Abstract
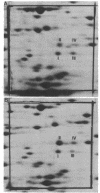
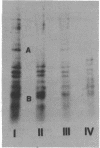
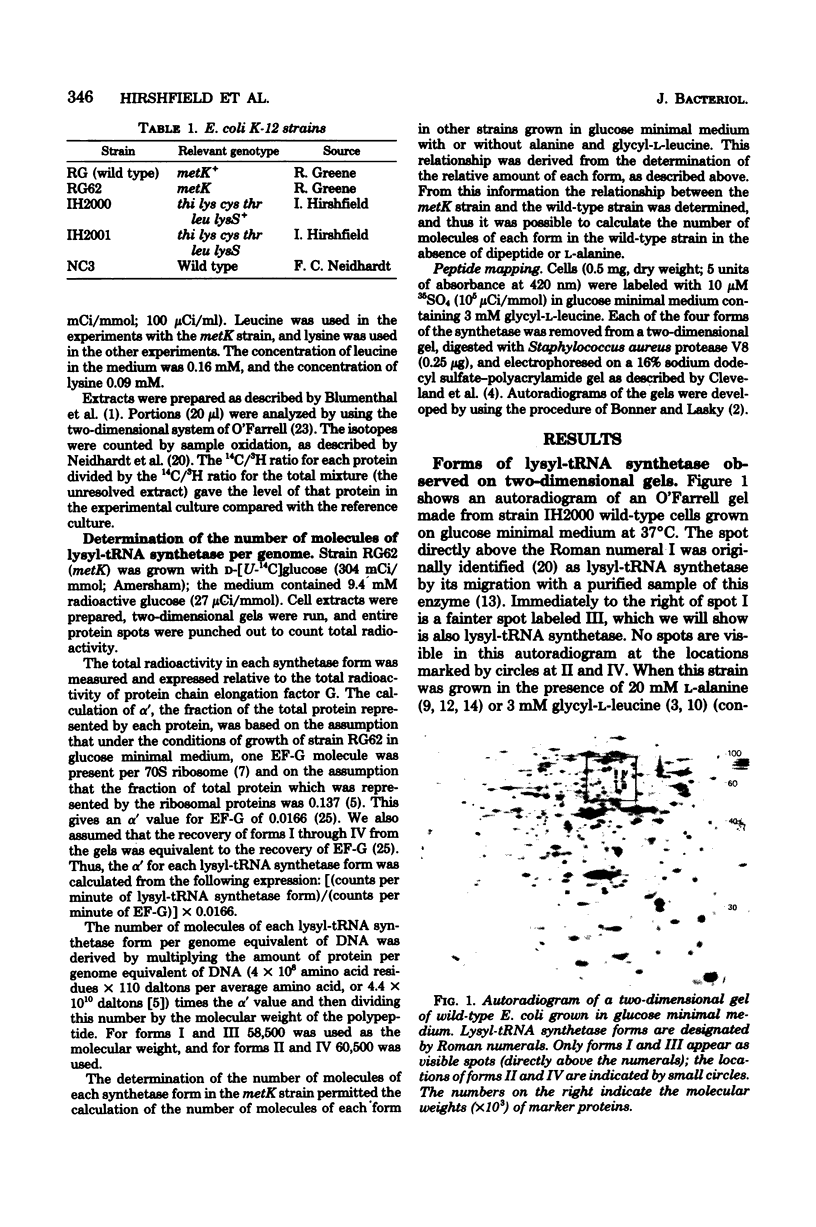
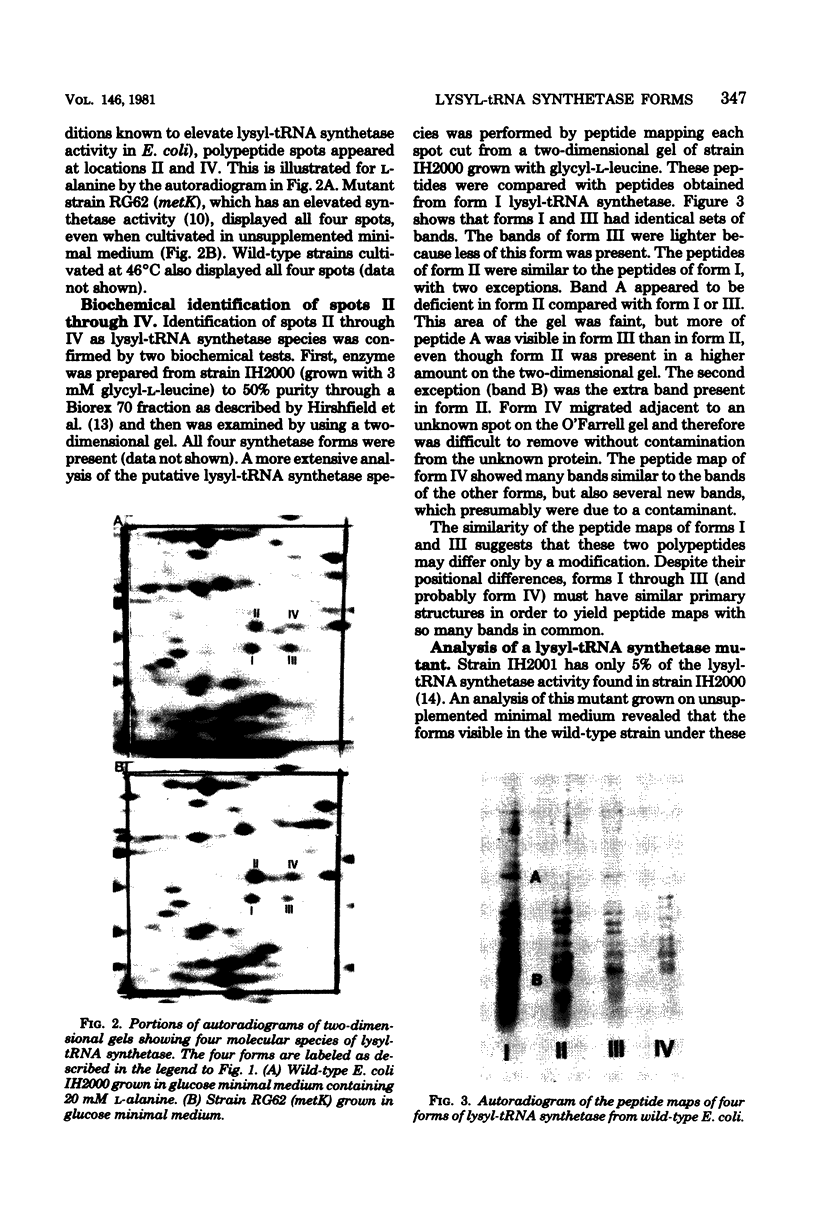
Lysyl-transfer ribonucleic acid synthetase (EC 6.1.1.6) was identified as four polypeptide spots after two-dimensional polyacrylamide gel electrophoresis of whole-cell lysates of Escherichia coli. Identification was made by migration with partially purified enzyme preparations, by peptide map patterns, by mutant analysis, and by correlation of spot intensities with changes in enzyme levels under different growth conditions. Wild-type cells growing at 37 degrees C in glucose minimal medium displayed the enzyme predominantly as two spots (spots I and III). Growth at 46 degrees C, growth in the presence of alanine or glycyl-L-leucine, or growth of a strain with a mutational deficiency in S-adenosylmethionine synthetase (metK) greatly increased the synthesis of two other spots (spots II and IV). Polypeptides I and III, but not polypeptides II and IV, had altered isoelectric points in a lysyl-transfer ribonucleic acid synthetase mutant. These data suggest that multiple forms of lysyl-transfer ribonucleic acid synthetase exist in vivo and that they may be encoded by more than one gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenthal R. M., Reeh S., Pedersen S. Regulation of transcription factor rho and the alpha subunit of RNA polymerase in Escherichia coli B/r. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2285–2288. doi: 10.1073/pnas.73.7.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buklad N. E., Sanborn D., Hirshfield I. N. Particular influence of leucine peptides on lysyl-transfer ribonucleic acid ligase formation in a mutant of Escherichia coli K-12. J Bacteriol. 1973 Dec;116(3):1477–1478. doi: 10.1128/jb.116.3.1477-1478.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Dennis P. P., Bremer H. Macromolecular composition during steady-state growth of Escherichia coli B-r. J Bacteriol. 1974 Jul;119(1):270–281. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano A. V. The elongation factor Tu coded by the tufA gene of Escherichia coli K-12 is almost identical to that coded by the tufB gene. J Biol Chem. 1977 Mar 25;252(6):2154–2157. [PubMed] [Google Scholar]
- Gordon J. Regulation of the in vivo synthesis of the polypeptide chain elongation factors in Escherichia coli. Biochemistry. 1970 Feb 17;9(4):912–917. doi: 10.1021/bi00806a028. [DOI] [PubMed] [Google Scholar]
- Greene R. C., Hunter J. S., Coch E. H. Properties of metK mutants of Escherichia coli K-12. J Bacteriol. 1973 Jul;115(1):57–67. doi: 10.1128/jb.115.1.57-67.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Liu C., Yeh F. M. Two modes of metabolic regulation of lysyl-transfer ribonucleic acid synthetase in Escherichia coli K-12. J Bacteriol. 1977 Aug;131(2):589–597. doi: 10.1128/jb.131.2.589-597.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirshfield I. N., Yeh F. M. An in vivo effect of the metabolites L-alanine and glycyl-L-leucine on the properties of the lysyl-tRNA synthetase from Escherichia coli K-12. II. Kinetic evidence. Biochim Biophys Acta. 1976 Jul 2;435(3):306–314. doi: 10.1016/0005-2787(76)90111-8. [DOI] [PubMed] [Google Scholar]
- Hirshfield I. N., Yeh F. M., Sawyer L. E. Metabolites influence control of lysine transfer ribonucleic acid synthetase formation in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1364–1367. doi: 10.1073/pnas.72.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Levinthal M., Williams L. S., Umbarger H. E. Role of threonine deaminase in the regulation of isoleucine and valine biosynthesis. Nat New Biol. 1973 Nov 21;246(151):65–68. doi: 10.1038/newbio246065a0. [DOI] [PubMed] [Google Scholar]
- McGinnis E., Williams L. S. Regulation of synthesis of the aminoacyl-transfer ribonucleic acid synthetases for the branched-chain amino acids of Escherichia coli. J Bacteriol. 1971 Oct;108(1):254–262. doi: 10.1128/jb.108.1.254-262.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Pedersen S., Reeh S. Chemical measurement of steady-state levels of ten aminoacyl-transfer ribonucleic acid synthetases in Escherichia coli. J Bacteriol. 1977 Jan;129(1):378–387. doi: 10.1128/jb.129.1.378-387.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neihardt F. C., Parker J., McKeever W. G. Function and regulation of aminoacyl-tRNA synthetases in prokaryotic and eukaryotic cells. Annu Rev Microbiol. 1975;29:215–250. doi: 10.1146/annurev.mi.29.100175.001243. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Williams L. S., Neidhardt F. C. Synthesis and inactivation of aminoacyl-transfer RNA synthetases during growth of Escherichia coli. J Mol Biol. 1969 Aug 14;43(3):529–550. doi: 10.1016/0022-2836(69)90357-x. [DOI] [PubMed] [Google Scholar]