Abstract
Hybrid or “recombinational” speciation refers to the origin of a new homoploid species via hybridization between chromosomally or genetically divergent parental species. Theory predicts that this mode of speciation is punctuated, but there has been little empirical evidence to support this claim. Here, we test the hypothesis of rapid hybrid speciation by estimating the sizes of parental species chromosomal blocks in Helianthus anomalus, a wild sunflower species derived via hybridization between H. annuus and H. petiolaris. Analysis of the frequency spectrum of parental species chromosomal blocks with respect to predictions based on R. A. Fisher’s [Fisher, R. A. (1953) Heredity 8, 187–197] junctions approach, suggests that H. anomalus arose rapidly, probably in fewer than 60 generations. This result is corroborated by independent lines of evidence demonstrating (i) a significant concordance between the genomes of H. anomalus and early generation H. annuus × H. petiolaris synthetic hybrids, and (ii) a rapid recovery of pollen fertility in these synthetic hybrid lineages. These results are not only consistent with theory but also provide a new and general method for estimating the tempo of hybrid speciation and dating the origin of hybrid zones.
A fundamental question in evolutionary biology is whether speciation is gradual or punctuated (1–5). This question has been difficult to evaluate critically because the evolutionary history of most plant and animal species is poorly known. However, it may be feasible to determine which modes of speciation are likely to be rapid and which ones are likely to occur gradually. For example, several recent theories of speciation suggest pathways by which new species might arise rapidly (6–9). One such pathway is hybrid or “recombinational” speciation, in which a new homoploid species arises via hybridization between chromosomally or genetically divergent parental species (10, 11). Simulation studies of recombinational speciation suggest that this mode is punctuated—long periods of hybrid zone stasis are followed by abrupt transitions in which parental species individuals are displaced rapidly by the hybrid neospecies (12). In this report, we test the hypothesis of rapid hybrid speciation by analyzing the sizes of parental species chromosomal blocks in Helianthus anomalus, a wild sunflower species thought to be derived via hybridization between H. annuus and H. petiolaris (13).
It is important to differentiate the process of hybrid speciation from the maintenance of stable hybrid zones. Stable hybrid zones are thought to be maintained by a balance between the dispersal of parental individuals into the hybrid zone and selection against unfit hybrid genotypes (14). By contrast, a hybrid species probably arises through a hybrid founder event, in which one or more early generation hybrids colonize a new locality and thus become spatially or ecologically isolated from the parental species (15, 16). In this scenario, early generation hybrids may give rise to a reproductively isolated recombinant lineage of high fitness rather than converging back toward one of the parental species via backcrossing. This second process is what we refer to as hybrid speciation, and it is the time taken to construct such a composite genome from the genetic alternatives present in an isolated hybrid population that we wish to gauge.
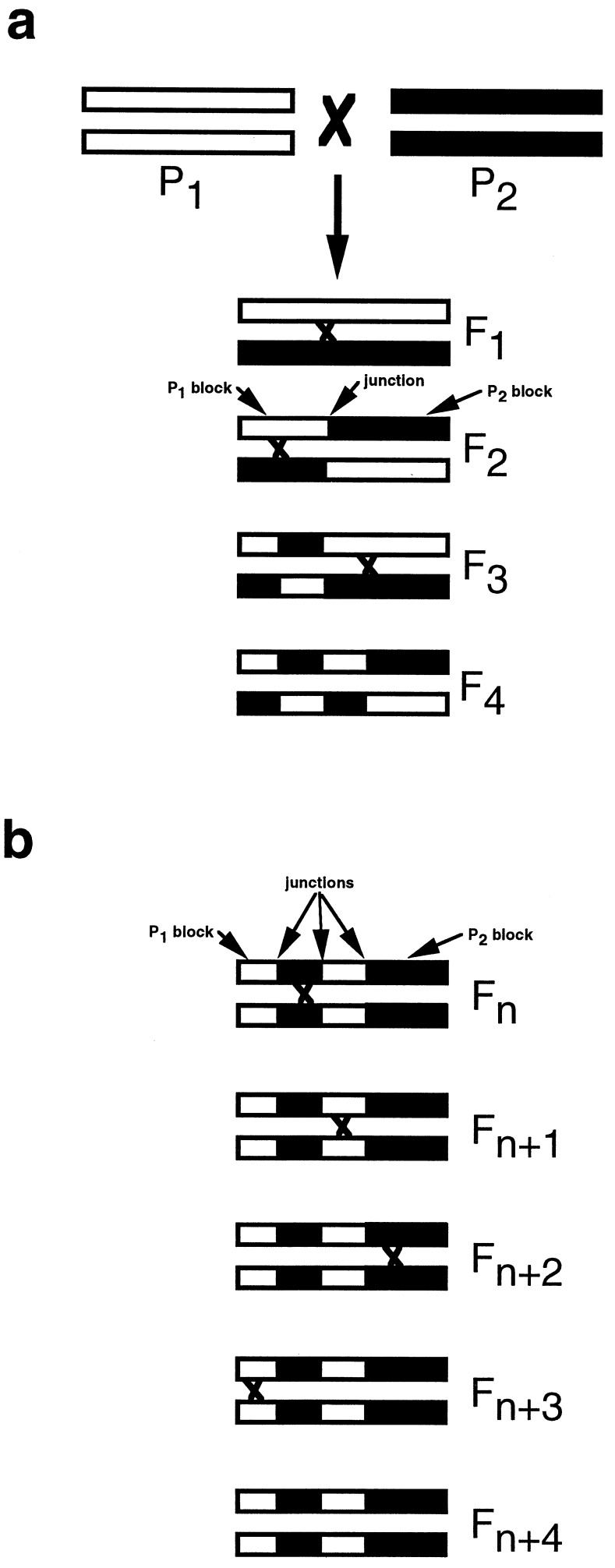
In a newly forming hybrid species, the sizes of parental species linkage blocks are expected to become progressively smaller over time due to recombination (17, 18) (Fig. 1a). However, continued reduction in block size will be countered by stabilization of the hybrid species’ genome; subsequent recombination will be among blocks derived from the same parental species (Fig. 1b). This analysis represents an empirical application of Fisher’s junctions approach (17), which tracks parental species blocks by monitoring recombination breakpoints or “junctions” between heterogeneous regions rather than all points on a genome. In the neutral case we can imagine a junctions “clock,” which is analogous to the molecular clock but slows over time as heterozygosity decreases due to drift. By comparing the frequency spectrum of observed parental species blocks with predictions based on computer simulations, we can estimate the number of generations required to stabilize the H. anomalus genome. Bear in mind that we are estimating the speed of hybrid speciation, not the age of the hybrid species.
Figure 1.
Illustration of the reduction and fixation of parental chromosomal blocks (black vs. white) over successive generations of hybridization. Chiasmata, and hence junction origin, are designated by “x”s between intra-generational chromosomes. (a) Hypothetical scenario demonstrating how parental species chromosomal block size decreases and junction number increases over successive generations of hybridization. (b) When genomic composition becomes fixed or stabilized, no further reduction in block size can occur, despite continued recombination in successive generations.
H. anomalus is a diploid (n = 17), outcrossing, annual sunflower restricted to xeric habitats in northern Arizona and southern Utah, USA, well within the range of its parental species, H. annuus and H. petiolaris (19). Although morphologically and ecologically distinct (19), it combines parental ribosomal DNA repeat units (13), allozymes (20), chloroplast DNA haplotypes (13, 20), randomly amplified polymorphic DNA (RAPD) markers (21), and amplified fragment length polymorphism (AFLP) markers (herein). The near absence of ribosomal DNA and chloroplast DNA sequence divergence between H. anomalus and its parents suggests a recent origin of the hybrid species, probably within the last 170,000 years (20). Reproductive isolation between H. anomalus and its parental species has been facilitated by rapid karyotypic evolution (22).
MATERIALS AND METHODS
Mapping.
A genetic linkage map based on 356 RAPD markers and one isozyme locus has been reported previously for H. anomalus (22). The map was generated by using 56 individuals derived from an intraspecific hybrid between two natural populations of H. anomalus (Mexican Water, AZ, ANO-1497 and Hanksville, UT, ANO-1506) crossed to an inbred sunflower line (CMS89). This design allowed the segregation of H. anomalus chromosomes to be monitored against a homozygous genetic background. This particular crossing design was chosen because of the need to map dominant RAPD and AFLP loci and because of the difficulty of producing F2s in self-incompatible wild sunflowers.
Here we report the placement of an additional 193 RAPD and 151 AFLP markers on the H. anomalus map. DNA isolations and RAPD methods are described elsewhere (21, 22). However, due to differences in base composition, the annealing temperature of RAPD primers 801–900 was increased from 36 to 52°C. AFLP methods followed standard protocols (23), except that primers were labeled with the fluorophore, Texas Red (Amersham), and polymorphisms were visualized by using a Hitachi FMBIO II Fluorescent Imaging Device (Tokyo). RAPD and AFLP primer sequences can be obtained on request from the authors.
Maps were developed with the computer program mapmaker (24) by using a logarithm of odds (LOD) score of 11.0 and a recombination limit of 0.15. Recombination values were converted to map distances by using the Haldane-mapping function. Potential scoring and ordering errors were detected by using the “genotype” command of mapmaker, and areas with apparent discrepancies were rescored or retested.
The parental species origins of markers mapped in the H. anomalus genome was determined by surveying five natural populations from each parental species: H. annuus (Siskiyou Co., CA, Rieseberg 101; Columbia Co., WA, Rieseberg 315; Pecos Co., TX, Rieseberg 1095; Keith Co., NE, Rieseberg 1238; and Cooper Co., MO, Soltis and Soltis s. n.), and H. petiolaris (Hildago Co., NM; Rieseberg 1087; Navaho Co., AZ, Rieseberg 1106; Cleveland Co., OK, Rieseberg 1224; Arthur Co., NE, Rieseberg 1243; and Roosevelt Co., NM, Seiler 1382). For Rieseberg 1238 and Rieseberg 1243, 15 individuals from each population were assayed. For the remaining eight populations, ten DNAs from each population were bulked and the eight bulked DNAs were assayed for the mapped H. anomalus markers. Only markers that were completely absent in one of the two parental species were considered species-specific.
Simulations.
As discussed earlier, it seems likely that H. anomalus arose in a hybrid founder population that was spatially or ecologically isolated from both parental species. Presumably, the founding population was fairly small and population expansion largely occurred after the H. anomalus genotype was stabilized. Otherwise H. anomalus would not be morphologically uniform throughout its range. We chose to model this scenario by simulating a hybrid swarm, starting with half and half H. annuus and H. petiolaris pure individuals, and allowing recombination to take its course in small, closed populations of n = 50, 100, 250, and 500 individuals. Differences between populations of 250 and 500 individuals were negligible, so there was no reason to examine larger populations. We also considered initiating the simulations with F1s or early generation hybrids but were concerned that this might lead to an underestimate of time to speciation.
Early generation hybrids between H. annuus and H. petiolaris have low fertility (19). Thus, a realistic model must include the effects of selection. Selection was modeled by genome-wide heterozygote disadvantage as described in Barton and Gale (25). Individual fitness is W(x) = 1 − s(4x [1 − x]), where x is the heterozygous proportion of the genome. Selection coefficients (s) of 0, 0.1, 0.2, and 0.4 were used. Genotypes with intermediate hybrid indices (such as that of H. anomalus) become increasingly rare in simulated populations when s > 0.4.
One thousand simulations were run for all combinations of s and N. In all simulations, generations were discrete and sexes were not distinguished, as is appropriate for self-incompatible, annual, hermaphrodites.
The core of the model was implemented by using a junctions simulation program previously developed in a study of multilocus clines (18). To analyze of the loss of variance due to inbreeding, Fisher (17) developed a representation of continuous genetic material in terms of the junctions along its length where material of different ancestry has come together as a result of recombination. A new junction is formed when a crossover occurs in a region for which the parent organism is heterozygous. The areas of genetic material between junctions are called blocks. Once produced, junctions are inherited like point mutations and may be lost or instead may increase to fixation. Recording the junctions produced between two original haplotypes allows complex offspring to be represented in a very concise way. Conventional simulations of L discrete loci for a population of N individuals are limited by both storage space and run-time requirements of order NL. If however we simulate a population by using the junction approach, we need only follow the fate of junctions produced, giving a maximum number of order NRt where R is the total map length. The number of loci considered is therefore no longer limiting, so this simulation approach is ideal for many problems involving linkage between many loci.
Given this background, the junctions approach seems highly suitable for the present study. If we consider secondary contact between two populations, which have diverged at many loci, then we can designate chromosomes from one population as type A, and from the other type B. The proportion of heterozygous material along a pair of chromosomes and block sizes can easily be calculated from the position of junctions along their length. Chromosomes are scaled to the map metric, so that recombination is uniform along their length, with a number of chiasmata drawn from a Poisson distribution with expectation R and assuming no interference. The size of blocks present in the simulated population were measured each generation as described below and compared with those found in H. anomalus.
Estimates of Block Sizes.
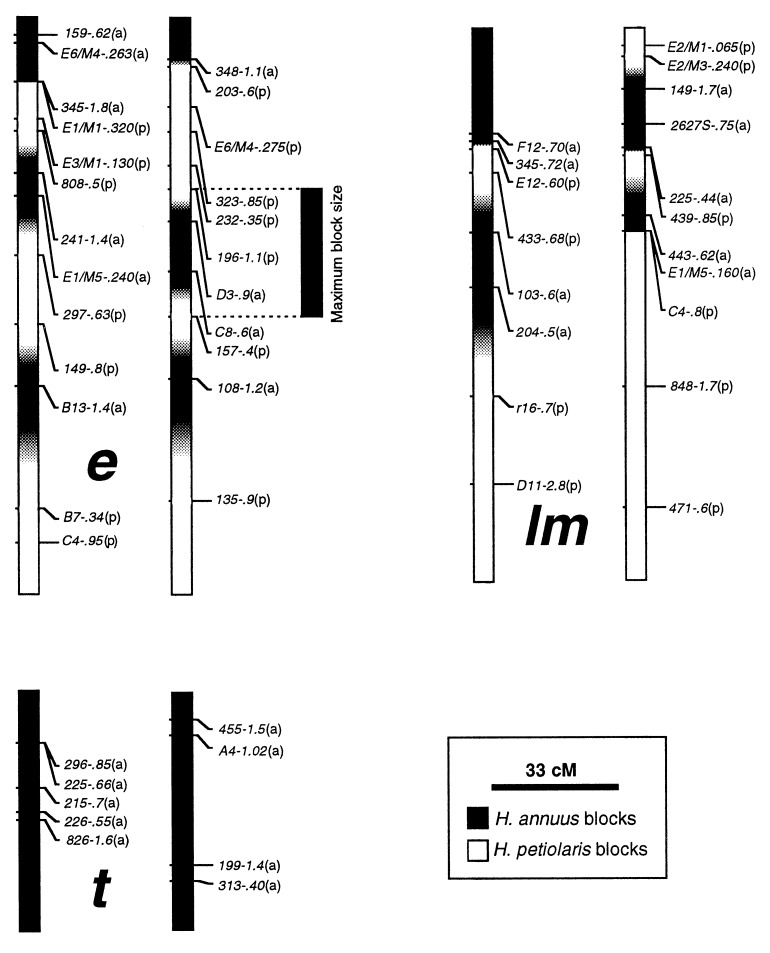
Even dense marker maps cannot fully reveal the underlying distribution of parental blocks because (i) the location of inferred junctions lying between markers of different ancestry cannot be identified precisely, and (ii) small blocks may not be detected due to gaps in marker distributions. For this reason, we have measured “maximum possible” block sizes, in which the maximum possible length of a parental block is the distance spanned by all consecutive markers from the same parental species, plus the distance to the nearest markers from the alternative species (Fig. 2). Blocks at the ends of linkage groups were not included because their maximum size cannot be estimated. Markers were placed on the simulated chromosomes at intervals drawn at random from a list of the intervals between markers on the H. anomalus map. The frequency spectrum (26) of maximum possible block lengths over the simulation data was calculated by using these markers precisely as they were used for the H. anomalus genome (the frequency spectrum shows the number of blocks in a length class, scaled by the span of that class).
Figure 2.
Selected linkage groups of H. anomalus. Each linkage is represented by two haplotypes, which are derived from the two natural populations of H. anomalus used to generate the mapping population. Large lower case letters between haplotypes designate linkage groups (22). Marker nomenclature includes, from left to right, the primer designation and the size in kilobases of the segregating fragments scored. Letters in parentheses after each marker indicate its parental species origin: a, H. annuus; p, H. petiolaris. Parental genomic composition is indicated by black and white blocks that span the distance between consecutive markers. Regions harboring recombination sites are indicated by a gray-scale, with the intensity of shading based on the genotype of the flanking markers. Maximum block size, which was used for analytical purposes, is illustrated by the H. annuus block to the right of linkage e.
Comparisons of Block Sizes.
We expect the number of blocks to increase exponentially with decreasing size, so exponentially decreasing size classes were used to evenly divide the number of blocks (18). A block is assigned to size class k if it is less than or equal to the kth class boundary, but greater than the (k + 1)th, where the kth boundary is (8/10)k, and 10 ≥ k ≥ 0. Thus size class zero runs from 1 to 8/10 (the parameter 8/10 was chosen because it leads to an even distribution of blocks within size classes for the anomalus data set). Because the genomic contributions of the parental species to the H. anomalus genome does not differ significantly from a 1:1 ratio, we assume that the hybrid species is derived from individuals with a hybrid index between 45 and 55%. So the observed data are compared with the frequency spectrum of blocks for simulated individuals within this group.
Comparisons of Genomic Composition.
Concordance in genomic composition between the two haplotypes comprising the H. anomalus genome was calculated by using the φ coefficient of association. The φ coefficient provides a measure of the degree of association between two properties (in this case, genomic composition of population haplotypes) and can vary from −1 to 1 with the positive or negative direction of the association indicated by the sign of the coefficient. Haplotype composition was analyzed as follows. Blocks between consecutive H. petiolaris markers, plus one-half the distance between terminal H. petiolaris markers to the nearest H. annuus markers, were considered H. petiolaris regions (1). Likewise, blocks between consecutive markers from H. annuus, plus one-half the distance between terminal H. annuus markers to the nearest H. petiolaris markers, were considered H. annuus genomic regions (0). Species-specific markers from one haplotype either matched (0,0 or 1,1) or did not match (0,1 or 1,0) the corresponding genomic region in the other haplotype.
RESULTS AND DISCUSSION
Because H. anomalus is diploid, each linkage group comprises two haplotypes (Fig. 2). These haplotypes represent the genomes of the two populations of H. anomalus used to generate the mapping population. Because the majority of markers employed for mapping are dominant, individual markers provide parentage information for one of the population haplotypes only. All haplotypes, with the exception of those comprising linkage group t, include chromosomal blocks derived from both H. annuus and H. petiolaris (Fig. 2; Table 1); all markers on linkage group t are derived from H. annuus. In addition, the genomic composition of haplotypes representing each linkage group are significantly associated (G = 22.71; df = 1; P < 0.0001; φ coefficient of association = 0.36). This is an expected result of drift in a recently formed hybrid species. We suspect that the haplotypes are identical in genome composition, and apparent differences (Fig. 2) are an artifact of limited marker resolution. Alternatively, some polymorphism for genomic composition may be maintained in H. anomalus. For example, drift may have led to the fixation of modest differences in genomic composition among geographically isolated populations of H. anomalus.
Table 1.
Locations in centiMorgans of parental species markers on H. anomalus linkage groups for each population haplotype (ANO-1497 and ANO-1506)
| Linkage a | Linkage b | Linkage c | Linkage d | Linkage e | Linkage f | Linkage gij | Linkage h | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 |
| 2.4a | 0.0a | 3.9a | 0.0p | 0.0p | 28.1a | 11.9p | 8p | 0.0a | 5.8a | 1.9a | 0.0a | 7.6p | 3.8p | 0.0p | 2.0p |
| 10.4a | 12.3a | 28.4a | 14.2a | 1.9p | 34a | 15.8p | 19.6a | 1.9a | 7.7p | 10.6a | 1.9a | 17.9p | 7.6p | 8.2a | 16.6a |
| 24.9a | 28.7a | 49.3p | 24.5a | 34a | 37.9a | 17.7a | 34.6a | 11.5a | 17.4p | 23.6p | 12.6p | 35.5a | 41.4a | 22.5a | 40.9a |
| 28.7ap | 41.3a | 55.2p | 34.3p | 41.8p | 47.7a | 35.5a | 36.4a | 11.5p | 23.2p | 39a | 36.2p | 49.1a | 45.2p | 30.6a | 51.1a |
| 55.2p | 47.2a | 63p | 61.1p | 59.7p | 51.6a | 40.3p | 80.9a | 20.3p | 31.2p | 46.7a | 40.9a | 59.4p | 63.3a | 54.9a | |
| 55.2p | 55.2p | 72.9p | 63a | 77.3p | 59.7p | 46.2p | 110.4p | 23.2p | 36.9p | 67p | 44.8a | 71.3ap | 71.3p | 56.8p | |
| 61p | 57.1a | 74.8p | 71p | 85.1a | 83.2p | 79a | 126.7a | 33.1a | 44.7a | 86.7a | 68.9a | 73.8p | 72.5p | ||
| 72.9a | 69a | 78.6a | 72.9a | 85.1a | 82.8a | 136.5p | 38.8a | 56.5a | 101.6a | 82.6p | 79.0p | 79.0a | |||
| 78.8a | 96.6p | 102.6a | 82.4a | 97.8p | 158.3a | 52.7p | 66.7p | 112.5p | 97.8p | 86.8a | 80.9a | ||||
| 86.8p | 95a | 119.8p | 172.3p | 68.6p | 81.2a | 97.8p | 94.9a | 101.8a | |||||||
| 91.7p | 130.6p | 83.1a | 109.3p | 119.1a | 111.1p | ||||||||||
| 138.6p | 111.2p | 126.9a | 125a | ||||||||||||
| 144.3a | 119.2p | 132a | 135.3p | ||||||||||||
| 154.5p | 146.4p | ||||||||||||||
| 168.5p | |||||||||||||||
| 176.3a | |||||||||||||||
| Linkage kj | Linkage lm | Linkage n | Linkage op | Linkage q | Linkage rs | Linkage u | Linkage vw | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 | 1497 | 1506 |
| 0.0p | 8.0p | 20.6a | 0.0p | 0.0a | 0.0a | 4.2p | 0.0p | 1.9p | 6.8p | 2.3a | 0.0a | 0.0p | 1.9p | 10.5a | 2a |
| 9.9p | 15.8p | 22.5a | 2.7p | 1.9a | 5.7a | 28.7p | 14.5p | 11.7p | 15.6a | 15.1p | 31.3a | 14.7a | 16.6p | 22.9p | 12.4p |
| 19.7p | 27.5a | 24.4p | 10.4a | 27.1p | 23.3p | 40.6a | 24.8a | 25.4p | 19.5p | 23.2p | 47.5a | 29.2a | 33.1a | 24.8p | 32.8p |
| 21.6p | 29.4p | 30.2p | 18.7a | 29.0a | 30.9p | 44.5a | 35.9p | 41.3a | 35.2a | 41.6a | 49.4p | 38.9p | 37p | 51.8a | 47.9a |
| 33.3a | 43.1p | 44.2a | 24.4a | 41.2p | 53.4a | 68.3p | 50.4a | 37.5a | 61a | 51.3a | 40.8a | 38.9a | 63.3a | 74.7a | |
| 33.3a | 57.0a | 56.8a | 26.3p | 51.5a | 53.4a | 86.5a | 58.5a | 66.9a | 57.2a | 42.7a | 48.6p | 76.6a | 101.9p | ||
| 39.2p | 89.7a | 81.9p | 40.4a | 53.4p | 55.3a | 62.4p | 86.7a | 59.1p | 66.3p | 57.9p | 124.6a | 114.5a | |||
| 51.1a | 95.6a | 102.2p | 44.2a | 55.3p | 67.0a | 68.3p | 96.5a | 66.9p | 70.9p | 79.3a | 129.1p | 114.5a | |||
| 51.1a | 112.6a | 44.3p | 61.2p | 70.9a | 76.3a | 71.5p | 74.8a | 90.8p | 136.9a | 122.4p | |||||
| 72.0a | 120.6a | 80.0p | 63.1a | 97.1p | 78.7a | 88.9p | 96.7a | 140.7a | 133p | ||||||
| 110.4a | 108.1p | 90.6p | 151.7a | 90.6a | 100.6p | 110.8a | 148.7a | 138.8p | |||||||
| 120.6p | 101.3a | 161.5p | 109.9p | 100.7a | 148.7a | 150.6a | |||||||||
| 122.5a | 119.8p | 174.7a | 152.5a | ||||||||||||
| 130.5a | 139.0p | 210.6a | 154.4a | ||||||||||||
| 141.4a | 155.6a | 180.7a | |||||||||||||
| 161.5p | 196.5a | ||||||||||||||
| 168.2p | 199.2a | ||||||||||||||
a, H. annuus; p, H. petiolaris
Linkage t is not shown since all of its markers are derived from H. annuus.
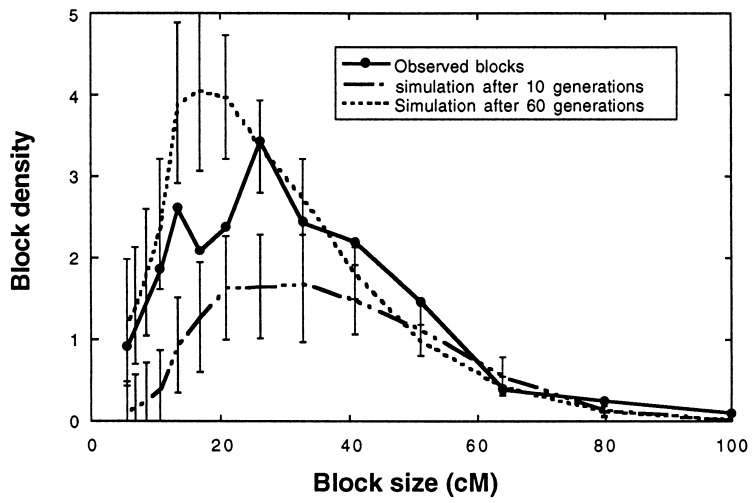
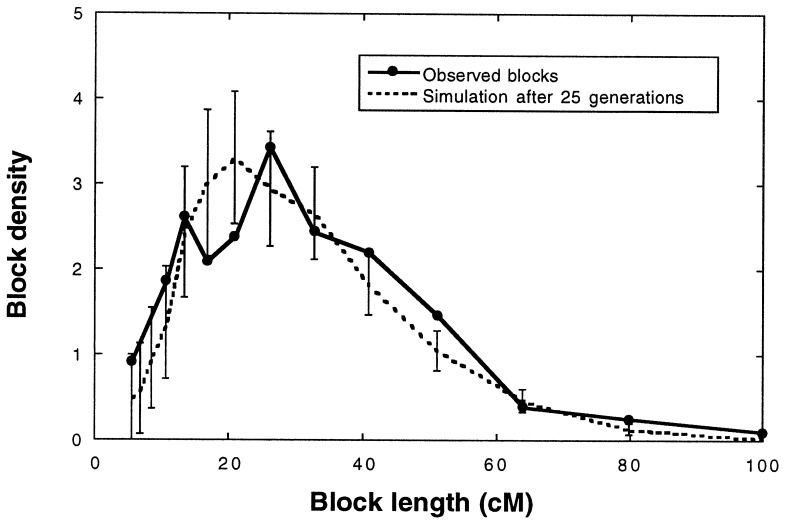
To estimate the number of generations of recombination required to achieve the present frequency spectrum of parental species chromosomal blocks in the H. anomalus genome, we compared observed block sizes with simulation results (Figs. 3–5). The results of this comparison suggest that between 10 and 60 generations of recombination would be required to stabilize the H. anomalus genome (Fig. 3), with the observed block sizes appearing most similar to those derived from simulations based on 25 generations of hybridization (Fig. 4). Because simulated block size distributions for different time intervals are most easily discriminated for blocks of intermediate size (between 13 and 23 cM), it is perhaps not surprising that it is for these size classes that the observed data fall both above the maximum 95% confidence interval for t = 10 generations and below the minimum 95% confidence interval for t = 60 generations (Fig. 3). For the remaining size classes, there was insufficient power in the data to discriminate between few and many generations of recombination.
Figure 3.
Comparison of the frequency spectra (26) of maximum possible parental species block sizes in the H. anomalus genome to those of simulation populations after 10 and 60 generations of hybridization (n = 500; s = 0). The frequency spectrum shows the number of blocks in a class (block density), scaled by the size of that class (see Materials and Methods) and is the standard way of representing the distribution of block sizes (18, 34). The increase in the area under the curve over time indicates the increasing degree to which the genome is broken up by junctions. (Error bars indicate 95% confidence intervals).
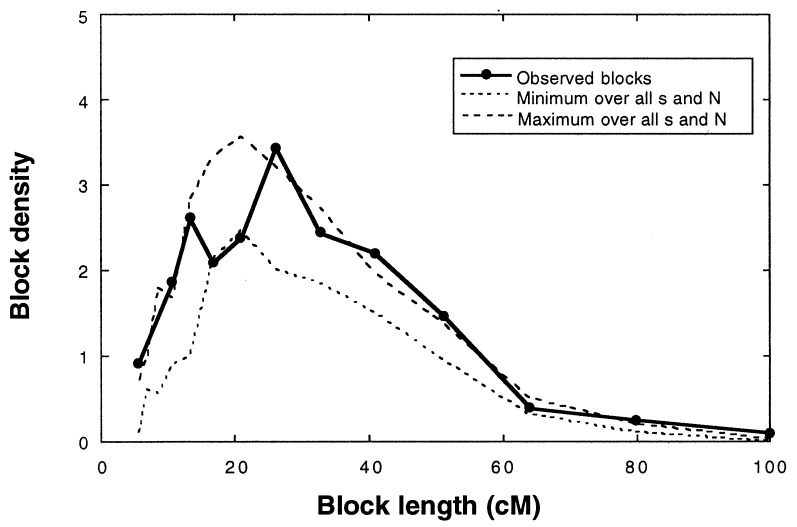
Figure 5.
Comparison of the frequency spectra of maximum possible parental species block sizes in the H. anomalus genome to those of simulation populations after 30 generations of hybridization (cf. Fig. 3). Dashed lines indicate the minimum and maximum average block sizes produced by any combination of population size (N) and selection coefficient (s). Note that variation in N and s has minimal effects on block sizes.
Figure 4.
Comparison of the frequency spectra of maximum possible parental species block sizes in the H. anomalus genome to those of simulation populations (cf. Fig. 3) after 25 generations of hybridization (population size = 500; selection coefficient = 0). (Error bars indicate 95% confidence intervals.)
The conclusion that H. anomalus arose rapidly is robust to variation in strength of selection and population size (Fig. 5). Extreme values for both parameters result in only minor changes in the expected distribution of block sizes (Fig. 5). Reproductive isolation appears to have accompanied genome stabilization in H. anomalus due to the sorting of parental chromosomal and genic sterility factors (22, 27). Thus, these time estimates should provide a valid measure of the number of generations required for the development of postmating reproductive barriers in H. anomalus.
Possibly, time of speciation is underestimated by our methods because recombination sometimes is reduced in distant hybrids (28, 29). Such an error would only, however, be of the same order as the change in recombination rate. For example, if a 25% reduction in recombination is assumed (i.e., 0.75 crossovers/chromosome/generation), such as has been observed for a rice interspecific cross (29), then our simulations would underestimate the time to speciation by 25%. The distribution of crossovers may vary between intraspecific and interspecific crosses as well, but this difference should not lead to biases in the sizes of blocks used in our analyses because they are based on the map of H. anomalus, not that of the parental species.
It also is possible that introgression with one or both parental species could contribute to the observed frequency spectrum. Possibly, some introgression occurred during the initial formation of the hybrid species because backcross hybrids are more fertile and easily produced than other hybrid genotypic classes (19). These early introgressions could lead to underestimates of the time to speciation and cannot be ruled out. More serious errors could result from sporadic recent introgressions. Natural hybridization between H. anomalus and its parents has not been reported, but the possibility of past episodes of hybridization cannot be dismissed. However, block sizes in H. anomalus do not differ significantly with regard to parental species origin or population haplotype. This symmetry seems unlikely if sporadic introgressions were important.
The rapid hybrid speciation suggested by this result is corroborated by independent lines of evidence from greenhouse-grown H. annuus × H. petiolaris hybrids. In an earlier report (27), the genomic composition of three independently generated synthetic hybrid lineages was compared with that of the hybrid species H. anomalus. All three hybrid lineages converged onto nearly identical gene combinations within five generations, and these proved statistically concordant with the genome of H. anomalus. Since then, we have added 185 species-specific markers to the H. anomalus map, strengthening this result (G = 80.10; df = 1; P < 0.0001; φ = 0.50). These data imply that although H. anomalus may have originated >100,000 years ago (20), its genome probably became stabilized in a small number of generations. A rapid transition also is suggested by analysis of pollen fertility in the synthetic hybrid lineages. Although F1s were mostly sterile (pollen fertility = 5.6 ± 2.21%), fertility was rapidly reestablished after only four generations of sib-mating or backcrossing (91.8 ± 4.47%), indicating that hybrid speciation could occur extremely rapidly under favorable conditions. Although the strong natural selection for fertility used to generate these lineages is predicted in nature, the low fertility of early generation hybrids could inhibit their establishment.
Rapid speciation may be the rule rather than the exception for recombinational species. Fertile, stable, hybrid neospecies have been experimentally synthesized in several plant genera, typically in 10 generations or fewer (10). These artificial neospecies are semisterile when crossed with their parental species, indicating that reproductive isolation can arise rapidly.
Recombinational speciation represents only one of several known or suspected pathways to rapid speciation (30). Others include allopolyploidy (8), changes in mating system (31), fixation of chromosomal rearrangements (9), and rapid adaptation to new habitats (32). Features shared by these mechanisms may facilitate rapid speciation. For example, both allopolyploidy and recombinational speciation involve hybridization, and rates of recombinational speciation are thought to be enhanced by chromosomal rearrangements, ecological divergence, and a shift to a selfing mating system (11, 12).
Analysis of parental species block sizes in hybrid zones or hybrid species has several general applications in addition to estimating the tempo of hybrid speciation. These include dating secondary contact of natural hybrid zones (18), estimating the number of genes affecting hybrid fitness, and measuring selection coefficients at these loci. Advances in molecular marker technology and laboratory automation (33) have only recently made collection of marker data of the necessary resolution feasible. Here, this quality of data has allowed the first direct application of Fisher’s junctions approach, an achievement which has significant potential for genomic analysis of experimental pedigrees, as well as natural hybrid populations and hybrid species. An analytical treatment extending the scope of our results is underway.
Acknowledgments
We thank S. Carney, R. Rieseberg, D. Wolf, and three anonymous reviewers for critical reading of the manuscript. This research was supported by a National Institutes of Health Genetics Training Grant (to M.C.U.) and by the National Science Foundation (L.H.R.).
ABBREVIATIONS
- RAPD
randomly amplified polymorphic DNA
- AFLP
amplified fragment length polymorphism
References
- 1. Morgan T H. The Physical Basis of Heredity. Philadelphia: Lippincott; 1919. [Google Scholar]
- 2.Goldschmidt R. The Material Basis of Evolution. New Haven, CT: Yale Univ. Press; 1940. [Google Scholar]
- 3.Eldredge N, Gould S J. In: Models in Paleobiology. Schopf T J M, editor. San Francisco: Freeman; 1972. pp. 82–115. [Google Scholar]
- 4.Charlesworth B, Lande R, Slatkin M. Evolution. 1982;36:474–498. doi: 10.1111/j.1558-5646.1982.tb05068.x. [DOI] [PubMed] [Google Scholar]
- 5.Rosenzweig M L. Science. 1997;277:1622–1623. [Google Scholar]
- 6.Mayr E. In: Evolution as a Process. Huxley J H, Hardy A C, Ford E B, editors. London: Allen & Unwin; 1954. pp. 157–180. [Google Scholar]
- 7.Carson H L. In: Population Biology and Evolution. Lewontin R C, editor. Syracuse, NY: Syracuse Univ. Press; 1968. pp. 123–137. [Google Scholar]
- 8.Lewis W. Polyploidy: Biological Relevance. New York: Plenum; 1980. [Google Scholar]
- 9.White M J D. Modes of Speciation. San Francisco: Freeman; 1978. [Google Scholar]
- 10.Grant V. Plant Speciation. New York: Columbia Univ. Press; 1971. [Google Scholar]
- 11.Templeton A R. Annu Rev Ecol Syst. 1981;12:23–48. [Google Scholar]
- 12.McCarthy E M, Asmussen M A, Anderson W W. Heredity. 1995;74:502–509. [Google Scholar]
- 13.Rieseberg L H. Am J Bot. 1991;78:1218–1237. [Google Scholar]
- 14.Barton N H, Hewitt G M. Annu Rev Ecol Syst. 1985;16:113–148. [Google Scholar]
- 15.Charlesworth D. Curr Biol. 1995;5:835–836. doi: 10.1016/s0960-9822(95)00166-7. [DOI] [PubMed] [Google Scholar]
- 16.Rieseberg L H. Annu Rev Ecol Syst. 1997;28:359–389. [Google Scholar]
- 17.Fisher R A. Heredity. 1953;8:187–197. [Google Scholar]
- 18.Baird S J E. Evolution. 1995;49:1038–1045. doi: 10.1111/j.1558-5646.1995.tb04431.x. [DOI] [PubMed] [Google Scholar]
- 19.Heiser C B, Smith D M, Clevenger S, Martin W C. Mem Torrey Bot Club. 1969;22:1–218. [Google Scholar]
- 20.Rieseberg L H, Beckstrom-Sternberg S, Liston A, Arias D. Syst Bot. 1991;16:50–76. [Google Scholar]
- 21.Rieseberg L H, Choi H-C, Chan R, Spore C. Heredity. 1993;70:285–293. [Google Scholar]
- 22.Rieseberg L H, Van Fossen C, Desrochers A M. Nature (London) 1995;375:313–316. [Google Scholar]
- 23.Travis S E, Maschinski J, Keim P. Mol Ecol. 1996;5:735–745. doi: 10.1111/j.1365-294x.1996.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 24.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 25.Barton N H, Gale K S. In: Hybrid Zones and the Evolutionary Process. Harrison R, editor. Oxford: Oxford Univ. Press; 1993. pp. 13–45. [Google Scholar]
- 26.Ewens W J. Mathematical Population Genetics. Berlin: Springer; 1979. [Google Scholar]
- 27.Rieseberg L H, Sinervo B, Linder C R, Ungerer M C, Arias D. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. [DOI] [PubMed] [Google Scholar]
- 28.Paterson A H, Damon S, Hewitt J D, Zamir D, Rabinowitch H D, Lincoln S E, Lander E S, Tanksley S D. Genetics. 1991;127:181–197. doi: 10.1093/genetics/127.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Causse M A, Fulton M T, Cho Y G, Ahn S N, Chunwongse J, Wu F S, Xiao J H, Ronald P C, Harrington S E, Second G, et al. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avise J C. Molecular Markers, Natural History and Evolution. New York: Chapman & Hall; 1994. [Google Scholar]
- 31.Gottlieb L D. Am J Bot. 1973;60:545–553. [Google Scholar]
- 32.Bradshaw A D. In: Ecological Genetics and Evolution. Creed R, editor. Oxford: Blackwell; 1972. pp. 20–50. [Google Scholar]
- 33.Karp A, Edwards K J, Bruford M, Funk S, Vosman B, Morgante M, Seberg O, Kremer A, Boursot P, Arctander P, et al. Nat Biotechnol. 1997;15:625–628. doi: 10.1038/nbt0797-625. [DOI] [PubMed] [Google Scholar]
- 34.Barton N H. Evolution. 1983;37:454–471. doi: 10.1111/j.1558-5646.1983.tb05563.x. [DOI] [PubMed] [Google Scholar]