Abstract
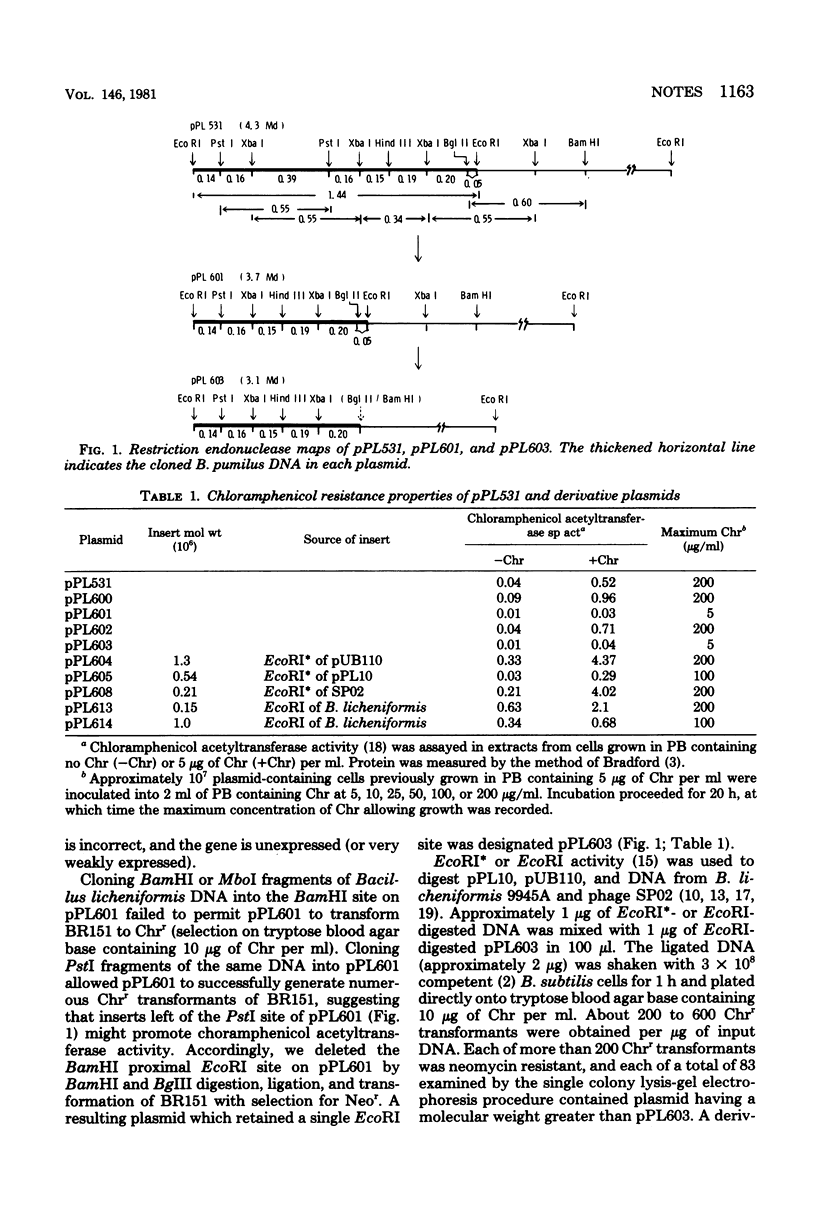
Plasmid pPL603 (3.1 megadaltons) specifies neomycin resistance in Bacillus subtilis and contains a structural gene for chloramphenicol acetyltransferase. Cells harboring the plasmid cannot grow on solid media containing 10 microgram of chloramphenicol per ml. Cloning EcoRI (or EcoRI)-generated fragments of deoxyribonucleic acid from several sources into the single EcoRI site in plasmid pPL603, with subsequent selection of transformants of media containing 10 micrograms of chloramphenicol per ml, permits the identification of restriction fragments that promote expression of the chloramphenicol acetyltransferase gene.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- An G., Friesen J. D. Plasmid vehicles for direct cloning of Escherichia coli promoters. J Bacteriol. 1979 Nov;140(2):400–407. doi: 10.1128/jb.140.2.400-407.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott K. F., Wilson G. A. Development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1967 Sep;94(3):562–570. doi: 10.1128/jb.94.3.562-570.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully D. F., Garro A. J. Expression of superinfection immunity to bacteriophage phi 105 by Bacillus subtilis cells carrying a plasmic chimera of pUB110 and EcoRI fragment F of phi 105 DNA. J Virol. 1980 Jun;34(3):789–791. doi: 10.1128/jvi.34.3.789-791.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill H. E., Whiteley H. R. Bacteriophages of Bacillus subtilis. Bacteriol Rev. 1975 Sep;39(3):257–315. doi: 10.1128/br.39.3.257-315.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Duvall E. J., Lovett P. S. Recombination between compatible plasmids containing homologous segments requires the Bacillus subtilis recE gene product. J Bacteriol. 1978 May;134(2):514–520. doi: 10.1128/jb.134.2.514-520.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Duvall E. J., Keggins K. M. Bacillus pumilus plasmid pPL10: properties and insertion into Bacillus subtilis 168 by transformation. J Bacteriol. 1976 Aug;127(2):817–828. doi: 10.1128/jb.127.2.817-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Polisky B., Greene P., Garfin D. E., McCarthy B. J., Goodman H. M., Boyer H. W. Specificity of substrate recognition by the EcoRI restriction endonuclease. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3310–3314. doi: 10.1073/pnas.72.9.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaie Y., Burtis K. C., Doi R. H. Purification and characterization of a kanamycin nucleotidyltransferase from plasmid pUB110-carrying cells of Bacillus subtilis. J Bacteriol. 1980 Mar;141(3):1178–1182. doi: 10.1128/jb.141.3.1178-1182.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher B. M., Law M. F., Garro A. J. Correlated genetic and EcoRI cleavage map of Bacillus subtilis bacteriophage phi105 DNA. J Virol. 1978 Oct;28(1):395–402. doi: 10.1128/jvi.28.1.395-402.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Graham S., Young F. E. Restriction-fragment map of the temperate Bacillus subtilis bacteriophage SPO2. Gene. 1979 Sep;7(1):51–68. doi: 10.1016/0378-1119(79)90042-8. [DOI] [PubMed] [Google Scholar]


