Abstract
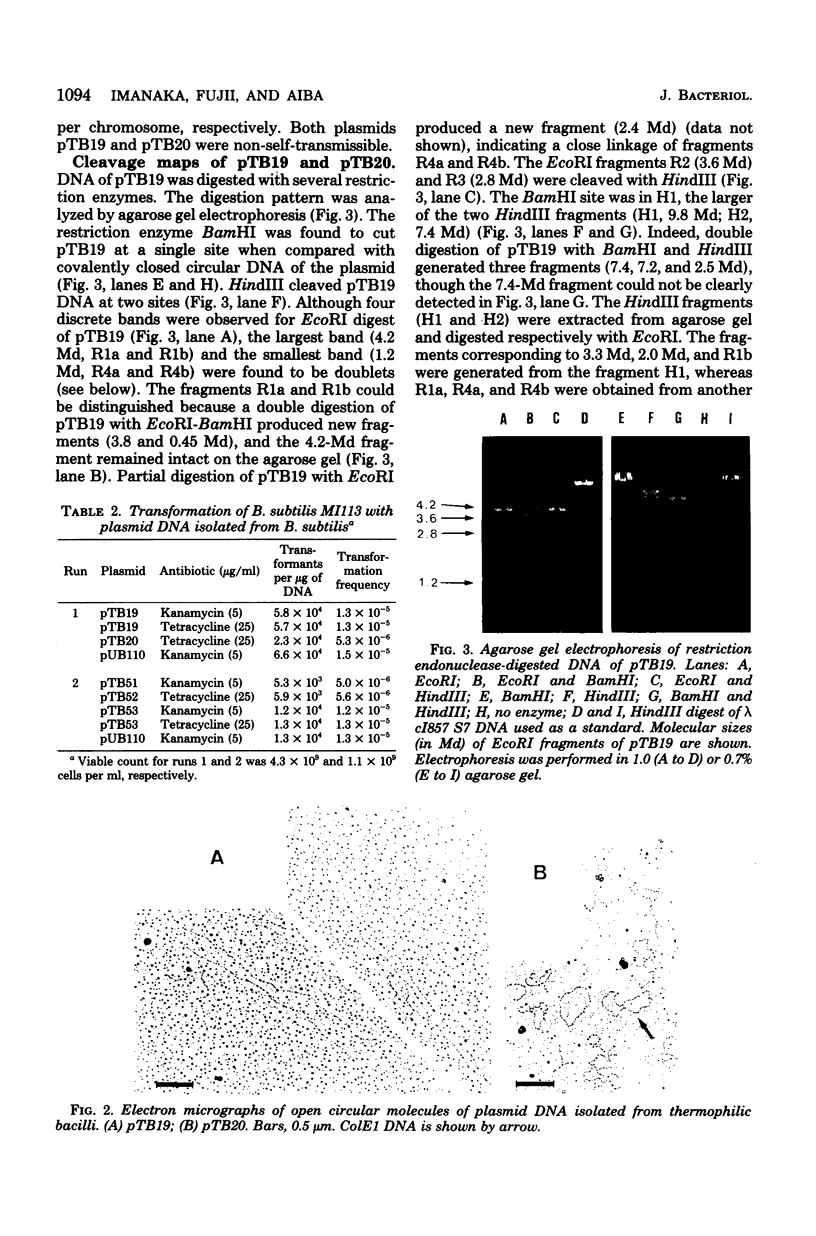
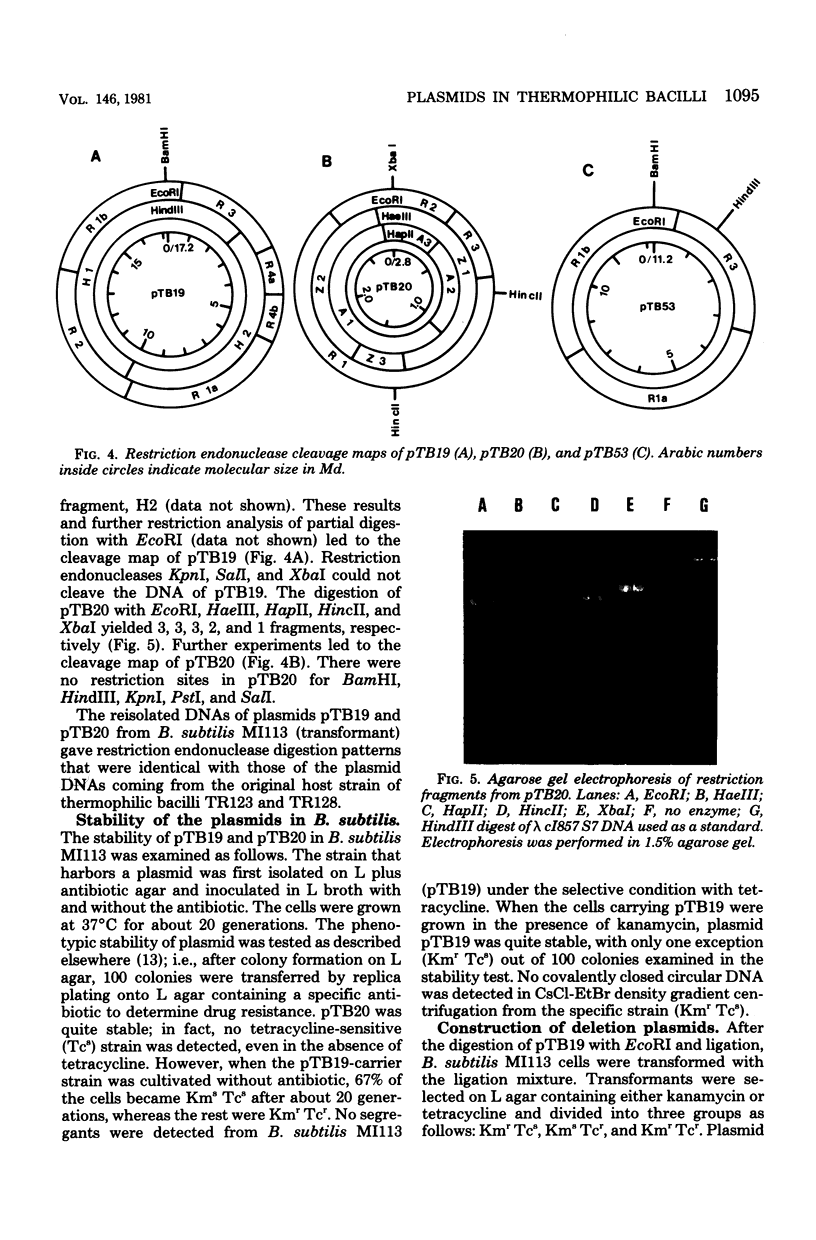
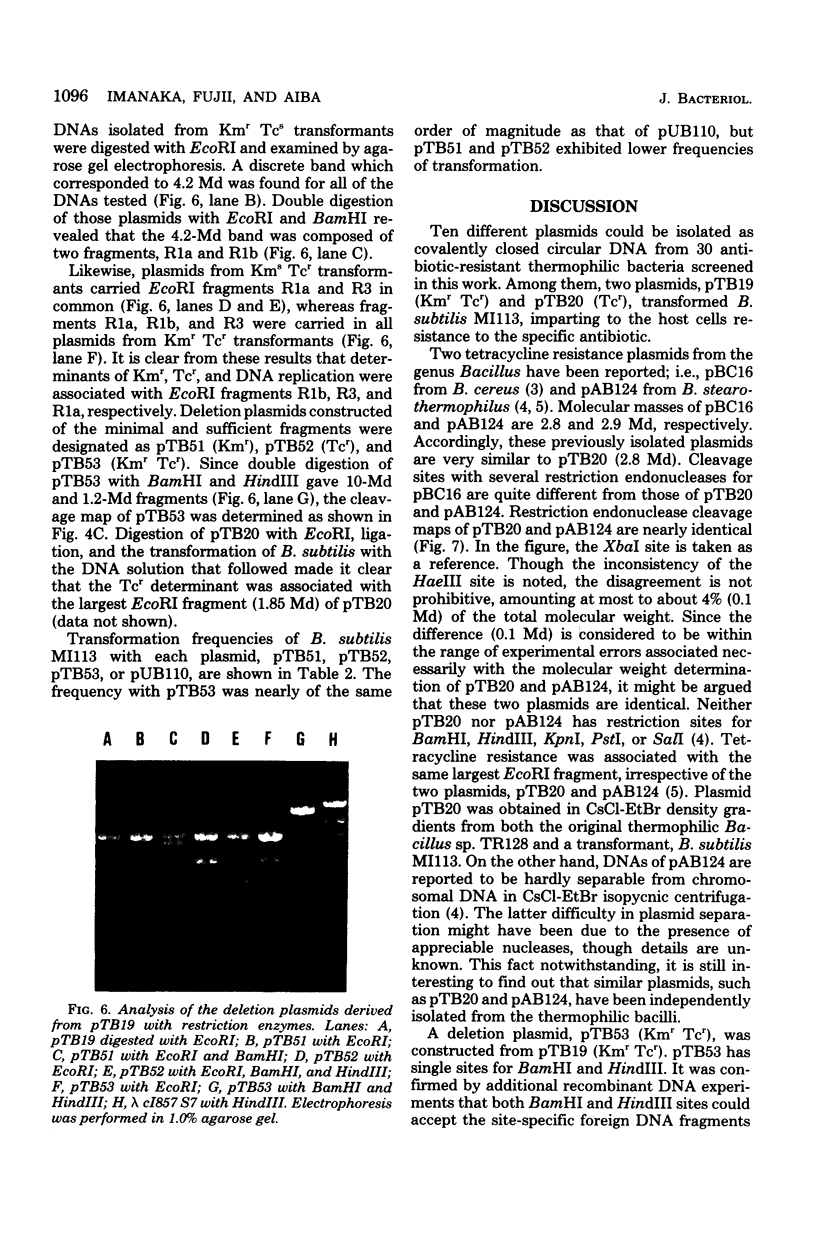
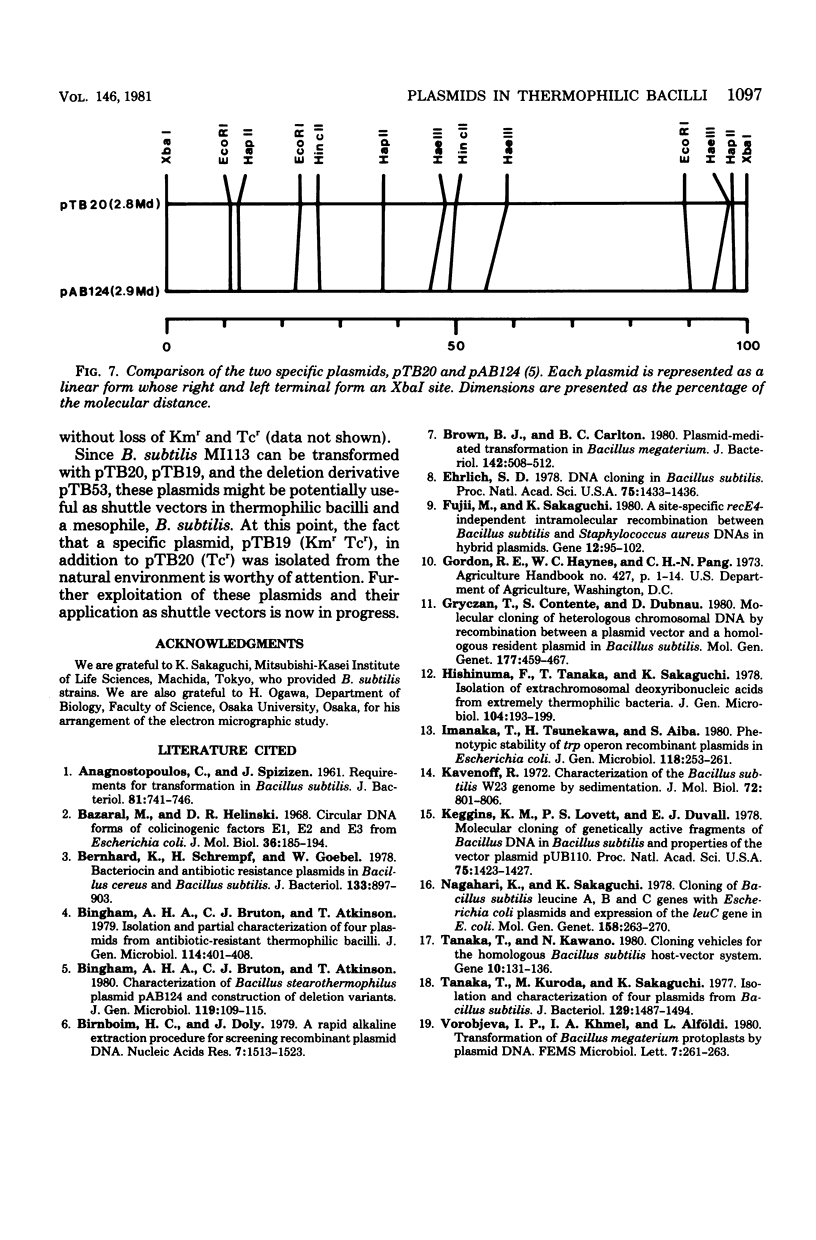
Ten plasmids were isolated as covalently closed circular deoxyribonucleic acid from antibiotic-resistant thermophilic bacteria. Of the 10 plasmids tested, 2 could transform Bacillus subtilis, yielding resistance to specific antibiotics. Plasmid pTB20 (2.8 X 10(6) daltons, approximately 24 copies per chromosome) specifies resistance to tetracycline (Tcr), whereas pTB19 (17.2 X 10(6) daltons, approximately 1 copy per chromosome) renders the host resistant to both kanamycin and tetracycline (KMrTcr). Three plasmids were not self-transmissible. The restriction endonuclease cleavage maps of the two plasmids, pTB19 and pTB20, were constructed. pTB19 and pTB20, both of which were originally isolated from thermophilic bacilli, were tested for stability in B. subtilis. Digestion of pTB19 followed by ligation yielded deletion plasmids pTB512 (Kmr), pTB52 (Tcr), and pTB53 (KmrTcr). Determinants of Kmr, Tcr, and DNA replication were associated with EcoRI fragments R1b (4.2 X 10(6) daltons), R3 (2.8 X 10(6) daltons), and R1a (4.2 X 10(6) daltons), respectively. Restriction endonuclease cleavage maps of pTB51, pTB52, and pTB53 were constructed. Tetracycline resistance of pTB20 was confirmed to be in the EcoRI fragment (1.85 X 10(6) daltons).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Bernhard K., Schrempf H., Goebel W. Bacteriocin and antibiotic resistance plasmids in Bacillus cereus and Bacillus subtilis. J Bacteriol. 1978 Feb;133(2):897–903. doi: 10.1128/jb.133.2.897-903.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. H., Bruton C. J., Atkinson T. Characterization of Bacillus stearothermophilus plasmid pab124 and construction of deletion variants. J Gen Microbiol. 1980 Jul;119(1):109–115. doi: 10.1099/00221287-119-1-109. [DOI] [PubMed] [Google Scholar]
- Bingham A. H., Bruton C. J., Atkinson T. Isolation and partial characterization of four plasmids from antibiotic-resistant thermophilic bacilli. J Gen Microbiol. 1979 Oct;114(2):401–408. doi: 10.1099/00221287-114-2-401. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B. J., Carlton B. C. Plasmid-mediated transformation in Bacillus megaterium. J Bacteriol. 1980 May;142(2):508–512. doi: 10.1128/jb.142.2.508-512.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Sakaguchi K. A site-specific recE4-independent intramolecular recombination between Bacillus subtilis and Staphylococcus aureus DNAs in hybrid plasmids. Gene. 1980 Dec;12(1-2):95–102. doi: 10.1016/0378-1119(80)90019-0. [DOI] [PubMed] [Google Scholar]
- Gryczan T., Contente S., Dubnau D. Molecular cloning of heterologous chromosomal DNA by recombination between a plasmid vector and a homologous resident plasmid in Bacillus subtilis. Mol Gen Genet. 1980 Feb;177(3):459–467. doi: 10.1007/BF00271485. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., Tanaka T., Sakaguchi K. Isolation of extrachromosomal deoxyribonucleic acids from extremely thermophilic bacteria. J Gen Microbiol. 1978 Feb;104(2):193–199. doi: 10.1099/00221287-104-2-193. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Tsunekawa H., Aiba S. Phenotypic stability of trp operon recombinant plasmids in Escherichia coli. J Gen Microbiol. 1980 May;118(1):253–261. doi: 10.1099/00221287-118-1-253. [DOI] [PubMed] [Google Scholar]
- Kavenoff R. Characterization of the Bacillus subtilis W23 genome by sedimentation. J Mol Biol. 1972 Dec 30;72(3):801–806. doi: 10.1016/0022-2836(72)90192-1. [DOI] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Duvall E. J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahari K., Sakaguchi K. Cloning of Bacillus subtilis leucina A, B and C genes with Escherichia coli plasmids and expression of the leuC gene in E. coli. Mol Gen Genet. 1978 Jan 17;158(3):263–270. doi: 10.1007/BF00267197. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kawano N. Cloning vehicles for the homologous Bacillus subtilis host-vector system. Gene. 1980 Jul;10(2):131–136. doi: 10.1016/0378-1119(80)90130-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kuroda M., Sakaguchi K. Isolation and characterization of four plasmids from Bacillus subtilis. J Bacteriol. 1977 Mar;129(3):1487–1494. doi: 10.1128/jb.129.3.1487-1494.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]