Abstract
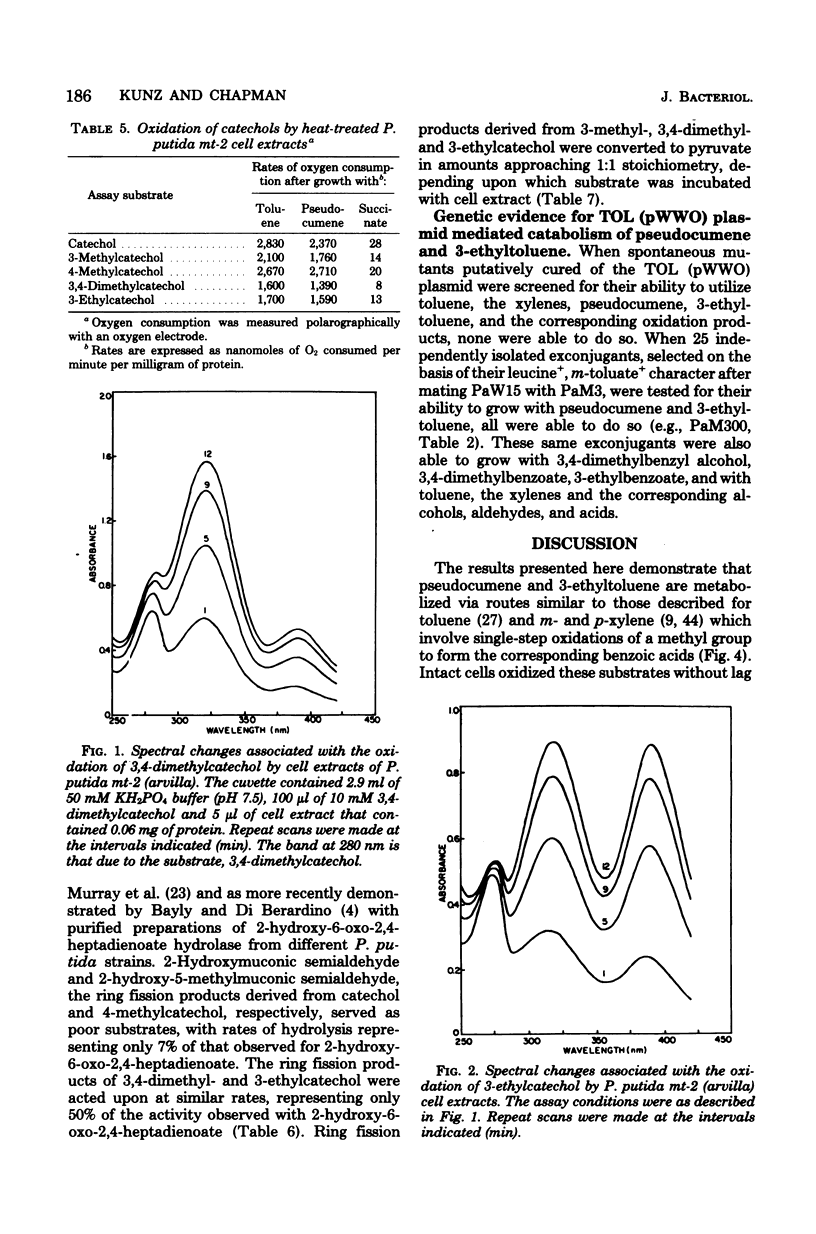
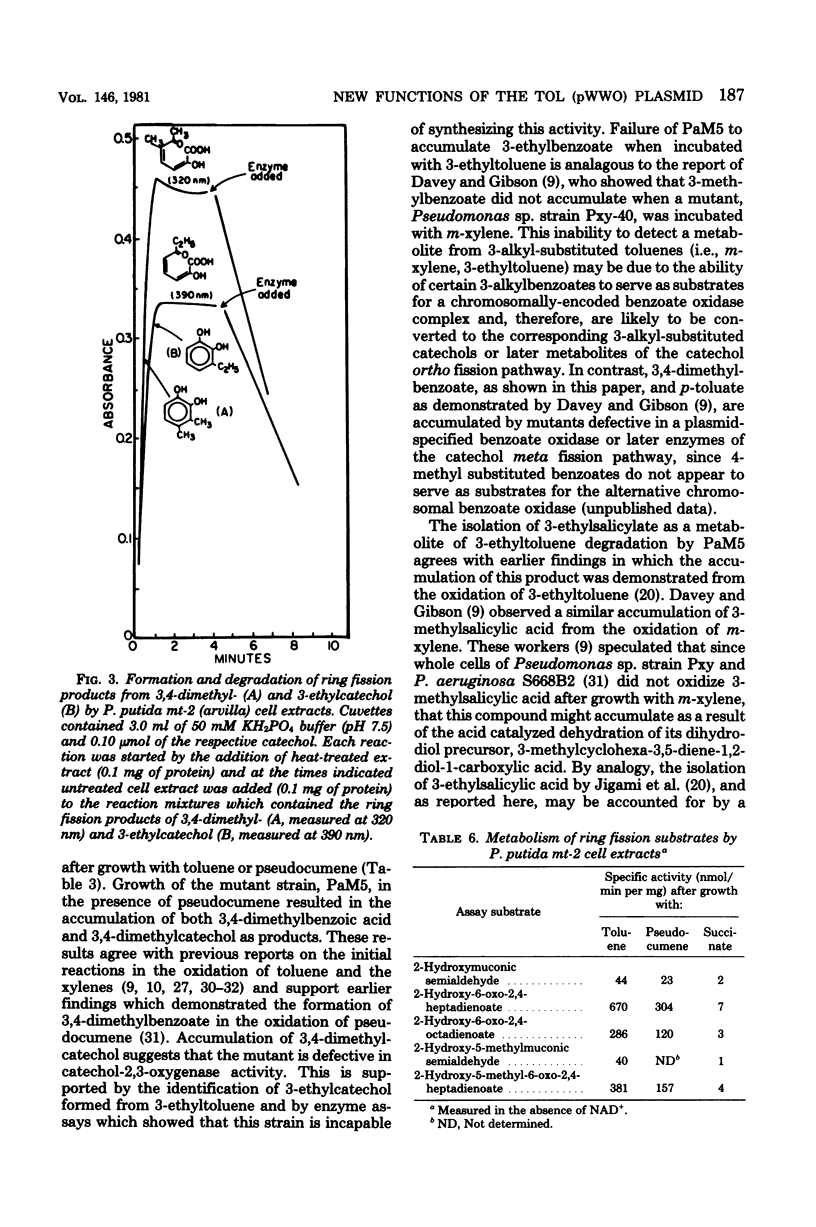
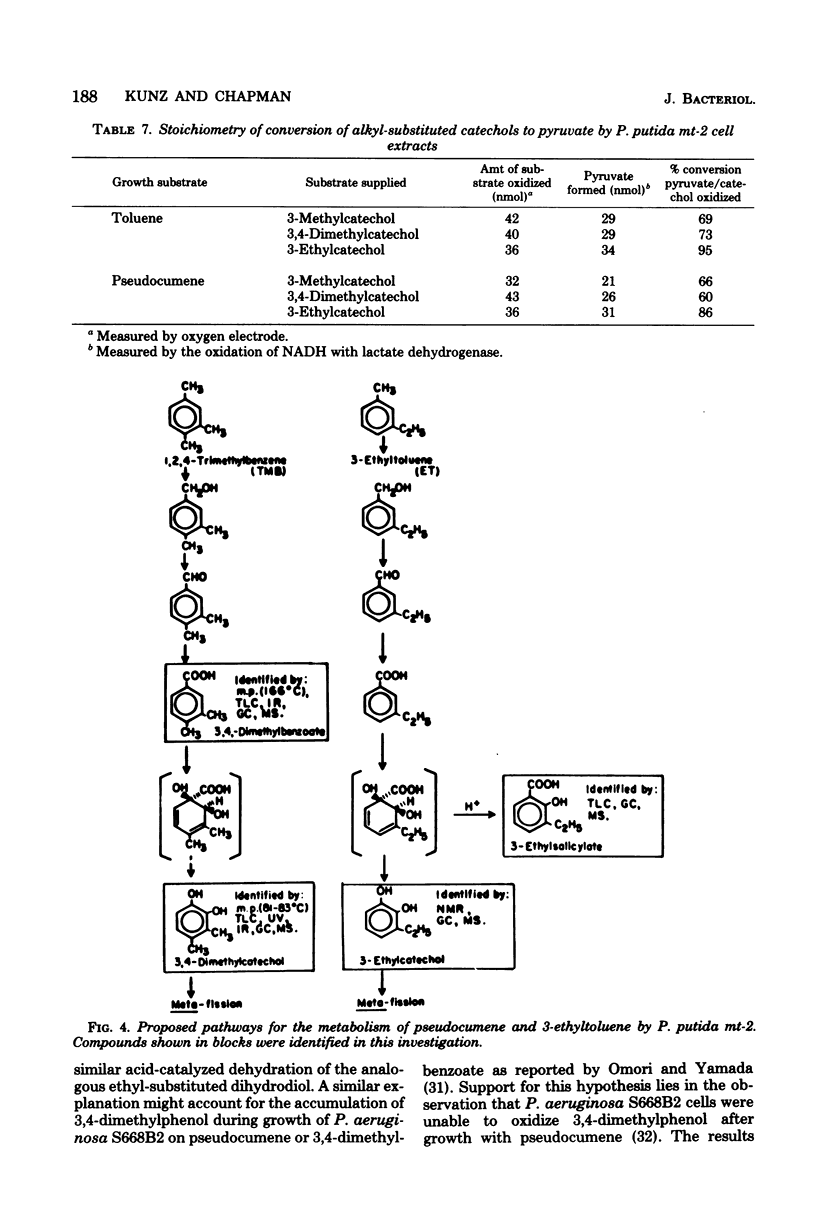
Pseudocumene (1,2,4-trimethylbenzene) and 3-ethyltoluene were found to serve as growth substrates for Pseudomonas putida (arvilla) mt-2, in addition to toluene, m-xylene, and p-xylene as previously described. Similar observations were made with several additional P. putida strains also capable of growth with toluene and the xylenes. Additional substrates which supported the growth of these organisms included 3,4-dimethylbenzyl alcohol, 3,4-dimethylbenzoate, and 3-ethylbenzoate. P. putida mt-2 cells grown either with toluene or pseudocumene rapidly oxidized toluene, pseudocumene, and 3-ethyltoluene as well as 3,4-dimethylbenzoate, 3-ethylbenzoate, 3,4-dimethylcatechol, and 3-ethylcatechol. Cell extracts from similarly grown P. putida mt-2 cells catalyzed a meta fission of 3,4-dimethylcatechol and 3-ethylcatechol to compounds having the spectral properties of 2-hydroxy-5-methyl-6-oxo-2,4-heptadienoate and 2-hydroxy-6-ox-2,4-octadienoate, respectively. The further metabolism of these intermediates was shown to be independent of oxidized nicotinamide adenine dinucleotide (NAD+) and resulted in the formation of essentially equimolar amounts of pyruvate, indicating that each ring fission product was degraded via the hydrolytic branch of the meta fission pathway. Treatment of cells with N-methyl-N'-nitro-N-nitrosoguanidine led to the isolation of a mutant, which when grown with succinate in the presence of pseudocumene or 3-ethyltoluene accumulated 3,4-dimethylcatechol or 3-ethylcatechol. Cells unable to utilize toluene, m-xylene, and p-xylene, obtained by growth in benzoate, also lost the ability to utilize pseudocumene and 3-ethyltoluene. The ability to utilize these substrates could be reacquired by incubation with a leucine auxotroph otherwise able to grow on all of the aromatic substrates.
Full text
PDF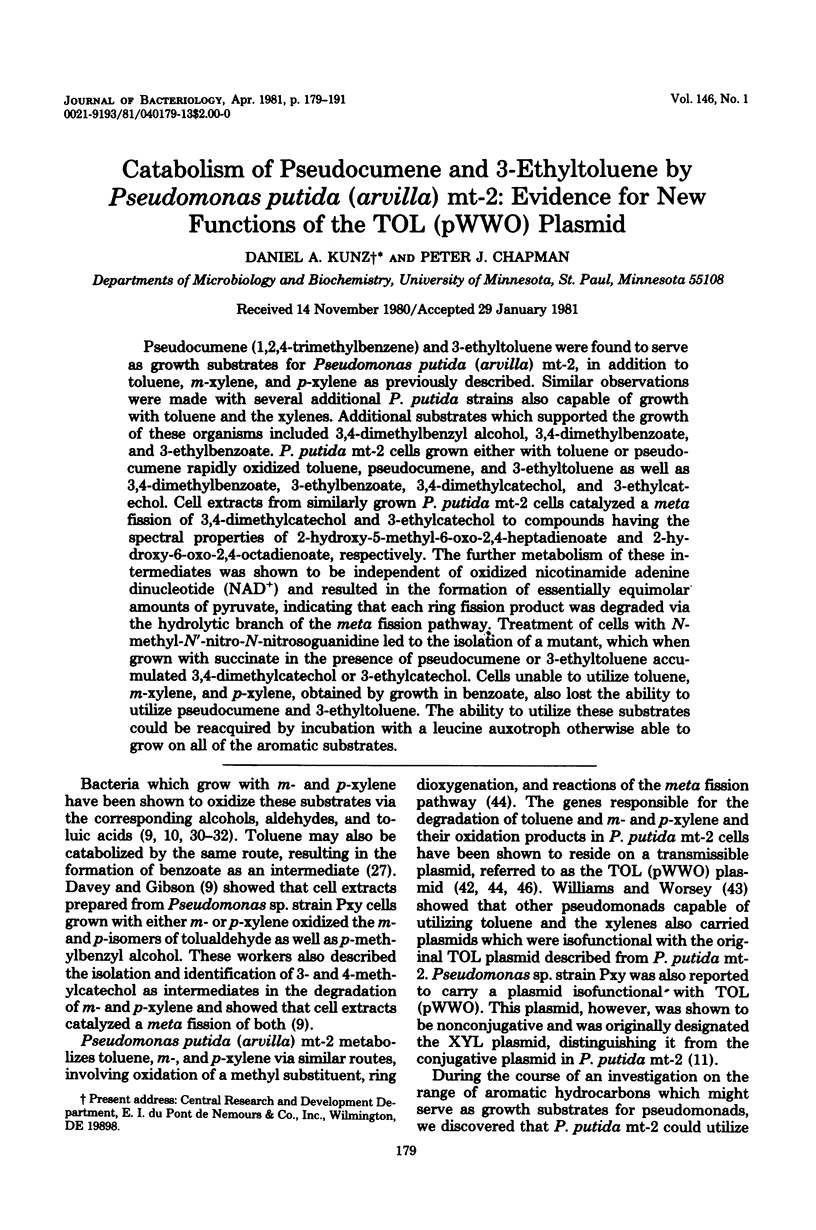









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayly R. C., Dagley S., Gibson D. T. The metabolism of cresols by species of Pseudomonas. Biochem J. 1966 Nov;101(2):293–301. doi: 10.1042/bj1010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., Dagley S. Oxoenoic acids as metabolites in the bacterial degradation of catechols. Biochem J. 1969 Feb;111(3):303–307. doi: 10.1042/bj1110303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., di Berardino D. Purification and properties of 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase from two strains of Pseudomonas putida. J Bacteriol. 1978 Apr;134(1):30–37. doi: 10.1128/jb.134.1.30-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAGLEY S., CHAPMAN P. J., GIBSON D. T., WOOD J. M. DEGRADATION OF THE BENZENE NUCLEUS BY BACTERIA. Nature. 1964 May 23;202:775–778. doi: 10.1038/202775a0. [DOI] [PubMed] [Google Scholar]
- DAGLEY S., GIBSON D. T. THE BACTERIAL DEGRADATION OF CATECHOL. Biochem J. 1965 May;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. F., Gibson D. T. Bacterial metabolism of para- and meta-xylene: oxidation of a methyl substituent. J Bacteriol. 1974 Sep;119(3):923–929. doi: 10.1128/jb.119.3.923-929.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. S., Hossler F. E., Stone R. W. Metabolism of p- and m-xylene by species of Pseudomonas. Can J Microbiol. 1968 Sep;14(9):1005–1009. doi: 10.1139/m68-166. [DOI] [PubMed] [Google Scholar]
- Friello D. A., Mylroie J. R., Gibson D. T., Rogers J. E., Chakrabarty A. M. XYL, a nonconjugative xylene-degradative plasmid in Pseudomonas Pxy. J Bacteriol. 1976 Sep;127(3):1217–1224. doi: 10.1128/jb.127.3.1217-1224.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D. T., Gschwendt B., Yeh W. K., Kobal V. M. Initial reactions in the oxidation of ethylbenzene by Pseudomonas putida. Biochemistry. 1973 Apr 10;12(8):1520–1528. doi: 10.1021/bi00732a008. [DOI] [PubMed] [Google Scholar]
- Gibson D. T., Hensley M., Yoshioka H., Mabry T. J. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry. 1970 Mar 31;9(7):1626–1630. doi: 10.1021/bi00809a023. [DOI] [PubMed] [Google Scholar]
- Gibson D. T. Initial reactions in the bacterial degradation of aromatic hydrocarbons. Zentralbl Bakteriol Orig B. 1976 Jul;162(1-2):157–168. [PubMed] [Google Scholar]
- HUGHES D. E. A press for disrupting bacteria and other micro-organisms. Br J Exp Pathol. 1951 Apr;32(2):97–109. [PMC free article] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Chapman P. J. Gentisic acid and its 3- and 4-methyl-substituted homologoues as intermediates in the bacterial degradation of m-cresol, 3,5-xylenol and 2,5-xylenol. Biochem J. 1971 Mar;122(1):19–28. doi: 10.1042/bj1220019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison V. W., Raymond R. L., Hudson J. O. Microbial Hydrocarbon Co-oxidation. III. Isolation and Characterization of an alpha, alpha'-Dimethyl-cis, cis-Muconic Acid-producing Strain of Nocardia corallina. Appl Microbiol. 1969 Jun;17(6):853–856. doi: 10.1128/am.17.6.853-856.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Fujisawa H., Nakazawa A., Nakazawa T., Kanetsuna F., Taniuchi H., Nozaki M., Hayaishi O. Studies on pyrocatechase. I. Purification and spectral properties. J Biol Chem. 1967 Jul 25;242(14):3270–3278. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- Murray K., Williams P. A. Role of catechol and the methylcatechols as inducers of aromatic metabolism in Pseudomonas putida. J Bacteriol. 1974 Mar;117(3):1153–1157. doi: 10.1128/jb.117.3.1153-1157.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NISHIZUKA Y., ICHIYAMA A., NAKAMURA S., HAYAISHI O. A new metabolic pathway of catechol. J Biol Chem. 1962 Jan;237:PC268–PC270. [PubMed] [Google Scholar]
- Nakazawa T., Yokota T. Benzoate metabolism in Pseudomonas putida(arvilla) mt-2: demonstration of two benzoate pathways. J Bacteriol. 1973 Jul;115(1):262–267. doi: 10.1128/jb.115.1.262-267.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki M., Kotani S., Ono K., Seno S. Metapyrocatechase. 3. Substrate specificity and mode of ring fission. Biochim Biophys Acta. 1970 Nov 11;220(2):213–223. doi: 10.1016/0005-2744(70)90007-0. [DOI] [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. 3. Enzymes of the catechol pathway. J Biol Chem. 1966 Aug 25;241(16):3795–3799. [PubMed] [Google Scholar]
- Ornston L. N. The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. IV. Regulation. J Biol Chem. 1966 Aug 25;241(16):3800–3810. [PubMed] [Google Scholar]
- Reiner A. M., Hegeman G. D. Metabolism of benzoic acid by bacteria. Accumulation of (-)-3,5-cyclohexadiene-1,2-diol-1-carboxylic acid by mutant strain of Alcaligenes eutrophus. Biochemistry. 1971 Jun 22;10(13):2530–2536. doi: 10.1021/bi00789a017. [DOI] [PubMed] [Google Scholar]
- Reiner A. M. Metabolism of benzoic acid by bacteria: 3,5-cyclohexadiene-1,2-diol-1-carboxylic acid is an intermediate in the formation of catechol. J Bacteriol. 1971 Oct;108(1):89–94. doi: 10.1128/jb.108.1.89-94.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Sparnins V. L., Chapman P. J., Dagley S. Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J Bacteriol. 1974 Oct;120(1):159–167. doi: 10.1128/jb.120.1.159-167.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. L., Dunn N. W. Transmissible plasmid coding for the degradation of benzoate and m-toluate in Pseudomonas arvilla mt-2. Genet Res. 1974 Apr;23(2):227–232. doi: 10.1017/s0016672300014853. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]



