Abstract
Myxococcus xanthus is a gliding bacterium that contains two motility systems: S-motility, powered by polar type IV pili, and A-motility, powered by uncharacterized motors and adhesion complexes. The localization and coordination of the two motility engines is essential for directed motility as cells move forward and reverse. During cell reversals, the polarity and localization of motility proteins are rapidly inverted, rendering this system a fascinating example of dynamic protein localization.
Introduction
Polarity of motility structures in bacteria is widespread. For example, Thiospirillum jenense produces a bundle of polar flagella required for locomotion and phototaxis. In contrast, motile cells of Caulobacter crescentus can swim in the direction of food by using a single polar flagellum; an alternative polar structure called the “stalk” allows the sessile form of this bacterium to adhere to a source of food. Other examples include Pseudomonas aeruginosa, which produces polar type IV pili required for twitching motility, and Listeria monocytogenes, which localizes ActA at the cell pole, nucleating host actin filaments to propel cells within the host cytoplasm. In this review, we will be focusing on polarity of motility structures in Myxococcus xanthus, which utilizes polar type IV pili and adhesion complexes for gliding motility. Of special interest in this system is that cells periodically reverse: during reversals, the leading cell pole becomes the lagging cell pole, necessitating very rapid re-localization and assembly of polar structures. Thus, the study of motility proteins in M. xanthus should yield important insights to the understanding of the dynamics and mechanism of protein localization in bacteria.
Motility plays a crucial role in the life–cycle of Myxococcus xanthus
M. xanthus is a Gram-negative soil bacterium that has a complex life cycle involving vegetative swarming and fruiting body formation. Motility is important for both of these functions. M. xanthus does not contain flagella and cannot swim in a liquid medium; it moves by gliding motility on solid surfaces. During vegetative growth, cells swarm to find prey bacteria or to gather nutrients in the environment. When M. xanthus cells are unable to find sufficient nutrients or in presence of prey [1], they enter a developmental process in which they aggregate in raised pigmented mounds, termed fruiting bodies. Within the fruiting bodies cells differentiate to form metabolically dormant spores [2].
M. xanthus cells utilize two genetically distinct systems for motility. The first system is called social (S)-motility and powered by the retraction of polar Type IV pili. The second system, called adventurous (A)-motility, is powered by not yet characterized engines involving focal adhesion sites (Fig 1A). The localization and coordination of the two motility engines is essential for bacteria to show directed motility (Fig 1B-C). M. xanthus cells move only on solid surfaces in the direction of the leading pole. Wild type vegetative cells reverse periodically, about every 6-8 min, so that the leading pole becomes the lagging pole. Directed movements occur because there is a bias in the timing of reversals. Thus cell reversals, like tumbling in the enteric bacteria, provides a mechanism for cells to reorient themselves in response to signals. How the components of the two engines of gliding motility, the S-engine and A-engine, re-orient themselves with opposing polarities as cells reverse is an important area of investigation.
Fig 1.
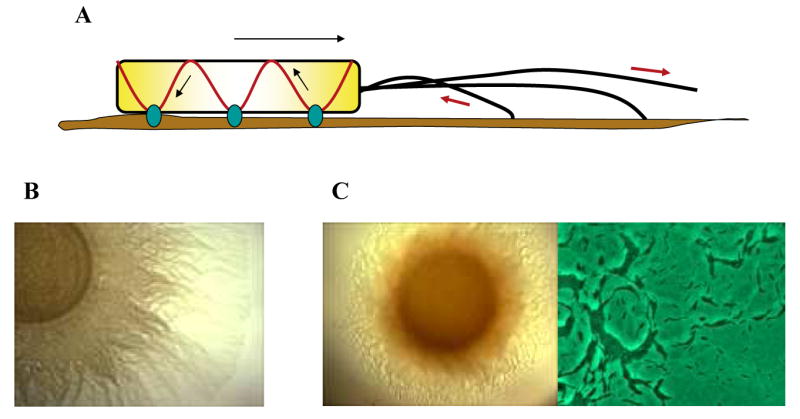
M. xanthus cells use two motility systems. (A) Schematic representation of A and S engine. The A engine includes membrane spanning adhesion sites (blue) connected to the cytoskeleton (red) and pushing against the substratum to moving the cell body forward. S-motility is powered by Type IV Pili localized at the leading pole and pulling the cell forward by attaching to the substrate or to another cell and then retracting. (B) S-motility assay of wild type cells plated on 0.5% agar. (C) A-motility assay of wild type cells plated on 1.5% agar. Many single cells are visible at the 20 times magnified edge of the colony.
Polar localization of type IV pili may require polar motility superstructures
Genetic and behavioral analyses have shown that S-motility requires type IV pili, lipopolysaccharide O-antigen, extracellular matrix polysaccharide (EPS, also known as fibrils), and FrzS [3,4••]. S-motility is similar to twitching motility in Pseudomonas aeruginosa [5], and is powered by retraction of the polar pili: pili are extruded from one cell pole, adhere to a surface or to another cell, and are then stimulated to retract [3]. Retraction of pili, triggered by specific attachment to polysaccharides or EPS, pulls cells in the direction of the sites of adhesion [3]. Retraction works as a signal for the production of EPS in a regulatory loop involving the dif pathway [6]. Thus, pili not only serve as the machinery that pulls cells forward, but likely also functions to sense the proximity of neighboring cells, and leads to the production of more EPS.
PilA is the major subunit of type IV pili and it localizes at the leading pole of moving cells. PilA is a soluble protein that is secreted through a channel in the outer membrane composed of PilQ [7]. Tgl is the lipoprotein necessary to assemble the PilQ secretin in M. xanthus. In the absence of Tgl, only the monomeric form of PilQ is found and cells are not able to secrete PilA, assemble pili or show S-motility. Interestingly, cells with a tgl mutation can be stimulated to assemble PilQ channels and pili after being mixed with wild type cells. The presence of Tgl in the tgl cells after stimulation suggests that transient contact and fusion between outer membranes of Tgl+ donor cells and Tgl- recipient cells can result in the physical transfer of Tgl protein from one cell to another [8••].
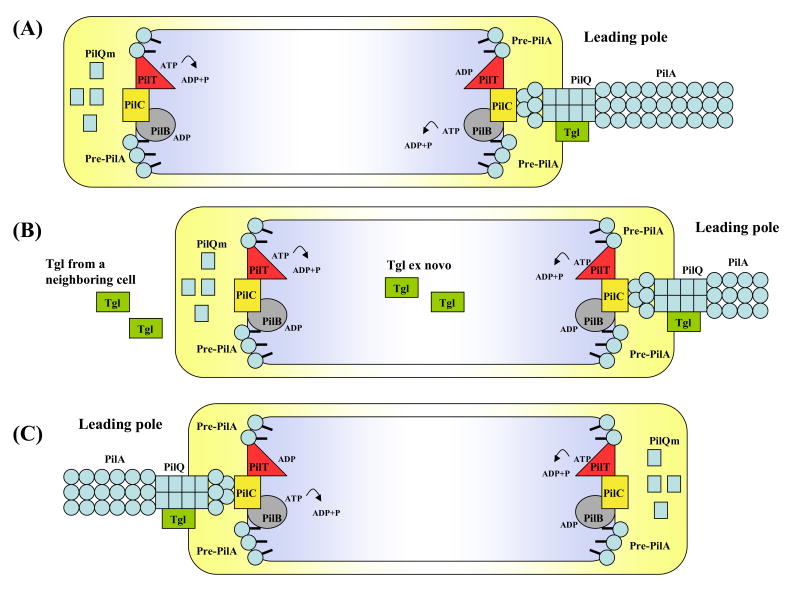
Tgl localizes at one pole, most likely the leading pole, since it is required for assembly of the PilQ channel and, consequently, PilA secretion and assembly [8••]. In contrast, PilQ is bipolar; its localization is Tgl independent. Thus only the leading pole should contain the assembled (functional) form of PilQ [9•]. As proposed in Fig 2, PilQ might be part of a superstructure (complex of motility proteins) present at both cell poles that includes the transporter, PilC, and the ATPases, PilB and PilT that are involved, respectively, in the assembly and retraction of pili. The leading pole superstructure should contain all S-motility components, while the lagging pole superstructure should lack PilA, Tgl and multimeric PilQ. When cells reverse and the lagging pole becomes the leading pole, Tgl and PilA need to be translocated and reassembled at the opposite cell pole; the translocation of Tgl may be facilitated by cell-cell contact. Tgl should activate the PilQ channel and induce transport and assembly of PilA filaments. The presence of a superstructure at both cell poles should facilitate rapid and efficient cell reversals. According to this model, a new motility superstructures need to be made only when cells divide.
Fig 2.
A model for the switching of polarity of Type IV pilus. A superstructure including PilB, PilT, PilC is present at both leading pole and lagging pole. The lipoprotein Tgl is only present at the leading pole and this causes the secretin PilQ to be disassembled at the lagging pole and assembled at the leading pole. The presence of PilQ channels allows the assembly of PilA filaments, from pre-pilin monomers (Pre-PilA), at the leading pole. Also PilB participates in the assembly of PilA by hydrolyzing ATP (A). During S-motility the retraction of TFP allows the cell to move forward. Disassembly of PilA filaments for TFP retraction is mediated by the ATPase activity of PilT (B). When the signal for a reversal occurs, Tgl molecules synthesized ex novo and/or taken from the external environment allow the formation of an active machinery at the new leading pole (C).
FrzS shows shifting polar localization as cells reverse
Another fascinating example of a protein involved in the reversal of cell polarity is FrzS. FrzS has an N-terminal pseudo-receiver domain, and an extended coiled-coil C-terminal domain. frzS mutants are defective in S-motility swarming [10]. FrzS preferentially accumulates at the leading (piliated) cell pole but it is also present at the lagging cell pole. During movements, the amount of FrzS at the leading pole decreases while FrzS at the lagging pole increases. When cells reverse, the remaining FrzS from the old leading pole rapidly shifts to the new leading pole. The frequency of FrzS oscillations is controlled by the Frz chemosensory system, which is essential for directed motility. Interestingly, mutants containing a deletion in the N-terminal pseudo-receiver domain of FrzS showed FrzS primarily localization to the lagging cell pole rather than the leading cell pole; in these mutants, upon cell reversals, FrzS is initially translocated to the leading cell pole, but is not retained there, causing FrzS to accumulate at the lagging cell pole. In contrast, mutants in a C-terminal region failed to dock at the lagging cell pole. FrzS mutants lacking both the pseudo-receiver domain and the C-terminal region localized the cryptic FrzS polypeptide in a filamentous structure along the cell body rather than at the cell poles. FrzS is hypothesized to be transported along a filament by way of the coiled-coil domain and delivered to the cell poles where it binds through the combined actions of the receiver domain and the C-terminal tail [11•].
A-motility proteins assemble in clusters at the leading cell pole
The A-motility system does not involve pili or other visible external structures, making it difficult to identify the motility engine. Recently, a “slime propulsion” model for A-motility was suggested to explain myxobacterial slime trails and polar nozzle-like structures attributed to A-motility [12]. This model was supported by calculations showing that the force generated by the hydration of slime within nozzles could push cells forward at the velocity observed for A-motility [12]. However, genetic and biochemical evidence supporting the slime propulsion model is weak. Furthermore, the gliding motor for A-motility is probably not focused at the cell poles, as suggested in the model, but distributed along the body of the cell ([13] and O. Sliusarenko, D. Zusman, and G. Oster, in press). In agreement with the idea of a distributed motor, Mignot et al. recently published a study about the localization of A-motility protein, AglZ [14••].
AglZ, discovered by the Hartzell laboratory, is an essential A-motility protein [15]. AglZ is structurally similar to FrzS in that it contains a N-terminal pseudo-receiver domain and a long C-terminal coiled-coil domain [15, 18]. To track the localization of AglZ in moving cells, M. xanthus strains were constructed that produced AglZ-YFP (Yellow fluorescent protein) [14••]. Fully motile cells showed AglZ-YFP to be present in clusters distributed in an ordered array spanning the cell length. As cells moved forward, these clusters maintained fixed positions with respect to the agar surface, rather than to their relative positions in the cell. Interestingly, the only AglZ-YFP cluster that moved relative to the cell body was the one located at the leading pole, suggesting that new clusters were assembled at the leading pole. Upon cellular reversal, AglZ localized rapidly to the new leading pole, and as cells began to move, localized to distributed clusters. These results suggest an alternative mechanism for A-motility whereby intracellular motor complexes that connect to membrane spanning adhesion complexes and to the cytoskeleton power motility by pushing against the substratum moving the cell body forward (Fig 1A). It is noteworthy that A and S motility systems do not show opposite polarity: during cellular reversals, proteins from both A- and S-motility systems are shifted to the new leading pole.
Frz chemosensory pathway regulates switching cell polarities
About every seven minutes on motility agar, M. xanthus cells switch their polarity and direction of movement [16]. The frequency and bias are regulated by the Frz chemosensory system in response to positive and negative stimulation. The Frz pathway is similar to the Che (chemotaxis) pathway from enteric bacteria except that instead of regulating the direction of rotation of flagella, it regulates cellular reversals (Fig 3). Regulating cell reversals is essential for directed cell movements in M. xanthus. The ability of cells to reverse direction allows cells to reorient themselves as part of a biased random walk, in much the same way that changing the rotation of flagella in the enteric bacteria causes tumbles, allowing cellular reorientations. Mutations in the Frz pathway generally diminish reversals, although some mutations cause hyper-reversals. When cells cannot regulate their reversal frequency, they lose coordination at the population level during A and S swarming, and fruiting body formation [19].
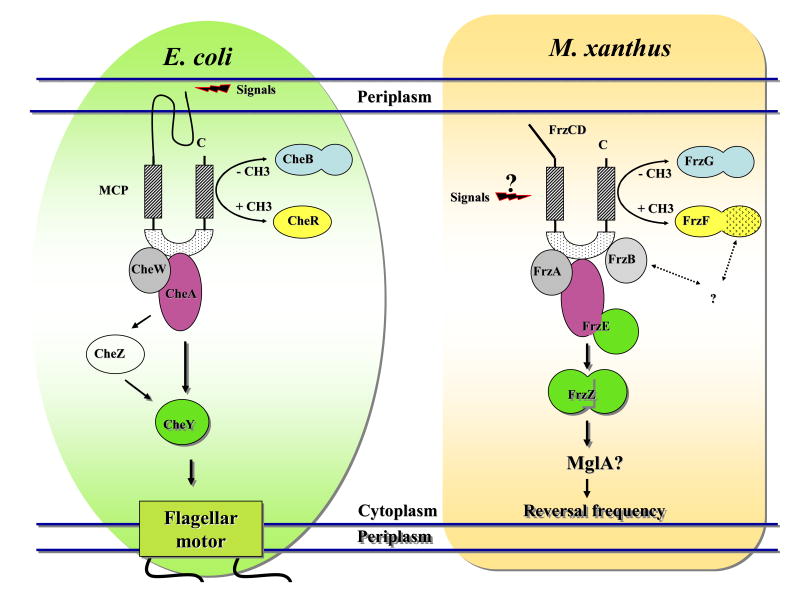
Fig 3.
Schematic representation of the chemotaxis system of E. coli and the Frz pathway of M. xanthus. The chemotaxis system of E. coli is composed of a transmembrane receptor (MCP) that receives signals from the periplasm; a methyltrasferase (CheR) and a methylesterase (CheB) that modulate the methylation state of the MCP; a histidine kinase (CheA) linked to the MCP through the helper protein CheW; a response regulator (CheY) that, once phosphorilated by CheA, can directly communicate with the flagellar motor; the phosphatase CheZ increases the spontaneous dephosphorilation of CheY to allow rapid signal termination. The stimulation from the periplasm leads, through the chemotaxis system, to the switch in the flagellar rotation so that E. coli cells can swim in the direction of nutrients. The Frz pathway of M. xanthus shares homology with the chemotaxis system of E. coli: FrzCD is a cytoplasmic MCP methylated and demethylated respectively by FrzF (CheR) and FrzG (CheB); FrzE is a fusion between a CheA (purple) and CheY (green) domain; FrzA and FrzB are CheW proteins; FrzZ is a fusion of two CheY domain. The Frz pathway regulates the frequency by which M. xanthus cells reverse the gliding direction to orientate in response to stimuli.
That the Frz pathway regulates reversals of S-motility was shown by Sun et al [17] who followed the retraction of pili in cells placed on glass slides in the presence of 1% methylcellulose. Under these conditions, the pili bind to the surface of the slide, tethering the cells. Cells remained bound to the surface for 7-8 min after which they are released as the pili are retracted. frz mutants that hypo or hyper-reverse remained tethered for extended or shortened periods, respectively. Unfortunately, the outputs that link the Frz pathway to the M. xanthus motility systems are unknown. However, recent findings from the study of AglZ and FrzS suggest that they may play a role in this process, since the oscillations of both proteins are regulated by the signaling activity of the Frz pathway. In frz hyporeversing strains, FrzS rarely switches from one end of the cell to the other and AglZ doesn't switch at all. On the other hand, in frz hypereversing strains FrzS and AglZ both show hyperoscillation [7, 18].
However, it is very unlikely that they are phosphorylated by the FrzE kinase, as both FrzS and AglZ lack the conserved aspartate residue that is phosphorylated in canonic receiver domains [11,18].
MglA: a possible coordinator of A- and S-motiltiy
mglA, the only known gene common to both A and S-motility [19], encodes a 22 KDa protein that shares similarities with small GTPases of the Ras family [24]. In eukaryotic cells, small GTPases act as regulatory proteins often by recruiting factors to their site of action. It is therefore possible that MglA acts to recruit factors that are important for reversal of the two motility systems. However, since mglA mutants are non-motile, it must function in recruiting other essential motility proteins as well. MglA is a good candidate for an output that links the Frz pathway to both the A and S motors. We speculate that MglA is activated by the Frz pathway to recruit molecules responsible for the switch of polarity in their site of action. MglA is essential for localization of both FrzS and AglZ: in an mglA mutant, FrzS remains localized to only one cell pole, while AglZ is no longer found in clusters but is diffused in the cytoplasm. Furthermore, protein interaction studies show that MglA can directly interact with both FrzS and AglZ (T. Mignot and D. Zusman, unpublished data). Clearly, additional experimental data are needed to elucidate the role of MglA in motility.
Conclusion
The dynamic localization of motility proteins in M. xanthus as cells move forward and reverse provides a powerful model system for the study of cell polarity and localization. The A- and S-motility systems each have many proteins that need to interact in specific ways to function. Some motility proteins are fixed in position while others, such as FrzS and AglZ, clearly shift their localization within seconds of receiving a reversal signal. FrzS, an S-motility protein, and AglZ, an A-motility protein, both move from the leading to the lagging pole utilizing different mechanisms. Nevertheless, the coordination of these various motility proteins is crucial for the two motility systems to function with the same directionality. These proteins are likely to interact with cytoskeletal proteins like MreB and with membrane proteins as they move from pole-to-pole within the cell. The study of these interactions should add new insights into the inner workings of bacterial cells.
Acknowledgments
We would like to thank T. Mignot, J. Berleman, J. Merlie, Y. Inclan, for critical comments on this manuscript and the entire Zusman lab for stimulating discussions. Research in our laboratory is funded by a grant from the National Institute of Health to DRZ (GM20509).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
Papers of particular interest, published within the annual period of review have being highlighted as:
• of special interest
•• of outstanding interest
- 1.Berleman JE, Kirby JR. Multicellular Development in Myxococcus xanthus Is Stimulated by Predator-Prey Interactions. J Bacteriol. 2007;189:5675–5682. doi: 10.1128/JB.00544-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimkets LJ. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A. 2003;100:5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4••.Mignot T, Merlie JP, Jr, Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 5.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black WP, Xu Q, Yang Z. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol Microbiol. 2006;61:447–456. doi: 10.1111/j.1365-2958.2006.05230.x. [DOI] [PubMed] [Google Scholar]
- 7.Wall D, Kolenbrander PE, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309:125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 9•.Nudleman E, Wall D, Kaiser D. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol Microbiol. 2006;60:16–29. doi: 10.1111/j.1365-2958.2006.05095.x. [DOI] [PubMed] [Google Scholar]
- 10.Ward MJ, Lew H, Zusman DR. Social motility in Myxococcus xanthus requires FrzS, a protein with an extensive coiled-coil domain. Mol Microbiol. 2000;37:1357–1371. doi: 10.1046/j.1365-2958.2000.02079.x. [DOI] [PubMed] [Google Scholar]
- 11.Mignot T, Merlie JP, Jr, Zusman DR. Two localization motifs mediate polar residence of FrzS during cell movement and reversals of Myxococcus xanthus. Mol Microbiol. 2007 doi: 10.1111/j.1365-2958.2007.05789.x. [DOI] [PubMed] [Google Scholar]
- 12.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 13.Sun H, Yang Z, Shi W. Effect of cellular filamentation on adventurous and social gliding motility of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1999;96:15178–15183. doi: 10.1073/pnas.96.26.15178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Mignot T, Shaevitz JW, Hartzell PL, Zusman DR. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang R, Bartle S, Otto R, Stassinopoulos A, Rogers M, Plamann L, Hartzell P. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J Bacteriol. 2004;186:6168–6178. doi: 10.1128/JB.186.18.6168-6178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackhart BD, Zusman DR. “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci U S A. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, Zusman DR, Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 18.Fraser JS, Merlie JP, Jr, Echols N, Weisfield SR, Mignot T, Wemmer DE, Zusman DR, Alber T. An atypical receiver domain controls the dynamic polar localization of the Myxococcus xanthus social motility protein FrzS. Mol Microbiol. 2007 doi: 10.1111/j.1365-2958.2007.05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]