998; Jin et al. 1998), it does not eliminate the possibility that MAD1 is hyperphosphorylated in mitosis. It is possible that in vertebrate systems, MAD1, MAD2, p55CDC, and hBUB1 act in parallel with hBUBR1 or through more complex interactions. Clearly, it will be important to clarify the biochemical relationship amongst these proteins to resolve whether they act together or along separate pathways in vertebrate cells.
Acknowledgments
The authors would like to thank B. Conner for excellent technical support, and Dr. M. Campbell (both from Fox Chase Cancer Center) for suggestions and comments.
This work was supported by United States Public Health Services grant GM44762: American Cancer Society, core grant CA06927, and an appropriation from the Commonwealth of Pennsylvania. T. Yen is supported by a Leukemia Society Scholar's Award. V. Sudakin is supported by a Human Frontier postdoctoral fellowship.
Footnotes
1.used in this paper: ACA, anticentromere autoimmune antibodies; APC, anaphase-promoting complex; CENP-E, centromere protein E; DAPI, 4′,6-diamidino-2-phenylindole
References
- Basu J., Logarinho E., Herrmann S., Bousbaa H., Li Z., Chan G.K., Yen T.J., Sunkel C.E., Goldberg M.L. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not Zw10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer C., Yu J., Riggins G.J., Willson J.K., Markowitz S.D., Kinzler K.W., Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- Chan G.K.T., Schaar B.T., Yen T.J. Characterization of the kinetochore-binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Waters J.C., Salmon E.D., Murray A.W. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chen R.H., Shevchenko A., Mann M., Murray A.W. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson I.A., Roth S., Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae . J. Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gately D.P., Hittle J.C., Chan G.K.T., Yen T.J. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol. Biol. Cell. 1998;9:2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen R.H., Murray A.W. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 1998;141:1193–1205. doi: 10.1083/jcb.141.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S.K., Quinn A.M. Protein kinase catalytic domain sequence databaseidentification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G., Murray A.W. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J. Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hunter T. A thousand and one protein kinases. Cell. 1987;50:823–829. doi: 10.1016/0092-8674(87)90509-5. [DOI] [PubMed] [Google Scholar]
- Hwang L.H., Lau L.F., Smith D.L., Mistrot C.A., Hardwick K.G., Hwang E.S., Amon A., Murray A.W. Budding yeast Cdc20a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Inoue S., Salmon E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 1995;6:1619–1640. doi: 10.1091/mbc.6.12.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski S.A., Chan G.K.T., Cooke C.A., Earnshaw W.C., Yen T.J. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Jin D.Y., Spencer F., Jeang K.T. Human T cell leukemia virus type 1 oncoprotein Tax targets the human mitotic checkpoint protein MAD1. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- Kallio M., Weinstein J., Daum J.R., Burke D.J., Gorbsky G.J. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J. Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Lin D.P., Matsumoto S., Kitazono A., Matsumoto T. Fission yeast Slp1an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- King R.W., Deshaies R.J., Peters J.M., Kirschner M.W. How proteolysis drives the cell cycle. Science. 1996;274:1652–1659. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A.W. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X., Nicklas R.B. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y., Benezra R. Identification of a human mitotic checkpoint genehsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li Y., Gorbea C., Mahaffey D., Rechsteiner M., Benezra R. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl. Acad. Sci. USA. 1997;94:12431–12436. doi: 10.1073/pnas.94.23.12431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R.B. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Rattner J.B., Lew J., Wang J.H. p34cdc2 kinase is localized to distinct domains within the mitotic apparatus. Cell Motil. Cytoskeleton. 1990;17:227–235. doi: 10.1002/cm.970170309. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Schultz A., Cole R., Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Cole R.W., Khodjakov A., Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Farr K.A., Hoyt M.A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K.T., Maddox P., Salmon E.D., Yen T.J. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro P.S., Vaisberg E., Hunt A.J., Tolwinski N.S., Whalen A.M., McIntosh J.R., Ahn N.G. Activation of the MKK/ERK pathway during somatic cell mitosisdirect interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J. Cell Biol. 1998;142:1533–1545. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V., Ganoth D., Dahan A., Heller H., Hershko J., Luca F.C., Ruderman J.V., Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell. 1995;6:185–197. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S.S., McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Ha E., McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1–related protein kinase. J. Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel J., Earnshaw W., Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Visintin R., Prinz S., Amon A. CDC20 and CDH1a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Waters J.C., Chen R.H., Murray A.W., Salmon E.D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J. Cell cycle-regulated expression, phosphorylation, and degradation of p55Cdc. A mammalian homolog of CDC20/Fizzy/slp1. J. Biol. Chem. 1997;272:28501–28511. doi: 10.1074/jbc.272.45.28501. [DOI] [PubMed] [Google Scholar]
- Weiss E., Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S., Cleveland D.W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yen T.J., Schaar B.T. Kinetochore functionmolecular motors, switches and gates. Curr. Opin. Cell Biol. 1996;8:381–388. doi: 10.1016/s0955-0674(96)80014-7. [DOI] [PubMed] [Google Scholar]
- Yu H., Peters J.M., King R.W., Page A.M., Hieter P., Kirschner M.W. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–1222. doi: 10.1126/science.279.5354.1219. [DOI] [PubMed] [Google Scholar]
- Zecevic M., Catling A.D., Eblen S.T., Renzi L., Hittle J.C., Yen T.J., Gorbsky G.J., Weber M.J. Active MAP kinase in mitosislocalization at kinetochores and association with the motor protein CENP-E. J. Cell Biol. 1998;142:1547–1558. doi: 10.1083/jcb.142.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]