Figure .
Vol. 169, No. 6, June 20, 2005. Pages 929–939.
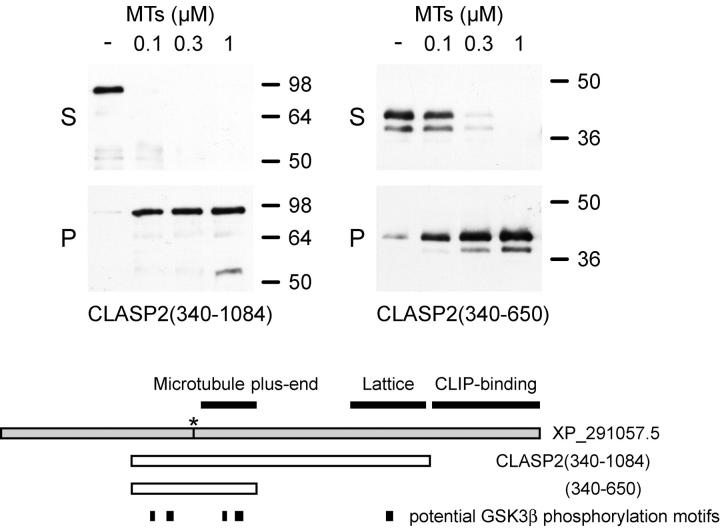
In Fig. 6 B of this article, we reported that bacterially expressed His6-CLASP2(340–875) does not bind microtubules in vitro. However, we found in subsequent experiments that the band on the immunoblot we interpreted as His6-CLASP2(340–875) is in reality a bacterial contamination of very similar molecular mass, which is unfortunately recognized by the antibody used in this experiment. We further found that we were not able to obtain sufficient amounts of nondegraded recombinant His6-CLASP2(340–875) from bacteria to accurately compare its microtubule binding to the longer construct. To resolve this, we instead cloned and expressed a shorter CLASP2 construct, His6-CLASP2(340–650), which contains the entire NH2-terminal region sufficient for plus end tracking in cells. We then repeated the microtubule-binding experiment and probed the immunoblot with a His-tag antibody (H-15; Santa Cruz Biotechnology, Inc.) and now find that both constructs, His6-CLASP2(340–1084), which associates with microtubule lattices in the cell periphery, and His6-CLASP2(340–650), which can only track plus ends in cells, bind to microtubules in vitro with similar high affinity (KD < 0.2 μM) within the accuracy of the microtubule pelleting assay. S, supernatant; P, pellet. Molecular masses are indicated in kilodaltons on the right. Thus, contrary to our earlier conclusion, the region between amino acids 875 and 1084 appears not to be directly required for high affinity microtubule binding. Interestingly, this also suggests that microtubule plus end tracking in cells is not due to an inherently lower affinity for microtubules as compared to classical lattice-binding microtubule-associated proteins. This does not affect any of the other results reported in this paper.