Abstract
Transcriptional regulation by members of the nuclear hormone receptor superfamily is a modular process requiring the mediation of distinct subclasses of coregulators. These subclasses include members of the steroid receptor coactivator-1 (SRC-1) coactivator family, p300/CBP and their associated proteins, such as p300/CBP-associated factor, human homologs of SWI/SNF proteins such as BRG-1, and the less well-characterized E3 ubiquitin-protein ligases E6 papillomavirus protein-associated protein and receptor-potentiating factor-1. Because functional studies indicate that these coregulators may form higher order complexes, we analyzed steady–state complexes of different coregulator subclasses in vivo. T47D and HeLa cell lysates were subjected to biochemical fractionation and screened by immunoblotting using coregulator-specific antibodies. We show that different subclasses of nuclear receptor coregulators exhibit distinct fractionation profiles. Furthermore, evidence is provided that SRC-1 family members may exist in vivo in heteromultimeric forms with each other. In addition, we demonstrate that liganded PR is present in stable complexes containing SRC-1 and transcription intermediary factor 2 (TIF2) in vivo. Our results suggest that the assembly of large, modular transcriptional complexes by recruitment of distinct subclasses of preformed coregulator subcomplexes may be involved in transcriptional regulation by activated nuclear receptors.
Members of the nuclear receptor family of ligand-inducible transcription factors activate transcription in response to their ligands via enhancer elements located in the promoters of target genes (1). Recently it has become clear that transactivation by these receptors is a modular process, requiring interaction with an array of cofactors capable of (i) modifying the chromatin structure of hormone-regulated promoters by intrinsic histone acetyltransferase (HAT) activities, (ii) mediating interactions between the receptors and other transcription factors, and (iii) directing assembly and stabilization of the transcriptional preinitiation complex. Several structurally distinct subclasses of nuclear receptor coregulators have been identified, including: members of the steroid receptor coactivator-1 (SRC-1) family, the cointegrators p300 and CBP and their associated proteins; mammalian homologs of yeast SWI/SNF proteins; and the less well characterized E3 ubiquitin-protein ligase coactivators.
Our laboratory initially cloned SRC-1 as a factor required for transactivation by nuclear receptors (2), and SRC-1 has been termed variously as p160/NCoA-1 (3), and ERAP-160 (4). The subsequent identification of two more members of the SRC-1 family, namely transcription intermediary factor-2 [TIF2/GRIP-1/SRC-2] (5–7), and p/CIP (8) [ACTR (9)/RAC-3 (10)/AIB-1 (11)/TRAM-1 (12)/SRC-3] established the existence of a class of structurally and functionally related nuclear receptor coactivators. Sequence alignment of the members of the SRC-1 family highlights the shared domain structure throughout and predicts common modes of action by the individual members. SRC-1 family members have C-terminal domains that contain HAT activity, suggesting that they modify chromatin (9, 13). The presence in their extreme N termini of a postulated multimerization motif, the Per-Arnt Sim/basic helix–loop–helix homology domain (14), implies that molecular interactions between SRC-1 family members and other Per-Arnt-Sim/basic helix–loop–helix homology domain proteins might be important for their function in vivo.
A class of coregulators structurally distinct from the SRC-1 family, the cointegrators, is defined by the functionally related proteins p300 and CBP. These proteins exhibit broad functional specificity in addition to extensive amino acid sequence identity (15, 16) and are proposed to function by adapting signaling pathways and integrating stimuli into an appropriate transcriptional response at a wide variety of promoters (3, 17). CBP synergizes with SRC-1 in the potentiation of estrogen receptor and progesterone receptor (PR)-dependent transactivation (18), indicating a role in nuclear receptor-dependent signaling. In addition, p300/CBP were among the first regulators of mammalian transcription in which HAT activity was identified (19). Furthermore, proteins such as the SRC-1 family member p/CIP (8) and the HAT protein p300/CBP-associated factor (PCAF) (20), first identified as binding partners of p300/CBP, have been characterized as nuclear receptor-associated proteins and coregulators in their own right (21, 22).
The SWI proteins were first identified as potentially important intermediates in nuclear receptor action when yeast strains bearing mutations in swi genes were found to be incapable of supporting glucocorticoid receptor-dependent transactivation (23). Subsequently, human SWI/SNF homologs were found to enhance the activation functions of glucocorticoid receptor (24) as well as estrogen receptor and retinoic acid receptor (25), and it has been shown that glucocorticoid receptor directs ligand-dependent nucleosomal remodeling activity of the SWI/SNF complex in yeast (26). The mammalian homologs of the closely related yeast swi2 and snf2 genes are termed brahma and brahma-related gene-1 (brg-1), respectively. BRG-1, the product of the brg-1 gene, has been shown to interact with glucocorticoid receptor in a ligand-dependent manner (27), suggesting that mammalian SWI/SNF proteins may be key elements in nuclear receptor action.
Another subclass of coregulators, relatively undefined functionally, but structurally distinct from those subclasses above, comprises the E3 ubiquitin-protein ligases receptor potentiating factor-1 (RPF-1) (28) and E6 papillomavirus protein-associated protein (E6-AP; Z.N., unpublished work). This subclass of coregulators differs from the SRC-1 family and the p300/CBP cointegrators in that they contain ubiquitin-protein ligase activity rather than HAT activity. They were initially identified as factors required for defining substrate specificity in proteolytic degradation by the proteosome system. The N-terminal receptor activation domains of E6-AP and RPF-1 are separable from their ubiquitin ligase domains that reside in their C-terminal HECT. In addition to these characterized subclasses of coregulators, a large number of receptor-interacting proteins have been identified, including RIP-140 (29), ARA-70 (30), Trip230 (31), and others.
Recently, attention has focused on mechanistic aspects of nuclear receptor coregulator function, in particular on the nature of the complexes that functional evidence indicates they potentially form. Liganded nuclear receptors are reported to recruit a variety of structurally diverse proteins: including SRC-1 family members SRC-1 (2), GRIP-1/TIF2/SRC-2 (5–7) and p/CIP/RAC3/AIB-1/ACTR/TRAM-1/SRC-3 (8–12); the cointegrators CBP and p300 (3, 32); PCAF (21, 22); human homologs of the yeast SWI/SNF proteins (27) as well as the E3 ubiquitin-protein ligase family members RPF-1 (28) and E6-AP (Z.N., unpublished work). In addition, multiple coregulator/coregulator interactions have been proposed, including p/CIP/CBP (8), CBP/PCAF (20), SRC-1/CBP (3), SRC-1/p300 (33), and SRC-1/PCAF (13). Viewed in their entirety, these individual observations raise questions concerning the steady–state organization of coregulators in the cell, as well as aspects of the nature, stability, and molecular relations of their putative complexes with activated nuclear receptors.
In light of these multiple reported interactions, we decided to address the steady–state relationships of multicoregulator transcriptional complexes in vivo by analyzing the biochemical fractionation profiles of coregulators representative of the different subclasses outlined above. We demonstrate that different subclasses of nuclear hormone receptor coregulators have distinct fractionation profiles. We suggest that at least two members of the SRC-1 coactivator family, SRC-1 and TIF2, can exist in stable complex(es) with each other in vivo. Furthermore, we provide evidence that PR interacts stably with complexes containing SRC-1 and TIF2 in a ligand-dependent manner. Our data suggest the existence of discrete, stable subcomplexes of different subclasses of coregulators that may facilitate the assembly of modular complexes required for transcriptional regulation by nuclear receptors.
MATERIALS AND METHODS
Cell Culture and Transient Transfections.
Cell lines were routinely maintained at 37°C/5% CO2 in DMEM (HeLa) or RPMI 1640 medium (T47D) supplemented with 5–10% charcoal-stripped fetal calf serum. Transfections were carried out using Lipofectin (Life Technologies, Gaithersburg, MD). pCR3.1-mCBP was constructed by inserting the BamHI-BamHI fragment of pRcRSV-mCBP8.0 into the corresponding site of pCR3.1 (Invitrogen). The construction of pCR3.1-E6-AP (Z.N., unpublished work), pCR3.1-hSRC-1A, and the reporter pPRE/GRE-E1b-Luc (21) have been described.
Gel Filtration.
Subconfluent T47D or HeLa cells were washed and harvested in PBS and lysed with a disposable manual homogenizer in 50 mM NaCl/5 mM KCl/20 mM Hepes, pH 7.5/1 mM EDTA/10% glycerol containing a mixture of protease inhibitors (Sigma), and supplemented with ligand where appropriate. After centrifugation, the supernatant was loaded on a Superose 6 gel filtration column (Pharmacia) preequilibrated with 150 mM NaCl/50 mM sodium phosphate, pH 7.0 (supplemented with ligand where appropriate), and controlled by an FPLC system (Pharmacia). For antibody shift experiments, clarified lysates were rocked for 90 min at 4°C with 1–2 μg of SRC-1 antibody and a 3- to 4-fold excess of rabbit anti-mouse IgG (Zymed).
Immunoblotting.
Immunoblotting was performed as described in Hanstein et al. (32). Commercially obtained antibodies used were anti-CBP (Upstate Biotechnologies, Lake Placid, NY), and anti-RNA polymerase II (RNA pol II) (Santa Cruz Biotechnology).
RESULTS
Subclasses of Nuclear Receptor Coregulators Exist in Primarily Distinct Complexes in Vivo.
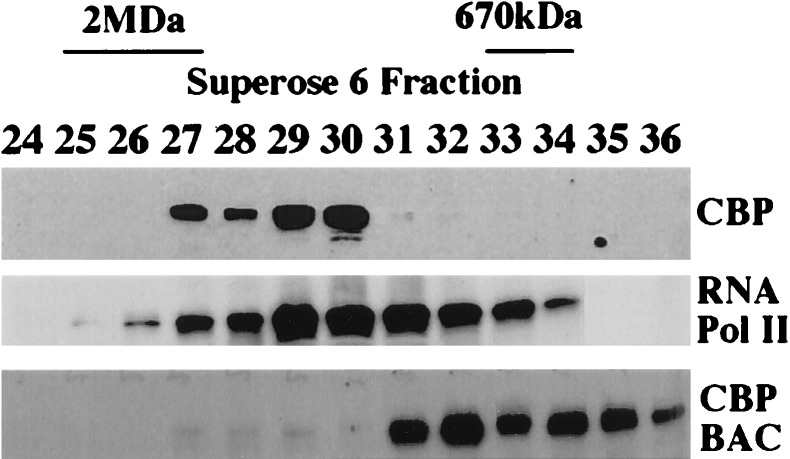
Our laboratory and others have previously shown that the functional interaction of nuclear hormone receptors with diverse subclasses of transcriptional coactivators is necessary for efficient receptor transactivation in vivo (2–3, 5–12, 27). Hypothesizing that such interactions might require the assembly of multiprotein complexes, we investigated the potential existence of nuclear hormone receptor coactivators in such complexes by biochemical fractionation of T47D and HeLa cell lysates, using a Superose 6 sizing column. Using antibodies against CBP and RNA pol II, we detected endogenous CBP and RNA pol II cofractionating in protein complexes of 1.5–2 MDa (Fig. 1), as estimated by Kee et al. (34). The elution profile of RNA pol II was much broader than that of CBP (Fig. 1; compare fractions 27–30 for CBP with fractions 26–34 for RNA pol II), also consistent with previous reports (34). We then compared the fractionation profile of endogenous CBP with that of purified baculovirus-expressed CBP, which elutes as an oligomer in distinct lower molecular size fractions (Fig. 1, CBP BAC fractions 31–36). This confirmed that CBP in T47D and HeLa cells forms high molecular weight multiprotein complexes in vivo, consistent with previous reports (34). In addition, the elution pattern of p300 in cell lysates closely resembled that of CBP, peaking in fractions containing complexes of 1.5–2 MDa, but more detectable in later fractions than CBP (Fig. 2, fractions 27–34).
Figure 1.
High molecular mass complexes contain CBP and RNA pol II. Fractionation of T47D lysate on a Superose 6 column was analyzed by immunoblot with CBP and RNA Pol II-specific antibodies (CBP and RNA pol II). Recombinant baculovirus-expressed CBP also was fractionated (CBP BAC). Indicated are elution peaks of molecular mass markers: mammalian SWI/SNF complex (≈2 MDa) and thyroglobulin (670 kDa). The void volume (4 MDa for globular proteins) was determined at fraction 20 by silver staining after fractionation of T47D cell lysate (data not shown).
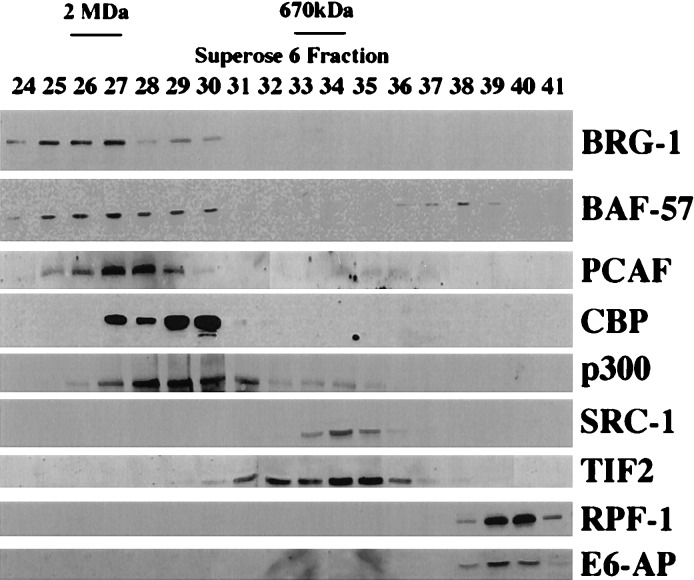
Figure 2.
Distinct steady–state fractionation profiles of different subclasses of nuclear receptor coregulators. T47D or HeLa cell lysate was fractionated on a Superose 6 column and subjected to immunoblot analysis by using coregulator-specific antibodies as indicated. Elution peaks of molecular mass standards are indicated. The relatively sharp elution peaks of SRC-1 and CBP were reproducible. No difference in fractionation pattern was observed between different cell lines.
We next compared the elution profiles of p300/CBP and RNA pol II with those of another class of nuclear receptor coregulators, the human homologs of the yeast SWI/SNF mediator complex proteins, which include BRG-1, the 220-kDa human homolog of yeast SWI2, and BAF-57, a 57 kDa-BRG-1-associated factor. These proteins exactly cofractionated in complexes of ≥2MDa (Fig. 2, peak fractions 25–27), consistent with previous estimates (35, 36). A distinct, second peak of BAF-57 was observed in later fractions (Fig. 2, peak fraction 38). Longer exposures of the BRG-1 immunoblots (data not shown) indicated that minor amounts of BRG-1 copurified with the peak fractions of SRC-1 and TIF2 (see below).
In the light of previous reports of an interaction between the cointegrators p300/CBP and PCAF (20), we next examined whether PCAF peaked in the same fractions in which p300/CBP peaked. PCAF was found to peak slightly earlier than the elution peaks of p300/CBP (Fig. 2, peak fractions 27–28), indicating that PCAF is not an exclusive binding partner of p300 and may form complexes with other proteins such as the human homologs of the SWI/SNF complex. A second minor pool of PCAF was observed (Fig. 2, fractions 35–37), which may represent partially dissociated PCAF complexes, or complexes with factors other than p300/CBP. These PCAF pools were variable in proportion between runs (data not shown), and exhibited the greatest variation of all coregulators analyzed.
Because several studies have suggested that SRC-1 may exist in complexes with CBP (3) (Fig. 2, elution peak fractions 29–30), p300 (33) (Fig. 2, elution peak fractions 28–30) and PCAF (13) (Fig. 2, major elution peak fractions 27–28), we next analyzed the elution profile of SRC-1 in relation to these proteins. Analysis of the fractionation pattern of SRC-1 showed that it peaked sharply in fractions containing protein complexes of an estimated 0.5–0.6 MDa (Fig. 2, fractions 33–35). Overlap between the elution patterns of SRC-1 and CBP was undetectable (Fig. 2), implying that these proteins may exist in distinct preformed complexes, contrary to previous reports (3). In contrast, the elution pattern of SRC-1 overlapped slightly with minor pools of p300 and PCAF (Fig. 2), suggesting that should stable complexes between SRC-1 and these coregulators exist, they represent only a small proportion of their respective cellular pools.
Monomeric SRC-1 was undetectable in cell lysates, suggesting that the kinetics of the complex formation strongly favor the sequestration of SRC-1 in these complexes, or that the free form is subject to rapid degradation. As a control, we fractionated baculovirus-expressed SRC-1 by Superose 6 gel filtration and found that it eluted exclusively in fractions 32–35 (data not shown), similar to its elution profile in cell lysate (Fig. 2, lanes 33–35) that might indicate homomultimerization of SRC-1, but also may be attributable to incomplete purification of recombinant SRC-1 from insect cell coregulators. Similar to its elution profile in T47D and HeLa cell lysate, no monomeric purified SRC-1 was detectable, further suggesting that the free form of SRC-1 may be kinetically unstable. We then examined the elution profile of a second member of the SRC-1 family, TIF2. TIF2 copurified with SRC-1, although its elution pattern was less defined and covered a wider range of fractions than SRC-1 (Fig. 2, fractions 31–36). No cross reactivity was observed between the SRC-1 antibody and TIF2 in immunoblots (not shown). The relatively broad elution profile of TIF2 suggests that it might form a greater variety of complexes than its family member SRC-1.
These initial observations suggested to us that different subclasses of coactivator involved in nuclear receptor transactivation might be sequestered in largely distinct complexes. To further test this hypothesis, we examined the elution profiles of two members of a less well-defined but functionally distinct subclass of nuclear hormone coregulators, the E3 ubiquitin-protein ligases RPF-1 and E6-AP. E6-AP and RPF-1 proteins were observed to copurify in complexes of 200–300 kDa and are distinct from all of the complexes previously observed (Fig. 2, fractions 38–41).
E6-AP and RPF-1 Synergistically Enhance PR Transactivation.
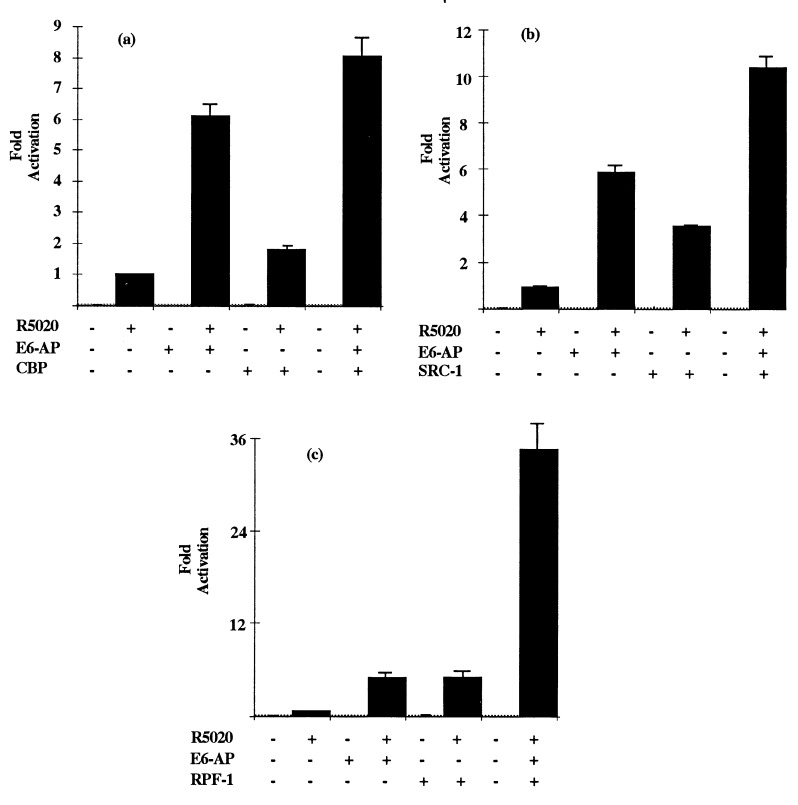
The copurification of E6-AP and RPF-1 by Superose gel filtration suggested to us that they might be present in common complexes. To test their possible functional interaction, we next examined whether these coactivators might synergistically enhance transactivation by PR. HeLa cells were transiently cotransfected with E6-AP/RPF-1, E6-AP/SRC-1, and E6-AP/CBP in a luciferase-based PR reporter assay (Fig. 3). Whereas the combinations of E6-AP/CBP (Fig. 3a) and E6-AP/SRC-1 (Fig. 3b) only additively enhanced PR transactivation, E6-AP and RPF-1 (Fig. 3c) synergistically enhanced PR transactivation.
Figure 3.
Synergistic enhancement of PR transactivation by E6-AP and RPF-1. HeLa cells were transiently transfected with 0.2 μg of PR-B expression plasmid and 1 μg of pPRE-E1b-Luc reporter in the presence and absence of 0.5 μg (total) of vectors expressing the indicated coactivators. The cells were treated with either vehicle only (−R5020) or 10nM R5020 (+). Data are expressed as the mean (± SD) of triplicate values.
Association of SRC-1 and TIF2 in a Single Complex in Vivo.
While the copurification of SRC-1 and TIF2 was evidence that they might form a complex in vivo (Fig. 2), we verified this by incubating cell lysate with anti-SRC-1 antibody and rabbit anti-mouse IgG before fractionation on the Superose 6 column. As anticipated, this resulted in a clear shift of SRC-1 immunoreactivity to fractions containing significantly larger protein complexes (compare Fig. 4 without anti SRC-1 antibody, fractions 33–35 and Fig. 4 with anti-SRC-1 antibody, fractions 28–32). Stripping and reprobing the same blot with anti-TIF2 antibody indicated a considerable shift of the immunoreactive TIF2 into fractions containing shifted SRC-1 (compare Fig. 4 without SRC-1 antibody, fractions 31–36 and Fig. 4 with anti-SRC-1 antibody, fractions 28–32). To demonstrate that the coeluting of TIF2 and shifted SRC-1 was not due to nonspecific primary or secondary antibody binding, the blot was stripped and reprobed with anti-CBP antibody demonstrating that CBP eluted in the same fractions irrespective of preincubation of lysate with SRC-1 antibody (data not shown). Because the monoclonal SRC-1 antibody does not cross react with TIF2, we take these results to indicate that TIF2 and SRC-1 can form common complexes. As shown earlier, the broader fractionation profile of TIF2 with respect to SRC-1 (Fig. 2) indicates that TIF2 likely also exists in complexes distinct from that which it forms with SRC-1. This is supported by the fact that, although incubation with SRC-1 antibody results in significant shift in the SRC-1 elution profile, a proportion of TIF2 is not shifted by anti-SRC-1 antibody (Fig. 4). Taken together, our results indicate that SRC-1 family members may associate with each other in heteromultimeric protein complexes.
Figure 4.
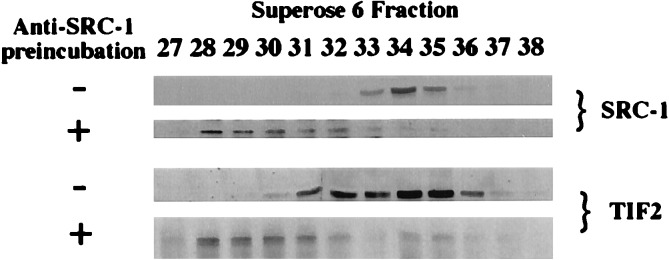
SRC-1 and TIF2 can form common complexes in vivo. SRC-1 complexes were collected by incubation with SRC-1 monoclonal antibody and polyclonal antimouse IgG and fractionated by gel filtration. Immunoblotting confirmed the shift of SRC-1 from its elution peak in the absence of preincubation with anti-SRC-1 antibody (−) to earlier fractions in the presence of anti-SRC-1 antibody (+). The relatively broad elution profile of shifted SRC-1 is most likely due to the heterogeneity of immune complexes formed in these fractions.
Liganded PR Recruits Preformed Complexes Containing SRC-1 and TIF2 in Vivo. To address the relationship of nuclear receptor with these coregulator complexes, we examined their relative migration patterns in the presence and absence of ligand.
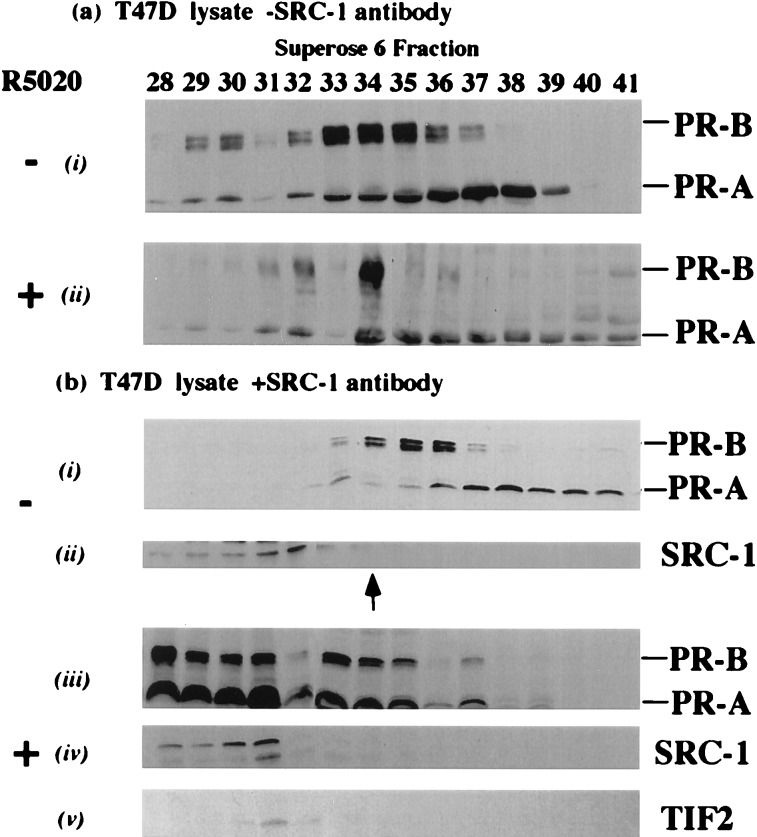
T47D cells were used for these experiments given their elevated endogenous levels of PR. Lysate from cells pretreated with vehicle or with hormone was subjected to fractionation on the Superose column. Unliganded PR A and B forms eluted in fractions containing protein complexes in the range of ≈500-kDa (Fig. 5a, i, lanes 32–39, longer exposure of 5a, i, lanes 32–41), masses consistent with previous reports (1, 37). In the presence of hormone, the liganded PR-B form copurified sharply with the elution peaks of SRC-1 and TIF2 (Fig. 5a, ii, lane 34; compare with Fig. 2, SRC-1 and TIF2). The liganded PR A form also coeluted with the peaks of SRC-1 and TIF2 but significant amounts did not (Fig. 5a, ii, lanes 36–41). Liganded PR was largely absent from fractions in which the majority of cellular p300/CBP eluted (compare Fig. 5a, ii with Fig. 2, p300/CBP).
Figure 5.
Liganded PR exists in stable complexes containing SRC-1 and TIF2 in vivo. (a) T47D cells were pretreated with vehicle (i) and with 1nM progesterone (ii) before fractionation and immunoblotting with PR antibody. (b) Cells were treated as above except lysate was incubated with anti-SRC-1 antibody, fractionated and immunoblotted for (i) PR, (ii) SRC-1, (iii) PR, (iv) SRC-1, and (v) TIF2. (The arrow indicates the peak of SRC-1 and TIF2 in the absence of preincubation with the SRC-1 antibody).
The presence of the liganded PR forms in fractions containing the peaks of SRC-1 and TIF2 was not conclusive evidence per se of an association of PR, SRC-1, and TIF2. To address more precisely the association of liganded PR with the SRC-1 and TIF2-containing complexes in vivo, we incubated SRC-1 antibody and polyclonal anti-mouse IgG with T47D lysates prepared from cells pretreated with and without hormone. After fractionation of T47D lysate preincubated with SRC-1 antibody, the elution pattern of the unliganded PR forms was largely unaltered (compare Fig. 5b, i with Fig. 5a, i), but SRC-1 was shifted to earlier fractions as predicted (Fig. 5b, ii, lanes 29–32). In contrast, after ligand treatment of T47D cells, preincubation of lysate with SRC-1 antibody resulted in the shifting of 60–70% of liganded PR A and B forms (Fig. 5b, iii, lanes 28–31) into fractions containing supershifted SRC-1 (Fig. 5b, iv, lanes 28–31) and TIF2 (Fig. 5b, v, lanes 30–31). The relatively broad elution profile of shifted liganded PR (compare Fig. 5b, iii with Fig. 5a, ii) is most likely due to the heterogeneity of immune complexes formed in these fractions. A significant proportion of liganded PR A and B forms was not shifted (Fig. 5b, iii), suggesting that liganded PR also may exist in complexes that do not bind SRC-1 antibody. Our data suggest that, in vivo, complexes containing SRC-1 and TIF2 associate stably with PR A and B forms in a ligand-dependent manner.
DISCUSSION
The formation of coregulatorsomes, or multicoregulator complexes, at hormone-regulated promoters has been widely postulated on the basis of multiple interactions between nuclear receptors and coregulators. Inferences as to the nature of the associations within these complexes have been founded largely on functional assays. In particular, the question has been raised of whether these coregulatorsomes associate in the steady–state or whether pools of specific precursor complexes exist. Our data provide direct evidence of the existence in vivo of stable subcomplexes of distinct nuclear receptor coregulator subclasses, possibly reflecting established functional differences between these subclasses of coregulators. We suggest that this physical partition of different subclasses of coactivators affords the potential for their efficient combinatorial assembly into higher order complexes. This is consistent with the functional data of Korzus et al. (38), which suggest that the requirement for maximal transcriptional activation at specific promoters may be a function of the existence of diverse groups of coactivator complexes. From our data, it is plausible that transient interactions between the stable subcomplexes we have observed would facilitate rearrangement of final coregulator complexes into multiple configurations.
One issue that is unclear from our data is whether the complexes we have observed represent component parts of larger transcriptional complexes, the kinetics of formation of which do not withstand our experimental conditions. Coimmunoprecipitation and in vitro experiments have detected interactions between SRC-1 and other subclasses of nuclear receptor coregulators such as p300 (32), CBP (3), and PCAF (13), as well as interactions between receptor and CBP (3), p300 (32), BRG-1(27), and PCAF (13). Our assay differs from these experiments in that we have been able to analyze multiple coregulator complexes in terms of the relative strengths of their steady–state interactions. In our assay, while SRC-1 was undetectable in fractions containing CBP (Fig. 2), we did observe some overlap of SRC-1 with minor pools of p300 and PCAF (Fig. 2). Interestingly, we also were able to copurify SRC-1 and small amounts of BRG-1 (Fig. 2), raising the possibility that these coregulators form stable steady–state complexes. Our data indicate however that putative complexes between SRC-1/BRG-1, SRC-1/p300, PR/BRG-1, and SRC-1/PCAF, in the steady–state of the cell, represent only small pools of the total amount of these proteins in the cell. In the context of our assay, it is possible that “final” transcriptional complexes are disrupted into the smaller, stable subcomplexes we have observed. However, we have reproduced the elution pattern of previously established complexes under our experimental conditions, such as the mammalian SWI/SNF complex (35, 36). Because we do not observe them under our conditions, final complexes comprised of different subcomplexes may be inherently labile and subject to rapid rearrangement, a plausible mechanism of fine control at transcriptionally active promoters. Additionally, we have not yet detected monomeric forms of coregulators in vivo, suggesting that an important mechanism of control of transcription may be the kinetic instability of the monomeric forms of coregulators.
The identification of the stable association of SRC-1 and TIF2 in a single complex, as well as the ability of SRC-1 to homomultimerize, suggests that protein-protein interactions between SRC-1 family members is important for their function in vivo. The sequence conservation between family members within the Per-Arnt-Sim/basic helix–loop–helix homology domains, taken together with our data, lends credence to the possibility that the Per-Arnt-Sim/basic helix–loop–helix homology domains mediate this interaction, but this is yet to be established. One consequence of this multimerization might be to increase the number of binding interfaces at which afferent signaling pathways might integrate with promoter-bound receptor.
The precise copurification of the functionally related coactivators E6-AP and RPF-1 in 200–300 kDa complexes is evidence that these proteins may form a stable complex in vivo. In light of the cooperative enhancement of PR transactivation by E6-AP and RPF-1, but not E6-AP/SRC-1 and E6-AP/CBP, we speculate that the putative physical association of E6-AP and RPF-1 in common complexes may be related to their synergism. Interestingly, SRC-1 and TIF2, while they can form common complexes, do not synergistically enhance transactivation by PR (data not shown). We suggest this anomaly is due to the fact that E6-AP and RPF-1 have different downstream targets, E6-AP being involved in p53 and HHR23A ubiquitination (39, 40), whereas RPF-1 is required for RNA pol II ubiquitination (41). Conversely, the HAT activities of SRC-1 and TIF2 probably have similar downstream chromatin targets and are likely to be redundant in cotransfection assays. Further studies are required to establish more clearly whether the mechanistic basis of the synergism of E6-AP and RPF-1 is related to their possible existence in a common complex.
Our demonstration of the ligand-dependent association of PR with the SRC-1/TIF2 complex is the first direct evidence that liganded PR associates stably with large coregulator complexes as a distinct step in transactivation in vivo. We have shown that unliganded PR forms stable complexes over the range of 400–500 kDa, consistent, within the error of the column, with previous estimates for unliganded PR complexes (1, 37). Liganded PR associates stably with similar sized complex(es) that contain SRC-1 and TIF2. The interaction between activated PR and SRC-1/TIF2 complexes that we have demonstrated is clearly a stable interaction in vivo, in comparison to any interaction with CBP or p300. Because liganded PR did not coelute with the major elution peaks of CBP or p300 in the context of our assay, we suggest that activated PR does not recruit these proteins in a stable complex. Rather, our data indicate that liganded PR associates stably with the major peaks of SRC-1 and TIF2, indicating that the complexes within these fractions may represent important fundamental intermediates in PR transactivation. Although our assay is not open to functional interpretation, it is possible that these stable PR/SRC-1/TIF2 complexes undergo relatively transient interactions with other subclasses of coregulators during transcriptional regulation. Our laboratory has suggested (42) that subsequent to formation of a stable committed complex, a “rapid-start” complex is assembled by liganded PR for subsequent rounds of transcription of a template. The relative stability of the liganded PR/SRC-1/TIF2 complexes, makes them plausible candidates for such a rapid-start complex. To further support such a notion, it has been shown that the functional requirement of p300 for estrogen receptor transactivation in vitro is reduced before transcriptional reinitiation (43), suggesting that the interaction of p300 with liganded estrogen receptor may be relatively transient. Future work will clarify the functional components of the complexes with which activated PR associates stably in vivo.
Transcriptional regulation by nuclear receptors is increasingly being seen as a modular process, involving multiple discrete steps, such as chromatin remodeling and recruitment of basal transcription factors (27, 43, 44). As a mechanistic basis for this, the multiple distinct subcomplexes we have identified here afford the possibility for their stepwise, sequential interactions with liganded receptor during transcriptional activation. A model based on our data (Fig. 6) suggests that hierarchical interactions, of varying stability, may contribute to transcriptional regulation by PR and coregulators. In our model, liganded PR, SRC-1, and TIF2 are present in comparatively stable core complexes that undergo relatively transient associations with other subcomplexes during transcriptional initiation. In support of such a notion, Fondell et al. (45) have identified a class of thyroid receptor-interacting proteins that copurify with constitutively liganded thyroid receptor. These thyroid receptor-interacting proteins are distinct from any coregulator class previously characterized and indicate that liganded receptor may undergo sequential interactions with different multiprotein complexes during transcriptional regulation. Future work will discern the functional significance of these and other complexes and their roles in regulation of gene expression by nuclear receptors.
Figure 6.
Mechanistic model for transcriptional activation by activated PR. The relative stability of the complexes between liganded PR and SRC-1/TIF2-containing subcomplexes suggests they may be important intermediates in PR transactivation. Interactions of SRC-1 with other subclasses of coregulators appear to be comparatively transient.
Acknowledgments
We thank members of the O’Malley laboratory for helpful discussions and critical reading, M. Burcin and J. Wong for recombinant proteins, D. Edwards for the E6-AP and SRC-1 antibodies, N. Weigel for the PR antibody, D. McDonnell for the RPF-1 antibody and RPF-1 construct, P. Chambon for the TIF2 antibody, R. Eckner for the p300 antibody, W. Wang for the BRG-1 and BAF-57 antibodies, Y. Nakatani for the PCAF antibody, and R. Goodman for the CBP construct. This work was supported by a National Institutes of Health grant.
ABBREVIATIONS
- SRC-1
steroid receptor coactivator-1
- CBP
CREB-binding protein
- BRG-1
product of the brg-1 gene
- E6-AP
E6 papillomavirus protein-associated protein
- RPF-1
receptor-potentiating factor-1
- TIF2
transcription intermediary factor 2
- PCAF
p300/CBP-associated factor
- RNA pol II
RNA polymerase II
- PR
progesterone receptor
- HAT
histone acetytransferase
References
- 1. Tsai M-J, O’Malley B W. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 3.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, et al. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 4.Halachmi S, Marden E, Martin G, Mackay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 5.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong H, Kohli K, Garabedian M J, Stallcup M R. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 8.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R, Schiltz L, Chakravarti D, Nash A, Nagy L, Privalsky M, Nakatani Y, Evans R. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Gomes P J, Chen J D. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 12.Takeshita A, Cardona G R, Koibuchi N, Suen C-S, Chin W W. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 13.Spencer T, Jenster G, Burcin M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Nature (London) 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 14.Lindebro M C, Poellinger L, Whitelaw M L. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 16.Eckner R, Ewen M, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–885. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 18.Smith C L, Onate S A, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 20.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 21.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M-J, O’Malley B W. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco J C, Minucci S, Lu J, Yang X-J, Walker K, Chen H, Evans R M, Nakatani Y, Ozato K. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 24.Muchardt C, Yaniv M. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiba H, Muramatsu M, Nomoto A, Kato H. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostlund-Farrants A K, Blomquist P, Kwon H, Wrange O. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fryer C J, Archer T K. Nature (London) 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 28.Imhof M O, McDonnell D P. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeh S, Chang C. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang K H, Chen Y M, Chen T T, Chou W H, Chen P L, Ma Y Y, Yangfeng T L, Leng X H, Tsai M-J, O’Malley B W, et al. Proc Natl Acad Sci USA. 1997;94:9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee B L, Arias J, Montminy M R. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, et al. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 37.Rehberger P, Rexin M, Gehring U. Proc Natl Acad Sci USA. 1992;89:8001–8005. doi: 10.1073/pnas.89.17.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 39.Huibrgetse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huibregtse J M, Scheffner M, Howley P M. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gavva N R, Gavva R, Ermekova K, Sudol M, Shen C K. J Biol Chem. 1997;272:24105–24108. doi: 10.1074/jbc.272.39.24105. [DOI] [PubMed] [Google Scholar]
- 42.Klein-Hitpass L, Tsai S Y, Weigel N L, Allan G F, Riley D, Rodriguez R, Schrader W T, Tsai M-J, O’Malley B W. Cell. 1990;60:247–257. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- 43.Kraus W L, Kadonaga J T. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong J, Shi Y B, Wolffe A P. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]