Abstract
Substrate-mediated gene delivery describes the immobilization of gene therapy vectors to a biomaterial, which enhances gene transfer by exposing adhered cells to elevated DNA concentrations within the local microenvironment. Surface chemistry has been shown to affect transfection by nonspecifically immobilized complexes using self-assembled monolayers (SAMs) of alkanethiols on gold. In this report, SAMs were again used to provide a controlled surface to investigate whether the presence of oligo(ethylene glycol) (EG) groups in a SAM could affect complex morphology and enhance transfection. EG groups were included at percentages that did not affect cell adhesion. Nonspecific complex immobilization to SAMs containing combinations of EG- and carboxylic acid-terminated alkanethiols resulted in substantially greater transfection than surfaces containing no EG groups or SAMs composed of EG groups combined with other functional groups. Enhancement in transfection levels could not be attributed to complex binding densities or release profiles. Atomic force microscopy imaging of immobilized complexes revealed that EG groups within SAMs affected complex size and appearance and could indicate the ability of these surfaces to preserve complex morphology upon binding. The ability to control the morphology of the immobilized complexes and influence transfection levels through surface chemistry could be translated to scaffolds for gene delivery in tissue engineering and diagnostic applications.
Keywords: Gene delivery, Reverse transfection, Atomic force microscopy (AFM), Self-assembled monolayers
1. Introduction
Developing systems capable of controlled and efficient gene transfer is a fundamental goal of biotechnology, with applications including functional genomics, gene therapy and tissue engineering. The primary challenge in applying gene delivery to these applications is inefficient delivery, with extracellular and intracellular barriers both limiting the efficiency. In non-viral approaches, plasmid is complexed with cationic lipids or polymers to facilitate transfection in vitro and in vivo [1–3]. Complexation can enhance interactions between positively charged DNA complexes and the negatively charged cellular membrane, in addition to providing stability against degradation [4] and facilitating intracellular trafficking.
Controlled release systems for DNA delivery have the potential to overcome extracellular barriers that limit gene transfer and enhance gene delivery relative to more traditional delivery methods [5]. These systems include delivery through polymeric release, in which the DNA is released from a polymer scaffold, or substrate-mediated delivery, in which DNA is retained at the surface of a substrate. Substrate-mediated delivery, also termed solid phase delivery or reverse transfection, involves the immobilization of DNA, complexed with nonviral vectors, to a biomaterial or substrate that supports cell adhesion [6]. Cells cultured on the substrate are exposed to elevated DNA concentrations within the local microenvironment, which enhances transfection.
DNA complexes can be immobilized on the substrate through specific or nonspecific interactions for delivery from the surface. Specific interactions can be introduced through complementary functional groups on the vector and surface, such as antigen–antibody or biotin–avidin [7–9]. Poly(L-lysine) (PLL) and polyethylenimine (PEI), modified with biotin residues, were complexed with DNA and bound to a neutravidin substrate [7,8], resulting in 100-fold increased transgene expression from the immobilized complexes relative to bolus delivery of complexes [7]. Plasmid DNA or DNA complexed with cationic polymers or lipids can also interact with substrates through nonspecific mechanisms [10–18]. Polyplexes and lipoplexes nonspecifically immobilized to substrates enhanced the extent of transgene expression in both cell lines and primary human-derived cells, along with an increased cellular viability [13]. This enhancement was dependent on the properties of both the complex (e.g. complexation agent, nitrogen to phosphate (N/P) ratio) and the substrate.
Surface chemistry has been shown to affect substrate-mediated delivery of nonspecifically immobilized complexes, impacting on both the initial binding and the subsequent transfection. Self-assembled monolayers (SAMs) of alkanethiols on gold were used to provide a flexible system for regulating surface chemistry [19–21] to examine complex–substrate interactions. DNA, complexed with cationic lipids, was immobilized through nonspecific mechanisms to SAMs presenting combinations of hydrophilic and charged (COO−), hydrophilic and uncharged (OH), and hydrophobic terminal functional groups (CH3) [15]. Surface hydrophilicity and ionization were found to mediate both DNA complex immobilization and transfection, but had no effect on complex release. The greatest amounts of binding and transfection were observed on surfaces presenting charged, hydrophilic groups, suggesting that electrostatic interactions allow for reversible interactions between the substrate and complexes and result in efficient gene delivery [22]. Hydrophobic substrates bound similar quantities of DNA as the hydrophilic surfaces, yet transfection was significantly reduced, suggesting the conformation of the DNA complexes may be altered upon binding to hydrophobic surfaces and result in low transfection levels [13,15].
However, as in traditional gene delivery approaches, further improvements are still needed for substrate-mediated gene delivery to address issues that limit gene transfer, including complex size stability, complex aggregation and strong interactions between the surface and complexes [9,13]. Polyethylene glycol (PEG), which has the monomeric repeat unit [–CH2–CH2–O]–, is widely used in drug and gene delivery and has been incorporated into DNA complexes of several cationic polymers, including polymethacrylate [23], PEI [24–32], PLL [33–35] and poly(amidoamine)s [36]. PEG reduces the surface charge of the complexes [25,26,31,34], which in turn reduces cytotoxicity [25,27,31]. The shielding effect of PEG also reduces the interaction between the complex and blood components (plasma proteins and erythrocytes) [25], and can prolong circulation of the complexes in the blood stream [25,32]. Furthermore, polyethylene glycosylation (PEGylation) can prevent salt-induced aggregation through steric stabilization [25,26,29–31,33,34]. Additionally, PEG is often used as a spacer for targeting ligands since the shielding effect of PEG is able to decrease nonspecific interactions with negatively charged cellular membranes, which results in reduction of nonspecific cellular uptake [37]. While some PEGylation strategies have had no effect on transfection efficiency in vitro [25,31,33] or in vivo [25], or even enhanced transfection [27,34], others have reported that PEGylation resulted in poor transfection [28,29,32], presumably due to interference with complexation [36]. These effects due to PEGylation have been associated with the extent of PEGylation, which may shield the surface charge [24,26], thus reducing cell binding and transfection, or alternatively, induce membrane leakage, resulting in enhanced cytoplasmic release [33,34].
In this report, SAMs of alkanethiols on gold were used to investigate substrate-mediated transfection by nonspecifically immobilized complexes on surfaces containing varying densities of PEG functional groups. We hypothesize that, rather than attach PEG to the complexes directly, its presence in a SAM could enhance substrate-mediated transfection by conveying the desired properties of PEG on gene delivery (reduced complex aggregation, complex size stability), and promote interactions between the complexes and cell membrane. SAMs presenting oligo(ethylene glycol) (EG) groups [38,39] have previously been used to resist protein adsorption according to the length of the EG chain and its percent composition within the monolayer [39,40], and are used here to modulate DNA complex adsorption for substrate-mediated gene delivery. In our studies, EG-terminated alkanethiols were incorporated into SAMs at concentrations that do not limit cell adhesion and combinations of EG- and COO−- terminated alkanethiols were examined for their ability to bind and release complexes and to subsequently support transfection. Complex morphology, a factor in gene delivery, was examined by atomic force microscopy (AFM) on these surfaces. The correlation between surface chemistry and morphology of immobilized complexes must be a design consideration for translating substrate-mediated gene delivery to biotechnology applications.
2. Material and methods
2.1. Gold slide preparation and monolayer self-assembly
Gold-coated glass slides were prepared using e-beam evaporation (Edwards Electron Beam Evaporator, Wilmington, MA), and consisted of a 5 nm titanium adhesion layer and 50 nm of gold. A diamond-tipped glass cutter was used to cut the gold-coated slides into smaller pieces that fit into standard 48-well tissue culture plates. Gold was prepared for SAM formation by extensive washing in acetone and ethanol, with subsequent drying under a stream of nitrogen. SAMs were formed by immersion of the clean gold substrates into 2 mM ethanolic solutions of alkanethiols for 18 h in the dark, under argon.
Monolayers were formed with combinations of four different alkanethiols (Table 1), including 1-decanethiol (DT10, CH3-terminated), 11-mercapto-1-undecanol (MUOH, OH-terminated), 11-mercaptoundecanoic acid (MUA, COO−-terminated) (Aldrich, St. Louis, MO) and HS(CH2)11(OCH2CH2)6OH, an EG-terminated alkanethiol (ProChimia, Gdansk, Poland). Alkanethiol solutions were freshly prepared in filtered, degassed ethanol. After monolayer formation, SAM samples were rinsed in pure ethanol and dried with nitrogen before further use.
Table 1.
Alkanethiols used for SAM formation
| Abbreviation | Terminal function group | Structure |
|---|---|---|

| ||
2.2. Cellular adhesion on SAMs
Cell morphology and adhesion studies were performed on SAMs with increasing percentages of EG-terminated alkanethiols in a background of MUA, prepared as described above. NIH/3T3 cells (ATCC; Manassas, VA), cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 1% sodium pyruvate, 1% penicillin–streptomycin, 1.5 g l−1 NaHCO3 and 10% fetal bovine serum, were seeded at a density of 15,000 cells per SAM. Cellular morphology and adhesion were analyzed after 24 and 48 h using phase microscopy. Adhesion was quantified by manually counting images obtained by phase microscopy, which were normalized to the associated surface area. Adhesion studies were also performed on SAMs containing only EG-terminated alkanethiols, with immobilized PEI–DNA complexes (as described below) or by coating the EG-SAMs with free PEI, at a concentration equal to that added to complexes of various N/Ps (see below).
2.3. DNA complex formation
Plasmids were purified from bacteria culture using Qiagen (Valencia, CA) reagents and stored in Tris–EDTA buffer solution (10 mM Tris, 1 mM EDTA, pH 7.4) at − 20 °C. Plasmid pEGFP-LUC encodes both the enhanced green fluorescent protein (EGFP) and firefly luciferase protein (LUC) under the direction of a CMV promoter, and was used for quantification of binding, release and transgene expression levels. Transfection efficiency was determined with plasmid pβGAL, which encodes for nuclear-targeted β-galactosidase under the direction of a CMV promoter. For DNA complex formation, branched PEI (25 kDa, Aldrich, St. Louis, MO) was diluted in 25 mM sodium phosphate buffer, pH 6.5, and then added dropwise to DNA in sodium phosphate buffer, vortexed for 10 s, and incubated for 15 min at room temperature. Complexes were formed at N/P ratios of 10 or 25, and 3 μg of DNA (300 μl final volume of complexes) was added to each SAM for binding, release and transfection studies.
2.4. Quantification of DNA complex immobilization and release on SAMs
Plasmid radiolabeled with [α-32P]dATP was used to measure the immobilization and release of DNA complexes on SAMs with varying amounts of EG-terminated alkanethiols in an MUA background. Briefly, a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NY) was used following the manufacturer’s protocol with minor modifications [8]. The labeled DNA was diluted with unlabeled DNA to a final concentration of 1% and this mixture was then used to form DNA complexes, as described above. After SAM preparation as described above, digital photographs of each sample were taken prior to complex immobilization and analyzed with ImageJ (NIH) to determine the area of each surface. SAMs were activated for complex immobilization by equilibration with 1× phosphate-buffered saline (PBS, pH 7.4) for 2 min, followed by a second incubation in MilliQ water for 10 min. Complexes were immobilized by incubation on these activated SAMs for 2 h. After the deposition period, the complex solution was removed and the SAMs were washed twice with PBS. The quantity of DNA immobilized was determined by immersing individual SAM samples in scintillation cocktail (5 ml of BioSafe II; Research Products International Corp., Mount Prospect, IL) for measurement with a scintillation counter. The counts were correlated to DNA mass using a standard curve. The density of DNA immobilized to each SAM sample was determined by normalizing the amount bound to area.
To determine the release profile, SAMs with immobilized DNA complexes were incubated with serum-containing cell growth media at 37°C in a humid chamber. At predetermined time points, half of the media was removed and replaced with fresh media. The activity of the collected sample was measured in a scintillation counter. At the final time point, the counts remaining on the SAM samples were also determined. The percentage of DNA released was calculated as the ratio of the cumulative counts released through a given time divided by the total counts initially on the substrate; thus, the release curves represent the percentage of DNA released relative to the initial amount bound to each surface.
2.5. Transfection on SAMs
Transfection studies were performed with NIH/3T3 cells, cultured as described above, on SAMs with varying amounts of EG-terminated alkanethiols, in backgrounds of MUA, MUOH or DT10, as well as SAMs containing combinations of MUA, MUOH and DT10. After complex formation and immobilization as described above, SAMs were immediately seeded with 15,000 cells in 48-well plates. Transfection was analyzed following a 48 h culture, characterized through the extent of transgene expression, which was quantified by measuring the luciferase activity using the Luciferase Assay System (Promega, Madison, WI). The luminometer (Turner Designs, Sunnyvale, CA) was set for a 3 s delay with an integration of the signal for 10 s. Luciferase activity (RLU) was normalized to the total protein amount determined with the BCA protein assay (Pierce, Rockford, IL). Transfection efficiency was also analyzed following a 48 h culture, characterized through the number of transfected cells using β-galactosidase expression visualized by staining with X-gal solution followed by imaging with a microscope (Leica, Bannockburn, IL) equipped with a color filter. The number of transfected cells was determined by counting five random fields on each SAM. The percentage of transfected cells was calculated as the ratio of the number of transfected cells divided by total cell number, determined by manual counting of phase images.
2.6. Atomic force microscopy of immobilized complexes
Atomic force microscopy was used to examine complex morphology. SAMs for AFM studies were prepared on gold-coated mica substrates (150 nm of gold; Agilent Technologies AFM, Tempe, AZ). SAM formation and complex formation and immobilization were performed as described above. After immobilization, surfaces were rinsed with 1× PBS, as described above, with two additional washes in MilliQ water, to remove any traces of salt on the surface. Samples were allowed to dry in air before imaging. AFM experiments were carried out with a DI Multimode AFM (Digital Instruments, Houston, TX) with a Type J scanner controlled with a NanoScope IIIa controller (Digital Instruments). Three images were collected in air at room temperature using contact AFM to obtain a representation for each surface. Silicon nitride cantilevers (Veeco, Santa Barbara, CA) with a spring constant of 0.12 N m−1 were used for collection of all images. Images were collected at 256 × 256 pixel resolution at a scan rate of <2 Hz. Initial image collection utilized the DI software, NanoScope 5.03, version r1 (Digital Instruments) and images were filtered using a flattening analysis, then further analyzed by WSxM, version 3.0 (Nanotec Electronica, Madrid, Spain) to generate height profiles and compare surface roughness using root-mean-square (RMS) calculations. Area–perimeter ratios analysis was performed with a height cut-off of 40 nm (for 0% EG, N/P 10 and 40% EG, N/P 25 conditions) in regions with large globular particles and of 7 and 15 nm (0% EG, N/P 10 and 40% EG, N/P 25, respectively) in regions without these large globular structures, which resulted in a range of ratios reported for these two conditions. For area–perimeter analysis on all other images (40% EG, N/P 10 and 0% EG, N/P 25), the height cut-off used was 7 nm.
2.7. Statistics
Statistical analysis was performed using JMP software (SAS Institute, Inc., Cary, NC). Comparative analyses were completed using one-way ANOVA with Tukey post-tests, at a 95% confidence level. Mean values with standard deviation are reported and all experiments were performed with at least three replicates.
3. Results and discussion
3.1. Cell adhesion on EG SAMs
The maximal amount of EG that could be incorporated into the monolayer without sacrificing cell morphology and adhesion (Fig. 1) was initially determined. SAMs were formed from solutions with increasing percentages of EG-terminated alkanethiols in a background of MUA (COO−). A monolayer containing no EG thiols resulted in a robust cell adhesion (Fig. 1a), similar to previous results on COO−-terminated SAMs [41,42]. Increasing the percentage of EG groups to 20 and 40% (Fig. 1b and c) did not appreciably affect the cell morphology or the cell monolayer. The mean cell density on surfaces with 0–40% EG groups was approximately 60,000 cells cm−2. Increasing the EG groups to 60 or 80% (Fig. 1d and e) altered the cell morphology, produced a more sporadic cell distribution on the surface with occasional cell patches (Fig. 1e) and resulted in a cell density twofold lower than surfaces with fewer EG groups. A SAM consisting of only EG groups (Fig. 1f) had no cells attached, which was expected due to the resistance of the surface to protein adsorption [39,40] that would be needed to support cellular adhesion. A complete elimination of cellular adhesion on SAMs containing 100% EG thiols indicates that the monolayers are robust. Our results are consistent with previous reports demonstrating that increasing the density of PEG groups decreases cell adhesion, due to the PEGylated surfaces’ increased ability to resist protein adsorption [43–46]. Finally, we note that increasing the EG content in solution results in an associated increase in the number of EG groups within the monolayer [38]. However, the surface concentration of EG groups is likely less than the solution concentration. Studies with mixed solutions of alkanethiols have indicated that the EG content on the surface may be reduced 3- to 30-fold relative to the solution content [38].
Fig. 1.
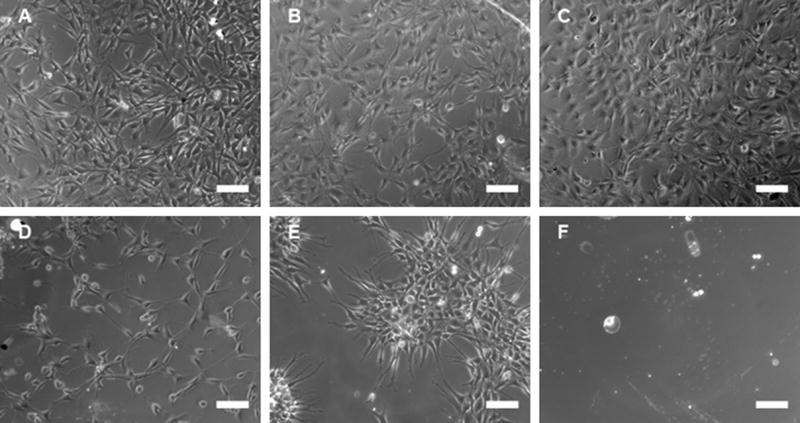
Cell adhesion on EG-containing SAMs. Images were captured 48 h after seeding cells on SAMs of (A) 0% EG/100% COO−, (B) 20% EG/80% COO−, (C) 40% EG/60% COO−, (D) 60% EG/40% COO−, (E) 80% EG/20% COO− and (F) 100% EG, all lacking immobilized DNA complexes. Scale bars correspond to 100 μm.
3.2. Quantification of complex immobilization
SAMs with varying densities of EG groups in a background of MUA (COO−) were subsequently employed to investigate the nonspecific immobilization of complexes. The amount of DNA immobilized to the surfaces did not vary with monolayer composition (Fig. 2), but was affected by the N/P ratio (Fig. 2). For an N/P of 10 (Fig. 2a), binding averaged approximately 0.35 μg cm−2, whereas complexes at N/P of 25 averaged higher binding densities, approximately 0.5 μg cm−2 (Fig. 2b). These amounts are similar to, or much greater than, previous reports of lipoplexes binding to SAMs [15,22], but are consistent with binding densities for PEI–DNA complexes on hydrophilic, serum-coated polystyrene substrates [13].
Fig. 2.
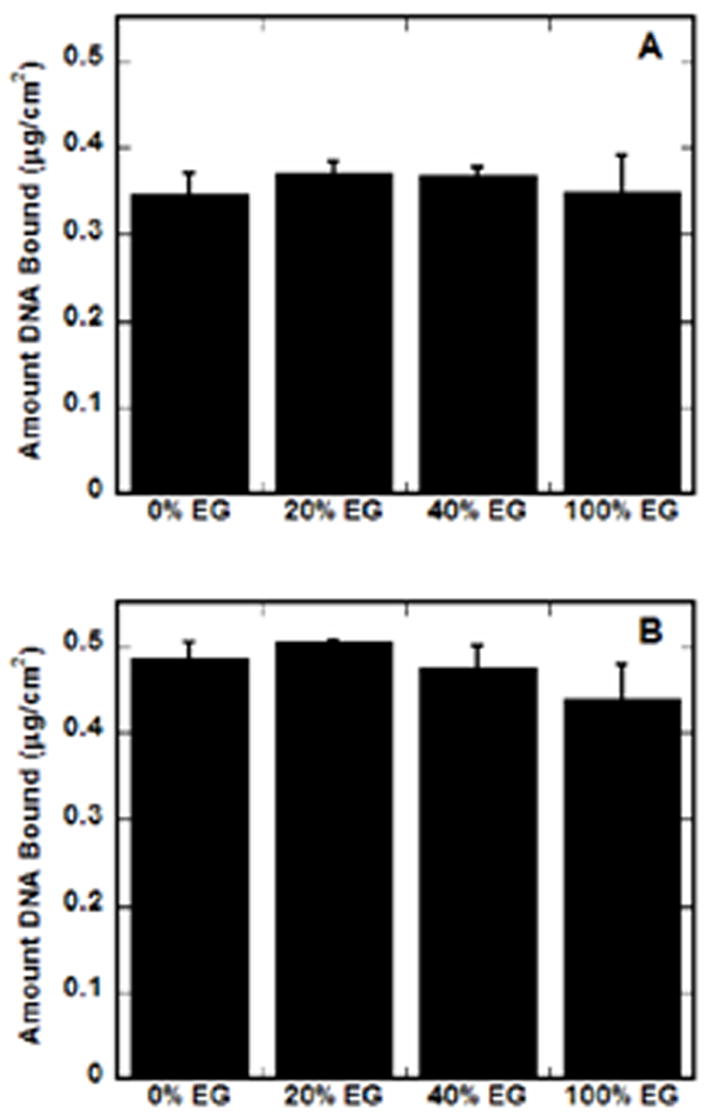
DNA complex immobilization on EG-containing SAMs. The amount of immobilized radiolableled DNA was determined for SAMs with increasing percentages of EG groups in a background of MUA (COO−), for complexes formed at N/P of 10 (A) and 25 (B). Values are reported as the mean ± SD.
Previous studies suggest that nonspecific DNA complex adsorption is mediated by at least two mechanisms: electrostatic and hydrophobic interactions [15]. Increasing the density of charged functional groups (COO−) in a background of uncharged groups (OH) increased complex immobilization, suggesting that electrostatic interactions play a major role in binding [15]. However, in this study, by decreasing the EG groups, and thus increasing the COO− background, no differences in binding were observed. The larger EG headgroups could be shielding the presentation of the COO− groups, similar to the shielding observed when PEG is incorporated into DNA–polymer complexes [26]. Shielding by the EG groups within the SAM could limit the electrostatic interactions between the carboxylic acids groups within the SAM and complexes and thus reduce the effect of charged functional groups on binding. Additionally, free PEI in the complex solution, which would be more abundant at an N/P of 25, could be binding to the SAM and changing the surface properties, eliminating differences between surfaces. Although these EG-modified SAMs have substantially reduced protein adsorption [39,40], presumably through steric stabilization and excluded volume, some adsorption of serum proteins does occur [47,48], and complexes may bind to these surfaces via similar mechanisms.
3.3. Quantification of complex release
Release studies were subsequently used to investigate the stability of the interaction between the complexes and surface (Fig. 3). Previous studies with complex adsorption to SAMs of varying surface chemistries [15,22] indicated no difference in the release rate or total amount of DNA released. For surfaces with increasing amounts of EG groups reported here, no significant differences in release rates or total amount of DNA released were observed (Fig. 3). For N/Ps of 10 (Fig. 3a) and 25 (Fig. 3b), most of the DNA release occurred by 24 h. After 8 days, less than 30% of the complexes were released, regardless of the composition of the SAM or complex. For each N/P condition, the mean amount of DNA released was slightly greater from SAMs containing only EG groups, which could result from the absence of electrostatic interactions between the complexes and these surfaces. For all surfaces, release rates are substantially lower than profiles previously determined for lipoplexes from SAMs [15], but similar to release curves obtained from PEI–DNA complexes on hydrophilic, serum-coated polystyrene substrates [13]. The presence of serum in the release media significantly enhances the release of nonspecifically immobilized complexes relative to incubation with PBS [13]. Our finding that release is independent of surface chemistry, both in this study and in previous reports [15], suggests that complex release from the substrate is mediated by competitive binding of serum components to the EG-containing SAMs, which can adsorb significant amounts of serum proteins [47].
Fig. 3.
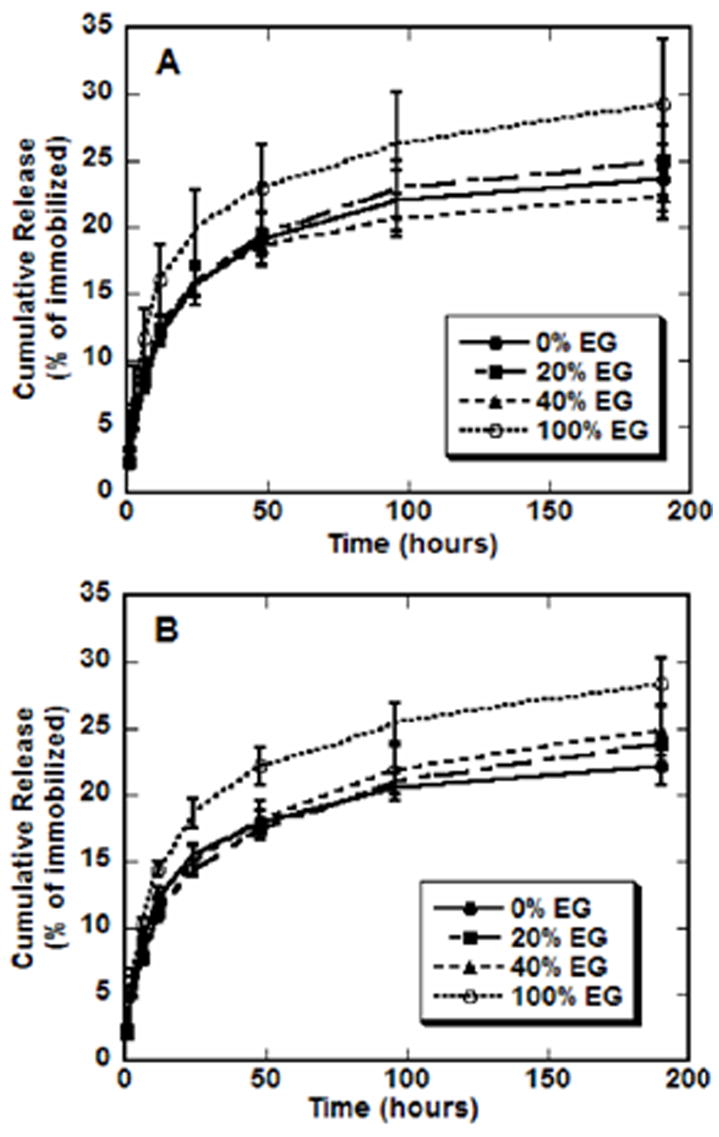
EG-containing SAMs and release. Radiolabeled DNA was used to quantify the amount of DNA released from each type of SAM (• 0% EG, ■ 20% EG, ▲ 40% EG, ○ 100% EG) into serum-containing media. Complexes formed at N/P 10 (A) and 25 (B). Values are reported as cumulative percentage released, reported as the mean ± SD at each time point.
3.4. Transfection on SAMS on gold
3.4.1. Transgene expression
SAMs with various densities of EG-terminated alkanethiols were explored to examine the ability of PEG on a surface to affect substrate-mediated gene delivery. While binding and release were not affected by the EG in the surface, expression levels increased with the percentage of EG alkanethiols in the SAM (Fig. 4) up to 40% EG. For the lower N/P (Fig. 4a), expression was significantly greater on 40% EG SAMs than surfaces containing no EG alkanethiols (p < 0.05) or surfaces containing only EG alkanethiols (p < 0.01); the latter observation presumably due to low cell binding. For complexes at the high N/P (Fig. 4b), transfection was again significantly greater on SAMs containing 40% EG than surfaces with no EG (p < 0.05); however, transfection was not statistically different between the 40% EG condition and SAMs with only EG alkanethiols, suggesting both significant cell adhesion and subsequent transfection on these surfaces. This issue of cell adhesion to complexes immobilized on EG-terminated alkanethiols is discussed in the next section. These findings, that increasing the percentage of EG groups and thus decreasing the percentage of carboxylic acid groups increases transfection, are in contrast to previous studies, where surfaces with 100% carboxylic acid functional groups resulted in highest transfection levels, presumably due to high binding efficiencies [15]. However, in the present study, the presence of EG in the surface did not affect complex immobilization, and therefore the amount of DNA bound did not contribute to enhanced transfection.
Fig. 4.
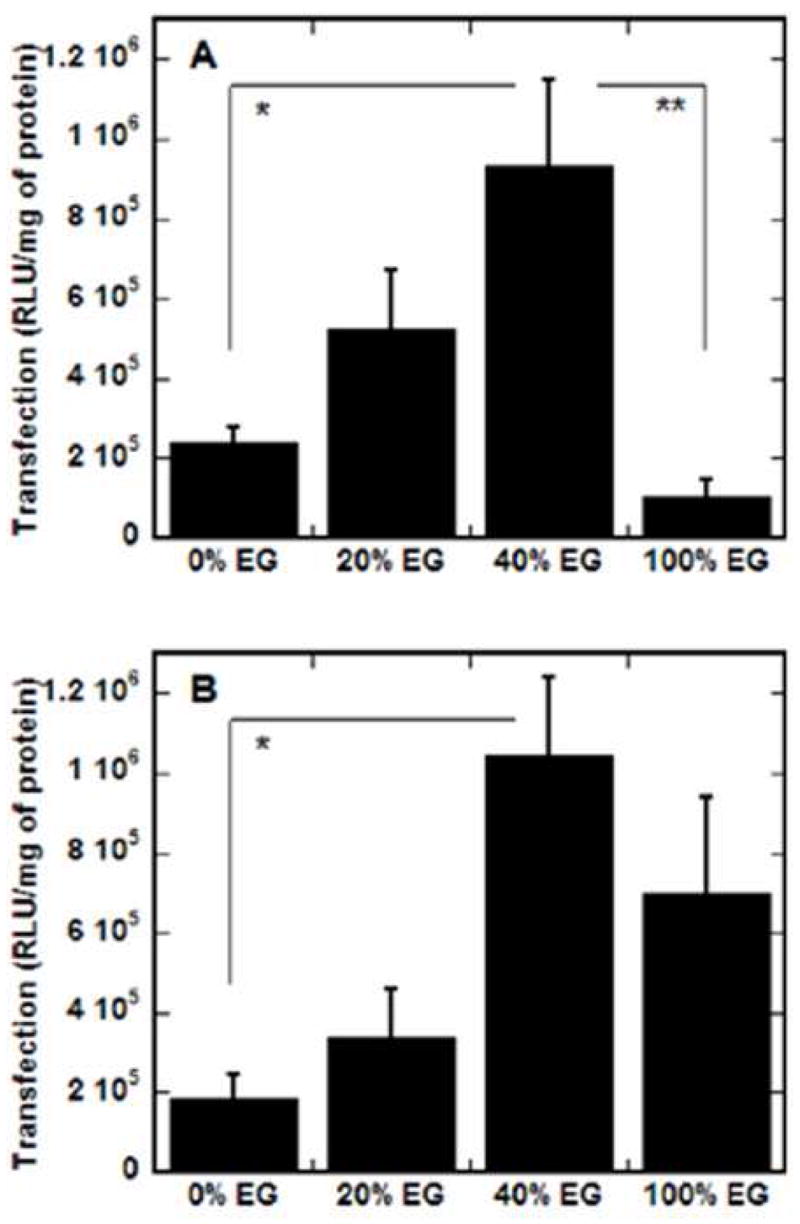
EG-containing SAMs and substrate-mediated transfection. SAMs were formed with increasing percentages of EG groups in a background of MUA (COO−) and transfection was assayed for complexes formed at N/P of 10 (A) and 25 (B) by normalizing luciferase levels to total protein amounts. Values are reported as the mean ± SD (*p < 0.05, **p < 0.01).
EG groups on the surface may enhance transfection through multiple mechanisms, including PEG–complex and PEG–cell interactions. PEG incorporated into DNA complexes of cationic polymers can reduce their surface charge [25,26,31,34], cytotoxicity [25,27,31] and interaction with blood components [25], as well as prevent aggregation through steric stabilization [25,26,29–31,33,34]. Because of its ability to modulate the properties of complexes, PEG has been reported to enhance transfection when attached to polymer–DNA complexes directly [24,27,33,34] or simply added to transfection media containing lipoplexes [49]. The enhancement of transfection by EG-containing SAMs could be attributed to the possible modulation of complex properties (surface charge, aggregation, complex–cell interaction), similar to the effects of PEG with soluble complexes, by mere association of the EG groups with the complexes on the surface. This enhancement of gene expression with surface PEG has similarly been observed with the addition of Pluronic, a block co-polymer of poly(ethylene oxide)–poly(propylene oxide)–poly(ethylene oxide), to microfluidic channels used for deposition of DNA complexes. Pluronics enhanced substrate-mediated transfection within these channels, possibly by preventing aggregation or enhancing complex association with the cells [18]. Therefore, the addition of PEG-like moieties to solutions of complexes during deposition could be an alternative method to enhance substrate-mediated gene delivery.
However, the association of the cell membrane with the SAMs containing EG groups could also contribute to an enhancement of transfection. PEG is known to induce association and fusion of phospholipid vesicles at high concentrations [50], widely used in the cell fusion required for formation of hybridomas. Ross and Hui have demonstrated that addition of PEG to transfection media containing lipoplexes increases the association of lipoplexes with the cell membrane up to 100-fold over controls without PEG [49]. They proposed that this enhanced association of lipoplexes with the cell could occur through a depletion effect, which produces an attractive osmotic force due to a PEG depleted region near the bilayer surface [49]. Thus, EG groups on the surface of the SAMs may be enhancing the interaction between the immobilized complexes and the cells through a similar depletion effect, which may promote internalization of the DNA complexes.
By incorporating PEG groups onto the SAM surface, complex properties and transfection can be enhanced, but the EG groups cannot interfere with complexation. PEGylation of the polymer prior to DNA association can interfere with complexation [36], which can reduce transfection [28,29,32]. Furthermore, in typical PEGylation strategies, the surface charge of polymer–DNA complexes decreases due to the shielding effect of PEG [26], with PEG side chains covering the surface of complexes and hindering the interaction between the complex and cell membrane. However, with EG present on the surface, complexes can be formed with the traditional procedures for subsequent immobilization. The presence of the EG groups on the surface should not interfere with cellular association of the complexes as previously reported [24,26], as the charge of the complexes would be unaffected by immobilization on EG-containing SAMs.
3.4.2. Cell adhesion on immobilized complexes
For complexes at the high N/P (Fig. 4b), high transfection levels on SAMs with only EG alkanethiols indicate cell adhesion on these surfaces. These surfaces had previously resisted cell adhesion (Fig. 1), presumably through resisting protein adsorption [39,40]. However, adhesion studies on SAMs containing only EG alkanethiols (100% EG) with immobilized complexes or PEI alone demonstrate significant cell adhesion (Fig. 5). PEI added alone at concentrations equivalent to N/P 10 (Fig. 5a) or N/P 25 (Fig. 5b) promoted cell adhesion, though cell monolayers were not as robust as those on surfaces with less EG alkanethiols and no PEI (Fig. 1). DNA complexes were also able to enhance cell adhesion, at N/P 10 (Fig. 5c) and N/P 25 (Fig. 5d). For high N/P, protein levels determined on the surface were low, indicating low cell numbers. Correspondingly low luciferase levels (data not shown), normalized to these low protein levels, result in robust transfection levels, and could give the appearance of enhancement of transfection on 100% EG surfaces (Fig. 4b).
Fig. 5.
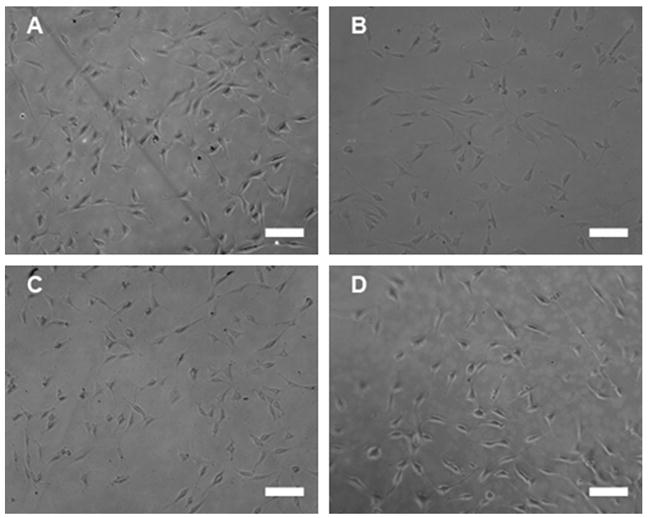
Cell adhesion on complexes immobilized on EG-containing SAMs. The morphology and adhesion of cells was observed 48 h after seeding cells on SAMs of (A) 100% EG with PEI concentration equivalent to N/P 10, (B) 100% EG with PEI concentration equivalent to N/P 25, (C) 100% EG with PEI–DNA complexes formed at N/P 10 and (D) 100% EG with PEI–DNA complexes formed at N/P. Scale bars correspond to 100 μm.
As complexes bind to surfaces through mechanisms similar to proteins [6,15], their binding on surfaces containing PEG groups would also be expected to be reduced. However, binding studies reveal that increasing the percentage of EG groups on the surface did not affect the amount of DNA complex immobilization (Fig. 2). The relatively high contact angles reported for EG-containing SAMs are consistent with an outer phase that exposes CH2 groups to solution [38]. PEG, while known as a hydrophilic polymer, is truly amphiphilic, with both hydrophilic and hydrophobic components, and this combination of properties is a key to its biocompatibility [51]. PEI molecules contain ethylene units along the backbone, and thus hydrophobic interactions between these ethylene units and the exposed CH2 groups of the EG-terminated alkanethiols could allow for immobilization of PEI–DNA complexes on EG-containing SAMs, similar to reports of DNA complex binding through hydrophobic interactions [15,52]. By binding to the SAMs containing EG alkanethiols, PEI–DNA complexes or free PEI could coat the surface, producing substrates with net positive charge, which could allow for protein immobilization through electrostatic interactions [53], and subsequent cell adhesion. This hypothesis agrees well with our transfection results, as complexes at high N/P (25) would have much more free PEI in solution than complexes at low N/P, which could further coat the surface and promote cell adhesion and thus transfection.
3.4.3. Transfection efficiency on SAMs
Transfection efficiency, reported as a percentage of transfected cells, was subsequently quantified for complexes immobilized on EG-containing SAMs (Fig. 6). β-Galactosidase expression was assayed at 48 h using phase microscopy. For both N/P ratios, the transfection efficiency increased as the percentage of EG groups within the SAMs increased, similar to the trend observed with the transfection levels. However, for both complex formulations, SAMs containing only EG groups resulted in the highest percentage of transfected cells. At low N/P (Fig. 6a), the transfection efficiency on 100% EG SAMs was statistically greater than surfaces containing 20% (p<0.05) or no EG groups (p<0.01). Additionally, 40% EG SAMs resulted in statistically greater transfection efficiency than 0% EG (p<0.05). For N/P of 25 (Fig. 6b), 100% EG SAMs supported the highest transfection efficiency, though the standard deviation was large and statistical tests did not indicate significant differences.
Fig. 6.
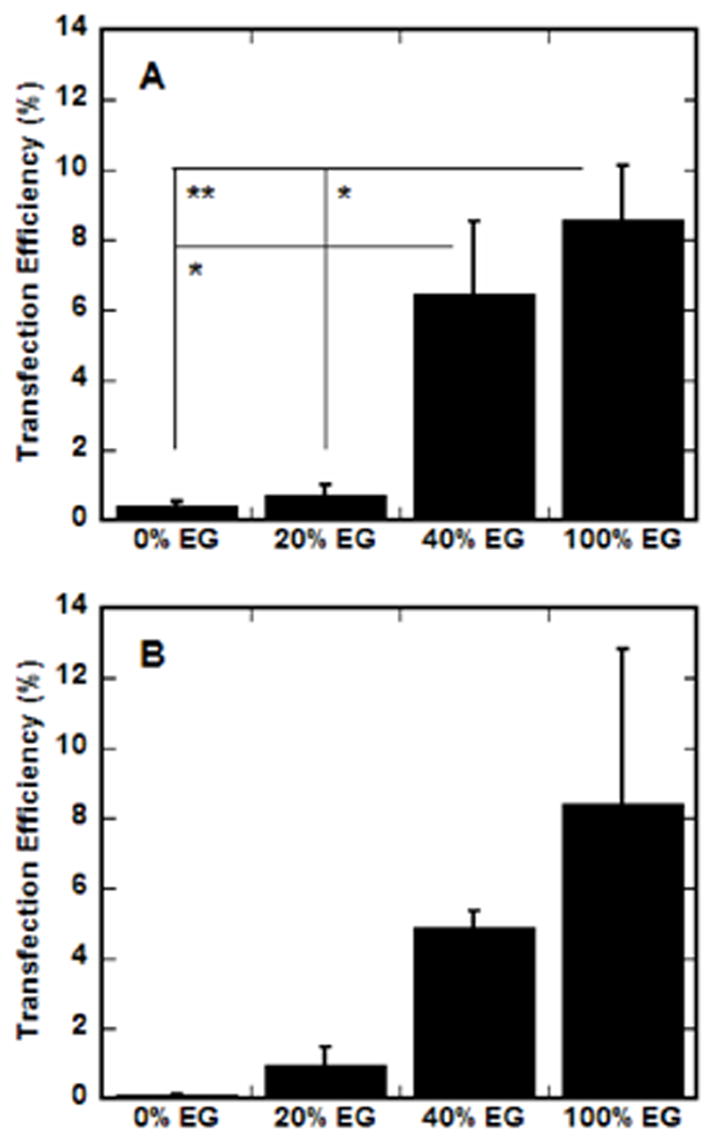
EG-containing SAMs and transfection efficiency. SAMs were formed with increasing percentages of EG groups in a background of MUA (COO−) and transfection efficiency was assayed for complexes formed at N/P of 10 (A) and 25 (B) by counting the number of cell expressing β-galactosidase and dividing by the total number of cells. Values are reported as the mean ± SD (*p < 0.05, **p < 0.01).
The transfection efficiency data indicate that more cells are transfected on surfaces containing more EG groups, but do not reflect that total cell numbers were reduced on the 100% EG SAMs. No trend could be determined between the number of cells on the surface (indirectly measured by BCA assay) and the percentage of EG in the monolayer with immobilized complexes (data not shown). For N/P 10, cell numbers were similar for all percentages of EG, but slightly higher for SAMs containing 20 and 100% PEG. For N/P of 25, cell numbers are nearly identical for all SAMs, except much lower for 100% EG (data not shown), consistent with our transfection efficiency results, which demonstrate few cells adhered (data not shown). Those cells that did adhere to these surfaces (100% EG, N/P 25) were more likely to be transfected than in the corresponding N/P 10 condition, resulting in huge variability in transfection efficiency evidenced by the large standard deviation (Fig. 6b).
3.4.4. Transfection enhancement specificity
SAMs containing combinations of alkanethiols with alternative functional groups were used to determine if the enhancement in transfection was specific to surfaces containing EG and COO− groups (Fig. 7). Alternative functional groups included OH and CH3 in backgrounds of 40% EG (Fig. 7a and b) and 60% COO− (Fig. 7c and d), as these compositions provide maximal expression levels and transfection efficiency with optimal cell adhesion and morphology. For complexes with N/P of 10 (Fig. 7a and c), surfaces containing the combination of 40% EG and 60% COO− groups resulted in statistically higher transfection than surfaces containing OH or CH3 groups in background of EG (p < 0.05) or COO− (p < 0.01). On SAMs with a background of 40% EG, complexes with the higher N/P of 25 (Figs. 7b) resulted in statistically higher transfection (p < 0.05) on surfaces with either hydrophilic functional group (COO− or OH). However, transfection by complexes with N/P 25 on SAMs with 60% COO− background (Fig. 7d) was statistically greater on surfaces with 40% EG than on SAMs with 40% OH (p < 0.05) or CH3 (p < 0.01). Therefore, the highest transgene expression occurs on SAMs with combinations of EG groups and carboxylic acids, which could indicate that charged functional groups contribute important properties within the monolayer, presumably providing reversible interactions between the substrate and complex that can also enhance transfection, similar to previous reports [15].
Fig. 7.
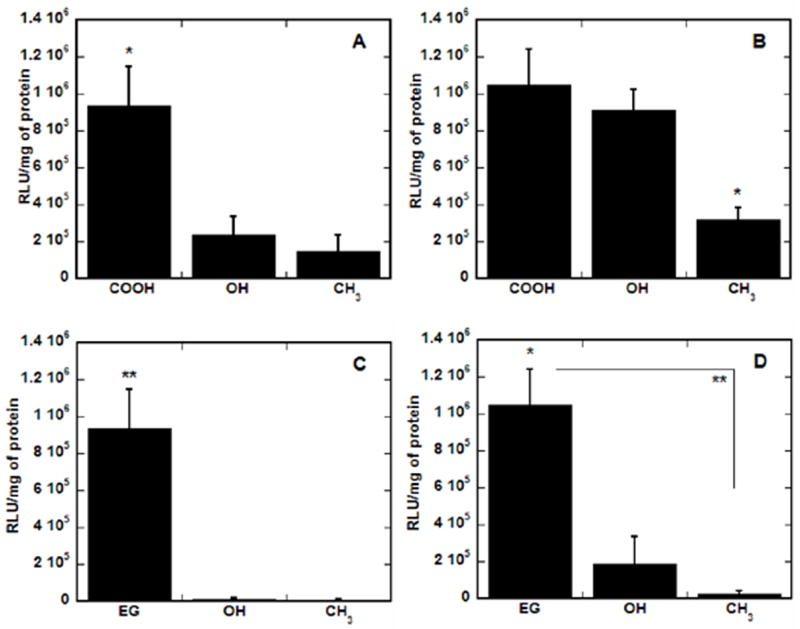
Substrate-mediated transfection on SAMs with backgrounds of EG and COO−. SAMs were formed with alkanethiols containing various terminal functional groups, including OH, CH3, EG and COO−, in backgrounds of 40% EG (A and B) and 60% COO− (C and D). Transfection was assayed for complexes formed at N/P of 10 (A and C) and 25 (B and D) by normalizing luciferase levels to total protein amounts. Values are reported as the mean ± SD (*p < 0.05, **p < 0.01).
3.5. Atomic force microscopy imaging of immobilized complexes
The enhancement in transfection levels and efficiency on SAMs containing EG groups cannot be attributed to binding densities of complexes or release profiles, thus the morphology of the complexes was examined using AFM. AFM has proven to be an excellent tool to image soft biological structures and can be performed after complex deposition on a surface, with high resolution. AFM images were obtained by scanning SAMs with immobilized complexes and representative images (Fig. 8a, c, e and g) revealed the morphology of PEI–DNA complexes on SAMs containing 0% EG (100% COO−) (Fig. 8a and e) and 40% EG (60% COO−) (Fig. 8c and g). On surfaces with no EG groups, complexes formed at N/P of 10 (Fig. 8a) exhibited large globular structures with varying heights (indicated by light intensity of image) and diameters, along with smaller structures throughout the background of the image. Further analysis of the height distribution of these complexes (Fig. 8b) reveals a rather large distribution of particles, with average heights of 20 nm (Table 2) and a mean roughness of 12 nm (Table 2). Note that images demonstrated two distinct regions: regions with large globular structures and regions with smaller, more uniform particles. Area–perimeter ratios were analyzed in each region, with ratios ranging from 337 to 1270 nm (Table 2). In contrast, complexes (N/P 10) on SAMs containing 40% EG (60% COO−) (Fig. 8c) had a very uniform morphology of small spherical particles evenly distributed across the surface. Analysis of the representative AFM image resulted in a narrow height distribution (Fig. 8d), with an average height of 8 nm, a mean roughness of 5 nm and an area–perimeter ratio of 100 nm (Table 2). These values are all much lower than for the corresponding 0% EG condition, indicating smaller, more uniform complexes on surfaces containing EG groups, which in turn resulted in higher transfection levels.
Fig. 8.
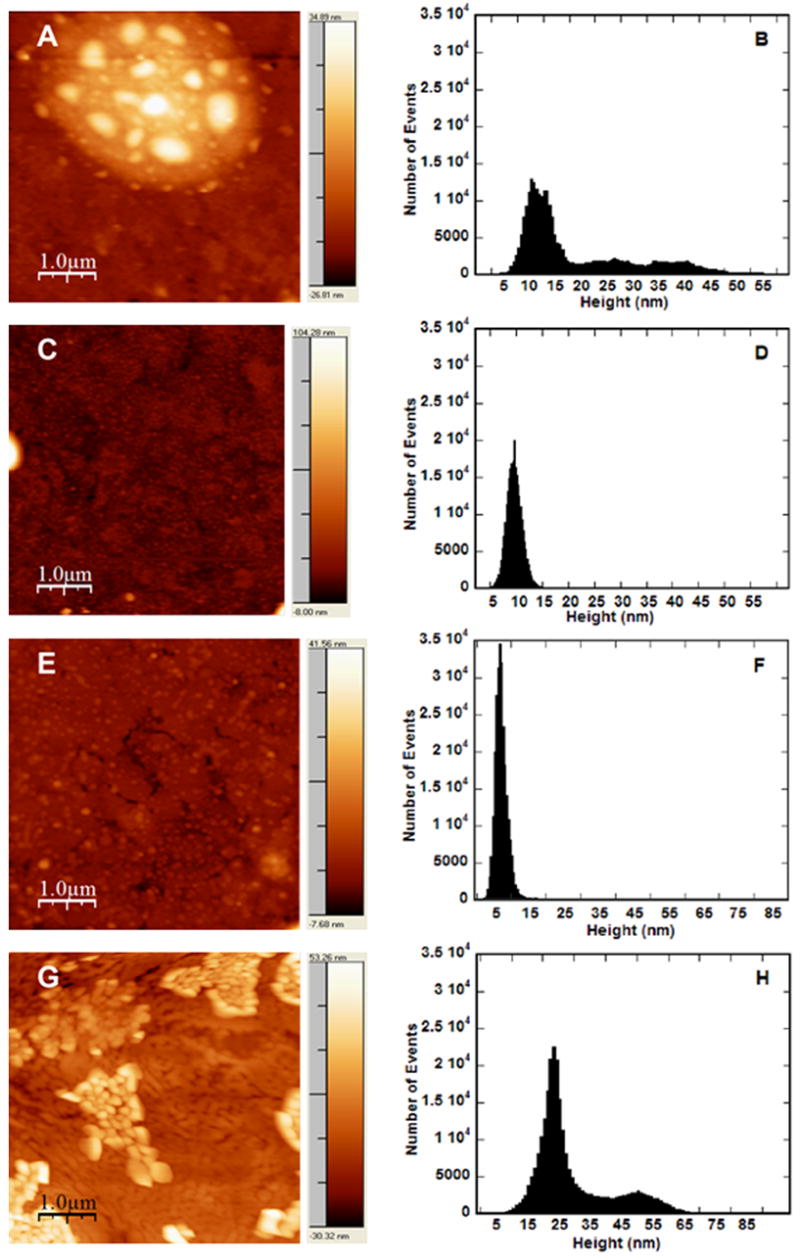
AFM images and analysis of complexes immobilized on SAMs. Complexes were formed at N/P of 10 (A–D) and N/P 25 (E–H) and immobilized on SAMs formed with 0% EG/100% COO− (A, B, E, F) or 40% EG/60% COO− (C, D, G, H). Pixel brightness in images (A, C, E, G) corresponds to particle height. Scale bars correspond to 1.0 μm. Height analysis histogram (B, D, F, H) reports analysis of image to left.
Table 2.
AFM analysis of PEI–DNA complexes on SAMs
For complexes formed at N/P of 25, the correlation between complex morphology and surface chemistry was reversed. On SAMs containing no EG-terminated alkanethiols (Fig. 8e), complexes exhibited a uniform morphology of small particles evenly distributed across the surface. Analysis of the representative AFM image resulted in a narrow height distribution (Fig. 8f), with an average height of 8 nm, a mean roughness of 2 nm and an area–perimeter ratio of 72 nm (Table 2), similar to complexes with N/P 10 on surfaces containing 40% EG (Fig. 8c). In contrast, complexes (N/P 25) on surfaces with 40% EG (60% COO−) (Fig. 8g) exhibited large aggregates of globular structures with varying heights and diameters. Further analysis of the height distribution of these complexes (Fig. 8h) reveals a rather large distribution of particles with two main populations of complexes, with an average height of 32 nm, a mean roughness of 12 nm and area–perimeter ratios ranging from 196 to 355 nm (Table 2), depending on the region analyzed. These values are higher than for the corresponding 0% EG condition, indicating smaller, more uniform complexes on surfaces containing no EG groups, but resulting in lower transfection levels.
Previous studies examining complexes using AFM, typically on mica substrates, reported a range of complex morphologies similar to the results presented here. PEI–DNA complexes had a disperse distribution of condensates with rounded, globular forms [54,55]. PEGylated PEI–DNA complexes analyzed by AFM had defined, spherical complexes [30–32], with less aggregation and smaller diameters than similar complexes without PEG, but were also demonstrated to be less uniform.
In the AFM images and analysis of PEI–DNA complexes formed at an N/P of 10, the presence of EG groups on the SAM reduced the average height and the area–perimeter ratio, and thus and the size of complexes (Fig. 8c vs. Fig. 8a). The surface PEG may function to decrease aggregation of the complexes on the surfaces, similar to studies that have demonstrated that PEGylation of polymer–DNA complexes can prevent salt-induced aggregation [25,26,29,34], to produce discrete particles with similar appearance with AFM [30,31,33]. Treatment of microfluidic channels with Pluronic has similarly demonstrated lower aggregation of complexes, evidenced by a homogeneous layer of deposited lipoplexes [18]. Alternatively, the presence of EG groups could be preventing complex unfolding, as conformational changes are widely reported at surfaces for other macromolecules [56–58]. This change in complex morphology on SAMs containing EG terminal functional groups correlates with high transfection levels and transfection efficiencies, and could indicate the ability of EG-containing SAMs to preserve complex morphology upon binding by limiting electrostatic interactions with the surface and reducing aggregation of the complexes through steric stabilization, or alternatively by preventing complex unfolding, which could also contribute to the large height and area–perimeter ratios of the complexes immobilized on surfaces without EG.
However, in the AFM images and analysis of PEI–DNA complexes formed at N/P of 25 (Fig. 8e and f), the presence of EG groups on the SAM leads to large particles that appear in clumps, suggesting the aggregation of complexes. The formation of these aggregates, which are morphologically different from the large, disperse globules observed for complexes of N/P of 10 on 0% EG SAMs yet produce high levels of expression, could be attributed to excess free PEI in the N/P 25 complexes, resulting in a layer of PEI on the surface prior to complex immobilization, which could alter the conformation of the complexes. Alternatively, these large aggregates could also be attributed, in part, to evaporation prior to AFM imaging. Though all samples were treated identically, drying has the potential to cause artifacts induced by the receding meniscus during drying [59]. Taken together, these AFM studies demonstrate how the design of both the surface and vector contribute to the morphology of immobilized complexes and how that, in turn, can affect substrate-mediated transfection.
Our observations with the adsorption of non-viral vectors are consistent with the mechanisms proposed for protein adsorption to surfaces. Protein adsorption has been described as occurring in three general steps: (i) partial dehydration of proteins and surfaces; (ii) redistribution of the groups at the interface; and (iii) conformational changes in the protein [60,61]. The functionality of the adsorbed proteins is determined by the adsorption profile (e.g. amount) and the protein bioactivity. PEGylation of surfaces typically increases hydrophilicity, which influences the quantity of protein bound and also its surface conformations [60,61]. In this report, non-viral vectors, which are a collection of DNA and cationic polymers rather than a single molecule, were adsorbed. Consistent with protein adsorption, we have previously demonstrated that hydrophilicity and surface charge influenced vector binding and transfection [15]. The addition of EG to these hydrophilic surfaces did not impact the quantity of bound complexes or their release, but influenced their conformation. The conformation of non-viral vectors has been a well-characterized factor that influences vector binding, internalization and intracellular trafficking [62], which contributes to the gene transfer efficiency. Thus, our results suggest that surface EG maintains or produces conformations that promote gene transfer more effectively.
4. Conclusions
Substrate-mediated delivery describes the immobilization of DNA, complexed with cationic lipids or polymers, to a biomaterial or substrate. Efficient delivery of DNA complexes from a surface is dependent on the interactions between the substrate and the complexes. SAMs presenting EG groups were used to investigate surfaces containing PEG-like moieties for complex binding, release, transfection and morphology. Nonspecific complex immobilization to SAMs containing combinations of EG- and COO−-terminated alkanethiols resulted in substantially greater transfection, which increased with increasing amounts of EG in the monolayer. These significant differences in transfection levels and efficiency on SAMs containing EG groups could not be attributed to binding densities of complexes or release profiles, nor could they be replicated in SAMs composed of EG-terminated alkanethiols combined with alkanethiols presenting other functional groups. This enhancement of transfection by EG-containing SAMS could be attributed to interactions through a proposed depletion mechanism [49], or by possible modulation of complex properties. We demonstrate that complex properties (surface charge, aggregation, complex–cell interaction) are modulated by mere association of the EG groups with the complexes. Atomic force microscopy imaging of immobilized complexes supports this hypothesis, revealing that the presence of PEG on the surface significantly affects the morphology of the complexes, which correlates with greater high transfection levels and transfection efficiencies. The ability to control the morphology of the immobilized complexes and thus influence transfection levels could be translated to scaffolds for gene delivery in tissue engineering applications [5,6], as well other applications of substrate-mediated gene delivery, including transfected cell arrays [63].
Acknowledgments
Support for this research was provided in part by grants from NIH (RO1 GM066830 to LDS, CMBD Training Grant to AKP) and NSF (Graduate Research Fellowship to AKP). Gold evaporation was performed at the Materials Processing and Crystal Growth core facility and the AFM measurements were performed at NIFTI/Nuance core facility, both at Northwestern University (Evanston, IL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9:1647. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- 2.Herweijer H, Wolff JA. Progress and prospects: naked DNA gene transfer and therapy. Gene Ther. 2003;10:453. doi: 10.1038/sj.gt.3301983. [DOI] [PubMed] [Google Scholar]
- 3.Segura T, Shea LD. Materials for non-viral gene delivery. In: Stupp S, editor. Ann Rev Mater Sci. Vol. 31. 2001. p. 25. Annual Reviews. [Google Scholar]
- 4.Ledley FD. Pharmaceutical approach to somatic gene therapy. Pharm Res. 1996;13:1595. doi: 10.1023/a:1016420102549. [DOI] [PubMed] [Google Scholar]
- 5.Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol Ther. 2004;10:19. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 6.Bengali Z, Shea LD. Gene delivery by immobilization to cell-adhesive substrates. MRS Bulletin. 2005;30:659. doi: 10.1557/mrs2005.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjug Chem. 2002;13:621. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- 8.Segura T, Volk MJ, Shea LD. Substrate-mediated DNA delivery: role of the cationic polymer structure and extent of modification. J Control Release. 2003;93:69. doi: 10.1016/j.jconrel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid–collagen hydrogels via a substrate-mediated approach. Biomaterials. 2005;26:1575. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielinska AU, Yen A, Wu HL, Zahos KM, Sun R, Weiner ND, Baker JR, Jr, Roessler BJ. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21:877. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- 11.Kneuer C, Sameti M, Bakowsky U, Schiestel T, Schirra H, Schmidt H, Lehr CM. A nonviral DNA delivery system based on surface modified silica-nanoparticles can efficiently transfect cells in vitro. Bioconjug Chem. 2000;11:926. doi: 10.1021/bc0000637. [DOI] [PubMed] [Google Scholar]
- 12.Manuel WS, Zheng JI, Hornsby PJ. Transfection by polyethyleneimine-coated microspheres. J Drug Target. 2001;9:15. doi: 10.3109/10611860108995629. [DOI] [PubMed] [Google Scholar]
- 13.Bengali Z, Pannier AK, Segura T, Anderson BC, Jang JH, Mustoe TA, Shea LD. Gene delivery through cell culture substrate adsorbed DNA complexes. Biotechnol Bioeng. 2005;90:290. doi: 10.1002/bit.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JT, Chua LS, Lynn DM. Multilayered thin films that sustain the release of functional DNA under physiological conditions. Langmuir. 2004;20:8015. doi: 10.1021/la048888i. [DOI] [PubMed] [Google Scholar]
- 15.Pannier AK, Anderson BC, Shea LD. Substrate-mediated delivery from self-assembled monolayers: effect of surface ionization, hydrophilicity, and patterning. Acta Biomaterialia. 2005;1:511. doi: 10.1016/j.actbio.2005.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korobko AV, Backendorf C, van der Maarel JR. Plasmid DNA encapsulation within cationic diblock copolymer vesicles for gene delivery. J Phys Chem B Condens Matter Mater Surf Interfaces Biophys. 2006;110:14550. doi: 10.1021/jp057363b. [DOI] [PubMed] [Google Scholar]
- 17.Reinisalo M, Urtti A, Honkakoski P. Freeze-drying of cationic polymer DNA complexes enables their long-term storage and reverse transfection of post-mitotic cells. J Control Release. 2006;110:437. doi: 10.1016/j.jconrel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Houchin-Ray T, Whittlesey KJ, Shea LD. Spatially patterned gene delivery for localized neuron survival and neurite extension. Mol Ther. 2007;15:705. doi: 10.1038/mt.sj.6300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulman A. Formation and structure of self-assembled monolayers. Chem Rev. 1996;96:1533. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 20.Sigal GB, Mrksich M, Whitesides GM. Effect of surface wettability on the adsorption of proteins and detergents. J Am Chem Soc. 1998;120:3464. [Google Scholar]
- 21.van der Veen M, Norde W, Stuart MC. Electrostatic interactions in protein adsorption probed by comparing lysozyme and succinylated lysozyme. Colloids and Surfaces B-Biointerfaces. 2004;35:33. doi: 10.1016/j.colsurfb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Yamauchi F, Kato K, Iwata H. Micropatterned, self-assembled monolayers for fabrication of transfected cell microarrays. Biochim Biophys Acta. 2004;1672:138. doi: 10.1016/j.bbagen.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Volcke C, Pirotton S, Grandfils C, Humbert C, Thiry PA, Ydens I, Dubois P, Raes M. Influence of DNA condensation state on transfection efficiency in DNA/polymer complexes: an AFM and DLS comparative study. J Biotechnol. 2006;125:11. doi: 10.1016/j.jbiotec.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3:E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin–PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6:595. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 26.Sung SJ, Min SH, Cho KY, Lee S, Min YJ, Yeom YI, Park JK. Effect of polyethylene glycol on gene delivery of polyethylenimine. Biol Pharm Bull. 2003;26:492. doi: 10.1248/bpb.26.492. [DOI] [PubMed] [Google Scholar]
- 27.Banerjee P, Weissleder R, Bogdanov A., Jr Linear polyethyleneimine grafted to a hyperbranched poly(ethylene glycol)-like core: a copolymer for gene delivery. Bioconjug Chem. 2006;17:125. doi: 10.1021/bc050083e. [DOI] [PubMed] [Google Scholar]
- 28.Erbacher P, Bettinger T, Belguise-Valladier P, Zou S, Coll JL, Behr JP, Remy JS. Transfection and physical properties of various saccharide, poly(ethylene glycol), and antibody-derivatized polyethylenimines (PEI) J Gene Med. 1999;1:210. doi: 10.1002/(SICI)1521-2254(199905/06)1:3<210::AID-JGM30>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 29.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur J Cell Biol. 2004;83:97. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 30.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Influence of polyethylene glycol chain length on the physicochemical and biological properties of poly(ethylene imine)-graft-poly(ethylene glycol) block copolymer/SiRNA polyplexes. Bioconjug Chem. 2006;17:1209. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 31.Petersen H, Fechner PM, Martin AL, Kunath K, Stolnik S, Roberts CJ, Fischer D, Davies MC, Kissel T. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug Chem. 2002;13:845. doi: 10.1021/bc025529v. [DOI] [PubMed] [Google Scholar]
- 32.Merdan T, Kunath K, Petersen H, Bakowsky U, Voigt KH, Kopecek J, Kissel T. PEGylation of poly(ethylene imine) affects stability of complexes with plasmid DNA under in vivo conditions in a dose-dependent manner after intravenous injection into mice. Bioconjug Chem. 2005;16:785. doi: 10.1021/bc049743q. [DOI] [PubMed] [Google Scholar]
- 33.Toncheva V, Wolfert MA, Dash PR, Oupicky D, Ulbrich K, Seymour LW, Schacht EH. Novel vectors for gene delivery formed by self-assembly of DNA with poly(L-lysine) grafted with hydrophilic polymers. Biochim Biophys Acta. 1998;1380:354. doi: 10.1016/s0304-4165(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 34.Mannisto M, Vanderkerken S, Toncheva V, Elomaa M, Ruponen M, Schacht E, Urtti A. Structure–activity relationships of poly(L-lysines): effects of pegylation and molecular shape on physicochemical and biological properties in gene delivery. J Control Release. 2002;83:169. doi: 10.1016/s0168-3659(02)00178-5. [DOI] [PubMed] [Google Scholar]
- 35.Guo Y, Sun Y, Li G, Xu Y. The molecular structures of poly(ethylene glycol)-modified nonviral gene delivery polyplexes. Mol Pharm. 2004;1:477. doi: 10.1021/mp0499205. [DOI] [PubMed] [Google Scholar]
- 36.Rackstraw BJ, Martin AL, Stolnik S, Roberts CJ, Garnett MC, Davies MC, Tendler SJ. Microscopic investigations into PEG-cationic polymer-induced DNA condensation. Langmuir. 2001;17:3185. [Google Scholar]
- 37.Lee M, Kim SW. Polyethylene glycol-conjugated copolymers for plasmid DNA delivery. Pharm Res. 2005;22:1. doi: 10.1007/s11095-004-9003-5. [DOI] [PubMed] [Google Scholar]
- 38.Pale-Grosdemange C, SImon ES, Prime KL, Whitesides GM. Formation of self-assembled monolayers by chemisorption of derivatives of oligo(ethylene glycol) of structure HS(CH2)11(OCH2CH2)mOH on gold. J Am Chem Soc. 1991;113:12. [Google Scholar]
- 39.Prime KL, Whitesides GM. Adsorption of proteins onto surfaces containing end-attached oligo(ethylene oxide): a model system using self-assembled monolayers. J Am Chem Soc. 1993;115:10714. [Google Scholar]
- 40.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. A survey of structure–property relationships of surfaces that resist the adsorption of protein. Langmuir. 2001;17:5605. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 41.Cooper E, Wiggs R, Hutt DA, Parker L, Leggett GJ, Parker TL. Rates of attachment of fibroblasts to self-assembled monolayers formed by the adsorption of alkylthiols onto gold surfaces. Journal of Materials Chemistry. 1997;7:435. [Google Scholar]
- 42.McClary KB, Ugarova T, Grainger DW. Modulating fibroblast adhesion, spreading, and proliferation using self-assembled monolayer films of alkylthiolates on gold. J Biomed Mater Res. 2000;50:428. doi: 10.1002/(sici)1097-4636(20000605)50:3<428::aid-jbm18>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 43.Deible CR, Petrosko P, Johnson PC, Beckman EJ, Russell AJ, Wagner WR. Molecular barriers to biomaterial thrombosis by modification of surface proteins with polyethylene glycol. Biomaterials. 1998;19:1885. doi: 10.1016/s0142-9612(98)00098-2. [DOI] [PubMed] [Google Scholar]
- 44.Jenney CR, Anderson JM. Effects of surface-coupled polyethylene oxide on human macrophage adhesion and foreign body giant cell formation in vitro. J Biomed Mater Res. 1999;44:206. doi: 10.1002/(sici)1097-4636(199902)44:2<206::aid-jbm11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 45.Jeon SI, Lee LH, Andrade JD, De Gennes PG. Protein–surface interactions in the presence of polyehtylene oxide. J Colloid Interface Sci. 1991;142:149. [Google Scholar]
- 46.McPherson T, Kidane A, Szleifer I, Park K. Prevention of protein adsorption by tethered poly(ethylene oxide) layers: experiments and single-chain mean-field analysis. Langmuir. 1998;14:176. [Google Scholar]
- 47.Falconnet D, Csucs G, Grandin HM, Textor M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials. 2006;27:3044. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 48.Ademovic Z, Holst B, Kahn RA, Jorring I, Brevig T, Wei J, Hou X, Winter-Jensen B, Kingshott P. The method of surface PEGylation influences leukocyte adhesion and activation. J Mater Sci Mater Med. 2006;17:203. doi: 10.1007/s10856-006-7306-2. [DOI] [PubMed] [Google Scholar]
- 49.Ross PC, Hui SW. Polyethylene glycol enhances lipoplex–cell association and lipofection. Biochim Biophys Acta. 1999;1421:273. doi: 10.1016/s0005-2736(99)00132-7. [DOI] [PubMed] [Google Scholar]
- 50.Yamazaki M, Ito T. Deformation and instability in membrane structure of phospholipid vesicles caused by osmophobic association: mechanical stress model for the mechanism of poly(ethylene glycol)-induced membrane fusion. Biochemistry. 1990;29:1309. doi: 10.1021/bi00457a029. [DOI] [PubMed] [Google Scholar]
- 51.Zhang F, Kang ET, Neoh KG, Huang W. Modification of gold surface by grafting of poly(ethylene glycol) for reduction in protein adsorption and platelet adhesion. J Biomater Sci Polym Ed. 2001;12:515. doi: 10.1163/156856201300194252. [DOI] [PubMed] [Google Scholar]
- 52.Delehanty JB, Shaffer KM, Lin B. A comparison of microscope slide substrates for use in transfected cell microarrays. Biosens Bioelectron. 2004;20:773. doi: 10.1016/j.bios.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 53.Wadu-Mesthrige K, Amro NA, Liu GY. Immobilization of proteins on self-assembled monolayers. Scanning. 2000;22:380. doi: 10.1002/sca.4950220607. [DOI] [PubMed] [Google Scholar]
- 54.Dunlap DD, Maggi A, Soria MR, Monaco L. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neu M, Sitterberg J, Bakowsky U, Kissel T. Stabilized nanocarriers for plasmids based upon cross-linked poly(ethylene imine) Biomacromolecules. 2006;7:3428. doi: 10.1021/bm060788z. [DOI] [PubMed] [Google Scholar]
- 56.Keselowsky BG, Collard DM, Garcia AJ. Surface chemistry modulates fibronectin conformation and directs integrin binding and specificity to control cell adhesion. J Biomed Mater Res A. 2003;66:247. doi: 10.1002/jbm.a.10537. [DOI] [PubMed] [Google Scholar]
- 57.Lan MA, Gersbach CA, Michael KE, Keselowsky BG, Garcia AJ. Myoblast proliferation and differentiation on fibronectin-coated self assembled monolayers presenting different surface chemistries. Biomaterials. 2005;26:4523. doi: 10.1016/j.biomaterials.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 58.Lee MH, Ducheyne P, Lynch L, Boettiger D, Composto RJ. Effect of biomaterial surface properties on fibronectin-alpha5beta1 integrin interaction and cellular attachment. Biomaterials. 2006;27:1907. doi: 10.1016/j.biomaterials.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 59.Hansma HG, Golan R, Hsieh W, Lollo CP, Mullen-Ley P, Kwoh D. DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res. 1998;26:2481. doi: 10.1093/nar/26.10.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial–cell interactions by adsorbed proteins: a review. Tissue Eng. 2005;11:1. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 61.Norde W, Lyklema J. Why proteins prefer interfaces. J Biomater Sci Polym Ed. 1991;2:183. doi: 10.1080/09205063.1991.9756659. [DOI] [PubMed] [Google Scholar]
- 62.Lin AJ, Slack NL, Ahmad A, Koltover I, George CX, Samuel CE, Safinya CR. Structure and structure-function studies of lipid/plasmid DNA complexes. J Drug Target. 2000;8:13. doi: 10.3109/10611860009009206. [DOI] [PubMed] [Google Scholar]
- 63.Pannier AK, Ariazi EA, Bellis AD, Bengali Z, Jordan VC, Shea LD. Bioluminescence imaging for assessment and normalization in transfected cell arrays. Biotechnol Bioeng. 2007 doi: 10.1002/bit.21477. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]


