Summary
A major goal of synthetic biology is to reprogram bacteria to carry out complex tasks, such as synthesizing and delivering drugs, and seeking and destroying environmental pollutants. Advances in molecular biology and bacterial genetics have made it straightforward to modify, insert, or delete genes in many bacterial strains, and advances in gene synthesis have opened the door to replacing entire genomes. However, rewriting the underlying genetic code is only part of the challenge of reprogramming cellular behavior. A remaining challenge is to control how and when the modified genes are expressed. Several recent studies have highlighted how synthetic riboswitches, which are RNA sequences that undergo a ligand-induced conformational change to alter gene expression, can be used to reprogram how bacteria respond to small molecules.
Introduction
One of the main goals of synthetic biology is to reprogram organisms to autonomously perform complex tasks. The revolution in genetics has provided the sequences of vast numbers of genes that carry out myriad functions, and many groups are beginning to view these gene sequences as snippets of code or “parts” that may be combined in new ways to reprogram how organisms behave.[1] Just as computer programs use conditional statements to determine if and when a command is executed, cells often apply conditional logic to determine whether particular genes are expressed. A classic example of conditional logic in E. coli involves the lac repressor protein, which induces expression of lactose-metabolizing genes in the lac operon only when lactose derivatives such as allolactose or IPTG are present.[2] Because of its simplicity, the lac repressor is one of the most commonly used ligand-inducible expression systems. However, for many applications in metabolic engineering and synthetic biology, it is desirable to have inducible expression systems that respond to specific small molecules in the engineered system, rather than an unrelated molecule such as IPTG. Such designer genetic control elements would allow the expression of genes in engineered metabolic pathways to be precisely choreographed to specific metabolite levels to minimize waste and improve yields, and would enable complex genetic programs to be executed at the desired time and place. Several groups have now put a new twist on a potentially ancient genetic regulatory system to create synthetic riboswitches that control gene expression in response to new ligands. Here we will describe how riboswitches function in bacteria, why synthetic riboswitches represent an attractive tool to control gene expression in bacteria, and some recent applications of synthetic riboswitches to reprogram cellular behavior. Our review will focus primarily on using small molecules and RNA to control gene expression in bacteria—an excellent review covering RNA-based synthetic biology with an emphasis on eukaryotic expression systems has appeared recently.[3]
Holes without Keys — How RNAs and Small Molecules Control Gene Expression in Prokaryotes
Throughout the 1990s, studies of the metabolic pathways for coenzyme B12 (AdoCbl) [4–6], methionine [7], and thiamine [8] in prokaryotes revealed a variety of conserved elements within the untranslated regions of various genes. Each of these elements, known as the B12 box [4–6], the S box [7], and the thi-box [8], shared two things in common. First, each element was important for the metabolite-dependent regulation of nearby genes. Second, each element was predicted to have two well-defined RNA secondary structures that could influence gene expression either transcriptionally by forming a terminator sequence, or post-transcriptionally by sequestering a potential ribosome binding site. While several groups postulated that metabolites might interact with the RNA to directly influence gene expression, it was difficult to rule out the presence of an additional “regulatory factor”, such as a protein, that could mediate the ligand-dependent changes in gene expression.[5,6,8] This situation changed when Breaker and coworkers reported that small molecules such as thiamine [9] and coenzyme B12 [10] could interact directly with mRNAs to control gene expression without the need for additional protein cofactors. The discovery that RNA switches, or riboswitches, could control gene expression launched a flurry of studies to determine how widely this mechanism is used in living systems. Since several reviews of riboswitches have appeared recently [11–14], we will focus on the common sensing mechanisms that may be engineered to control gene expression in bacteria.
Riboswitch Architecture and Mechanisms of Action
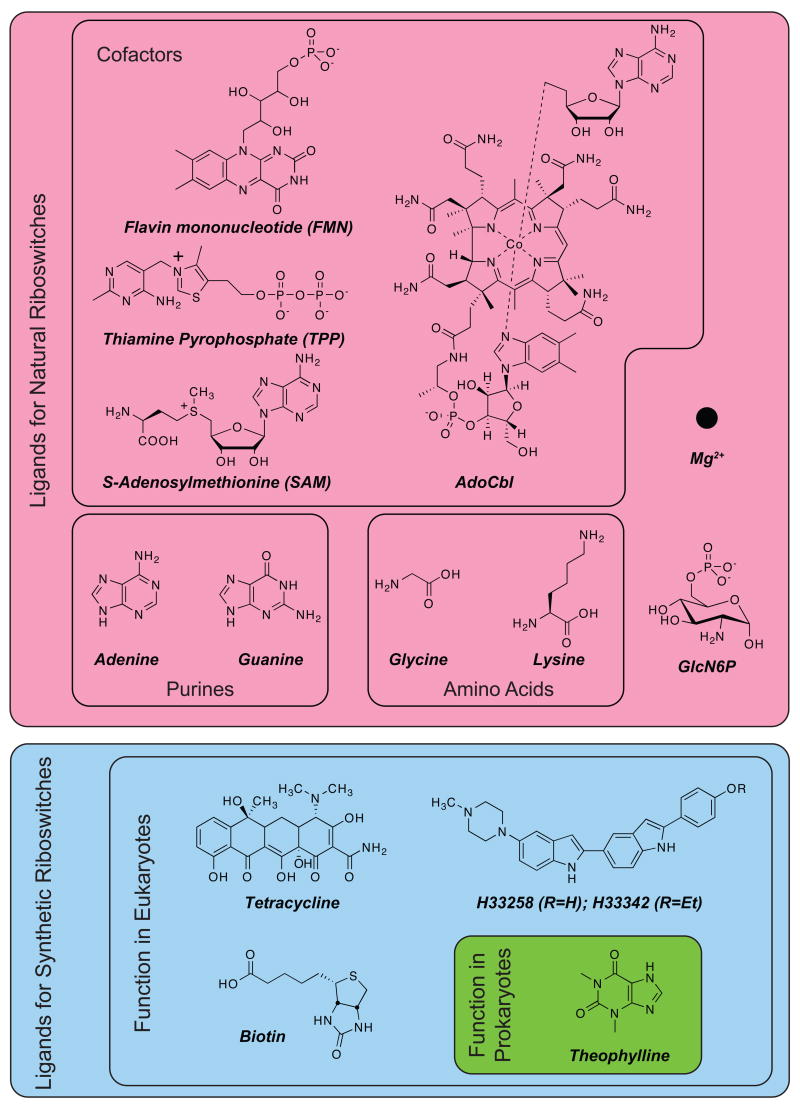
Riboswitches are typically comprised of two domains, an aptamer domain that recognizes the ligand, and an expression platform that couples ligand binding to a change in gene expression. Natural riboswitches recognize many different classes of compounds (Figure 1) that range from elaborately structured cofactors such as coenzyme B12 [10,15] and flavin mononucleotide (FMN) [16], to simpler structures such as purines (adenine [17] and guanine [18,19]) and amino acids (glycine [20] and lysine [21,22]), down to one of the simplest structures, the Mg2+ ion [23,24]. In addition to binding their cognate ligands tightly, natural aptamers often distinguish between closely related structures, such as S-adenosylmethionine (SAM) and S-adenosylhomocysteine (SAH), which differ by only a methyl group [25,26], and thiamine pyrophosphate and thiamine monophosphate, which differ by a single phosphate group [9,27–30]. This discriminatory power of aptamers allows riboswitches to differentiate between reactants and products along metabolic pathways (e.g. SAM and SAH) and may be particularly useful in creating synthetic riboswitches that can detect the products of biocatalytic reactions in vivo.
Figure 1.
Examples of ligands for natural and synthetic riboswitches.
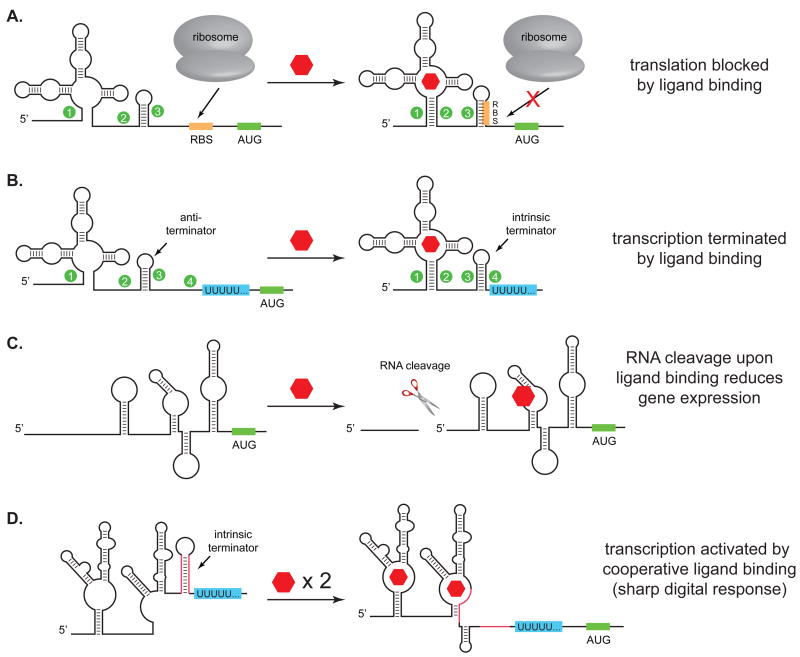
For a riboswitch to function, ligand binding must be coupled to some change in cell behavior. Riboswitches use a variety of different mechanisms to enhance or repress gene expression at either the transcriptional- or post-transcriptional level (Figure 2). One of the simplest post-transcriptional mechanisms operates at the level of mRNA translation. In most riboswitches that act at the translational level, ligand binding causes the mRNA to undergo a conformational shift that sequesters the ribosome-binding site (RBS), which represses translation of the downstream gene (Figure 2A). Although natural riboswitches typically repress, rather than activate translation, this is likely the result of evolutionary pressure to down-regulate the expression of metabolic genes when ligands are plentiful, rather than any limitation on the mechanism. As we’ll see, synthetic riboswitches often control gene expression by activating protein translation.
Figure 2.
Examples of riboswitch mechanisms of action. A. Binding of ligand induces a conformational change that blocks the ribosome binding site (RBS) and reduces translation. The inverse of this mechanism can also operate, where binding of ligand reveals the RBS to activate translation. B. Binding of ligand induces formation of an intrinsic transcriptional terminator, reducing gene expression. The inverse of this mechanism can activate gene expression. C. Binding of ligand induces RNA cleavage, reducing gene expression, presumably by destabilizing the mRNA. This mechanism operates in the glmS riboswitch. D. Two aptamers can be used in tandem to activate gene expression by disrupting a transcriptional terminator. In the case of the glycine riboswitch, ligand binding is cooperative, leading to a sharp increase in gene expression over a narrow concentration range.
In addition to controlling translation, riboswitches can also control transcription (Figure 2B). This typically occurs when a ligand binds to an elongating mRNA during transcription and causes either the formation of a terminator structure that stops transcription, or the disruption of a terminator structure, which allows transcription to continue. In the FMN, SAM, and guanine riboswitches, ligand binding shifts the conformation from a structure that presents an anti-terminator to one that displays an intrinsic transcriptional terminator, thus repressing the expression of the downstream genes in the presence of the ligand. Interestingly, the adenine riboswitch, which has a structure very similar to the guanine riboswitch, activates transcription upon binding adenine.1[17,18,31] That two similar aptamers can act to either activate or repress transcription highlights the independence of the aptamer and the expression platform and underscores the flexibility of the riboswitch architecture.
More recently, riboswitches that act through more complex mechanisms have been reported.[20,32,33] For example, the glmS riboswitch functions as a ligand-inducible ribozyme that controls gene expression in Gram-positive bacteria.[33] The glmS mRNA encodes the enzyme glutamine-fructose-6-phosphate amidotransferase, which catalyzes the synthesis of glucosamine-6-phosphate (GlcN6P). When concentrations of GlcN6P are low, the glmS mRNA is translated normally, which increases the production of GlcN6P. When the concentration of GlcN6P rises above a certain threshold, GlcN6P binds to the glmS mRNA and initiates a self-cleavage reaction that reduces gene expression, presumably by rendering the mRNA more sensitive to intracellular proteases (Figure 2C).[33] Despite being one of the larger riboswitches, glmS is well-characterized structurally in both the presence and absence of ligand.[34–40] These studies revealed that unlike many riboswitches, which undergo extensive conformational changes upon ligand binding, the glmS ribozyme does not change its structure significantly upon ligand binding. Rather, the ligand appears to function as a cofactor that increases the cleavage rate by ~100,000-fold.[33,34,38,39]
The glycine riboswitch provides another example of how riboswitches can perform more complex functions.[20] The glycine riboswitch controls the transcription of the gcvT operon, which encodes a glycine cleavage pathway in B. subtilis. When glycine is scarce, this pathway is repressed. When glycine is abundant, the operon is transcribed, leading to glycine cleavage. Unlike most riboswitches, the glycine riboswitch employs not one, but two aptamers, which bind glycine cooperatively to activate gene expression over a narrow concentration range (Figure 2D). The cooperative nature of ligand binding allows the glycine riboswitch to produce a more ‘digital’ response [20], rather than a graded ‘analog’ response to the ligand concentration, demonstrating that riboswitches may be finely tuned for specific applications.
The metE (SAM-AdoCbl) riboswitch from B. clausii also takes advantage of two aptamers, but unlike the glycine riboswitch, each aptamer recognizes a different ligand.[32] The first aptamer is capable of binding S-adenosylmethionine, while the second can bind adenosylcobalamin. Each aptamer resides upstream of an intrinsic transcriptional terminator, and each can independently repress transcription of the downstream metE gene in the presence of its cognate ligand. One possible explanation for the existence of this dual aptamer system is that B. clausii has two different enzymes that catalyze the conversion of homocysteine to methionine (metE and metH), and a third enzyme (metK) that converts methionine to SAM.[32] While all of these genes contain a SAM riboswitch that represses their transcription when SAM is abundant, metE also contains an AdoCbl riboswitch that represses transcription when AdoCbl is abundant. This transfers the burden of methionine production to metH, which uses the AdoCbl derivative methylcobalamin as a cofactor to produce methionine more efficiently than metE. Formally, the metE riboswitch acts as a ‘NOR’ logic gate, in which either of two inputs can produce the same output, in this case, repression of transcription.[32] The ability to implement elements of Boolean logic at the molecular scale not only has important implications for the regulation of cell metabolism, but also for creating designer control elements to reprogram cell behavior.
Taken together, these systems show that Nature has evolved a variety of sensing mechanisms that provide the underpinnings of conditional logic to control how cells function. By combining these motifs and principles with well-established in vitro selection methods, the stage is set to develop designer genetic control elements that will allow us to reprogram cell metabolism and behavior.
Applied Studies Revealed Hints to How Riboswitches Might Function, and How they Might be Reengineered
The discovery of how living systems employ the riboswitch control mechanism was, in many ways, a great surprise and undoubtedly, there are many more surprises to come. However, many key aspects of how riboswitches might function had already been established using synthetic systems. The clearest example was the discovery that, starting from a large pool of RNA, in vitro selection (or SELEX) could selectively amplify sequences that could tightly and specifically bind to small molecules.[41,42] These ‘aptamers’ could bind a variety of ligands (including many that were later shown to be ligands for riboswitches, such as FMN) with high affinity and, in some cases, outstanding selectivity against structurally related compounds.[43] In addition to selecting for ligand binding, many groups were able to demonstrate that aptamers could be incorporated into allosteric switches that could regulate other processes, such as ligand-regulated RNA cleavage [44] — these studies nicely presaged the discovery of the glmS riboswitch that employs ligand-dependent RNA cleavage, albeit by a slightly different mechanism.
Perhaps the biggest hint of what was to come was provided by Werstuck and Green [45], who, in an effort to control ligand-dependent gene expression in eukaryotes, selected aptamers that bound the Hoescht dyes H33258 and H33343 (Figure 1). They subsequently cloned these aptamers into the 5′-UTR of a β-galactosidase reporter gene and demonstrated ligand-dependent repression of translation in Chinese hamster ovary (CHO) cells. Although the mechanisms of prokaryotic and eukaryotic gene expression differ considerably, this study established that all of the pieces of what would later become known as a riboswitch (aptamer, expression platform, gene) could function together in an intact cell.[45] Their work was followed by a number of studies that used different ligand/aptamer combinations to control eukaryotic gene expression.[3,46–48] This work in eukaryotes not only set the stage for the discovery of natural riboswitches, it suggested the tantalizing possibility of using in vitro selection to create new aptamers that could control gene expression in response to nearly any small molecule.
Using Synthetic Riboswitches to Reprogram Bacteria
Given the prevalence of riboswitches in bacteria, it is perhaps surprising that there are relatively few examples of synthetic riboswitches in prokaryotes. The first bacterial synthetic riboswitch was reported by Suess and coworkers[49], who incorporated the theophylline aptamer into a designed helix in the 5′-UTR of a reporter gene. The helix was designed to undergo a 1 nt shift upon ligand binding to reveal a ribosome binding site, which would enhance translation of the downstream gene. When this synthetic riboswitch was expressed in the Gram-positive bacterium B. subtilis, addition of theophylline caused a dose-dependent increase in gene expression.[49] Because the authors used the riboswitch to activate a repressor protein, the net effect was an 8.8-fold decrease in expression of a second protein that was under the control of the repressor.[49] Soon thereafter, Desai et al. reported a theophylline-sensitive synthetic riboswitch that activated gene expression in the Gram-negative bacterium E. coli.[50] Unlike the riboswitch described by Suess et al. [49], their riboswitch was not designed to operate by any specific base-pairing mechanism. Rather, they reasoned that simply increasing the strength of the secondary structure near the ribosome binding site upon ligand binding might be sufficient to repress translation in a manner analogous to how increasing base-pairing near the RBS reduces gene expression. Surprisingly, the riboswitch did not repress translation upon the addition of ligand, it actually activated translation of several reporter genes, including β-galactosidase and the antibiotic resistance gene chloramphenicol acetyltransferase (cat).[50] By activating antibiotic resistance in a ligand-dependent fashion, the authors demonstrated that cell survival could be coupled to the presence of a non-metabolite.[50] The ability to use synthetic riboswitches to detect the production of small molecules opens the door to creating powerful genetic selections to guide directed evolution experiments.
While the riboswitches created by Desai et al. [50] were useful, they suffered from several drawbacks: they showed background expression in the absence of the ligand, the activation of expression was modest (~8-fold), and the mechanism of action was unclear. To address these issues, Lynch et al. [51] developed a high-throughput screen to identify riboswitches that display extremely low background expression in the absence of the ligand, and robust activation of gene expression (>35-fold) in its presence. Sequence analysis, covariant mutagenesis, in vivo kinetic assays, and in vivo RNA footprinting experiments also allowed the authors to determine the mechanisms of action of the new switches, and revealed that switching is kinetically controlled and likely occurs co-transcriptionally. This study also demonstrated that it is relatively straightforward to discover new synthetic riboswitches starting with a single aptamer and an efficient screen.[51] Efficient methods to rapidly convert aptamers into synthetic riboswitches should lower the barrier to creating designer genetic control elements for reprogramming bacteria.
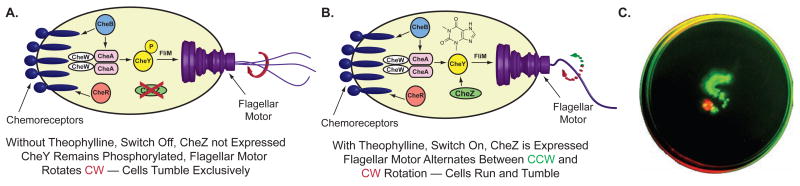
In an example of using synthetic riboswitches to reprogram bacteria, Topp and Gallivan asked whether a riboswitch could induce bacteria to follow a new small molecule that was not normally a chemoattractant.[52] They reasoned that such cells could be engineered to seek and destroy pollutants in the environment, or to seek out disease sites and synthesize and release therapeutics. By using a theophylline-sensitive synthetic riboswitch to activate the translation of a key gene in the chemotaxis pathway, the authors showed that they could induce cell motility in a theophylline-dependent fashion.[52] The reprogrammed cells could precisely follow a theophylline-containing path (Figure 3), while avoiding a path containing caffeine, and could also respond to ligand gradients by migrating toward increasing theophylline concentrations. By using video-microscopy to track the motion of individual E. coli in liquid media, the authors demonstrated that this behavior was primarily dictated by the fraction of the cells that were motile at a given theophylline concentration.[52] This study demonstrated how a synthetic riboswitch could provide a complex phenotype that would be difficult to produce using protein engineering.
Figure 3.
Reprogramming bacteria to follow a small molecule. A. In the absence of the cheZ gene, E. coli tumble in place and are non-motile. B. Activation of cheZ translation using a theophylline-sensitive synthetic riboswitch allows the cells to become motile. C. E. coli containing the synthetic riboswitch (green) are plated at the top of an S-shaped path containing theophylline on semi-solid agar. These cells migrate exclusively along the path. Cells lacking the synthetic riboswitch and the cheZ gene (red) are plated at the bottom of the path and are non-motile.
Conclusions
We are still at the earliest stages of reprogramming bacteria using synthetic riboswitches. From studies of natural and synthetic systems it is clear that riboswitches can control gene expression in response to a great variety of ligands using a number of different mechanisms. These mechanisms can operate either transcriptionally or post-transcriptionally, can activate or repress gene expression, can perform Boolean logic, and can respond in ‘digital’ or ‘analog’ fashions. The tremendous power of established in vitro selection techniques provides a means to discover new aptamers that can bind to nearly any molecule that is capable of interacting with an RNA. Both rational design and in vivo screening methods can convert new aptamers into riboswitches that function in cells.
Despite these advances, most synthetic riboswitches have thus far used just a handful of aptamers that had been previously selected for another purpose. While these early studies provide proof-of-principle results, in the coming years it will be important to demonstrate the generality of these methods. Specifically, this will require selecting aptamers that respond to new ligands and using these to create synthetic riboswitches that can reprogram cell behavior. Fortunately, each step in the process has been validated, suggesting that the time is right to begin reprogramming bacteria with small molecules and RNA.
Acknowledgments
I wish to thank the members of my laboratory for their outstanding contributions and for their helpful comments on this manuscript. This work was supported by the NIH (GM074070). J.P.G. is a Beckman Young Investigator and a Camille Dreyfus Teacher-Scholar.
Footnotes
With a single C->U mutation at position 74, the guanine riboswitch becomes sensitive to adenine and insensitive to guanine, as would be predicted by a Watson-Crick base-pair between the ligand and the base at position 74.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert W, Muller-Hill B. Isolation of the Lac repressor. Proc Natl Acad Sci USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. • A nice overview of synthetic biology of RNA that includes extensive examples of eukaryotic gene regulatory systems. [DOI] [PubMed] [Google Scholar]
- 4.Ravnum S, Andersson DI. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol Microbiol. 1997;23:35–42. doi: 10.1046/j.1365-2958.1997.1761543.x. [DOI] [PubMed] [Google Scholar]
- 5.Ravnum S, Andersson DI. An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cob mRNA of Salmonella typhimurium. Mol Microbiol. 2001;39:1585–1594. doi: 10.1046/j.1365-2958.2001.02346.x. [DOI] [PubMed] [Google Scholar]
- 6.Nou X, Kadner RJ. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc Natl Acad Sci USA. 2000;97:7190–7195. doi: 10.1073/pnas.130013897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 8.Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci USA. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 10.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic Control by a Metabolite Binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 11.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Winkler WC. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr Opin Chem Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Grundy FJ, Henkin TM. From ribosome to riboswitch: Control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–338. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 14.Coppins RL, Hall KB, Groisman EA. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176–181. doi: 10.1016/j.mib.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucl Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 18.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 20.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 21.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 22.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 25.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 26.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 27.Edwards TE, Ferre-D'Amare AR. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thore S, Leibundgut M, Ban NN. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. • The first crystal structure of a riboswitch from eukaryotes. [DOI] [PubMed] [Google Scholar]
- 30.Miranda-Rios J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15:259–265. doi: 10.1016/j.str.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. •• This paper shows that tandem riboswitches that respond to different ligands (SAM and AdoCbl) can act together to perform logical functions in living cells. [DOI] [PubMed] [Google Scholar]
- 33.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 34.McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson SR, Been MD. A pseudoknot in the 3' non-core region of the glmS ribozyme enhances self-cleavage activity. RNA. 2005;11:1788–1794. doi: 10.1261/rna.2203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hampel KJ, Tinsley MM. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- 37.Jansen JA, McCarthy TJ, Soukup GA, Soukup JK. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat Struct Mol Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- 38.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. •• The first crystal structure of the glmS riboswitch, which functions as a ribozyme. The structure reveals mechanistic insight into how the switch functions. [DOI] [PubMed] [Google Scholar]
- 39.Roth A, Nahvi A, Lee M, Jona I, Breaker RR. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the glmS ribozyme bound to its catalytic factor. Chem Biol. 2007;14:95–103. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 42.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 43.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 44.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werstuck G, Green MR. Controlling gene expression in living cells through small molecule-RNA interactions. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 46.Grate D, Wilson C. Inducible regulation of the S. cerevisiae cell cycle mediated by an RNA aptamer-ligand complex. Bioorg Med Chem. 2001;9:2565. doi: 10.1016/s0968-0896(01)00031-1. [DOI] [PubMed] [Google Scholar]
- 47.Harvey I, Garneau P, Pelletier J. Inhibition of translation by RNA-small molecule interactions. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucl Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucl Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J Am Chem Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 51.Lynch SA, Desai SK, Sajja HK, Gallivan JP. A high-throughput screen for synthetic riboswitches reveals mechanistic insights into their function. Chem Biol. 2007;14:173–184. doi: 10.1016/j.chembiol.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. •• An example of how a synthetic riboswitch can reprogram cells to have a complex phenotype — following a new small molecule. This work also shows how synthetic riboswitches can help solve problems that would be difficult to address using protein engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]