Abstract
Commitment to the T and NKT cell lineages is determined during αβ T cell receptor (TCR)-mediated interactions of common precursors with ligand-expressing cells in the thymus. Whereas mainstream thymocyte precursors recognize Major Histocompatibility Complex (MHC) ligands expressed by stromal cells, NKT precursors interact with CD1d ligands expressed by cortical thymocytes. Here, we demonstrate that such homotypic T-T interactions generate ‘second signals’ mediated by the cooperative engagement of the homophilic receptors Ly108 and SLAM and downstream recruitment of the adaptor SAP and the Src kinase Fyn, which are essential for the lineage expansion and differentiation of the NKT lineage. These receptor interactions are required during TCR engagement and therefore only occur when selecting ligands are presented by thymocytes rather than epithelial cells which do not express Ly108 or SLAM. Thus, the topology of NKT ligand recognition determines the availability of a co-signaling pathway that is essential for NKT lineage development.
INTRODUCTION
NKT cells constitute a separate lineage of innate-like T lymphocytes involved in various infectious, autoimmune, allergic and cancerous conditions (Bendelac et al., 2006; Godfrey et al., 2004). They emerge from the thymus as memory/effector cells which explosively release Th1 and Th2 cytokines and chemokines upon recognition, through conserved semi-invariant αβ TCRs, of glycolipid ligands presented by the MHC-like molecule CD1d. There is growing evidence indicating that differential signaling associated with TCR recognition of ligands at the CD4+CD8+ double-positive (DP) stage of thymic development underlies the divergence between NKT cells, mainstream CD4 and CD8 T cells, regulatory T cells and CD8αα cells (Benlagha et al., 2005; Gapin et al., 2001; Jordan et al., 2001; Leishman et al., 2002; Yamagata et al., 2004). However, the specific nature of these differences and their molecular basis have remained elusive.
NKT cells exhibit autoreactivity to CD1d-expressing cortical thymocytes (Bendelac, 1995) and the dominant subset expressing Vα14-Jα18/Vβ8, 7, 2 TCRs (Park et al., 2001) recognizes endogenous ligands such as iGb3 as weak agonists (Schumann et al., 2006; Zhou et al., 2004). By contrast, conventional T cells recognize thymic peptides presented by MHC proteins as partial agonists (Hogquist et al., 1994). Importantly, the development of NKT cells is unique, as it relies on ligand expression by cortical thymocytes (Coles and Raulet, 2000; Schumann et al., 2005; Wei et al., 2005). Indeed, CD1d expression by cortical thymocytes alone was both required and sufficient in vivo for positive selection, lineage expansion and differentiation into the NKT lineage. This could impact their lineage differentiation because some of the signals emanating from such homotypic thymocyte-thymocyte interactions during TCR recognition of ligand likely differ from those associated with heterotypic thymocyte-stromal cell interactions. Interestingly, ectopic expression of MHC class II by the thymocytes of mice expressing a CIITA transgene driven by a CD4 promoter resulted in the selection of CD4 T cells expressing a memory/effector differentiation similar to NKT cells (Choi et al., 2005; Li et al., 2005). Furthermore, mice lacking Tec kinases developed memory/effector CD8 T cells that also resembled NKT cells in their expression of NK lineage markers and in their dependence on MHC class I ligand expression by bone marrow rather than epithelial cells (Atherly et al., 2006; Broussard et al., 2006). Together, these observations suggest that unidentified signals provided by bone marrow-derived selecting cell-types may contribute to the differentiation of memory/effector lineages (Locksley, 2002).
Indirect evidence suggests that members of the SLAM family of receptors may be involved in this process. These proteins, which are all encoded in the Slam locus, are mostly homotypic self-associating receptors expressed by cells of hemopoietic origin (Engel et al., 2003; Veillette, 2006). They have recently emerged as TCR-dependent or -independent regulators of adhesion and cellular activation during interactions between mature T, B, macrophage and dendritic cells and control several aspects of innate and adaptive responses as well as chronic diseases (Cannons et al., 2006; Crotty et al., 2003; Howie et al., 2005; Kumar et al., 2006; Wang et al., 2004). Despite intricate patterns of expression of several of the SLAM family receptors during hematopoiesis (Kiel et al., 2005) and thymopoiesis, their role in lymphocyte development has not been demonstrated. In T cells and thymocytes, SLAM receptor-initiated signaling is mediated in part by SLAM-associated protein (SAP, also called SH2D1A), an adaptor that recruits Src kinase Fyn, which in turn phosphorylates SLAM receptors which then serve as a docking site for a set of signaling molecules (Engel et al., 2003; Veillette, 2006). Mice lacking Fyn or SAP exhibited severe NKT cell defects as did humans with the X-linked lymphoproliferative (XLP) syndrome associated with SAP mutation (Chung et al., 2005; Eberl et al., 1999; Gadue et al., 1999; Nichols et al., 2005; Pasquier et al., 2005). Based on these observations, it could be envisioned that a costimulatory pathway involving SAP and Fyn is recruited by SLAM family receptors at some critical stage of NKT cell development (Borowski and Bendelac, 2005). However, in mice lacking the SLAM family receptors Ly108 (NTBA), SLAM (CD150) or Ly9 (CD229), NKT cell development seemed unperturbed (Graham et al., 2006; Howie et al., 2005; Wang et al., 2004). Other reports have suggested that Fyn could be activated downstream of TCR signaling (Hermiston et al., 2002) and that SAP could function without binding Fyn (Cannons et al., 2006; Chan et al., 2003; Gu et al., 2006; Howie et al., 2002; Simarro et al., 2004), raising the possibility that Fyn and SAP mediate in part separate pathways contributing to NKT cell development which may be independent of SLAM-family receptors. Thus, the precise pathways recruiting SAP and Fyn during NKT cell development remain enigmatic. In addition, the developmental stages at which they are required have not been characterized. Contrasting with the hypothesis that SAP and Fyn costimulate TCR signaling, the apparent restoration of NKT cell development in Fyn-/- mice by transgenic expression of the canonical Vα14-Jα18 TCR α chain, has suggested that Fyn acted at early stages of development, upstream of TCR expression and therefore prior to thymic positive selection (Gadue et al., 2004).
Here, we use genetic and bone marrow chimera approaches to provide evidence for a linear pathway of signaling that is initiated by homophilic self association of SLAM and Ly108 expressed by cortical thymocytes and is propagated by SAP and Fyn. Importantly, we show that this pathway is recruited during the TCR signaling events driving the positive selection of NKT precursors. Furthermore, we demonstrate that, while unessential for positive selection, SLAM and Ly108 signaling critically controls the characteristic expansion and differentiation of the NKT lineage that follows thymic selection.
RESULTS
NKT cell developmental arrest in the absence of SAP and Fyn
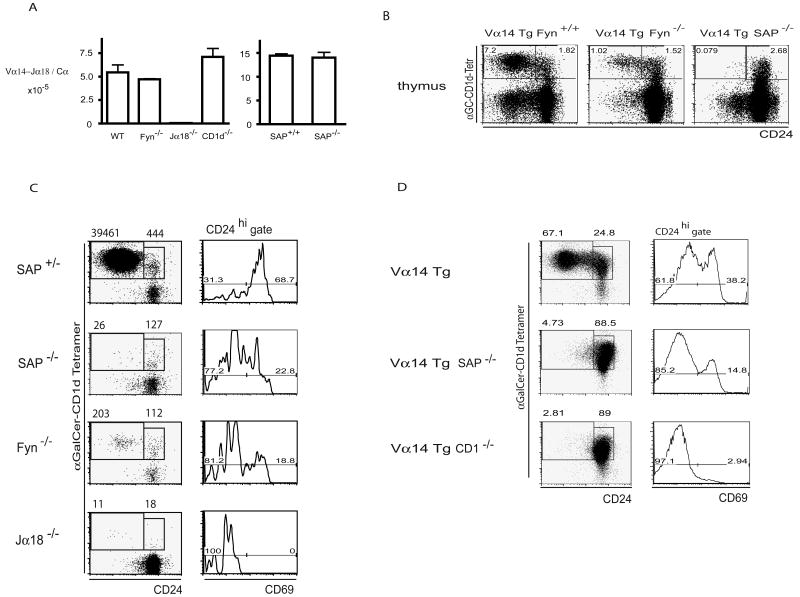
We first examined whether the NKT cell defects in Fyn or SAP mutants preceded or followed Vα14-Jα18 TCR expression (Gadue et al., 2004). We found identical frequencies of canonical Vα14-Jα18 rearrangements in sorted DP thymocytes from wild-type, SAP or Fyn deficient mice (Fig. 1A), ruling out a role of SAP or Fyn in the generation of the canonical NKT TCR α chain. We then directly analyzed the NKT cell developmental defects in different Fyn and SAP deficient mice. In Vα14-Jα18 transgenic mice, Fyn ablation reduced by 7-fold the frequency of mature NKT thymocytes identified by CD1d-αGalCer tetramers and low level of the heat stable antigen CD24 (Fig. 1B). Furthermore, ablation of SAP, which in SLAM receptor signaling functions upstream of Fyn, induced a nearly complete block in the development of mature NKT cells. The less severe phenotype of Fyn mutants might suggest compensation by other Src kinases, such as Lck (Simarro et al., 2004). To extend these observations to non-transgenic thymocytes and further characterize the arrested stage, we stained rare developmental intermediates with allophycocyanin (APC)-conjugated CD1d-αGalCer tetramers and enriched them using anti-APC paramagnetic beads. This enrichment procedure allows the unambiguous detection by FACS of the few hundred CD24high Tetramerhigh CD69high NKT lineage precursors that are present in the normal thymus and represent the stage that immediately follows positive selection (Benlagha et al., 2005). As shown in Fig. 1C, NKT cell development was arrested at the CD24high Tetramerhigh CD69high stage in both SAP- and Fyn-deficient thymocytes. The developmental block in Fyn-deficient mice again appeared to be leaky, because rare cells (<1% of wild type) reached the mature CD24low stage. The arrested cells were reduced in numbers compared with wild-type cells and they expressed CD69, a marker of positive selection, albeit at a 3-fold reduced frequency. A similar arrest at the CD24high Tetramerhigh CD69high stage following positive selection by CD1d ligand was observed in Vα14-Jα18 transgenic thymocytes lacking SAP (Fig. 1D). Together, these results demonstrate that the NKT cell developmental arrest in SAP- or Fyn-deficient mice occurs not only after TCR expression but also during or just after positive selection.
Figure 1. NKT cell developmental arrest in SAP-/- and Fyn-/- mice.
A. Conserved frequency of Vα14-Jα18 rearrangements in Fyn and SAP-deficient thymocytes. Quantitative RT-PCR of canonical Vα14-Jα18 rearrangements and Cα in sorted DP thymocytes from WT, Fyn-/-, Jα18-/-, CD1-/- (left) and in DP thymocytes from CD1-/-SAP+/+ and CD1-/- SAP-/- mice (right). SAP-/- mice were crossed onto a CD1-/- background to eliminate any contamination of DP thymocyte by mature NKT cells. The values shown are ratios between Vα14-Jα18 and Cα transcripts. B. Vα14-Jα18 transgenic thymocytes on a Fyn+/+, Fyn-/- or SAP-/- background were stained with CD1d-αGalCer tetramers and CD24 to enumerate the immature CD24high and mature CD24low stages. Cell frequencies are indicated in the corresponding gates. C. Left column, CD1d-αGalCer tetramer positive thymocytes were enriched by autoMACS from pools of thymi obtained from 2 week-old SAP+/- and SAP-/- littermates, Fyn-/- and Jα18-/- mice prior to FACS analysis with CD24. Numbers in the CD1d-αGalCer+ CD24high and CD24low gates represent absolute cell numbers recovered from 2 pooled thymi. Right column, gated CD1d-αGalCer+ CD24high cells were analyzed for CD69 expression. Percentages are indicated over corresponding brackets. Similar results were obtained in three independent experiments. D. Stage of NKT cell developmental arrest in the Vα14 Tg SAP-/- thymus. CD1d-αGalCer tetramer+ thymocytes were enriched using paramagnetic beads and submitted to FACS analysis as in 1C.
Thymic expression pattern of SLAM family receptors
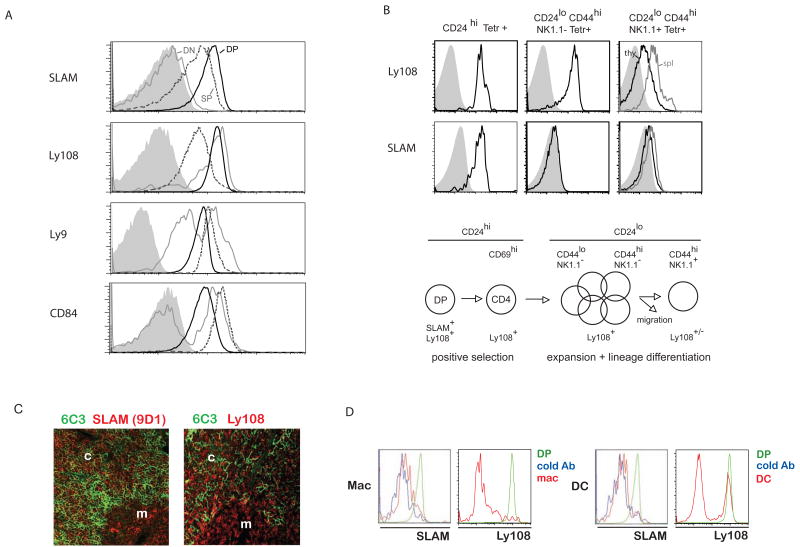
We next addressed the question of whether and which SLAM family members might be involved in NKT cell development. The Slam locus comprises genes encoding several SAP binding family members that are conserved in mouse and human, including SLAM (CD150), 2B4 (CD244), CD84, Ly108 (NTBA), Ly9 (CD229) (Engel et al., 2003; Veillette, 2006). Using monoclonal antibodies, we found that 2B4 was not expressed on mouse thymocytes (not shown). Of the remaining receptors, SLAM and Ly108 displayed highest expression on DP cortical thymocytes with conserved or decreased expression on mature single-positive (SP) thymocytes, a pattern similar to CD1d (Roark et al., 1998), whereas Ly9 and CD84 had lower expression on DP than SP cells (Fig. 2A). Developing NKT thymocytes had a similar pattern of expression with SLAM rapidly downregulated after the DP stage whereas Ly108 expression persisted longer, until the CD24low stage (Fig. 2B). Furthermore, SLAM and Ly108 were conspicuously absent from other thymic cell-types, including epithelial cells, dendritic cells and macrophages, with the exception of a subset of CD11c+ cells expressing Ly108 (Fig. 2C-D).
Figure 2. Expression pattern of SLAM family members on thymocytes and NKT cells.
A. Double negative (DN), double positive (DP) and mature single positive (SP) thymocytes from B6 mice (or BALB/c for Ly9) were analyzed for expression levels of SLAM family receptors. DP are represented by a solid black line, SP a dashed dark grey line and DN by a light grey line as indicated. Shaded profiles represent isotype controls or staining of thymocytes lacking the corresponding gene or epitope. B. SLAM and Ly108 expression at different stages of NKT thymocyte development. Left panels, immature CD24high; middle panels, mature CD24lowCD44highNK1.1-; right panels, terminally differentiated CD24lowCD44highNK1.1+ cells in the thymus (black) and spleen (grey). The findings are summarized in the NKT cell developmental chart below the FACS panels. C. Immunohistochemical staining shows abundant SLAM and Ly108 expression in cortical and medullary cells (mostly T lineage) but not on the cortical epithelial cells. D. Flow cytometry analysis of thymic CD11b+ macrophages and CD11c+ dendritic cells (red) compared with DP thymocytes (green). The blue profile corresponds to a staining with biotin-conjugated antibody after incubation with excess unconjugated antibody (negative control).
NKT cells in SLAM- or Ly108-deficient mice
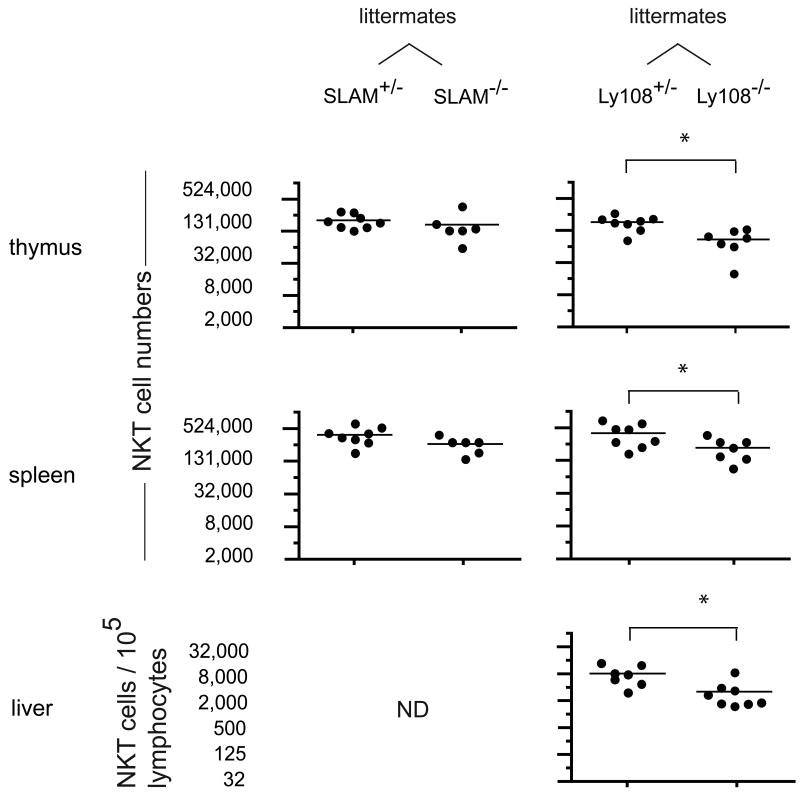
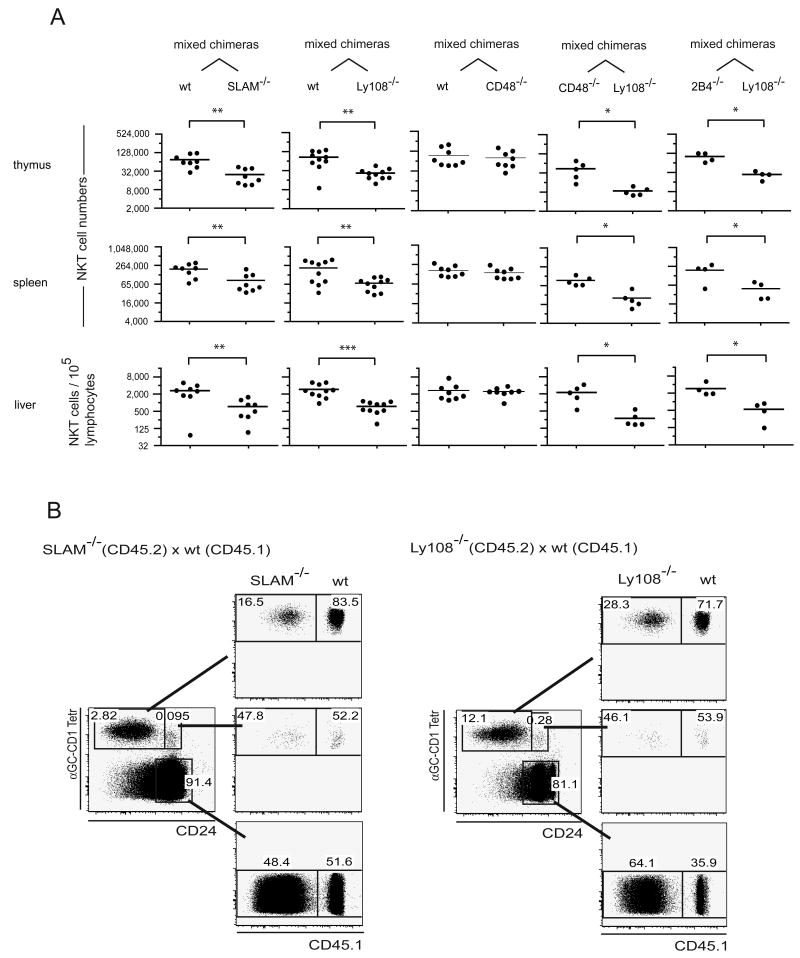
Using flow cytometry, we compared the thymus, spleen and liver lymphocytes of Ly108 and SLAM +/- and -/- littermate pairs and found little difference in NKT cell numbers in most cases. These results are consistent with previous reports of Ly108 (Howie et al., 2005), SLAM (Wang et al., 2004) or Ly9 (Graham et al., 2006) deficient mice. The compilation of data did suggest a modest reduction (∼50%), in the case of Ly108-/- mice (Fig. 3). To reveal latent, intrinsic NKT cell defects associated with these mutations, we generated radiation chimeras where lethally irradiated Jα18-deficient hosts were competitively reconstituted with a 1:1 mixture of wild-type and SLAM or Ly108 deficient bone marrow (bm) cells expressing different marker alleles of CD45 for convenient FACS identification. In all these chimeras, the KO:WT ratio of thymocytes and splenocytes ranged between 0.3 and 0.7, demonstrating similar capacity of WT and KO bone marrow cells for general lymphocyte reconstitution. Significant NKT cell decreases of ∼50-75% on average were observed for both the SLAM and the Ly108-deficient populations compared with wild type (Fig. 4A). Because the SLAM-deficient and the Ly108-deficient bone marrows used in these chimeras were from mice that had been originally derived from embryonic stem cells of 129 origin and were subsequently backcrossed to B6, the possibility existed that these relatively modest differences resulted from some heterogeneity between the Slam locus of 129 and B6. We thus generated control mixed chimeras using ‘neutral’ mutations of 2B4 or its ligand CD48. Both proteins are encoded in the same Slam locus but they should not affect NKT cell development because, whereas CD48 is highly expressed by all immature and mature thymocytes, 2B4 is not expressed in the thymus or during NKT cell development (data not shown). The 2B4 mutants were derived from embryonic stem cells of B6 origin and the CD48 mutants were derived from embryonic stem cells of 129 origin and backcrossed onto the B6 genetic background. In such control (2B4-/- + Ly108-/-) or (CD48-/- + Ly108-/-) mixed chimeras, the development of NKT cells lacking Ly108 was as defective relative to NKT cells lacking 2B4 or CD48, as it was defective relative to wild type NKT cells in the (wt + Ly108-/-) mixed chimeras (Fig. 4A). An additional set of control (wt + CD48-/-) chimeras directly demonstrated that the Slam locus from 129 did not contribute to the NKT cell defects observed in SLAM-/- or Ly108-/- mice (Fig 4A).
Figure 3. Individual contributions of SLAM and Ly108 to NKT cell development.
Scatter plots showing absolute numbers (log2 scale) of CD1d-αGalCer positive cells in individual thymi, spleens and livers of 3-5 week-old SLAM +/- and -/- littermates and Ly108 +/- and -/- littermates. Because of variations in liver lymphocyte recovery between experiments, liver NKT cell numbers are enumerated per 105 lymphocytes. Liver NKT cells were not examined in SLAM -/- mice (ND, not determined). * denotes statistical significance (p<0.05) using an unpaired t test.
Figure 4. NKT cell development in competition chimeras.
A. 1:1 mixtures of WT + SLAM-/-, WT + Ly108-/-, WT + CD48-/-, 2B4-/-+ Ly108-/-, CD48-/-+ Ly108-/- bone marrow were injected into lethally irradiated Jα18-/- hosts and chimeras were analyzed at 6-8 weeks. Absolute NKT cell numbers were calculated for each CD45 allele-marked compartment, then divided by the number of lymphocytes in the corresponding CD45 allele-marked fraction and multiplied by the total number of lymphocytes to adjust for differences between the ratio of reconstitution by the two bone marrows which ranged between 0.3 and 0.7. *, ** and *** denote statistical significance (p<0.05, p<0.01 and p<0.001 respectively) using a t test for paired comparisons (2 experiments are pooled as they showed similar results (ANOVA)). B. FACS analysis of MACS-enriched CD1d-αGalCer tetramer+ CD24high and CD24low cells originating from the wt and mutant (CD45 allele-marked) bone marrows. The chimeric distribution of the tetramer-negative thymocytes is also shown (bottom dot plots).
A more detailed analysis of the NKT cell developmental stages in these mixed chimeras revealed a clear defect at the transition between the CD24high and CD24low stages in the SLAM- or Ly108-deficient cells compared with wild-type (Fig. 4B). This transition corresponds to the phase of expansion of the NKT cell lineage after positive selection has occurred.
SLAM/Ly108 double deficiency
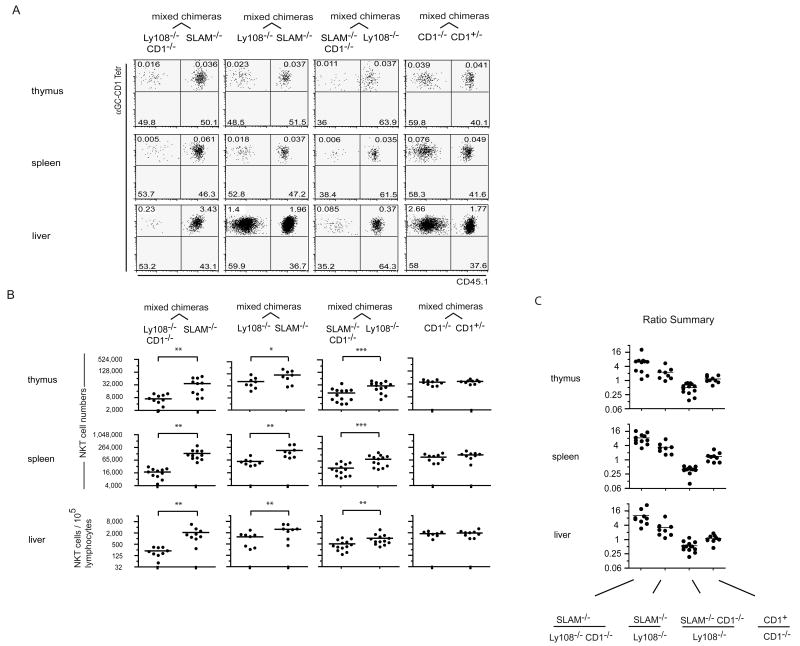
Owing to their overlapping expression pattern and their shared usage of SAP and Fyn for intracellular signaling, SLAM, Ly108 and some of the other SLAM-family receptors might be partly redundant in the thymus as well as in peripheral immune responses. Because they are encoded in the same locus, however, double or triple mutant mice could not be generated by breeding single mutants. Instead, we reconstituted lethally irradiated Jα18-deficient hosts with a 1:1 mixture of CD45 allele-marked CD1d-deficient bm that lacked Ly108 (Ly108-/- CD1-/-) and CD1d-sufficient bm that lacked SLAM (SLAM-/-). In these chimeras, NKT precursors originating from the CD1-/- compartment lacked Ly108 and were forced to engage their TCR on CD1d-expressing thymocytes that lacked SLAM, creating a special ‘double mutant’ situation during these cell encounters because neither SLAM not Ly108 could engage in homophilic self-interactions (Fig. 5, top). These Ly108-/- CD1-/- precursors, however, were able to receive SLAM signals from bystander thymocytes in trans. In contrast, the SLAM-/- cells had to interact with other SLAM-/- cells for their TCR to see CD1d, creating a single mutant situation (Fig. 5, bottom). Fig. 6A-B (1st column) shows that in these chimeras the ‘double mutant’ cells generated on average 6 to 10 times less NKT cells than the single mutants in the thymus and peripheral tissues. Control chimeras expressing a 1:1 mixture of wild type and CD1-/- bone marrow demonstrated that the absence of CD1d on half of the thymocytes did not interfere with NKT cell development (Fig. 6A-B, 4th column). Notably, in these mixed chimeras as in those shown in Fig. 4A comparing CD48-/- and Ly108-/-, the NKT precursors shared the same Slam locus of 129 origin and differed only with respect to the Ly108 and SLAM mutations. Taking into account the fact that the SLAM-/- cells, against which the Ly108-/- CD1-/- cells were competing, were themselves impaired 2-fold when compared with wild-type (Fig. 4), the findings suggest that the combined interruption of homophilic SLAM/SLAM and Ly108/Ly108 interactions between the CD1d-presenting thymocyte and the NKT precursor results in a 12-20 fold reduction in NKT cell development which cannot be rescued by ‘bystander’ interactions with other thymocytes expressing these receptors. As expected from previous studies of SAP-/- and Fyn-/- mice, the development of conventional CD4 and CD8 T cells was not altered in the mixed chimeras (not shown).
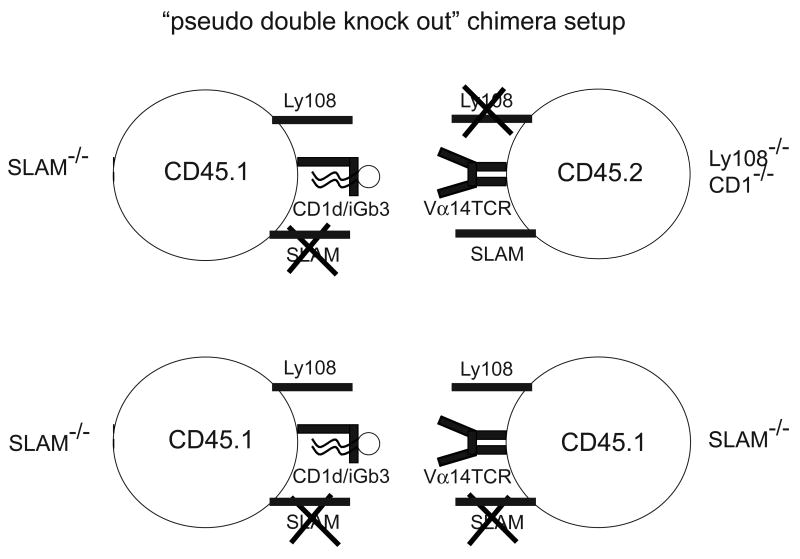
Figure 5. Design of the pseudo-double KO chimeras.
Cell interaction schemes illustrating the functional double deficiency created in the CD1-/- compartment of mixed chimeras of the SLAM-/- + Ly108-/-CD1-/- (top) vs the single deficiency in the CD1+/+ compartment (bottom). The Ly108-/-CD1-/- NKT precursors must interact with SLAM-/- thymocytes, the sole source of CD1d ligands, functionally removing both SLAM and Ly108 signals during TCR engagement (top), whereas the SLAM-/- NKT precursors must interact with SLAM-/- thymocytes, creating a single KO situation (bottom).
Figure 6. NKT cell developmental block in pseudo-double KO chimeras.
A. Mixed radiation bone marrow chimeras as indicated. FACS dot plots are gated on CD1d-αGC+ NKT lineage cells and show expression of the CD45.1 allelic marker in tetramer-positive and negative cells in different tissues, as indicated. B. Summary scatter plots show NKT cell numbers in different tissues and in CD45 allele-marked compartments of individual mixed chimeras. *, ** and *** denote statistical significance (p<0.05, p<0.01 and p<0.001 respectively) using a t test for paired comparisons of the two hemopoietic compartments in individual mixed chimeras. Results are pooled from 2-3 sets of chimeras showing similar results (ANOVA). C. Calculated ratios of NKT cells found in the two bone marrow-derived compartments of individual chimeras.
A second set of mixed chimeras was constructed using a 1:1 mixture of SLAM-/- and Ly108-/- bone marrow cells. In these chimeras, a NKT precursor lacking SLAM, for example, will be deprived of SLAM interactions permanently but will also miss Ly108 interactions in 50% of its cellular interactions because the other half of the thymocytes lack Ly108. Interestingly, these competitive chimeras suggested that the impact of Ly108 might be more important than that of SLAM, because its absence resulted in 2-3 fold fewer NKT cells on average than that of SLAM (Fig. 6A-B, 2nd column). This may be due to the persisting expression of Ly108 after the DP stage (Fig. 2B), as it is known that NKT precursors continue to engage their TCR for an extended period of time until terminal maturation into NK1.1+ cells (McNab et al., 2005). In addition, statistical comparison between the two sets of mixed chimeras in the 1st and 2nd columns of Fig. 6A-B showed that the lack of CD1d in the Ly108-/- compartment further increased the magnitude of the NKT cell defects by comparison with the SLAM-/- compartment in different tissues by ∼3 fold on average (Fig. 6C) (p<0.001, ANOVA), demonstrating that functionally relevant self-interactions between SLAM family members occurred at the same time as TCR engagement and with the same CD1d-expressing thymocyte. This conclusion was further supported by reverse chimeras made of a 1:1 mixture of Ly108-/- and SLAM-/- CD1-/- bone marrows (Fig. 6A-B, 3rd column) where the functionally double mutant SLAM-/- CD1-/- population now generated less NKT cells than the single mutant Ly108-/- population. Statistical comparison of the two sets of mixed chimeras in the 2nd and 3rd columns of Fig. 6A-B showed that the ablation of CD1d in the SLAM-/- cell population, which functionally changes these cells into double mutants, increased the magnitude of its NKT cell defect by 6 fold on average in different tissues (Fig 6C) (p<0.001, ANOVA). The ratios of NKT cells developing in the different compartments of individual mixed chimeras are summarized in Fig. 6C.
DISCUSSION
TCR and CD4/CD8 coreceptor signaling events are central to the selection and lineage differentiation of thymocytes. However, the characterization of new lineages recognizing ligands presented by cells as divergent from thymic epithelial cells as cortical thymocytes has suggested that unidentified, cell-type specific signals might also decisively impact the selection and differentiation events. The present study identified two members of the SLAM family of receptors, SLAM and Ly108, that are highly expressed by cortical thymocytes and control the transition between positive selection and the subsequent expansion and differentiation of the NKT cell lineage. NKT precursors lacking either SLAM or Ly108 exhibited only modest defects. However, in mixed bone marrow chimeras designed to abrogate both SLAM and Ly108 signaling during CD1d ligand recognition in the thymus, a stronger developmental arrest was observed, demonstrating that SLAM and Ly108 exerted essential and intrinsic, but partly redundant effects through homophilic self association across the cell-cell synapse. The demonstration that SLAM and Ly108 could exert partially redundant effects is consistent with their shared recruitment of SAP and Fyn, and may suggest an explanation for the past failure to ascribe the multiple phenotypes associated with the SAP or Fyn deficiencies to individual members of the SLAM family of receptors (Veillette, 2006). Deficiencies of SLAM, Ly108 or their intracellular signaling partners Fyn and SAP affected NKT cell development with increasing severity. It is possible therefore that the residual NKT cells developing in mixed SLAM+Ly108 “pseudo-double KO” chimeras have signaled through other, less prominent SLAM family receptors such as Ly9, CD84 or CRACC (CD319) for example. Alternatively, bystander engagement of SLAM and Ly108 by 3rd party cortical thymocytes (not directly involved in TCR/CD1d ligand interactions) might have provided the required signals, albeit inefficiently. This issue will be resolved when double and triple mutant of the SLAM family of receptors become available. All mutations impacted the same stage of NKT cell development, further supporting a linear model of signaling through SLAM family receptors and intracellular SAP and Fyn. Thus, whereas CD24highCD69highTet+ cells, representing the stage just following positive selection, were clearly identified, the subsequent intrathymic expansion and differentiation of NKT cells was invariably defective. These results stand in contrast with a previous conclusion that Fyn acted upstream of TCR expression based on the apparent restoration of NKT cells in Fyn-/- mice upon expression of a Vα14-Jα18 transgene (Gadue et al., 2004). The reasons for this discrepancy are unclear, but our own experiments clearly demonstrated that NKT cell development was severely quantitatively impaired in Vα14-Jα18 transgenic mice lacking either Fyn or SAP. One possibility is that overexpression of the TCR transgene combined with the leakiness of the Fyn mutation might have resulted in the apparent ‘rescue’ of NKT cells in the discordant report. Another, non-exclusive possibility is that the ‘rescued’ cells corresponded to a subset of tetramer-positive, CD1d-independent, non-NKT lineage cells that is found in these transgenics, which used a prematurely expressed Vα11 promoter (Bendelac et al., 1996; Wei et al., 2006), but is not present in our new transgenic lines which use a CD4 promoter. In any case, the present combined results render the previous conclusion that Fyn was involved prior to TCR engagement speculative and, instead, support a unified model where the bifurcation of the NKT lineage is driven by the peculiar signals emanating through SLAM family receptors, SAP and Fyn at the intercellular synapse formed during the thymic selection events. These findings help focus the search for the mechanisms of developmental arrest during the events associated CD1d ligand recognition rather than during some defective TCR arrangement or lineage precommitment. The arrested cells did not exhibit signs of increased apoptosis and could not be rescued by a Bcl-xL transgene driven by a proximal Lck promoter (data not shown), suggesting that, rather than a mere lack of a survival signal, a defect in positive selection, a block in proliferation or an increased negative selection might also underlie the developmental defect.
Signaling through SAP and Fyn is reported to activate SHIP1, Dok1/2 and RasGap and inhibit Ras signaling (Veillette, 2006). Fyn also activates the NF-kB pathway (Cannons et al., 2004) whose importance for NKT cell development, particularly for survival, is well established (Schmidt-Supprian et al., 2004; Sivakumar et al., 2003; Stanic et al., 2004). The coordinated expression of CD1d and SLAM-family members in mouse and human thymocytes, segregated from MHC class I and class II in thymic stromal cells, favors the spatial and temporal co-signaling of SLAM family members along with the TCR during NKT cell development. Notably, because Ly108 and SLAM signaling appears to be required concomitantly with TCR engagement, it would not be available to mainstream thymocytes as they interact with MHC ligands on stromal cells (which do not express these SLAM family members). Thus, our findings suggest that the peculiar thymic expression patterns of MHC, CD1d and SLAM family glycoproteins in mouse and human dictate alternative lineage fates. The topology of ligand expression defines therefore a specialized niche that provides the necessary signals for divergent lineage differentiation.
Interestingly, recent reports have suggested that inactivation of the Tec kinases, which signal downstream of the TCR, induced a shift from the conventional T cell to a memory/effector NK1.1+ ‘innate-like’ T cell lineage through the induction of eomesodermin and the IL-2 receptor β chain (Atherly et al., 2006; Broussard et al., 2006), a striking parallel to the induction of T-bet, a homolog of eomosodermin, and IL-2 receptor β chain required for the NKT cell lineage (Intlekofer et al., 2005; Matsuda et al., 2006; Townsend et al., 2004).
Development of these MHC-restricted innate-like T cells required MHC class I expression on bone marrow-derived rather than epithelial cells. Likewise, normal or transgenic expression by thymocytes of MHC class Ib or MHC class II, respectively, resulted in the development of memory/effector type T cells (Choi et al., 2005; Li et al., 2005; Urdahl et al., 2002). These intriguing findings may indicate that SLAM family receptor signaling is generally involved in the formation of memory/effector innate-like lineages by bone marrow-derived thymocytes and suggest novel mechanisms of lymphocyte lineage instruction based not only on TCR/coreceptor signals but also on accessory signals specifically provided by different antigen-presenting cell types.
Notably, polymorphism at the SLAM locus, in particular the differential expression of Ly108 splice variants with differential signaling properties, is associated with autoimmunity and lupus (Kumar et al., 2006; Wandstrat et al., 2004). The absence of SLAM expression on thymic DP cells was recently reported in NOD mice, a phenotype correlating with homozygosity for the SLAMNOD locus in genetic analysis (Jordan et al., 2007). The link between SLAM and variations in NKT cell numbers remains correlative but it is consistent with a previous mapping of NKT cell deficiency in this autoimmune strain identifying two independent loci, Nkt1 and Nkt2 (Jordan et al., 2007; Rocha-Campos et al., 2006), since the Slam locus is a candidate for Nkt1. Although Nkt1 congenic NOD mice did not show amelioration of diabetes (Rocha-Campos et al., 2006), the NKT cell increase in these mice was modest and the role of NKT cells in type I diabetes remains intriguing (Delovitch and Singh, 1997).
Although previous research emphasized the role of SLAM family receptors in multiple forms of innate and adaptive immune responses, our findings reveal a new essential role of the SLAM family of receptors in lymphocyte development and further suggest a more general mechanism of T cell lineage differentiation through topologically segregated expression of TCR ligands and costimulatory receptors. The results also strengthen the emerging concept that polymorphism of the Slam locus may underlie important genetic variations in the development or function of entire populations of immune cells to fine-tune the balance between the effector and regulatory pathways that control infection and autoimmunity.
METHODS
Mice
SLAM-/- (Wang et al., 2004) mice were used after 8 backcrosses to C57BL/6, Ly108-/- (Howie et al., 2005) (lacking exons 2-3 encoding the entire extracellular domain) after 6 backcrosses, CD48-/- after 12 backcrosses, SAP-/- (Wu et al., 2001) and Fyn-/- (Stein et al., 1992) after 10 backcrosses, Jα18-/- (Cui et al., 1997) and CD1d-/- (Carnaud et al., 1999) mice after >12 backcrosses. 2B4-/- were on a pure C57BL/6 background (Vaidya et al., 2005). C57BL/6 (CD45.2), B6.SJL-Ptprca Pep3b/BoyJ (CD45.1). We verified that SLAM-/-, Ly108-/- and CD48-/- mice shared the same SLAM locus of 129 origin by using strain-specific antibodies against Ly9 and 2B4 and by genotyping for a SNP rs315321197 located at the other end of the locus (data not shown). To generate the Vα14-Jα18 transgenic mice, the pre-rearranged Vα14-Jα18 TCR α chain cDNA of the DN32.D3 hybridoma (Bendelac et al., 1996) was inserted into the SalI site of a plasmid containing the minimal CD4 promoter/enhancer and the intronic silencer (Sawada et al., 1994). The linearized (via Not I) construct was injected into fertilized C57BL/6 oocytes, and the injected oocytes were implanted into pseudopregnant CD-1 (VAF+) outbred female mice. Transgenic mice were screened using PCR (forward primer: 5′-TGT AGG CTC AGA TTC CCA ACC-3′; reverse primer: 5′-GAG GAT GGA GCT TGG GAG TCA GG-3′) and were bred onto various gene deficient backgrounds. All mouse work was done in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Chicago.
Antibodies and flow cytometry
Anti-Ly108 (clone 13G3) was generated from the spleen of a Ly108-/- mouse immunized with wild type thymocytes and is described further elsewhere (S.R. et al., manuscript in preparation) and anti-CD84 mAb (a generous gift from Pablo Engel) was reported (Romero et al., 2005). CD1d-αGalCer tetramers were prepared as previously described (Benlagha et al., 2000). Fluorochrome labeled monoclonal antibodies (clone indicated in parentheses) against SLAM (9D1), CD229.1/Ly9.1 (30C7), Ep-CAM (G8.8), Ly51 (6C3), NK1.1 (PK136), CD44 (IM7), CD24 (M1/69), B220 (RA3-6B2), CD8α (53–6.7), CD4 (RM4-5), CD45 (30-F11), CD11c (HL3), CD11b (M1/70), CD1d (1B1), CD45.1 (A20), rat κ light chain (MRK1), and were purchased from eBioscience or BD Biosciences. The FITC-conjugated anti-Syrian and Armenian hamster IgG cocktail (G70-204, G94-56) was purchased from BD Biosciences. For NKT cell enrichment, thymocytes were stained with APC-conjugated CD1d-αGalCer tetramers, bound to anti-APC paramagnetic beads and positively selected with an autoMACS (Miltenyi biotech) as described (Benlagha et al., 2005). All steps were performed at ice-cold temperature. Samples were analyzed on BD Canto or LSRII flow cytometers. Dead cells were excluded with DAPI staining and doublets by gating on FSC and SSC area, height and width.
Generation of mixed bone marrow chimeras
Four to eight-week old Jα18-/- mice were subjected to 900 Rads irradiation using a gamma cell 40 irradiator with a cesium source. Four to eight hours later, irradiated mice were injected intravenously with 5×106-107 bone marrow cells isolated from femurs of donor mice, and depleted of T cells by magnetic cell sorting (autoMACS, Miltenyi biotech) using CD3-PE antibody (eBiosciences) and anti-PE beads. Mice were analyzed six to eight weeks after reconstitution.
Quantitative real-time PCR
Canonical Vα14-Jα18
CD4+CD8+CD24hi thymocytes were sorted from C57Bl/6, CD1d-/-, SAP-/-, Fyn-/- and Jα18-/- mice on FACSAria (BD Biosciences) and MoFlo (Dako Cytomation) cell sorters. Total RNA of 5 × 105 sorted thymocytes was extracted into 10μl and treated with DNase1 using the RNAqueous Micro Kit (Ambion). 8μl of each DNase1-treated RNA sample was primed with oligo-dT and reverse transcribed into a 20μl volume using the SuperScript III Platinum Two-Step qRT-PCR kit (Invitrogen). Vα14 and Cα-specific primers, as well as the Cα-specific Taqman probe were previously described 6. A probe specific for the sequence spanning the NKT clonotypic Vα14-Jα18 junction (5′-FAM-CTGTGTGGTGGGCGATA-MGBNFQ-3′) was purchased from Applied Biosystems. PCR reactions were run in an ABI Prism 7700 Sequence Detector (Applied Biosystems). Each duplicate reaction contained 5μl cDNA, 0.2μM each primer and Taqman probe, and Platinum Quantitative PCR Supermix-UDG with ROX reference dye (Invitrogen) to a final volume of 50μl. Initial 2 minutes holds at 50°C and 95°C were followed by 50 cycles consisting of 15 seconds at 95°C and 45 seconds at 60°C. Amplification was analyzed using ABI Prism Sequence Detection Software Version 1.9.1 (Applied Biosystems) and the Standard Curve Method described in the ABI Prism 7700 Sequence Detection System User Bulletin #2. 10-fold serial dilutions of a plasmid containing a rearranged Vα14 TCRα cDNA were used for Vα14 and Cα standard curve generation.
Immunohistochemistry
Thymi from four-week-old C57Bl/6 mice were rinsed with PBS and frozen in OCT freezing medium (Tissue-Tek, Sakura Finetek) by flotation on 2-methylbutane (Sigma-Aldrich) over dry ice. Frozen thymi were stored at −80°C. Thymic sections were cut at 6mm on a LEICA microtome, fixed with aceton for 5 min and stained with purified anti-SLAM (9D1) and a Cy3 TSA amplification system (Perkin Elmer) or biotinylated anti-Ly108 (13G3) followed by Streptavidin-Rhodamin (Molecular Probes) and FITC conjugated anti-Ly51 (6C3), (a marker for cortical epithelium). Images were captured on a Leica SP2 A OBS Laser Scanning Confocal and a Zeiss Axiovert 200 immunofluorescence microscope, and processed using LCS Leica Confocal Software, Openlab software and Adobe Photoshop.
Appendix 1. SLAM locus analysis.
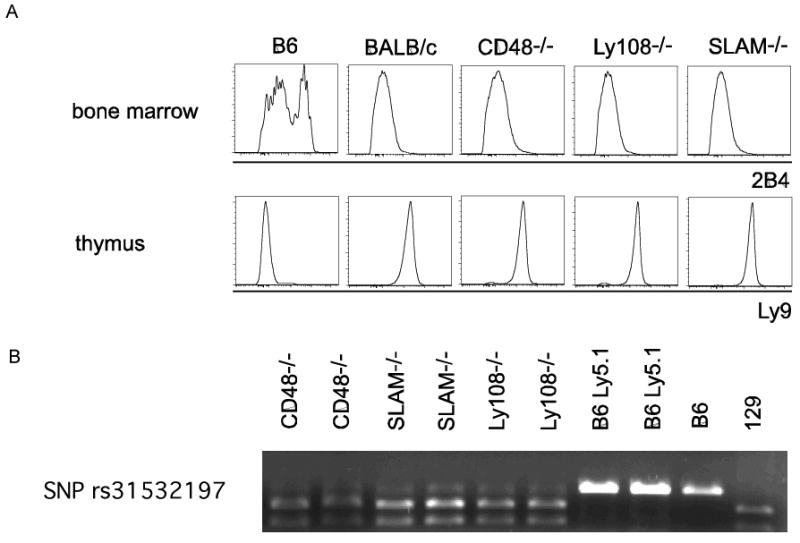
A. Strain-specific antibody staining. Flow cytometric analysis of bone marrow with antibody clone 2B4 (anti CD244.2) and thymi with antibody clone 30C7 (anti Ly9) of indicated mouse strains. B. Expression of SNP rs31532197 downstream of Ly108. PCR amplification of genomic DNA from indicated strains using primers Forward - GCCTTTCATCTTGGGTTTCA Reverse - AGCTGGGGAAAGGTAAGGAG followed by BSSII digest.
Acknowledgments
We thank members of the Bendelac and Terhorst laboratories for help and advice, Vanja Dukic for help with statistical analysis, Kelly Hudspeth for technical help, Arlene Sharpe and Vinay Kumar for providing CD48 and 2B4 deficient mice, Alexander Chervonsky for helpful comments on the manuscript. A.B. is a Howard Hughes Medical Institute Investigator. This work was supported by NIH RO1 AI038339 (AB) and DK073339 (CT). SR is supported by a research fellowship from the Crohn's and Colitis Foundation of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The tec family tyrosine kinases itk and rlk regulate the development of conventional CD8(+) T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Hunziker RD, Lantz O. Increased interleukin 4 and immunoglobulin E production in transgenic mice overexpressing NK1 T cells. J Exp Med. 1996;184:1285–1293. doi: 10.1084/jem.184.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The Biology of NKT Cells. Annu Rev Immunol. 2006 doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages in thymic NKT cell development. Journal of Experimental Medicine. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski C, Bendelac A. Signaling for NKT cell development: the SAP-Fyn connection. Journal of Experimental Medicine. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broussard C, Fleischecker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8(+) T cell lineages in mice deficient for the tec kinases itk and rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL. SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL. SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med. 2006;203:1551–1565. doi: 10.1084/jem.20052097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnaud C, Lee D, Donnars O, Park SH, Beavis A, Koezuka Y, Bendelac A. Cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol [Cutting Edge] 1999;163:4647–4650. [PubMed] [Google Scholar]
- Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ. SAP couples Fyn to SLAM immune receptors. Nat Cell Biol. 2003;5:155–160. doi: 10.1038/ncb920. [DOI] [PubMed] [Google Scholar]
- Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol. 2000;164:2412–2418. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R. SAP is required for generating long-term humoral immunity. Nature. 2003;421:282–287. doi: 10.1038/nature01318. [DOI] [PubMed] [Google Scholar]
- Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- Delovitch TL, Singh B. The nonobese diabetic mouse as a model of autoimmune diabetes: immune dysregulation gets the NOD. Immunity. 1997;7:727–738. doi: 10.1016/s1074-7613(00)80392-1. published erratum appears in Immunity 1998 Apr;8(4):531. [DOI] [PubMed] [Google Scholar]
- Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- Engel P, Eck MJ, Terhorst C. The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat Rev Immunol. 2003;3:813–821. doi: 10.1038/nri1202. [DOI] [PubMed] [Google Scholar]
- Gadue P, Morton N, Stein PL. The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadue P, Yin L, Jain S, Stein PL. Restoration of NK T cell development in fyn-mutant mice by a TCR reveals a requirement for Fyn during early NK T cell ontogeny. J Immunol. 2004;172:6093–6100. doi: 10.4049/jimmunol.172.10.6093. [DOI] [PubMed] [Google Scholar]
- Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- Graham DB, Bell MP, McCausland MM, Huntoon CJ, van Deursen J, Faubion WA, Crotty S, McKean DJ. Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with SLAM- and SAP-deficient mice. J Immunol. 2006;176:291–300. doi: 10.4049/jimmunol.176.1.291. [DOI] [PubMed] [Google Scholar]
- Gu C, Tangye SG, Sun X, Luo Y, Lin Z, Wu J. The X-linked lymphoproliferative disease gene product SAP associates with PAK-interacting exchange factor and participates in T cell activation. Proc Natl Acad Sci U S A. 2006;103:14447–14452. doi: 10.1073/pnas.0606624103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston ML, Xu Z, Majeti R, Weiss A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J Clin Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C. Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions. J Immunol. 2005;174:5931–5935. doi: 10.4049/jimmunol.174.10.5931. [DOI] [PubMed] [Google Scholar]
- Howie D, Simarro M, Sayos J, Guirado M, Sancho J, Terhorst C. Molecular dissection of the signaling and costimulatory functions of CD150 (SLAM): CD150/SAP binding and CD150-mediated costimulation. Blood. 2002;99:957–965. doi: 10.1182/blood.v99.3.957. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- Jordan MS, Boesteanu A, Reed AJ, Petrone AL, Holenbeck AE, Lerman MA, Naji A, Caton AJ. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 2006;312:1665–1669. doi: 10.1126/science.1125893. [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Gapin L, Capone M, Palmer E, MacDonald HR, Kronenberg M, Cheroutre H. Precursors of functional MHC class I- or class II-restricted CD8alphaalpha(+) T cells are positively selected in the thymus by agonist self-peptides. Immunity. 2002;16:355–364. doi: 10.1016/s1074-7613(02)00284-4. [DOI] [PubMed] [Google Scholar]
- Li W, Kim MG, Gourley TS, McCarthy BP, Sant'Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–386. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Locksley RM. Blood lines. Nat Immunol. 2002;3:705–706. doi: 10.1038/ni0802-705. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab FW, Berzins SP, Pellicci DG, Kyparissoudis K, Field K, Smyth MJ, Godfrey DI. The influence of CD1d in postselection NKT cell maturation and homeostasis. J Immunol. 2005;175:3762–3768. doi: 10.4049/jimmunol.175.6.3762. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d-restricted repertoire is dominated by a few autoreactive t cell receptor families. J Exp Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–3127. [PubMed] [Google Scholar]
- Rocha-Campos AC, Melki R, Zhu R, Deruytter N, Damotte D, Dy M, Herbelin A, Garchon HJ. Genetic and functional analysis of the Nkt1 locus using congenic NOD mice: improved Valpha14-NKT cell performance but failure to protect against type 1 diabetes. Diabetes. 2006;55:1163–1170. doi: 10.2337/diabetes.55.04.06.db05-0908. [DOI] [PubMed] [Google Scholar]
- Romero X, Zapater N, Calvo M, Kalko SG, de la Fuente MA, Tovar V, Ockeloen C, Pizcueta P, Engel P. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J Immunol. 2005;174:7033–7042. doi: 10.4049/jimmunol.174.11.7033. [DOI] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci U S A. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann J, Mycko MP, Dellabona P, Casorati G, MacDonald HR. Cutting edge: influence of the TCR Vbeta domain on the selection of semi-invariant NKT cells by endogenous ligands. J Immunol. 2006;176:2064–2068. doi: 10.4049/jimmunol.176.4.2064. [DOI] [PubMed] [Google Scholar]
- Schumann J, Pittoni P, Tonti E, Macdonald HR, Dellabona P, Casorati G. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J Immunol. 2005;175:7303–7310. doi: 10.4049/jimmunol.175.11.7303. [DOI] [PubMed] [Google Scholar]
- Simarro M, Lanyi A, Howie D, Poy F, Bruggeman J, Choi M, Sumegi J, Eck MJ, Terhorst C. SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. Int Immunol. 2004;16:727–736. doi: 10.1093/intimm/dxh074. [DOI] [PubMed] [Google Scholar]
- Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, Joyce S. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004;172:2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- Stein PL, Lee HM, Rich S, Soriano P. pp59fyn mutant mice display differential signaling in thymocytes and peripheral T cells. Cell. 1992;70:741–750. doi: 10.1016/0092-8674(92)90308-y. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- Urdahl KB, Sun JC, Bevan MJ. Positive selection of MHC class Ib-restricted CD8(+) T cells on hematopoietic cells. Nat Immunol. 2002;3:772–779. doi: 10.1038/ni814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya SV, Stepp SE, McNerney ME, Lee JK, Bennett M, Lee KM, Stewart CL, Kumar V, Mathew PA. Targeted disruption of the 2B4 gene in mice reveals an in vivo role of 2B4 (CD244) in the rejection of B16 melanoma cells. J Immunol. 2005;174:800–807. doi: 10.4049/jimmunol.174.2.800. [DOI] [PubMed] [Google Scholar]
- Veillette A. Immune regulation by SLAM family receptors and SAP-related adaptors. Nat Rev Immunol. 2006;6:56–66. doi: 10.1038/nri1761. [DOI] [PubMed] [Google Scholar]
- Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr, Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DG, Curran SA, Savage PB, Teyton L, Bendelac A. Mechanisms imposing the Vbeta bias of Valpha14 natural killer T cells and consequences for microbial glycolipid recognition. J Exp Med. 2006;203:1197–1207. doi: 10.1084/jem.20060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DG, Lee H, Park SH, Beaudoin L, Teyton L, Lehuen A, Bendelac A. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, et al. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol. 2001;2:410–414. doi: 10.1038/87713. [DOI] [PubMed] [Google Scholar]
- Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]