Abstract
Cytokinesis completes cell division and partitions the contents of one cell to the two daughter cells. Here we characterize CAR-1, a predicted RNA binding protein that is implicated in cytokinesis. CAR-1 localizes to germline-specific RNA-containing particles and copurifies with the essential RNA helicase, CGH-1, in an RNA-dependent fashion. The atypical Sm domain of CAR-1, which directly binds RNA, is dispensable for CAR-1 localization, but is critical for its function. Inhibition of CAR-1 by RNA-mediated depletion or mutation results in a specific defect in embryonic cytokinesis. This cytokinesis failure likely results from an anaphase spindle defect in which interzonal microtubule bundles that recruit Aurora B kinase and the kinesin, ZEN-4, fail to form between the separating chromosomes. Depletion of CGH-1 results in sterility, but partially depleted worms produce embryos that exhibit the CAR-1–depletion phenotype. Cumulatively, our results suggest that CAR-1 functions with CGH-1 to regulate a specific set of maternally loaded RNAs that is required for anaphase spindle structure and cytokinesis.
Introduction
Cytokinesis completes cell division by creating membranous barriers that partition the cytoplasm of one cell to form two topologically distinct daughter cells. In animal cells, cytokinesis requires continuous interplay between the microtubule and cortical acto-myosin cytoskeletons (Straight and Field, 2000; Glotzer, 2003; Robinson and Spudich, 2004). After chromosome segregation, an array of interzonal microtubule bundles forms between the segregated chromosomes. Concurrently, a cortical acto-myosin–based contractile ring assembles and constricts, and changes the shape of the cell to facilitate division. As cytokinesis proceeds, the interzonal microtubule bundles compact to form the spindle midbody, and the contractile ring constricts around this structure. The midbody is believed to direct localized membrane fusion that generates the two topologically distinct daughter cells (Finger and White, 2002; Schweitzer and D'Souza-Schorey, 2004).
The embryo of the nematode Caenorhabditis elegans recently emerged as a powerful system for studying cell division. In C. elegans, RNA-mediated interference (RNAi) can be used to generate oocytes that are depleted of targeted essential proteins in a process that does not depend on intrinsic protein turnover (Oegema and Hyman, 2005). The first mitotic division of the depleted embryos can be monitored after fertilization. Several genome-wide RNAi-based screens have identified the set of nonredundant genes that is required for embryonic viability (Gunsalus and Piano, 2005). Additional screens in which embryos that were depleted of each of these gene products were imaged by differential interference contrast (DIC) microscopy defined further the subset of these genes that have essential roles in cell division (Gönczy et al., 2000; Piano et al., 2000; Zipperlen et al., 2001; Sönnichsen et al., 2005). This approach successfully identified most proteins that were known to function during cytokinesis. A small number of new gene products also were identified, including one that corresponds to an uncharacterized, potential RNA-binding protein (Y18D10a.17) that we named CAR-1 (see Results section).
Sequence analysis showed that CAR-1 is a member of the Scd6 family of proteins. Scd6 family members contain several distinct sequence features, including regions of low complexity that are enriched in charged residues (e.g., RS and RG motifs) that are typical of RNA-binding proteins (Dreyfuss et al., 1993), and a recently defined FDF domain of unknown function (Anantharaman and Aravind, 2004). All Scd6 family members also have a divergent Sm domain at their NH2-termini (Anantharaman and Aravind, 2004). Sm and Sm-like domains are present in a variety of proteins that have been implicated in RNA metabolism, including small nuclear ribonucleoprotein particles that contribute to pre-mRNA splicing, RNA processing, and telomere replication (for review see Kambach et. al., 1999), and protein complexes that are implicated in mRNA decapping and degradation (for review see Coller and Parker, 2004). Sm domains typically associate with other Sm domains to form heptameric or hexameric toroids that bind to short uracil-rich stretches of RNA (Toro et al., 2001; Thore et al., 2003). However, the sequence divergence within the atypical Sm domain that is found in Scd6 family members is predicted to confer unique RNA binding properties to this protein family (Anantharaman and Aravind, 2004).
Because the sequence features of the conserved Scd6 protein family were recognized only recently, their function remains unclear. To better understand the function of Scd6 family proteins, and how depletion of a predicted RNA-binding protein results in a cytokinesis defect, we characterized the C. elegans Scd6 homologue, CAR-1. We show that CAR-1 is a component of a multiprotein complex that also contains the DEAD box RNA helicase, CGH-1, and a Y-box–containing protein, CEY-2. CAR-1 and CGH-1 localize to RNA-containing P-granules that concentrate in the germline precursors, and to smaller cytoplasmic particles that are present in the gonad and in all cells of early embryos. Depletion of CAR-1 results in a specific defect in the microtubule cytoskeleton that becomes pronounced after anaphase onset, when assembly of interzonal microtubule bundles is impaired severely and cytokinesis fails. Cumulatively, our results suggest that CAR-1 functions with CGH-1 to regulate a specific set of RNAs that is required for anaphase spindle structure and cytokinesis during early embryogenesis.
Results
Maternally loaded CAR-1 is essential for cytokinesis during early embryogenesis
RNAi-based functional genomic screens of C. elegans, in which embryos that were laid by RNA-treated worms were imaged by DIC microscopy, identified several genes that are required for cytokinesis (Gönczy et al., 2000; Piano et al., 2000; Zipperlen et al., 2001; Sönnichsen et al., 2005). One of these, Y18D10a.17, was a previously uncharacterized, but widely conserved, 340-aa protein containing a predicted RGG box and an atypical Sm domain, two motifs that are found commonly in RNA-binding proteins (Fig. 1 A). Consistent with the sequence predictions, purified GST fusions with the Sm domain and the RGG box bound to immobilized RNA (poly(U)-sepharose) beads (Fig. S1; available at http://www.jcb.org/cgi/content/full/jcb.200506124/DC1). Based on the primary sequence features and depletion phenotype, we and other investigators have named this gene car-1, for cytokinesis/apoptosis/RNA.
Figure 1.
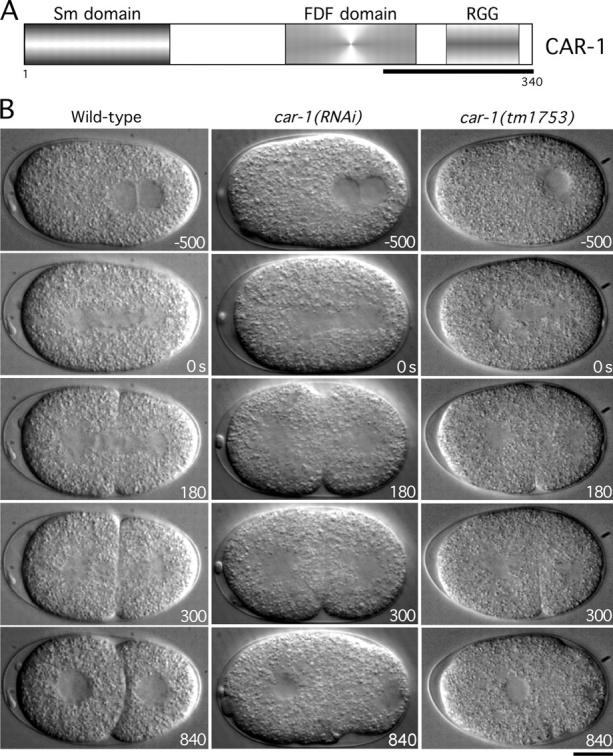
Cytokinesis fails in car- 1 mutant embryos. (A) Schematic illustration of the domain structure of CAR-1. Polyclonal antibodies were generated against the underlined region. (B) Selected panels from time-lapse DIC sequences of wild-type embryos (left column), car-1(RNAi) embryos (middle column), and car-1(tm1753) mutant embryos (right column). Time in seconds after apparent chromosome alignment is indicated in the lower right corner of each panel. (See also Videos 1–3). Bar, 10 μm.
To study its role in cytokinesis, we injected hermaphrodites with double-stranded RNA (dsRNA) against car-1, and analyzed embryos that were laid by the injected mothers, which are depleted of maternally loaded CAR-1 protein. To ensure specificity, dsRNAs against three different regions of the car-1 gene were tested (Table S1 A; available at http://www.jcb.org/cgi/content/full/jcb.200506124/DC1). 45 h after injection, when Western blotting revealed that CAR-1 was >95% depleted (e.g., see Fig. 3 D), >99% embryonic lethality was observed (e.g., see Fig. 3 F). Analysis of the depleted embryos by DIC confirmed the cytokinesis defect that was reported by Zipperlen and coworkers (2001) (Fig. 1 B; see Videos 1 and 2). In a minority of cases (n = 9/50), the first cytokinesis succeeded, but subsequent divisions failed, which accounted for the penetrant embryonic lethality.
Figure 3.
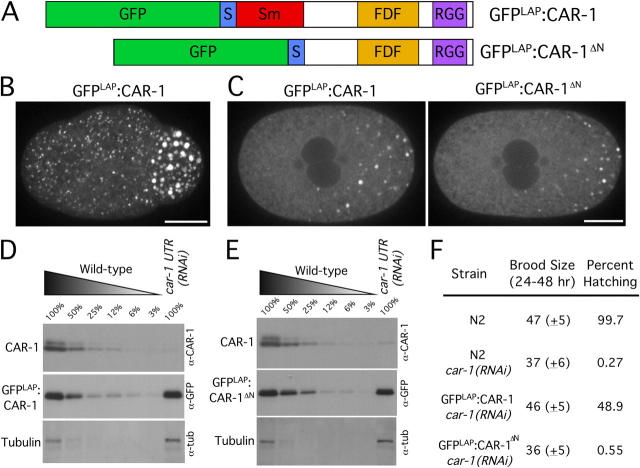
The atypical Sm domain is not required for CAR-1 localization but is essential for its function. (A) Schematic illustrations of the domain structure of two fusion proteins whose functionality was compared. A GFP-containing localization and purification (GFPLAP) tag was fused to full-length CAR-1 (top) or a truncated version of CAR-1 lacking the NH2-terminal Sm domain (bottom). The S-peptide sequence, a component of the GFPLAP tag used for biochemical purification, also is indicated. (B) Projected three-dimensional dataset of a living wild-type embryo at the four-cell stage expressing GFPLAP:CAR-1. Bar, 10 μm (see also Video 4). (C) Single section spinning disc confocal images of living prometaphase embryos expressing GFPLAP:CAR-1 (left) or GFPLAP:CAR-1ΔN (right) are shown after depletion of endogenous CAR-1 using car-1 3′UTR RNAi. Bar, 10 μm. Western blots of extracts prepared from GFPLAP:CAR-1–expressing worms (D) or GFPLAP:CAR-1ΔN–expressing worms (E) that have been depleted specifically of endogenous CAR-1 by RNAi against the car-1 3′UTR. Serial dilutions of extracts prepared from untreated worms expressing GFPLAP:CAR-1 or GFPLAP:CAR-1ΔN were loaded to quantify depletion levels. (F) Wild-type (N2), GFPLAP:CAR-1–expressing, and GFPLAP:CAR-1ΔN–expressing hermaphrodites that were injected with dsRNA targeted against the car-1 3′UTR to deplete the endogenous protein were scored for brood size and embryonic lethality.
To determine if CAR-1 function also is required at other stages of development, we analyzed a deletion allele of car-1. The deletion tm1753, which became available from the Japanese National Bioresource Project during the course of this work, is predicted to delete the COOH-terminal 225 amino acids of CAR-1, including the RGG box and the FDF domain. Homozygous car-1(tm1753) mutant embryos laid by heterozygous mothers contain maternally loaded CAR-1 that allows them to progress through the early stages of embryogenesis normally. Surprisingly, homozygous car-1(tm1753) mutant embryos hatched and developed through the larval stages to form apparently normal adult hermaphrodites. However, homozygous adult car-1(tm1753) hermaphrodites laid significantly fewer embryos than did the wild type (22 ± 6 in 24 h at 20°C vs. 48 ± 5 for wild-type; n = 5), and all of these embryos failed to hatch. Embryos laid by homozygous car-1 mutant mothers exhibited a cytokinesis defect identical to that observed in embryos that were depleted of CAR-1 by RNAi (Fig. 1 C; Video 1; Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200506124/DC1). These results suggest that our RNAi phenotype corresponds to a complete loss of maternally supplied CAR-1. In addition, we conclude that although zygotic transcription of car-1 is not required for development to adulthood, maternally loaded CAR-1 is essential for cytokinesis during early embryogenesis.
CAR-1 localizes to RNA-containing particles
To determine how depletion of CAR-1 leads to a defect in embryonic cytokinesis, we examined its localization. An affinity-purified antibody to the COOH terminus of CAR-1 (Fig. 1 A) detected two closely spaced bands on Western blots that were reduced by >95% by RNAi of car-1 (see Fig. 3 D). CAR-1 localized to cytoplasmic particles whose size and distribution varied during the early embryonic divisions (Fig. 2 A). Particles were not detected in car-1(RNAi) embryos (Fig. 2 B), which confirmed the specificity of the localization. From the latter half of the first division onward, CAR-1 localized prominently to large particles that were similar in size and distribution to P-granules, which are enriched in the germline precursors and contain poly(A)+ RNAs and several proteins that are predicted to bind RNA (Strome and Wood, 1982; Seydoux and Fire, 1994). By performing immunofluorescence in a strain expressing GFP:PGL-1 (Cheeks et al., 2004), we confirmed that a subset of CAR-1 colocalizes with PGL-1 to P-granules (Fig. 2). Depletion of CAR-1 did not affect P-granule formation or distribution (Fig. 2 B), which indicated that CAR-1 is not necessary for either of these events.
Figure 2.
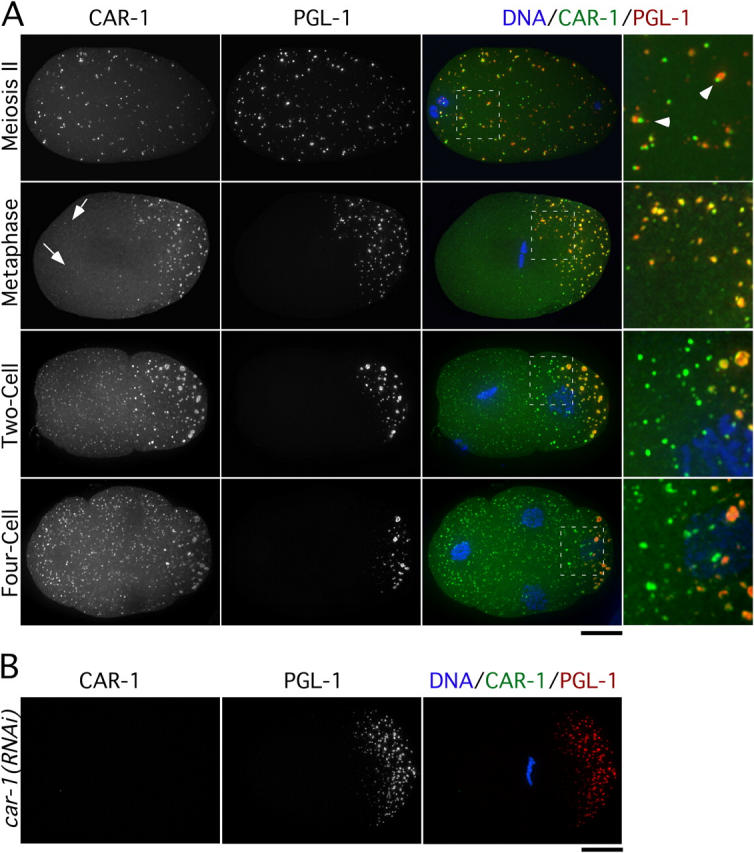
CAR-1 localizes to P-granules and additional smaller cytoplasmic particles. (A) Projected three-dimensional datasets of fixed embryos at the indicated cell cycle stages stained for CAR-1 (left column) and PGL-1 (middle column). Merged images with CAR-1 in green, PGL-1 in red, and DNA in blue also are shown. Panels in the right column are of the boxed area magnified 3× relative to the adjacent images. Arrowheads point to examples of juxtaposed CAR-1– and PGL-1–containing particles during meiosis II. Arrows identify smaller cytoplasmic CAR-1–containing particles that lack PGL-1. Bar, 10 μm. (B) No CAR-1 staining is detected in car-1(RNAi) embryos. Bar, 10 μm.
A more detailed comparison revealed a dynamic nature to the particulate localization of CAR-1 and PGL-1. In embryos in which the female pronucleus was completing meiosis, both proteins localized to numerous small particles that were distributed throughout the embryo (Fig. 2 A). However, although particles that contained the two markers were juxtaposed, they were not coincident (Fig. 2 A). By metaphase of the first mitotic division, most particulate CAR-1 colocalized with PGL-1 in large P-granules in the embryo posterior. However, a few smaller particles containing CAR-1, but not PGL-1, remained scattered throughout the embryo (Fig. 2 A). In two- and four-cell–stage embryos, CAR-1 continued to colocalize with P-granules in the germline precursors, but numerous additional smaller CAR-1–containing particles also were evident throughout the cytoplasm of all cells (Fig. 2). These smaller CAR-1–containing particles were not detected after the 50–100-cell stage (unpublished data).
To examine CAR-1 particle dynamics directly, we filmed embryos expressing a functional GFPLAP:CAR-1 fusion (see Fig. 3 B, D, and E and Video 4). The formation of the small GFPLAP:CAR-1–containing particles in both daughter cells after completion of the first cytokinesis was particularly prominent in these sequences. The number of small particles increased as embryos proceeded into the four- and eight-cell stages, but the particles were not detected in older embryos. In summary, consistent with the RNA-binding motifs in its primary sequence, CAR-1 localizes to P-granules that are partitioned to the germline precursors and to smaller particles that are present in all cells of early embryos.
The atypical Sm domain of CAR-1 is essential for function but not localization
CAR-1 and all other Scd6 family proteins contain an atypical Sm domain at their NH2 terminus. To determine whether this RNA-binding domain is important for CAR-1 function, we generated integrated strains expressing fusions of GFPLAP with full-length CAR-1 or CAR-1 lacking the atypical Sm domain (CAR-1ΔN;Fig. 3, A–C), and used a dsRNA against the car-1 3′UTR to deplete endogenous CAR-1 specifically in embryos from both strains. The transgenes expressing the GFP fusions use the pie-1 3′UTR, and therefore, are insensitive to the car-1 3′UTR dsRNA. Western blotting confirmed that car-1 3′UTR RNAi resulted in >95% depletion of endogenous CAR-1 without significantly altering the levels of the GFP fusions (Fig. 3, D and E). Expression of full-length GFPLAP:CAR-1 rescued the viability of CAR-1–depleted embryos to ∼50% (Fig. 3 F), and their ability to complete cytokinesis (n = 21/22 one-cell–stage embryos completed cytokinesis; not depicted). In contrast, GFPLAP:CAR-1ΔN failed to rescue the viability of CAR-1–depleted embryos (Fig. 3 F) or their ability to complete cytokinesis (n = 18/24 one-cell–stage embryos did not complete cytokinesis; not depicted). Thus, the Sm domain of CAR-1 is essential for its function.
Surprisingly, after depletion of endogenous CAR-1, GFPLAP: CAR-1ΔN localized identically to full-length GFPLAP:CAR-1 (Fig. 3 C). This result indicates that the CAR-1 Sm domain is dispensable for its localization to P-granules and smaller particles. Together with the fact that PGL-1 targets normally in CAR-1–depleted embryos (Fig. 2 B), this result indicates that inhibition of CAR-1 function does not prevent assembly of P-granules or the smaller particles that are present in all cells.
CAR-1 associates with the essential RNA helicase CGH-1 and the Y-box–containing protein CEY-2
The above results suggested that CAR-1 interacts with other RNA-binding proteins to execute its function in cytokinesis. To identify such proteins, we used a tandem affinity purification scheme (Cheeseman et al., 2004). Protein complexes containing GFPLAP:CAR-1 were purified by immunoprecipitation with antibodies to GFP, released by cleavage with the tobacco etch virus protease, and reisolated by binding to S-protein agarose. Proteins eluted from the S-protein agarose with urea were analyzed by solution mass spectrometry. Under these stringent purification conditions, we obtained significant sequence coverage for CAR-1 and two additional proteins, the RNA helicase, CGH-1, and the Y-box domain–containing protein, CEY-2, which also is predicted to bind RNA (Fig. 4, A and B).
Figure 4.
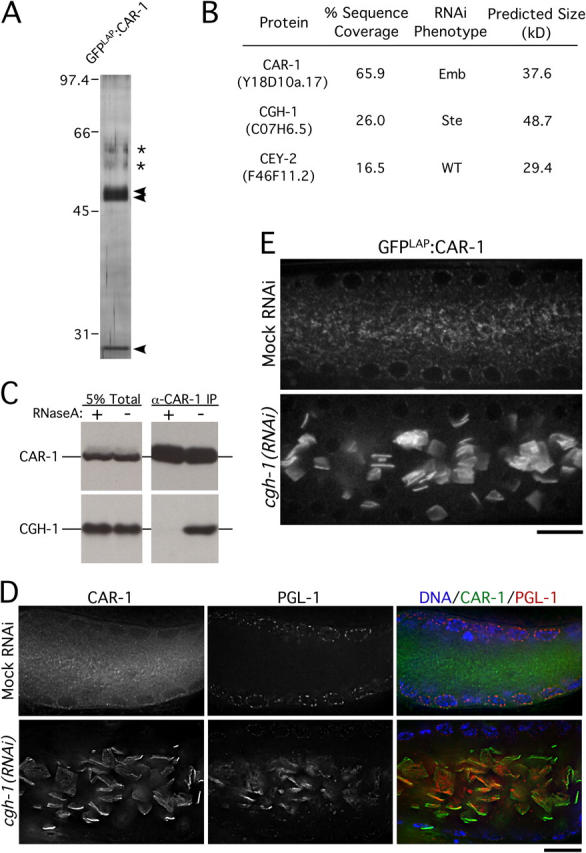
CAR-1 copurifies with two widely conserved RNA-binding proteins. (A) Silver stained gel of proteins eluted from S-protein agarose after tandem affinity purification of GFPLAP:CAR-1. Three distinct bands (arrowheads) are present. The two bands labeled with asterisks are contaminants (keratins). (B) Table showing the three proteins identified by solution mass spectrometry. The percent sequence coverage, RNAi phenotype, and molecular weight of each protein is shown. (C) Western blots of CAR-1 immunoprecipitates in the presence (+) or absence (−) of RNaseA, probed with the indicated antibodies. (D) Single sections from deconvolved three-dimensional widefield images of fixed gonads stained for CAR-1 (left column) and PGL-1 (middle column). Merged images with DNA staining (blue) are shown in the right column. Bar, 20 μm. (E) Single section spinning disc confocal imaging of gonads (middle section) from living wild-type or CGH-1–depleted hermaphrodites expressing GFPLAP:CAR-1. Bar, 10 μm.
Like CAR-1, CGH-1 and CEY-2 are conserved widely (Sommerville and Ladomery, 1996; Tanner and Linder, 2001). Consistent with their biochemical association, CGH-1 localizes to P-granules and smaller cytoplasmic particles in early embryos in a pattern similar to that of CAR-1 (Navarro et al., 2001) (Fig. S2). However, worms depleted of CGH-1 by RNAi or harboring a cgh-1 deletion (ok492) exhibit penetrant sterility (Navarro et al., 2001) (unpublished data). This phenotype is not observed in CAR-1–depleted worms or the car-1(tm1753) mutant, which suggests that CGH-1 performs multiple functions, of which only a subset requires CAR-1. In contrast to cgh-1 and car-1, a strain harboring a cey-2 deletion allele (ok902) did not exhibit any phenotype. Consequently, we focused our subsequent analysis on the relationship between CAR-1 and CGH-1.
To determine if the interaction between CAR-1 and CGH-1 requires RNA, we used antibodies to immunoprecipitate CAR-1 from extracts after incubation in the presence or absence of RNaseA. Whereas CGH-1 was detected in CAR-1 immunoprecipitates from control extracts, no CGH-1 was present after RNase treatment (Fig. 4 C). We conclude that CAR-1 is a component of an RNase-sensitive, multiprotein complex of conserved RNA-binding proteins.
CGH-1 controls the localization of CAR-1
To explore the functional relationship between CAR-1 and CGH-1, we determined the effect of depleting each protein on the localization of the other. Because depletion of CGH-1 results in sterility, we examined CAR-1 localization in the syncytial gonad of depleted worms. The gonad is composed of a cylindrical shell of meiotic nuclei that surrounds a common cytoplasmic core called the rachis. CAR-1 weakly colocalized with PGL-1 to P-granules, which surround the nuclei in the central portion of the gonad. However, most CAR-1 was present in smaller granules—distributed throughout the cytoplasm of the rachis—that did not contain PGL-1 (Fig. 4, D and E).
In CGH-1–depleted worms, the localization of CAR-1 in the gonad was perturbed dramatically. CAR-1 still weakly localized to P-granules around the nuclei, but the small CAR-1–containing particles were absent. Instead, CAR-1 accumulated in large, bar-shaped structures in the center of the rachis that appeared to form sheets (Fig. 4 E). This result was confirmed using a strain harboring a cgh-1 deletion allele (ok492; not depicted); an identical reorganization was observed in living CGH-1–depleted worms expressing GFP:CAR-1 (Fig. 4 F). The localization of GFP:PGL-1 also was altered after depletion of CGH-1. Like CAR-1, GFP:PGL-1 was still detected in P-granules, but it also was associated prominently with the aberrant CAR-1–containing structures in the rachis. In contrast to depletion of CGH-1, inhibition of CAR-1 did not affect the targeting of CGH-1 or PGL-1 to either type of granule. Similarly, depletion of CEY-2 or PGL-1 did not affect the targeting of CAR-1 or CGH-1. These results suggest that CGH-1 controls the formation of the small CAR-1–containing particles, whereas CAR-1 is not required for the formation of either particle type (Table I; see also Figs. 2 B and 3 C).
Table I. Localization dependencies of RNA-binding proteins.
| Mutant/protein | CAR-1 | CGH-1 | PGL-1 |
|---|---|---|---|
| car-1(tm1753) | - | Normal | Untested |
| car-1(RNAi) | - | Normal | Normal |
| cgh-1(ok494) | Bar-like structures | - | Untested |
| cgh-1(RNAi) | Bar-like structures | - | Bar-like structuresa |
| cey-2(ok902) | Normal | Normal | Untested |
| pgl-1(bn102) | Normal | Normal | - |
Gonad and embryo localizations of CAR-1, CGH-1, and PGL-1 in mutant worms. For cgh-1(RNAi) and cgh-1(ok492), localization was performed only in gonads due to sterility.
In addition to localizing to bar-like structures, a significant fraction of PGL-1 remained concentrated on P-granules in cgh-1 (RNAi) gonads. Localization of PGL-1 was performed using a GFP fusion to PGL-1.
Cleavage furrow dynamics are disrupted in CAR-1–depleted embryos
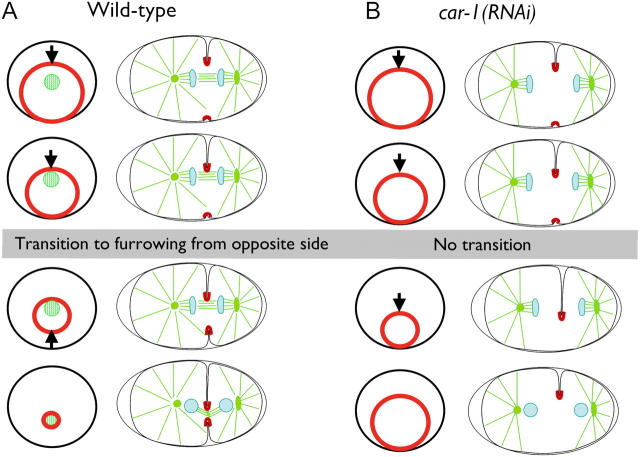
To analyze the cytokinesis defect in CAR-1–depleted embryos, we constructed a C. elegans strain expressing a fusion of GFP with a pleckstrin homology (PH) domain derived from mammalian PLC1δ1 to monitor cleavage furrow ingression (Fig. 5). The PLC1δ1 PH domain binds with high affinity to a phosphoinositide lipid (PI4,5P2) that is generated on the plasma membrane in most cell types (for review see Hurley and Meyer, 2001). Imaging of GFP:PHPLC1δ1 in wild-type embryos revealed a striking asymmetry in cleavage furrow ingression. A primary furrow was observed initially coming in from one side of the embryo (yellow line on kymograph in Fig. 5 C; Video 5). As the primary furrow ceased its ingression, a secondary furrow began to come in from the opposite side of the embryo (pink line on kymograph in Fig. 5 C). Simultaneous visualization of microtubules and the plasma membrane (Video 6) revealed that the secondary furrow began to ingress when the primary furrow came into contact with the interzonal microtubule bundles that form between the separating chromosomes (see Fig. 9 for a schematic of this transition). After ingression of the secondary furrow slows, a small gap remains that gradually closes (Fig. 5, B and C). This asymmetric ingression is not a consequence of embryo compression (unpublished data).
Figure 5.
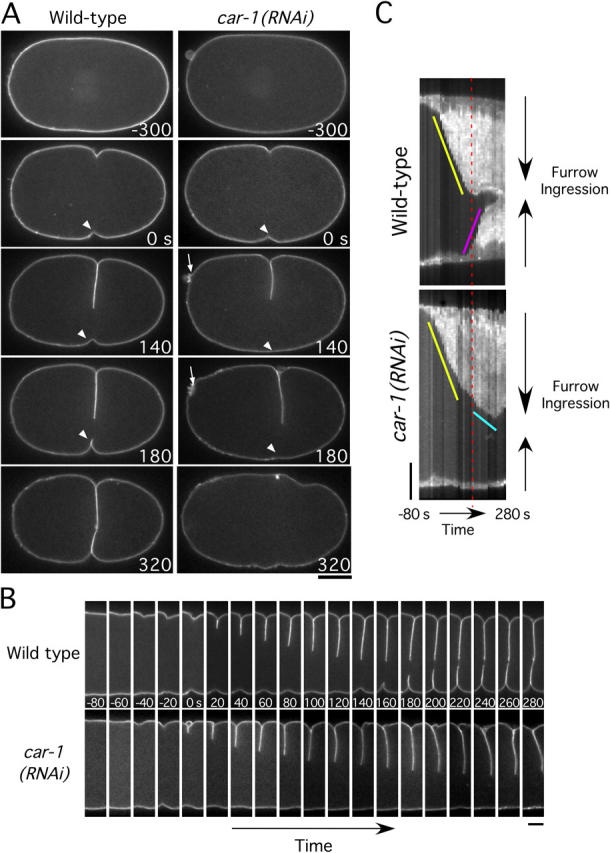
Disruption of cleavage furrow ingression in CAR-1–depleted embryos. (A) Selected panels from time-lapse sequences of wild-type (left column) and CAR-1–depleted (right column) embryos expressing GFP:PHPLC1δ1, which localizes to the plasma membrane. Time in seconds after initiation of furrowing is indicated in the lower right corner of each panel. (See also Video 5.) Bar, 10 μm. Arrows highlight the failure of polar body extrusion. Arrowheads point to secondary furrow, which fails to ingress in CAR-1–depleted embryos. (B) Same as in A, except only a 7.3-μm wide vertical section containing the furrow is shown for each time point. Bar, 5 μm. (C) Kymographs of the regions shown in B, comparing furrow ingression in wild-type and CAR-1–depleted embryos (see Methods and materials for details on kymograph construction). The slope of the yellow line indicates the initial rate of primary furrow ingression, which is identical in wild-type and CAR-1–depleted embryos. In wild-type embryos, the primary furrow encounters the midbody and ceases to ingress (time indicated by red dashed line). At a similar time in CAR-1–depleted embryos, the primary furrow slows (cyan line) but does not stop. In wild-type embryos, a secondary furrow begins to ingress from the opposite side of the embryo (pink line) as the primary furrow ceases its inward movement. In CAR-1–depleted embryos, no ingression of a secondary furrow is observed. Bar, 5 μm.
Figure 9.
Schematic illustrating the asymmetric furrow ingression in wild-type and car-1(RNA i) embryos. End-on and side views are shown. (A) The contractile ring is positioned asymmetrically within the circumference of the cell equator. In a side view, a primary furrow is observed coming in from one side of the embryo. Contact between the primary furrow and the interzonal microtubule bundles between the separated chromosomes triggers the other side of the contractile ring (the secondary furrow) to pull away from the edge of the embryo, and the contractile ring closes down around the compacted interzonal microtubule bundles (the midbody). (B) In car-1(RNAi) embryos, the primary furrow ingresses, but fails to come into contact with interzonal microtubule bundles. This prevents the structural transition that allows the contractile ring to pull away from the side of the embryo opposite the primary furrow. The furrow fails to close completely and cytokinesis fails.
In CAR-1–depleted embryos, the primary furrow initially ingressed at a rate similar to that in wild-type embryos, which indicated that there is no defect in its assembly or constriction. However, it failed to stop during the time interval in which it normally would contact the interzonal microtubules. Instead, the primary furrow continued to ingress at a slower rate (Fig. 5 C, cyan line) and began to move laterally within the embryo (Video 5). This lateral instability never is observed in wild-type embryos (Fig. 5 B, compare 100-s and 280-s panels). Strikingly, ingression of the secondary furrow from the opposite side of the embryo was blocked completely in CAR-1–depleted embryos. These observations suggest that a defect in the structure of the anaphase spindle, rather than in contractile ring assembly or constriction, underlies the cytokinesis failure in CAR-1–depleted embryos.
CAR-1–depleted embryos exhibit a pronounced defect in anaphase spindle structure
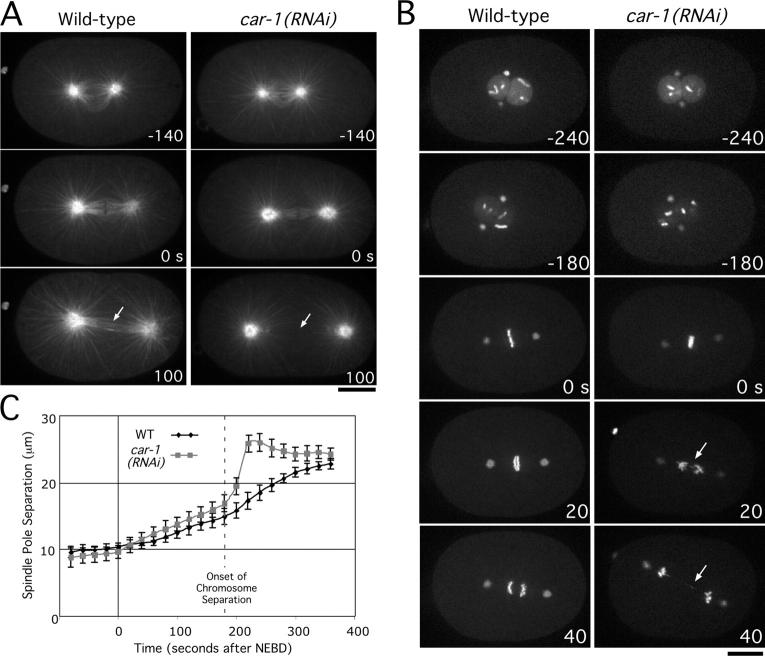
To analyze spindle structure directly, we examined embryos expressing GFP:α-tubulin or coexpressing GFP-histone H2B and GFP:γ-tubulin (Fig. 6 A; Video 7). Although spindle length before anaphase was not affected appreciably by CAR-1 depletion, subtle defects in spindle structure during mitosis and meiosis were evident before the onset of chromosome segregation (Fig. S3). Consistent with this, lagging chromosomes and chromosome bridges were observed frequently in CAR-1–depleted embryos (n = 45/50) (Fig. 6 B, 20/40-s panels; Video 8). At anaphase onset, a dramatic defect was apparent in CAR-1–depleted embryos; the spindle poles separated abruptly and prematurely, and the interzonal microtubule bundles that normally form between the separating chromosome masses were not detectable (Fig. 6 A, arrows in 100-s panel). Average plots of spindle pole separation versus time confirmed this reproducible anaphase “spindle snapping” defect (Fig. 6 C). These results indicate that CAR-1 is required for assembly of the anaphase spindle; this may explain the nature of the cytokinesis defect that is observed in CAR-1–depleted embryos.
Figure 6.
CAR-1 depletion results in a pronounced defect in anaphase spindle structure. (A) Selected panels from time-lapse sequences of wild-type (left column) and CAR-1–depleted (right column) embryos expressing GFP:α-tubulin. Time in seconds after chromosome alignment is indicated in the lower right corner of each panel. Arrows indicate the region between the segregated chromosomes where interzonal microtubule bundles normally form (see also Video 7). Bar, 10 μm. (B) Selected panels from time-lapse sequences of wild-type (left column) and CAR-1–depleted (right column) embryos expressing GFP:histone H2B and GFP:γ-tubulin. Time in seconds after chromosome alignment is indicated in the lower right corner of each panel. Arrows point to chromosome bridges that become evident after anaphase onset in CAR-1–depleted embryos. (See also Video 8.) Bar, 10 μm. (C) The distance between spindle poles was tracked for 15 wild-type (WT) and 19 CAR-1–depleted embryos imaged as in A. Average pole-to-pole distance is plotted versus time after NEBD (nuclear envelope breakdown). Error bars represent the SEM with a confidence interval of 0.95. For reference, the time of onset of chromosome segregation is indicated. Embryos in which extrusion of the second polar body failed and extra chromatin was present in the mitotic spindle (n = 6/50) were not used for the analysis of spindle length.
Targeting of the chromosomal passenger Aurora B and the centralspindlin kinesin, ZEN-4, is disrupted in CAR-1–depleted embryos
In CAR-1–depleted embryos, interzonal microtubule bundles are absent or reduced significantly. This defect is specific, because it is not seen after a variety of perturbations that destabilize the microtubule cytoskeleton, such as nocodazole treatment or depletion of the microtubule-stabilizing protein, ZYG-9 (Albertson, 1984; Matthews et al., 1998). The formation of interzonal microtubule bundles requires two protein complexes: the chromosomal passenger complex, which includes the Aurora B kinase, AIR-2 (reviewed in Vagnarelli and Earnshaw, 2004), and centralspindlin, a complex that contains the plus-end–directed kinesin, ZEN-4 (for review see Glotzer, 2003). Both complexes are recruited to interzonal microtubule bundles as they form, and therefore, are excellent markers for these structures. To examine the dynamics of both complexes, we used strains expressing GFP fusions with AIR-2 and ZEN-4. In wild-type embryos, GFP:AIR-2 localizes to chromosomes at metaphase (Fig. 7 A, 0-s panel). Following chromosome separation, GFP:AIR-2 is found on chromosomes and interzonal microtubule bundles. In CAR-1–depleted embryos, GFP:AIR-2 was present on mitotic chromosomes as in the wild type. However, after chromosome separation, GFP:AIR-2 remained exclusively localized to chromosomes (Fig. 7A). Similarly, GFP:ZEN-4, which normally accumulates on interzonal microtubule bundles, was essentially absent in CAR-1–depleted embryos (Fig. S4). Identical results were obtained from fixed samples using antibodies to the endogenous proteins (Fig. 7, B and C). The presence of normal levels of Aurora B on chromosomes, and of ZEN-4 on meiotic midbody microtubules in CAR-1–depleted embryos (Fig. 7 C), suggests that that the failure of interzonal microtubule bundle formation in CAR-1–depleted embryos is not due to destabilization of Aurora B or ZEN-4.
Figure 7.
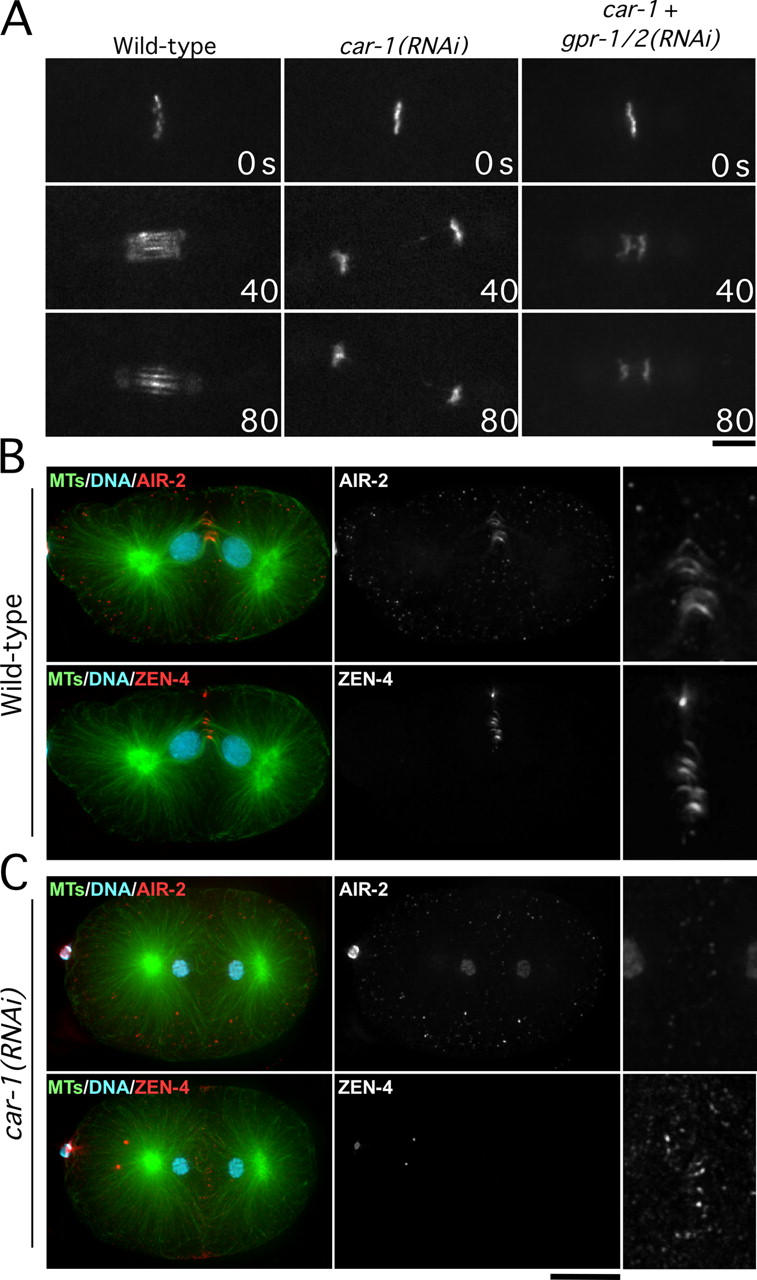
AIR-2 and ZEN-4 fail to target to interzonal microtubule bundles in CAR-1–depleted embryos. Simultaneous depletion of GPR-1/2, which inhibits pulling forces on the spindle poles and prevents spindle snapping, does not rescue this defect. (A) Selected images from time-lapse sequences of wild-type, car-1(RNAi), and car-1/gpr-1,2 double RNAi embryos expressing GFP:AIR-2. Only the region of the spindle is shown. Time after chromosome alignment (in seconds) is indicated in the lower right corner of each panel. Bar, 5 μm. Projected three-dimensional datasets of fixed wild-type (B) and CAR-1–depleted (C) embryos stained for DNA, microtubules, AIR-2, and ZEN-4. High magnification panels in the right column are magnified 2.5× relative to the adjacent images. The ZEN-4 high magnification image is 10× overexposed. Bar, 10 μm.
The failure of cytokinesis and the loss of AIR-2 and ZEN-4 targeting to interzonal microtubules might be secondary consequences that result from premature elongation of the anaphase spindle. To explore this possibility, we prevented premature spindle elongation by inactivating the cortical forces that normally pull on the asters to position the spindle within the embryo. We used a dsRNA that depletes two highly homologous G-protein regulators, GPR-1 and GPR-2, which are required for the production of astral pulling forces (Willard et al., 2004). Codepletion of CAR-1 and GPR-1/2 prevented the premature spindle elongation that is observed in embryos that are depleted of CAR-1 alone, but recruitment of AIR-2 and ZEN-4 to microtubule bundles between the segregating chromosomes remained dramatically reduced and cytokinesis still failed (Fig. 7 A and Fig. S4). Based on these results, we conclude that the failure to form interzonal microtubule bundles that can recruit AIR-2 and ZEN-4 is not a secondary consequence of premature spindle elongation, but is a primary defect in CAR-1–depleted embryos.
Partial depletion of CGH-1 phenocopies CAR-1 inhibition
CAR-1 associates with the helicase, CGH-1, whose inhibition results in penetrant sterility. To address whether CGH-1 function is required for embryonic cytokinesis, we analyzed partial CGH-1 depletions. 24 h after injection of a dsRNA targeting cgh-1, all worms ceased embryo production. However, 22–24 h after injection, several worms were able to fertilize up to two oocytes. The resulting embryos were osmotically sensitive, but could be imaged in utero or by using specialized media to provide osmotic support (see Materials and methods). Strikingly, the defects observed in partial CGH-1–depleted embryos were almost identical to those in CAR-1–depleted embryos (Fig. 8). Cytokinesis failed in most partial CGH-1–depleted embryos (n = 11/19), and imaging of the microtubule cytoskeleton confirmed the absence of interzonal microtubule bundles (Fig. 8 B). In addition, AIR-2 accumulated on mitotic chromosomes but failed to target to interzonal microtubules (Fig. 8 C). Consistent with the analysis in gonads (Fig. 4, D and E), localization of CAR-1 was perturbed severely in partial cgh-1(RNAi) embryos. Most of the CAR-1 accumulated in small, bar-like structures that did not migrate to the posterior during the first mitotic division (Video 9).
Figure 8.
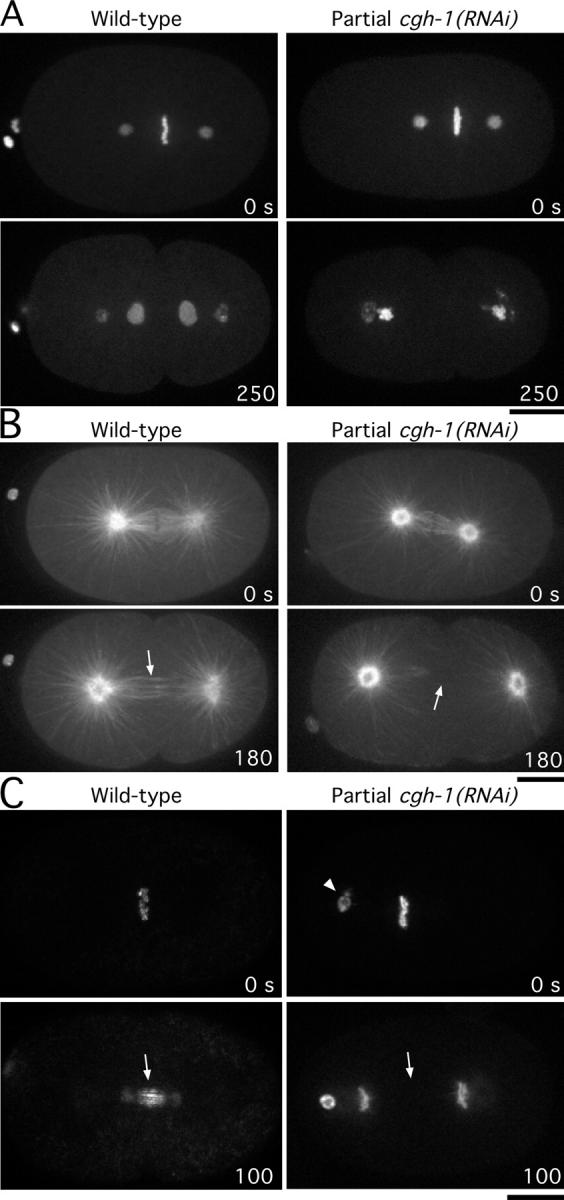
Partial depletion of CGH-1 phenocopies depletion of CAR-1. Selected panels from time-lapse sequence of wild-type (left column) and CGH-1 partially depleted (right column) embryos expressing GFP:histone H2B and GFP:γ-tubulin (A), GFP:α-tubulin (B), and GFP:AIR-2 (C). Arrows in B highlight the presence or absence of interzonal microtubules. Arrows in C highlight the presence or absence of AIR-2 on interzonal microtubules; the arrowhead points to polar body chromatin that failed to be extruded properly. Time in seconds after chromosome alignment is indicated in the lower right corner of each panel. Bar, 10 μm.
These results suggest that CGH-1 and CAR-1 coordinately regulate anaphase spindle structure during embryonic cytokinesis, and support the idea that CAR-1 is required for only a subset of CGH-1 functions.
General inhibition of translation does not phenocopy CAR-1 and partial CGH-1 depletions
CGH-1 has been proposed to function in the regulation of mRNA translation (Navarro et al., 2001). To determine whether the observed cytokinesis defect in CAR-1 and partial CGH-1 depletions could arise from a general defect in the translation of mRNA in the gonad and early embryo, we examined whether loss of interzonal microtubules was a consequence of inhibiting translation. Depletions of six different ribosomal subunits (RPS-1, -3, -5, -11, -18, and -23) each resulted in penetrant sterility (unpublished data). Consequently, we performed partial depletions of each ribosomal subunit, and examined the localization of GFP:AIR-2 in the final few embryos produced before onset of sterility. In every case, GFP:AIR-2 accumulated on interzonal microtubule bundles (Fig. S4 and not depicted). Defects in cytokinesis were not observed after partial depletion of ribosomal subunits, although embryos were osmotically sensitive and exhibited other pleiotropic phenotypes, consistent with those previously reported for general inhibition of translation (Gönczy et al., 2000). These results suggest that the cytokinesis defect after CAR-1 or partial CGH-1 depletion is not a consequence of generally inhibiting translation in the gonad and during early embryogenesis.
Discussion
Maternally supplied CAR-1 is required for cytokinesis in the early C. elegans embryo
The sequence features that define the Scd6 family of proteins suggest a role in RNA regulation (Anantharaman and Aravind, 2004). However, the function of members of this protein family has been unclear. Here we show that inhibition of the C. elegans Scd6 family member, CAR-1, results in a dramatic defect in anaphase spindle structure and failure to complete cytokinesis during early embryogenesis. Embryos, either depleted of >95% CAR-1 protein by RNAi or laid by homozygous car-1(tm1753) mutant mothers, exhibit an identical defect. This suggests that car-1(tm1753)—which deletes two thirds of the coding region—is null for car-1 function. car-1(tm1753) behaves like a maternal effect mutation; homozygous mutants develop to adulthood but lay inviable embryos. The maternal effect nature of the car-1 deletion, together with the functionally important RNA-binding motifs in the CAR-1 sequence, its association with conserved RNA-binding proteins, and its localization to P-granules and smaller cytoplasmic particles in the gonad and early embryo, is consistent with a model in which CAR-1 functions to regulate a specific set of maternally supplied RNAs.
CAR-1 forms a conserved complex with a DEAD box RNA helicase and a Y-box protein
CAR-1 copurifies with the essential DEAD box RNA helicase, CGH-1, and the Y-box protein, CEY-2. In Drosophila, a maternally expressed protein complex containing the CGH-1 homologue, Me31B, and a Y-box protein was described (Wilhelm et al., 2000; Nakamura et al., 2001). The CAR-1 homologue in Drosophila, trailerhitch, is a component of this complex (Wilhelm, J., personal communication). The amphibian homologues of CGH-1 and CAR-1, Xp54 and RAP55, also were enriched in mRNP particles that were isolated from oocytes (Ladomery et al., 1997; Lieb et al., 1998). This suggests that the regulation of maternally supplied RNA, by a complex containing a Scd6 family protein and a DEAD box helicase, during oogenesis and early embryogenesis is conserved among metazoans.
Partial depletion of CGH-1 phenocopies depletion of CAR-1, which supports the idea that these two proteins function together. However, the CGH-1 depletion and deletion phenotypes are more severe than that of CAR-1. Adults that are homozygous for a deletion in car-1 lay embryos that fail to complete cytokinesis, whereas adults that are homozygous for a deletion in cgh-1 are sterile. Consistent with the difference in phenotypic severity, depletion of CAR-1 does not disrupt the localization of CGH-1, PGL-1, or a nonfunctional CAR-1 truncation lacking the Sm domain. These data indicate that although CAR-1 may function in the context of P-granules or smaller cytoplasmic particles, it is not required for their formation. In contrast, depletion of CGH-1 results in the loss of the small CAR-1–containing particles, and causes CAR-1 and PGL-1 to accumulate in aberrant sheet-like structures in gonad. The discrepancy between our data, and a previous study that reported that PGL-1 targeting was normal after RNAi of cgh-1 (Navarro et al., 2001), probably is due to the more severe nature of our perturbation. The cgh-1(RNAi) worms that were examined previously were the final surviving F1 progeny of hermaphrodites that were injected with dsRNA against cgh-1. In contrast, we analyzed the gonads of hermaphrodites that were injected as L4 larvae.
CAR-1/CGH-1: a link between maternal RNA regulation and anaphase spindle structure
In Xenopus extracts, RNPs containing the mRNA export factor, Rae1, concentrate around chromatin and in the centers of the centrosomal microtubule asters, where they have been proposed to have a direct, translation-independent role in spindle assembly (Blower et al., 2005). Although, we cannot rule out a direct role for RNAs regulated by CAR-1/CGH-1 in spindle structure, the fact that CAR-1/CGH-1–containing particles do not concentrate in the vicinity of the spindle makes this unlikely. Instead, we favor the idea that CAR-1 and CGH-1 modulate spindle dynamics indirectly by regulating the translation of a specific set of maternally supplied mRNAs in the gonad and early embryo.
Why is the function of CAR-1/CGH-1 restricted to the gonad and early embryo? The C. elegans gonad is a syncytium, lined by ∼800 nuclei, that progresses in an orderly fashion from a distal mitotic zone, through the various stages of meiotic prophase, and ultimately become packaged in oocytes. During this process, the meiotic nuclei also serve a nurse function, and supply the common cytoplasm with mRNA (Gumienny et al., 1999). Oocytes containing maternally supplied protein and mRNA bud off the tip of the gonad and are fertilized to form embryos. We speculate that spatial and temporal control of translation is important to maintain the local environments in the different regions of the gonad and early embryo. Regulation of maternally supplied RNA would be critical until the embryonic cells begin to transcribe their own genome—approximately the time that CAR-1/CGH-1–containing particles can no longer be detected in embryos. Because the dynamics of cytoskeletal polymers are very sensitive to the concentration of their regulators, it seems possible that the anaphase spindle defect in CAR-1–depleted embryos results from the translational misregulation of a component of the microtubule cytoskeleton.
A role in translational competence would be similar to that proposed previously for the RNA binding protein, CPEB, the inhibition of which results in defects in spindle structure in Xenopus oocytes (Groisman et al., 2000). Homologues of CPEB regulate the translational competence of mRNAs in oocytes and neurons by promoting the polyadenylation and activation of mRNAs that are stored in a deadenylated, translationally silenced state (de Moor et al., 2005). A possible functional connection with CAR-1 is suggested by the fact that clam CPEB/p82 has been copurified with the CGH-1 homologue, p47 (Minshall et al., 2001). However, because we did not isolate any of the C. elegans homologues of CPEB in our CAR-1 purification, and inhibition of cpb-3, the closest C. elegans CPEB homologue, does not result in embryonic lethality (Luitjens et al., 2000), additional experiments are needed to test this hypothesis.
A role for CAR-1 and its associated helicase, CGH-1, in regulating the translational competence of maternal RNAs in the gonad and early embryo is consistent with other studies. The Xenopus and Drosophila homologues of CGH-1 were proposed to function in maintaining maternally loaded RNAs in a translationally repressed state until they are needed during the early rapid embryonic divisions (Ladomery et al., 1997; Minshall et al., 2001; Nakamura et al., 2001). If CAR-1 represses translation, the cytokinesis defect in car-1–inhibited embryos could result from the inappropriate translation of a specific set of maternally contributed RNAs. Alternatively, CAR-1 could play a role in activating the translation of a subset of maternally loaded RNAs. In this scenario, depletion of critical target RNAs might recapitulate the CAR-1 depletion phenotype. Identification of the relevant targets is necessary to understand further the connection between this conserved RNP complex and embryonic cytokinesis.
A novel role for interzonal microtubule bundles during asymmetric cytokinesis
Our analysis of furrow dynamics using a GFP-fusion that specifically localizes to the plasma membrane highlights the asymmetric nature of cytokinesis in wild-type C. elegans embryos (see Fig. 9 for schematic). When viewed in cross section, a primary furrow first comes in from one side of the embryo (Fig. 9 A; top panels). When the primary furrow encounters the interzonal microtubule bundles between the separating chromosomes, a transition occurs in which ingression of the primary furrow slows, and a secondary furrow begins to come in from the opposite side of the embryo (Fig. 9 A; bottom panels). In CAR-1–depleted embryos, which lack interzonal microtubule bundles, a primary furrow initiates ingression from one side of the embryo with a rate similar to the wild type. However, the transition to ingression from the opposite side of the embryo never occurs (Fig. 9 B). Consequently, the contractile ring frequently fails to close and the furrow regresses.
Asymmetric furrowing could be due to two sequential signaling events. Recent work suggests that in C. elegans, the microtubule asters and the spindle midzone send sequential signals to position the cleavage furrow (Bringmann and Hyman, 2005). It is possible that an early signal from the asters initiates furrowing from one side, and a late signal from the spindle midzone causes a furrow to come in from the opposite side. Alternatively, our data support the idea that interzonal microtubule bundles promote an important structural transition. Perhaps resistance from physical contact with interzonal microtubule bundles prevents further ingression from one side. Continued constriction of the contractile ring would cause a furrow to come in from the opposite side (Fig. 9). In this way, the interzonal microtubule bundles, anchored between the two centrosomal microtubule asters, would position the contractile ring within the division plane. We speculate that the asymmetry of this process may help to constrain furrowing to a single plane at the spindle equator. In large embryonic cells with rapid cell cycles, an asymmetric, step-wise mechanism would ensure the timely completion of cell division.
Materials and methods
Worm strains and DNA manipulations
GFP and GFPLAP fusion constructs were generated in pAZ132, a plasmid expressing GFP under the control of the pie-1 promoter, or pIC26, a plasmid expressing GFP followed by a cleavage site for the tobacco etch virus protease and S-peptide, also under the pie-1 promoter. The unspliced genomic locus for Y18D10a.17 (CAR-1) and B0207.4 (AIR-2) and the spliced coding region of M03D4.1 (ZEN-4) were cloned into pIC26 after SpeI digestion. To generate a construct to express GFP:PHPLC1δ1, the PH domain (aa 1–175) from PLC1δ1 (accession no. P10688) was cloned into pAZ132 after SpeI digestion. To generate the truncated form of Y18D10a.17 (CAR-1ΔN), the unspliced genomic locus of Y18D10a.17 lacking exon 1, intron 1, and the first 51 nucleotides of exon 2 was cloned into pIC26 after SpeI digestion. Constructs were integrated into DP38 (unc-119 (ed3)) using a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad Laboratories) (Praitis et al., 2001). Worm strains expressing GFP fusion of α-tubulin, γ-tubulin, histone H2B, and PGL-1 were described previously (Oegema et al., 2001; Cheeks et al., 2004). All strains used are listed in Table SI B.
dsRNA and antibody production
Oligonucleotides that were used for double-stranded (dsRNA) production are listed in Table SI A. DNA templates for dsRNA synthesis were amplified from N2 genomic DNA. To generate antibodies directed against CAR-1, the final 300 nucleotides of Y18D10a.17 were amplified from cDNA yk729c6 (obtained from Y. Kohara, National Institute of Genetics, Mishima, Japan) and inserted into pGEX6P-1 (GE Healthcare). The purified GST fusion protein was outsourced for injection into rabbits (Covance), and affinity purification of α–CAR-1 antibodies was performed as described previously, after removal of the GST by cleavage of the antigen with Prescission protease (Desai et al., 2003). Purified antibodies were labeled directly with a Cy3 fluorescent dye as described (Francis-Lang et al., 1999).
RNAi, blotting of dsRNA-injected worms, and brood size determination
L4 hermaphrodites were injected with dsRNA and were incubated at 20°C for 45 h before analysis. For partial depletions, L4 hermaphrodites were incubated for 22–24 h at 20°C after injection of dsRNA. Immunoblotting after RNAi was performed as described (Desai et al., 2003). To determine brood size, injected or control worms were moved every 12 h to individual plates, and the total number of embryos laid on the plate was counted for each period. For each time interval, percent hatching was determined by calculating the number of viable L1 progeny and dividing by the total number of embryos on that plate.
Microscopy and kymograph construction
For analysis of fixed samples mounted in PPDM (90% glycerol, 0.5% p-phenylenediamine, 20 mM Tris-HCl pH 8.8), images were acquired on a DeltaVision deconvolution Olympus IX70 microscope (Applied Precision) equipped with a CoolSnap CCD camera (Roper Scientific) at 20°C using a 100×, 1.35 NA Olympus U-Planapo oil objective lens. Immunofluorescence of fixed embryos was performed as described (Desai et al., 2003), using the following rabbit antibodies at a concentration of 1 μg/ml: α–CAR-1 (Cy3-labeled; described above); α–AIR-2 (Cy-5 labeled; generated against a GST fusion to the full-length protein); α–ZEN-4 (Cy-5 labeled; generated against a GST fusion to the COOH-terminal 108 aa); the mouse monoclonal antibody DM1α (Oregon green 488–labeled; Sigma-Aldrich); the goat polyclonal GFP antibody (Oregon green 488–labeled; generated against a 6x-histidine fusion to the full- length protein); and the unlabeled rat CGH-1 antibody (JDCR5; a gift of K. Blackwell, Joslin Diabetes Center, Boston, MA). For analysis of gonads, the tails of adult hermaphrodites were amputated in 5% sucrose and 100 mM NaCl to extrude the gonads. Fixation and immunofluorescence on gonads was performed as described for embryos. For live analysis, embryos were mounted on agarose pads as described previously (Oegema et al., 2001), and imaged on a spinning disc confocal microscope (Nikon Eclipse TE2000-E) equipped with a Hamamatsu Orca-ER CCD camera at 20°C using a Nikon 60×, 1.4 NA Planapo oil objective lens. For osmotically sensitive embryos and embryos imaged in the absence of compression, filming was performed in a depression slide containing meiosis media (25 mM Hepes at pH 7.4, 60% Leibowitz L-15 Media, 20% FBS, 500 μg/ml inulin) and sealed with petroleum jelly. Analysis of spindle pole separation, spindle microtubule density, and furrow movement was performed using Metamorph software.
Kymographs were constructed by compressing the image of the furrow region from each time point (same region as in Fig. 5 B) to a single vertical line, in which the maximum intensity along the x-axis of each original image is displayed for each point along the y-axis. The vertical strips for sequential time points are laid adjacent to each other so that time increases from left to right along the x-axis.
GFPLAP purification, mass spectrometry, and immunoprecipitation
Adult hermaphrodites expressing GFPLAP:CAR-1 were grown synchronously in liquid culture (Cheeseman et al., 2004), washed in lysis buffer (50 mM Hepes at pH 7.4, 1 mM EDTA, 1 mM MgCl2, 100 mM KCl, and 10% glycerol), and drop frozen in liquid N2. Extracts were generated, and CAR-1 interacting proteins were isolated as described (Cheeseman et al., 2004). Mass spectrometry was performed as described (Cheeseman et al., 2004), using the most recent version of the predicted C. elegans proteins (Wormpep111).
Immunoprecipitations were performed as described previously (Desai et al., 2003), except that extracts were incubated for 20 min in the presence or absence of 5 μg/ml RNaseA at room temperature before immunoprecipitation.
Online supplemental material
Fig. S1 shows that both the CAR-1 Sm domain and RGG box bind to RNA-coated beads. Fig. S2 shows that CAR-1 and CGH-1 both localize to P-granules and smaller cytoplasmic particles. Fig. S3 depicts how microtubule density is reduced in the region between the centrosome and the chromosomes in metaphase spindles from CAR-1–depleted embryos. Fig. S4 shows the absence of inter-zonal microtubules following car-1(RNAi) results in a defect in ZEN-4 localization. Video 1: the first mitotic division of a wild-type C. elegans embryo filmed using DIC microscopy. Video 2: the first mitotic division of an embryo laid by a car-1(RNAi) hermaphrodite (RNA #194) filmed by DIC microscopy. Video 3: the first mitotic division of an embryo laid by a homozygous car-1(tm1753) hermaphrodite filmed by DIC microscopy. Video 4: the first seven mitotic divisions of an embryo expressing GFPLAP:CAR-1 filmed by spinning disk confocal microscopy. Video 5: the first mitotic division of a wild-type and a car-1(RNAi) (dsRNA #194) embryo expressing GFP:PHPLC1δ1 filmed by spinning disk confocal microscopy. Video 6: the first mitotic division of a wild-type embryo expressing both GFP:α-tubulin and GFP:PHPLC1δ1 filmed by spinning disk confocal microscopy. Video 7: the first mitotic division of a wild-type and a car-1(RNAi) embryo (RNA #194) expressing GFP:α-tubulin filmed by spinning disk confocal microscopy. Video 8: the first mitotic division of a wild-type and a car-1(RNAi) embryo (RNA #194) expressing GFP:γ-tubulin and GFP:histone H2B filmed by spinning disk confocal microscopy. Video 9: the first mitotic division of a partial cgh-1(RNAi) embryo (RNA #273) expressing GFPLAP:CAR-1.Table S1: dsRNAs and worm strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200506124/DC1.
Acknowledgments
We are grateful to P. Maddox for assistance with microscopy, I. Cheeseman for helpful advice on isolating protein complexes, Y. Kohara for gene-specific cDNAs, the Caenorhabditis Genetics Center (University of Minnesota) and the National Bioresource Project for C. elegans (Tokyo, Japan) for mutant strains, K. Blackwell for CGH-1 antibodies, L. Lewellyn for technical assistance, and B. Weaver for useful discussions. We also thank J. Wilhelm, K. Blackwell, P. Boag, J. White, and J. Squirrel for useful discussions and communicating results before publication.
A. Audhya is a Helen Hay Whitney Postdoctoral Fellow. A.S. Maddox is a fellow of the Giannini Family Foundation. K. Oegema is a Pew Scholar in the Biomedical Sciences. A. Desai is a Damon-Runyon Scholar. This work was supported by funding from the Ludwig Institute for Cancer Research to K. Oegema and A. Desai, and National Institutes of Health grant RR11823 that funds the Yeast Resource Center.
Abbreviations used in this paper: dsRNA, double-stranded RNA; DIC, differential interference contrast; RNAi, RNA-mediated interference.
References
- Albertson, D. 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101:61–72. [DOI] [PubMed] [Google Scholar]
- Anantharaman, V., and L. Aravind. 2004. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 16:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., M. Nachury, R. Heald, and K. Weis. 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 121:223–234. [DOI] [PubMed] [Google Scholar]
- Bringmann, H., and A.A. Hyman. 2005. A cytokinesis furrow is positioned by two consecutive signals. Nature. 436:731–734. [DOI] [PubMed] [Google Scholar]
- Cheeks, R.J., J.C. Canman, W.N. Gabriel, N. Meyer, S. Strome, and B. Goldstein. 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr. Biol. 14:851–862. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Niessen, S. Anderson, F. Hyndman, J.R. Yates III, K. Oegema, and A. Desai. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861–890. [DOI] [PubMed] [Google Scholar]
- de Moor, C.H., H. Meijer, and S. Lissenden. 2005. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 16:49–58. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Rybina, T. Muller-Reichert, A. Shevchenko, A. Shevchenko, A.A. Hyman, and K. Oegema. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., M.J. Matunis, S. Pinol-Roma, and C.G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289–321. [DOI] [PubMed] [Google Scholar]
- Finger, F.P., and J.G. White. 2002. Fusion and fission: membrane trafficking in animal cytokinesis. Cell. 108:727–730. [DOI] [PubMed] [Google Scholar]
- Francis-Lang, H., J. Minden, W. Sullivan, and K. Oegema. 1999. Live confocal analysis with fluorescently labeled proteins. Methods Mol. Biol. 122:223–239. [DOI] [PubMed] [Google Scholar]
- Glotzer, M. 2003. Cytokinesis: progress on all fronts. Curr. Opin. Cell Biol. 15:684–690. [DOI] [PubMed] [Google Scholar]
- Gönczy, P., C. Echeverri, K. Oegema, A. Coulson, S.J. Jones, R.R. Copley, J. Duperon, J. Oegema, M. Brehm, E. Cassin, et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 408:331–336. [DOI] [PubMed] [Google Scholar]
- Groisman, I., Y. Huang, R. Mendez, Q. Cao, W. Theurkauf, and J.D. Richter. 2000. CPEB, Maskin, and Cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 103:435–447. [DOI] [PubMed] [Google Scholar]
- Gumienny, T.L., E. Lambie, E. Hartwieg, H.R. Horvitz, and M.O. Hengartner. 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 126:1011–1022. [DOI] [PubMed] [Google Scholar]
- Gunsalus, K.C., and F. Piano. 2005. RNAi as a tool to study cell biology: building the genome-phenome bridge. Curr. Opin. Cell Biol. 17:3–8. [DOI] [PubMed] [Google Scholar]
- Hurley, J.H., and T. Meyer. 2001. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 13:146–152. [DOI] [PubMed] [Google Scholar]
- Kambach, C., S. Walke, and K. Nagai. 1999. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9:222–230. [DOI] [PubMed] [Google Scholar]
- Ladomery, M., E. Wade, and J. Sommerville. 1997. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 25:965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb, B., M. Carl, R. Hock, D. Gebauer, and U. Scheer. 1998. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp. Cell Res. 245:272–281. [DOI] [PubMed] [Google Scholar]
- Luitjens, C., M. Gallegos, B. Kraemer, J. Kimble, and M. Wickens. 2000. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14:2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, L.R., P. Carter, D. Thierry-Mieg, and K. Kemphues. 1998. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141:1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall, N., G. Thom, and N. Standart. 2001. A conserved role of a DEAD box helicase in mRNA masking. RNA. 7:1728–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., R. Amikura, K. Hanyu, and S. Kobayashi. 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 128:3233–3242. [DOI] [PubMed] [Google Scholar]
- Navarro, R.E., E.Y. Shim, Y. Kohara, A. Singson, and T.K. Blackwell. 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 128:3221–3232. [DOI] [PubMed] [Google Scholar]
- Oegema, K., and A.A. Hyman. 2005. Cell division. In Wormbook, ed. The C. elegans Research Community. Available at: http://www.wormbook.org (accessed June 15, 2005).
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano, F., A.J. Schetter, M. Mangone, L. Stein, and K.J. Kemphues. 2000. RNAi analysis of genes expressed in the ovary of Caenorhabditis elegans. Curr. Biol. 10:1619–1622. [DOI] [PubMed] [Google Scholar]
- Praitis, V., E. Casey, D. Collar, and J. Austin. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 157:1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.N., and J.A. Spudich. 2004. Mechanics and regulation of cytokinesis. Curr. Opin. Cell Biol. 16:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, J.K., and C. D'Souza-Schorey. 2004. Finishing the job: cytoskeletal and membrane events bring cytokinesis to an end. Exp. Cell Res. 295:1–8. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and A. Fire. 1994. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development. 120:2823–2834. [DOI] [PubMed] [Google Scholar]
- Sommerville, J., and M. Ladomery. 1996. Transcription and masking of mRNA in germ cells: involvement of Y-box proteins. Chromosoma. 104:469–478. [DOI] [PubMed] [Google Scholar]
- Sönnichsen, B., L.B. Koski, A. Walsh, P. Marschall, B. Neumann, M. Brehm, A.M. Alleaume, J. Artelt, P. Bettencourt, E. Cassin, et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 434:462–469. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., and C.M. Field. 2000. Microtubules, membranes and cytokinesis. Curr. Biol. 10:R760–R770. [DOI] [PubMed] [Google Scholar]
- Strome, S., and W.B. Wood. 1982. Immunofluorescence visualization of germ-line specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 79:1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, N.K., and P. Linder. 2001. DexD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 8:251–262. [DOI] [PubMed] [Google Scholar]
- Thore, S., C. Mayer, C. Sauter, S. Weeks, and D. Suck. 2003. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 278:1239–1247. [DOI] [PubMed] [Google Scholar]
- Toro, I., S. Thore, C. Mayer, J. Basquin, B. Seraphin, and D. Suck. 2001. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 20:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli, P., and W.C. Earnshaw. 2004. Chromosomal passengers: the four dimensional regulation of mitotic events. Chromosoma. 113:211–222. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., J. Mansfield, N. Hom-Booher, S. Wang, C.W. Turck, T. Hazelrigg, and R.D. Vale. 2000. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J. Cell Biol. 148:427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard, F.S., R.J. Kimple, and D.P. Siderovski. 2004. Return of the GDI: the GoLoco motif in cell division. Annu. Rev. Biochem. 73:925–951. [DOI] [PubMed] [Google Scholar]
- Zipperlen, P., A.G. Fraser, R.S. Kamath, M. Martinez-Campos, and J. Ahringer. 2001. Roles for 147 embryonic lethal genes on C. elegans chromosome I identified by RNA interference and video microscopy. EMBO J. 20:3984–3992. [DOI] [PMC free article] [PubMed] [Google Scholar]