Abstract
Nuclear pore complexes (NPCs) govern macromolecular transport between the nucleus and cytoplasm and serve as key positional markers within the nucleus. Several protein components of yeast NPCs have been implicated in the epigenetic control of gene expression. Among these, Nup2p is unique as it transiently associates with NPCs and, when artificially tethered to DNA, can prevent the spread of transcriptional activation or repression between flanking genes, a function termed boundary activity. To understand this function of Nup2p, we investigated the interactions of Nup2p with other proteins and with DNA using immunopurifications coupled with mass spectrometry and microarray analyses. These data combined with functional assays of boundary activity and epigenetic variegation suggest that Nup2p and the Ran guanylyl-nucleotide exchange factor, Prp20p, interact at specific chromatin regions and enable the NPC to play an active role in chromatin organization by facilitating the transition of chromatin between activity states.
Introduction
Nuclear pore complexes (NPCs) govern communication between the nucleus and cytoplasm (for reviews see Wente, 2000; Rout and Aitchison, 2001); but, evidence also supports a direct role for NPCs in the structural organization of the nucleus and the chromatin contained within it. In interphase, DNA is divided into silenced heterochromatin and actively transcribed euchromatin, which predominate at the nuclear periphery and intranuclear regions, respectively (Cockell and Gasser, 1999). Euchromatic channels also penetrate into regions of the nuclear periphery proximal to NPCs (Fawcett et al., 1994), raising the possibility that NPCs are intimately involved in the division of chromatin into active and silent states.
Saccharomyces cerevisiae, is an excellent model to elucidate cellular mechanisms of genome organization as, although they lack observable heterochromatin, they possess several heterochromatin-like silenced regions, including subtelomeric regions (Gottschling et al., 1990; Fourel et al., 1999) and the tandem rDNA repeats (Smith and Boeke, 1997). In diploid yeast, the 64 telomeres localize to <10 foci at the nuclear periphery (Gotta et al., 1996), and this localization correlates with the silenced state of subtelomerically encoded reporters (Maillet et al., 2001). Moreover, this effect appears to be a result of peripheral localization, as artificially tethering a reporter gene to the nuclear envelope (NE) is sufficient to induce the silenced state (Andrulis et al., 1998). The precise mechanism for this position-based silencing is not known, but a protein network beneath the NE comprised of Mlp1p and Mlp2p has been implicated in the organization of functional nuclear subcompartments, the maintenance of gene expression states, and telomeric clustering (Galy et al., 2000; Feuerbach et al., 2002). The presumed role of NPCs in this function is to provide a peripheral anchor for the Mlp proteins. Yeast that lack both Mlp proteins or certain NPC components, collectively termed nucleoporins or nups, exhibit aberrant telomere localization and transcriptional activation of subtelomerically encoded reporter genes, suggesting that NPCs are—at least indirectly—involved in these functions (Galy et al., 2000; Feuerbach et al., 2002). However, mlp mutants exhibit defects in both NE morphology and nucleocytoplasmic transport (Strambio-de-Castillia et al., 1999) and, given that contradictory results regarding their roles in telomeric anchoring have been reported (Hediger et al., 2002a,b), this model remains controversial.
The best studied examples of silenced chromatin in yeast are the subtelomeric HML and HMR mating type loci (Loo and Rine, 1994), which are encoded on the left and right arms of chromosome 3, respectively. The examination of these regions has revealed another function for NPCs in chromatin organization. Active or silenced DNA is partitioned along chromosomes by insulators or boundary elements, defined by their ability to buffer the spread of transcriptional activation and repression between flanking chromatin regions (Gerasimova and Corces, 2001; Pai and Corces, 2002). In yeast, boundary elements flank the silenced mating type loci to prevent the spread of silencing into adjacent chromatin (Donze et al., 1999; Donze and Kamakaka, 2001). Boundaries also exist at additional subtelomeric regions (Fourel et al., 1999), and it is likely that additional boundaries exist elsewhere in the genome (Fourel et al., 2004). Using an engineered, partially derepressed HML locus, Ishii et al. (2002) made the surprising observation that physically tethering certain components of the nucleocytoplasmic transport machinery to chromatin allows the formation of chromosome boundaries, a function they termed boundary activity (BA). Central to NPC-mediated BA was Nup2p, as it was the sole nucleoporin shown to exhibit BA and was required for the BA of the other nucleocytoplasmic transport factors identified.
Although the boundary trap assay revealed a potentially exciting new function of NPCs, several questions remained, including whether these data reflect an endogenous mechanism of NPC-mediated BA and, if so, what provides the physical link to DNA and why is BA unique to Nup2p among nucleoporins. Nup2p is also unique with respect to its association at the NPC, as it transcends the classical division between the mobile and stationary phases of the transport apparatus, moving on and off the NPC in a Ran-dependent manner (Denning et al., 2001; Dilworth et al., 2001). To further investigate the functions of Nup2p and its potential role in endogenous NPC-mediated BA, we performed proteomic, transcriptomic, and genetic analyses that implicate Nup2p in the maintenance of gene expression states and telomeric silencing. Furthermore, these studies reveal that Prp20p, the chromatin-associated Ran/Gsp1p guanylyl-nucleotide exchange factor, which performs an essential role in nucleocytoplasmic transport, also functions in NPC-mediated BA and, thus, can provide a mechanism to physically link NPCs with chromatin. Interestingly, we also show that Nup60p, the nup responsible for anchoring Nup2p and the Mlp proteins to the NPC (Feuerbach et al., 2002), is required for Nup2p-dependent BA, suggesting that peripheral silencing and Nup2p-mediated BA are functionally linked. Given these data and the mobile characteristics of Nup2p, we propose a dynamic model of NPC-mediated BA in which the NPC functions as a nexus through which Nup2p facilitates the passage of chromatin between transcriptionally distinct nuclear subcompartments.
Results
The link between NPCs and chromatin
NPC-mediated BA implies an (at least indirect) connection between Nup2p and DNA (Ishii et al., 2002). Although we and others have reported that Nup2p interacts primarily with Nup60p and the karyopherins, Kap60p and Kap95p (Denning et al., 2001; Dilworth et al., 2001), there is also indication that Nup2p may interact with Prp20p, which is reported to play a role in nuclear organization and bind to chromatin (Aebi et al., 1990). However, a link between Nup2p and Prp20p is not well established. Indeed, although Denning et al. (2001) cite in vitro methods indicating that full-length Nup2p and Prp20p bind directly to one another, the data were not shown and another group, using similar methods, reported that these proteins do not interact (Solsbacher et al., 2000). In support of a potential link, our own investigations revealed that if not tethered to the NPC through Nup60p, at restrictive temperatures, mutations in PRP20 (prp20-7) lead to a mislocalization of Nup2p to the cytoplasm (Dilworth et al., 2001). Interestingly, the mutant version of Prp20p also moves to the cytoplasm at the restrictive temperature (Amberg et al., 1993). Because this interaction is potentially a critical link between Nup2p and DNA, we sought to examine this connection by a variety of experimental approaches.
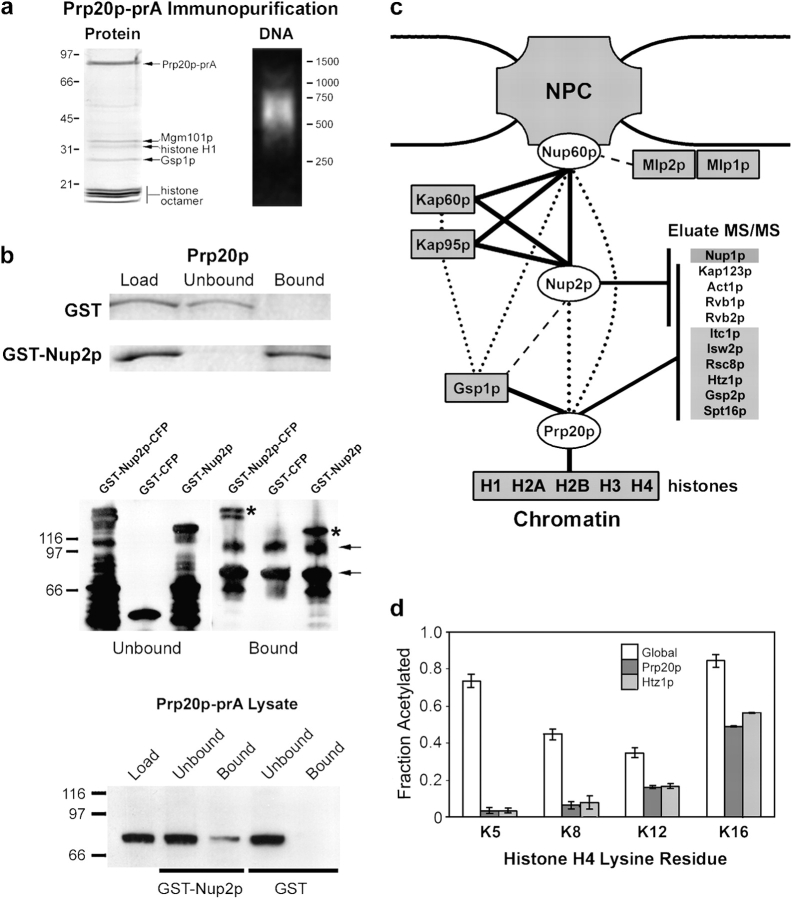
First, it was established that Prp20p binds avidly to nucleosomal DNA as detected by ethidium bromide staining of eluates immunopurified from whole-cell lysates and separated by agarose gel electrophoresis (Fig. 1 a). This was in contrast to Nup2p, and the majority of yeast proteins that, using similar methods, do not yield readily detectable DNA. To address the interaction between Nup2p and Prp20p (Fig. 1 b), we first performed solution binding assays using recombinant proteins to confirm that these proteins do indeed interact directly in vitro. Second, we used whole-cell lysates to immobilize the Prp20p–nucleosome complex on beads and then assessed the ability of this complex to bind to bacterially expressed and purified Nup2p. The presence of Nup2p in the bound fractions suggests that Nup2p can interact with Prp20p in this context, which is analogous to chromatin-bound Prp20p. With the third experimental approach, we showed that beads coated with GST-Nup2p, but not GST alone, were able to capture Prp20p from yeast whole-cell extracts. Importantly, none of these experiments exclude the possibility that, rather than associate with nucleosome-bound Prp20p, Nup2p binds instead to a small pool of free Prp20p. To address this uncertainty, we resolved bound and unbound protein mixtures over 10–70% glycerol gradients. Nup2p was detected in the highest glycerol densities only when the Prp20p–nucleosome complex was present (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200509061/DC1). Together with the observation that all detectable Prp20p was found in the high density fractions, the data support the existence of a large DNA–protein complex with which Nup2p can associate. When these data are combined with previously published physical and yeast two-hybrid interactions involving Nup2p, Nup60p, and Prp20p (Dingwall et al., 1995; Rexach and Blobel, 1995; Denning et al., 2001; Feuerbach et al., 2002), they suggest that the Nup2p–Prp20p interaction provides a link between the NPC and chromatin (Fig. 1 c). As a final test of an in vivo interaction between these proteins, we immunopurified Nup2p-prA from formaldehyde cross-linked cells that coexpressed myc-tagged Prp20p. In this case, we were able to detect the myc epitope in Nup2p eluates by immunoblotting using a monoclonal myc antibody and by mass spectrometry (Fig. S2). Together, these data support an in vivo interaction between Nup2p and Prp20p.
Figure 1.
Prp20p and Nup2p interact with chromatin remodeling factors. (a) Prp20p is nucleosome associated. (Left) Prp20p-prA was immunopurified from yeast whole-cell lysates and abundant copurifying proteins were identified by MS analysis of gel slices. Components of the histone octamer, H2A, H2B, H3, and H4, were present as well as the linker histone, H1 and Ran/Gsp1p. The presence of Mgm101p is not specific (see text). (Right) Prp20p-prA eluates contain DNA. Eluates were resolved by agarose gel electrophoresis and visualized by ethidium bromide staining. Prp20p-prA associated DNA is ∼600 bp in length due to chromatin shearing during the cell lysis procedures. (b) The interaction between Nup2p and the Prp20p–nucleosome complex can be reconstituted in vitro. (Top) Bacterially expressed and purified Prp20p was incubated with glutathione resin coated with GST or GST-Nup2p. Unbound and bound proteins were resolved by SDS-PAGE and visualized with Coomassie blue. (Middle) The Prp20p–nucleosome complex was bound to IgG-coated magnetic beads and then incubated with bacterially expressed and purified GST-Nup2p-CFP, GST-CFP, or GST-Nup2p. Immunoblotting for GST revealed that although GST alone could not bind to the Prp20p–nucleosome complex, GST chimeras containing Nup2p bound efficiently (asterisks). Arrows indicate nonspecific immunoreactive proteins. (Bottom) Glutathione resin coated with GST-Nup2p, but not GST alone, was able to capture Prp20p-prA from yeast extracts. (c) Interactions between the NPC and chromatin. Proteins present at Coomassie blue–detectable levels in Nup2p, Nup60p, and/or Prp20p immunopurifications are connected by solid black lines. Other known physical and yeast two-hybrid interactions are shown by dotted and dashed lines, respectively (Dingwall et al., 1995; Rexach and Blobel, 1995; Denning et al., 2001; Feuerbach et al., 2002). High coverage tandem MS (MS/MS) of immunopurification eluates from Nup2p, Prp20p, and Nup60p,as well as Kap95p and Nup49p (as controls) was also performed. The inset list (Eluate MS/MS) shows proteins present exclusively in Nup2p eluates (top) or Prp20p eluates (bottom) and those present in both eluates (middle). (d) Nucleosomes associated with Prp20p and Htz1p possess unique acetylation patterns suggestive of boundary chromatin. The acetylation levels of residues K5, K8, K12, and K16 of histone H4 were quantified by mass spectrometry for global (white), Prp20p-associated (dark gray), and Htz1p-associated (light gray) nucleosomes.
To further investigate the hypothesis that Prp20p can serve as a chromatin anchor for Nup2p to elicit BA, we performed high coverage tandem mass spectrometry on trypsin-digested eluates from Nup2p, Prp20p, Nup60p, Kap95p, and Nup49p immunopurifications. This approach does not depend on Coomassie blue visualization of copurifying proteins and is, therefore, capable of identifying proteins present at low (substoichiometric) levels (Marelli et al., 2004). Relevant proteins absent in Nup60p, Kap95p, and Nup49p eluates but present specifically in Nup2p eluates, Prp20p eluates, or both Nup2p and Prp20p eluates are shown in Fig. 1 c (inset table). The identification of Nup1p and Nup60p in Nup2p eluates underscores the sensitivity of this technique: all three nups are located at the nuclear basket, yet our past methodologies failed to detect these proteins with Coomassie blue in Nup2p immunopurifications (Dilworth et al., 2001). In support of the association of Nup2p and Prp20p with chromatin, both proteins yielded Rvb1p, Rvb2p, and Act1p, which are components of several DNA remodeling complexes, including the recently identified SWR-C complex (Krogan et al., 2003). Several additional proteins involved in chromatin remodeling were detected specifically with Prp20p. Among these, the histone 2A variant, Htz1p, has been shown to be recruited to chromatin by the SWR-C complex, and it is believed to act as a boundary factor in yeast (Adam et al., 2001; Meneghini et al., 2003). Interestingly, Prp20p not only copurified with the Ran homologue Gsp1p, but also with the nonessential counterpart, Gsp2p. Notably, Gsp2p, but not Gsp1p, was identified along with Nup2p in the boundary trap screen performed by Laemmli's group (Ishii et al., 2002). These data lend credence to hypothesis that Prp20p is involved in endogenous boundary function.
Histone modification patterns of Prp20p-associated nucleosomes are typical of neither classically silent nor active chromatin
Active and silent chromatin are associated with nucleosomes that exhibit unique histone acetylation patterns, which can be quantified (Smith et al., 2003), and methylation patterns that can be analyzed qualitatively, by mass spectrometry (van Leeuwen et al., 2002). In particular, hypoacetylation at lysine residues K5, K8, K12, and K16 of histone H4 correlates with silent DNA (Kurdistani et al., 2004), and hypomethylation at residue K79 of histone H3 has been postulated to predominate in this minor fraction of yeast chromatin (van Leeuwen et al., 2002). We investigated the modification states present in Prp20p-associated histones. Because of the known role of Htz1p in chromatin remodeling, we performed the same analysis with Htz1p-associated histones. Htz1p and Prp20p were immunopurified from cell extracts in the presence of butyric acid to inhibit deacetylation and we compared the acetylation and methylation patterns of histones in these eluates to those of histones derived from whole-cell lysates. Htz1p and Prp20p yielded similar profiles (Fig. 1 d): relative to bulk histones, histones associated with Prp20p and Htz1p were highly hypoacetylated at residues K5 and K8 of histone H4 (10–20% of bulk histone H4 levels at this residue), suggestive of heterochromatin. However, in Prp20p and Htz1p eluates, histone H4 was only modestly hypoacetylated at residues K12 and K16 (50–60% of bulk histone H4 levels at this residue) and histone H3 exhibited a high level of methylation at residue K79 of histone H3 (∼80% cumulative mono-, di-, and tri-methylated at K79; unpublished data), which together are suggestive of active chromatin (van Leeuwen et al., 2002; Kurdistani et al., 2004). This mixed phenotype, being characteristic of neither silent nor active DNA (Tackett et al., 2005), is consistent with Prp20p and Htz1p binding to DNA regions near chromosome boundaries, where active and silent DNA converge and, furthermore, supports the premise that Htz1p and Prp20p bind to similar chromatin regions. Alternatively, these results could be explained if these proteins interact with several species of differentially modified histones; however, we believe this to be unlikely given that the modification patterns observed for Prp20p and Htz1p are strikingly similar to those obtained for Dpb4p, a protein localized to chromatin boundaries and involved in their maintenance (Iida and Araki, 2004; Tackett et al., 2005).
Prp20p harbors BA
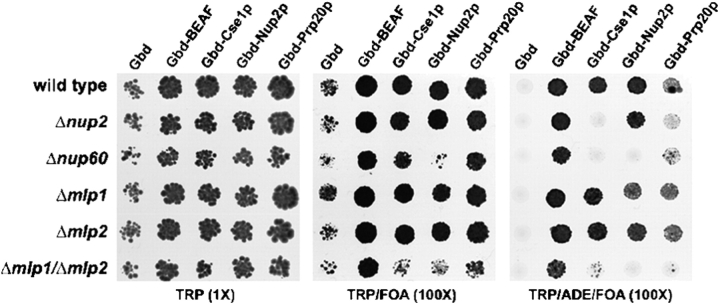
Our analysis of interactions made by Nup2p led us to hypothesize that Nup2p BA is mediated by its interaction on the one hand with Nup60p at the NPC, and on the other hand with Prp20p on chromatin. Using the boundary trap assay (Ishii et al., 2002), we examined if Nup2p-mediated BA was dependent on NPC association, and whether Prp20p possesses its own BA. In this assay, two genes (URA3 and ADE2) are placed within a partially derepressed HML locus with the ADE2 gene flanked by DNA sequences that bind Gal4p (UASg sequence). Boundary activity, such as that exhibited by the control Drosophila protein BEAF, is detected by the ability of proteins fused to the DNA binding domain of Gal4p (Gbd) to allow the expression of the ADE2 marker while maintaining the adjacent URA3 gene in an “OFF” state. This activity is interpreted as a selective insulation of the ADE2 gene from the surrounding silenced chromatin resulting from the binding of the Gbd chimera to the UASg sites flanking ADE2 (Ishii et al., 2002). The BA of Nup2p (and Cse1p) was strictly dependent on its ability to bind to the NPC through Nup60p (Fig. 2). We also found that Prp20p elicited BA at frequencies comparable to those of Gbd-Nup2p and Gbd-Cse1p and, in addition, full activity required Nup2p and Nup60p. It should be noted, however, that a low amount of BA was still detected for Gbd-Prp20p in the absence of Nup2p or Nup60p, which we interpret to be due to the ability of Prp20p to bind to several other components of the nucleocytoplasmic machinery (Floer et al., 1997; Noguchi et al., 1997; Solsbacher et al., 2000; Akhtar et al., 2001), which could offer alternative (albeit less efficient) associations with the NPC. In further support of the pivotal role of Nup2p in BA, neither Nup60p nor Htz1p were active in the boundary trap assay (unpublished data), indicating that the ability to bind to Prp20p is not sufficient to confer BA, and confirming that BA is not a general feature of nups (Ishii et al., 2002).
Figure 2.
The boundary trap assay confirms the links between the NPC and chromatin-bound Prp20p. The ability of Nup2p to bind to the NPC through Nup60p is required for BA. The boundary trap strain, KIY54, and isogenic Δnup2, Δnup60, Δmlp1, Δmlp2, and Δmlp1/Δmlp2 derivatives expressing plasmids encoding the Gal4 DNA binding domain alone, Gbd (pGBC11) or fused to the COOH-terminal portion of the Drosophila BEAF protein, Gbd-BEAF (pGBC11-BEAF-C), a GFP-tagged portion of Cse1p, Gbd-Cse1p (pGBC11-CSE1[474–960]-GFP), full-length Nup2p, Gbd-Nup2p (pGBC12-NUP2[1–720]) or GFP-tagged Prp20p, Gbd-Prp20p-GFP (pGBC12-PRP20-GFP) were serially spotted onto CSM-TRP (T), CSM-TRP+FOA (TF) and CSM-TRP-ADE+FOA (TAF) to assess boundary function. Cells lacking Nup60p were defective in their ability to silence the URA3 reporter, indicated by a reduced viability on media containing 5-FOA. This phenotype was also shared by two independently isolated double-mutant strains lacking both of the Mlp proteins. BA is indicated by growth on media lacking adenine and containing 5-FOA (TAF). A plasmid encoding only Gbd failed to elicit BA and the positive control fusion, Gbd-BEAF, exhibited BA in all genotypes tested. The BA of Gbd-Cse1p was dependent on Nup2p (Ishii et al., 2002), and BA of both Gbd-Cse1p and Gbd-Nup2p was absent in Δnup60 mutants. Double-mutant Δmlp1/Δmlp2 strains exhibited reduced BA. Gdb-Prp20p possesses BA in wild-type and single Mlp mutant strains at frequencies comparable to transport factors (Ishii et al., 2002). Deletion of NUP2 or NUP60 or both MLP1 and MLP2 resulted in dramatically reduced Gbd-Prp20p BA.
In light of the evidence linking Nup60p with Mlp-dependent silencing, we investigated the requirement of Mlp1p and Mlp2p for BA. Although single MLP1 or MLP2 null mutants showed no defects, we observed a significant decrease in the BA of Gbd-Cse1p, Gbd-Nup2p, and Gbd-Prp20p, but not Gbd-BEAF, in mlp double mutant strains (Fig. 2), suggesting that although not essential for BA, the integrity of the Mlp structure associated with the distal regions of NPC is important for full function. However, we note that even though it was observed in two independently derived double mutant strains, the Δmlp1/Δmlp2 defect was only partially penetrant (∼80%; unpublished data); that is, double-mutant strains occasionally exhibited near normal levels of BA, which can explain why it was initially reported that BA was not dependent on MLP1 and MLP2 (Ishii et al., 2002).
Endogenous NPC-mediated boundary function
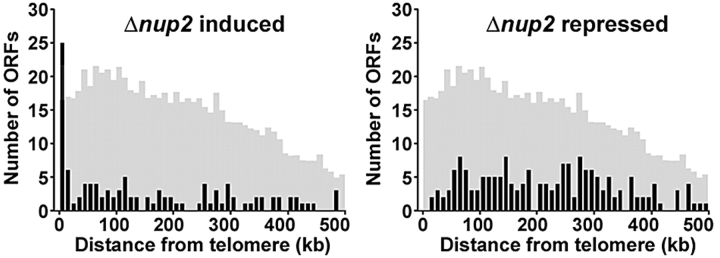
The physical connections between Nup2p, Prp20p, chromatin-modifying proteins, and atypically modified nucleosomes suggest that Nup2p can act as an endogenous boundary factor. Therefore, we predicted that boundaries in Δnup2 cells would not be stably maintained and these cells would exhibit unique transcriptional profiles relative to their wild-type counterparts. To test this hypothesis, we determined the global steady-state mRNA levels in logarithmically growing wild-type and nup2 null cells using DNA microarrays (representing 6,271 yeast ORFs) (Ideker et al., 2001; Smith et al., 2002) and analyzed the chromosomal locations of the top 5% (313) most significantly induced (123) or repressed (190) ORFs in Δnup2 cells. These experiments revealed a striking bias of Δnup2-induced ORFs to subtelomeric regions, whereas repressed ORFs tended toward intrachromosomal sites (Fig. 3). This pattern suggests a steady-state alleviation of telomeric repression in cells lacking Nup2p, supporting a role for Nup2p in maintaining chromosomal boundaries in these regions. We note this phenotype might also arise if Nup2p, like Nup60p, plays a direct role in maintaining the Mlp peripheral silencing apparatus; however, these possibilities are not mutually exclusive and, unlike NUP60, deletion of NUP2 does not alter the localization of Mlp1p nor Mlp2p (unpublished data).
Figure 3.
Genes exhibiting aberrant expression in cells lacking Nup2p map to distinct chromosome regions. For the top 5% of significant Δnup2 induced or repressed ORFs, the distance from each ORF to the nearest telomere was determined. These distances were grouped into 10-kb bins and plotted as a function of telomeric distance. These plots reveal an enrichment of Δnup2-induced ORFs at subtelomeric regions, as 25% of induced ORFs reside within 20 kb of a chromosome end. Only 1% of significantly repressed ORFs were within this distance. The shaded histograms indicate the distribution of telomeric distances for all ORFs plotted at 1/8 scale on the y-axis. Statistical comparison of the Δnup2-induced and repressed distributions to the profile for all ORFs using a two-sample Kolmogorov-Smirnov test confirms that the induced profile is unique (P = 0.0000386), but the repressed profile is not (P = 0.287).
Interestingly, an opposite telomeric bias was observed in Δhtz1 mutants (Meneghini et al., 2003; Mizuguchi et al., 2004), which exhibited subtelomeric enrichment of repressed genes. Htz1p functions in maintaining chromosomal boundaries by promoting the spread of activation near these regions (Adam et al., 2001); thus, our data suggest an opposing role for Nup2p at boundaries—either promoting repression or antagonizing activation. Importantly, the reciprocal nature of the transcriptional profiles of Δnup2 and Δhtz1 cells together with the activity of Nup2p in the boundary trap assay suggest that the bias of induced ORFs to telomeric regions is a consequence of a role for Nup2p in chromatin organization, rather than an indirect manifestation of weak nucleocytoplasmic transport defects (Booth et al., 1999; Solsbacher et al., 2000).
We also note that the magnitude of the expression changes for most of the aberrantly expressed ORFs was less than twofold. This is expected if Nup2p is involved in boundary maintenance rather than absolutely required for the formation of boundaries. As such, we would predict that only a subset of cells in a given Δnup2 population would experience a stochastic breakdown of boundaries (or alleviation of repression) in subtelomeric regions, leading to the slight overall increase in gene expression observed.
The localization of Prp20p and Nup2p along chromatin correlates with ORFs induced in Δnup2 cells
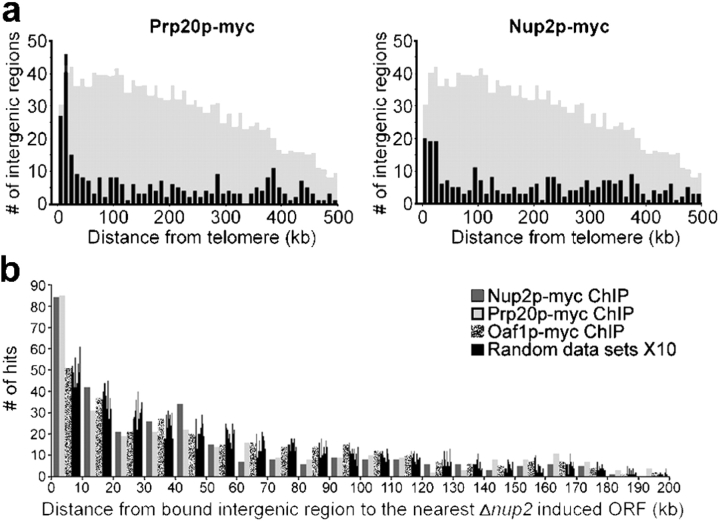
To further examine the role of Prp20p and Nup2p in endogenous NPC-mediated BA, we performed genome localization studies by chromatin immunopurification microarray (ChIP-CHIP) using myc-epitope tagged Prp20p and Nup2p and microarrays containing 6,081 yeast intergenic regions (Ren et al., 2000). Because the ChIP-CHIP procedure involves cross-linking proteins to DNA followed by PCR amplification of bait-associated DNA fragments, we were able to perform these experiments with myc-tagged Nup2p despite the inability of this protein to stably associate with DNA during standard immunopurification procedures (Nup2p-myc yielded ∼50-fold less DNA than Prp20p-myc as determined by SyBr Green fluorimetry; unpublished data). Strikingly similar to the telomeric bias of induced ORFs observed in Δnup2 cells, the top 5% of Prp20p- and Nup2p-enriched intergenic regions (304 targets) clustered together and enriched near telomeres (compare Fig. 4 a with Fig. 3). This telomerically biased distribution was not observed with numerous transcription factors that we have investigated (unpublished data).
Figure 4.
DNA regions bound by Prp20p and Nup2p enrich near telomeres and lie in close proximity to ORFs induced in cells lacking Nup2p. (a) Histograms of minimal telomeric distance for the top 5% of significantly enriched intergenic regions bound by Prp20p and Nup2p reveal a telomeric enrichment similar to that observed for ORFs induced in cells lacking Nup2p (see Fig. 3). The shaded histograms represent the distribution of all intergenic regions shown at 1/4 scale on the y-axis. The Prp20p and Nup2p profiles are significantly distinct from the distribution of all intergenic regions (P < 0.000001 and P = 0.000367, respectively). In contrast, the profile of the transcription factor Oaf1p displayed no significant enrichment relative to all intergenic regions (P = 0.368; not depicted). (b) Chromosomal proximity of transcriptionally induced ORFs in Δnup2 cells and ChIP-CHIP enriched intergenic regions. The distance between each enriched intergenic region and the nearest Δnup2-induced ORF was determined for Nup2p, Prp20p, Oaf1p, and 10 randomized datasets. Intergenic regions bound by Nup2p and Prp20p are found much closer to ORFs induced in cells lacking Nup2p, relative to the Oaf1p or randomized datasets, as evidenced by the high number of Nup2p and Prp20p enriched intergenic regions found within 10 kb of Δnup2 ORFs. The Kolmogorov-Smirnov test reveals significant differences between the Oaf1p profile and those obtained with Prp20p and Nup2p (P = 0.0282 and P = 0.00102, respectively).
The Nup2p and Prp20p ChIP-CHIP datasets exhibited a 16% exact match correlation (48 out of 304) for the top 5% most significantly enriched intergenic regions. The probability of this overlap occurring by chance alone is 3.1 × 10−13 (calculated by hypergeometric distribution analysis) (Smith et al., 2002), which supports the hypothesis that Nup2p and Prp20p bind to chromatin at similar regions. In contrast to our ChIP-CHIP studies, a genome localization analysis of several nucleocytoplasmic transport machinery components, which included Nup2p and Prp20p, revealed no obvious enrichment at telomeres (not depicted; Casolari et al., 2004). However, given that Casolari et al. (2004) performed their experiments with ORF spotted microarrays while we used arrays containing intergenic regions, these datasets cannot be directly compared with one another as they are representative of distinct chromatin environments (coding vs. noncoding DNA), which might provide an explanation of the observed discrepancy (Hanlon and Lieb, 2004). Indeed, ORF spotted microarrays also did not detect a telomeric bias for the Mlp proteins (Casolari et al., 2004), which might be expected given that others have shown that the Mlp proteins tether telomeres to the nuclear periphery through an interaction with Yku70p, which associates with chromosome ends (Galy et al., 2000). However, using intergenic microarrays we did observe telomeric enrichment for Mlp1p and Mlp2p ChIP-CHIP datasets (unpublished data), suggesting that although these discrepancies remain unresolved, the experimental protocols target different aspects of protein function. Given that it was more recently shown that chromatin can be anchored to the nuclear periphery by both DNA- and RNA-dependent mechanisms (Casolari et al., 2005), one possibility is that the use of ORF spotted microarrays biases toward detection of transcriptionally dependent RNA anchors, whereas intergenic arrays bias toward detection of DNA-dependent anchors, such as those thought to be used in NPC-dependent chromatin boundaries.
To test for a correlation between our Δnup2 mRNA expression microarrays and results from our ChIP-CHIP studies, we calculated the chromosomal distance between each Nup2p or Prp20p ChIP-CHIP enriched intergenic region and the closest Δnup2 induced or repressed ORF. Relative to ChIP-CHIP results using an unrelated control protein, Oaf1p, as well as randomized datasets, Nup2p and Prp20p ChIP-CHIP enriched intergenic sites lie in close proximity to Δnup2 induced ORFs (Fig. 4 b), whereas comparisons made to Δnup2 repressed ORFs did not reveal a bias toward close proximity (not depicted).
Genetic interactions support a link between Prp20p, Nup2p, Nup60p, and Htz1p
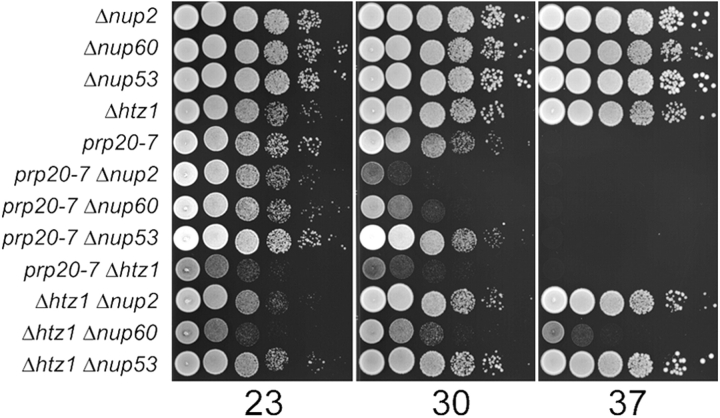
NUP2 has previously been shown to interact genetically with PRP20 and NUP60 (Booth et al., 1999; Dilworth et al., 2001). Given the requirement of Nup60p for the NPC association of Nup2p and the telomeric biases of aberrantly expressed ORFs in Δnup2 and Δhtz1 cells, we tested for genetic interactions between these genes. The growth rates of strains harboring single- and double-mutant combinations of NUP2, NUP60, PRP20, and HTZ1 at various temperatures were tested. NUP53 (Marelli et al., 1998) was included as a negative control. As shown in Fig. 5, the temperature-sensitive allele of PRP20, prp20-7, conferred increased sensitivity when combined with null mutations of NUP2, NUP60, or HTZ1, as these double-mutant strains, but not double-mutants involving NUP53, grew poorly at 30°C. As a confirmation of specificity, we obtained similar results using another temperature-sensitive allele of PRP20, srm1-1 (Clark and Sprague, 1989), with the sole exception that the genetic interaction with HTZ1 was very weak in the srm1-1 background (unpublished data). In addition, we detected HTZ1-NUP2 and HTZ1-NUP60 genetic interactions, which support functional relationships between Nup2p-interacting proteins and the activity of Htz1p. We note, however, that the HTZ1-NUP2 interaction is rather weak as it is readily evident only at 23°C, showing only a modest growth defect at 30°C and none at 37°C.
Figure 5.
Genetic interactions support links between NUP 2, NUP60, PRP20, and HTZ1. Growth rate analysis of Δhtz1, Δnup2, Δnup60, Δnup53, and prp20-7 single-mutant and relevant double-mutant strains at 23, 30, and 37°C. Double-mutant prp20-7 Δhtz1, Δhtz1 Δnup2, Δhtz1 Δnup60, prp20-7 Δnup2, and prp20-7 Δnup60 strains all exhibited more severe growth defects than those detected in their parental strains, whereas double-mutant combinations involving deletion of NUP53 revealed no genetic interactions.
Nup2p is required for the efficient maintenance of telomeric silencing
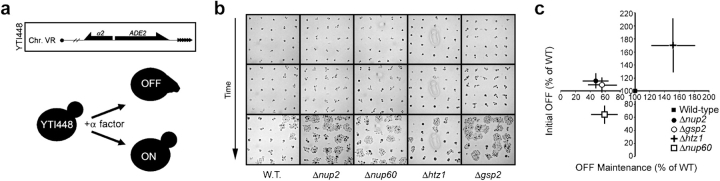
Genes within subtelomeric regions are capable of epigenetically switching between transcriptionally active (“ON”) and silent (“OFF”) states (Gottschling et al., 1990). Our transcriptomic data suggest that Nup2p plays a role in the maintenance of subtelomeric gene expression states. To test this, we used a single-cell telomeric silencing assay that detects expression variegation in individual cells based on their response to mating pheromone (Iida and Araki, 2004). When grown in the presence of α-factor, the vast majority of yeast a-type cells (MATa) arrest in G1 before START and form pseudopod-like projections, termed shmoos. Diploid a/α cells (MATa/MATα) and α cells (MATα) are insensitive to α-factor and therefore continue to divide in its presence. However, if haploid MATa cells express the normally silenced α2 gene, they are insensitive to α-factor. Exploiting this phenomenon, Iida and Araki (2004) incorporated a subtelomeric copy of the α2 gene in a MATa background, allowing the variegated expression of the α2 gene to be determined in single cells by their response to α-factor—cells in the ON state are insensitive and continue to divide, whereas cells in the OFF state arrest (Fig. 6 a). Consequently, by monitoring the ability of individual cells to maintain the arrested or insensitive phenotype over a time course, the OFF to ON switching rate can be determined (Fig. 6 b).
Figure 6.
Loss of Nup2p or Gsp2p results in subtelomeric gene silencing defects that are distinct from those observed for strains lacking Nup60p or Htz1p. (a) The expression status of a telomerically encoded α2 reporter gene was assayed in single cells by monitoring the response of cells to the α-factor. Cells that do not express α2 (OFF) respond to α-factor arrest in G1 and shmoo; cells expressing α2 (ON) continue to bud and divide. The initial OFF proportion was determined by scoring cells after a 4-h treatment with α-factor. (b) Determination of OFF maintenance ratio. α2 OFF maintenance was assayed by monitoring α-factor arrested cells over time. Cells that have switched to the ON state give rise to microcolonies, whereas stably arrested cells do not divide. (c) The wild-type normalized initial OFF ratios (y-axis) and OFF maintenance ratios (x-axis) were plotted for each strain (error bars indicate the SD for three independent experiments). Strains lacking Nup2p or Gsp2p exhibited very similar phenotypes (marginally increased initial OFF ratios [y ≥ 1], and poor OFF state maintenance [x < 1]). Cells lacking Nup60p exhibited a steady-state defect in the establishment of the OFF state and an inability to maintain the OFF state (x and y < 1), whereas cells lacking Htz1p showed the opposite phenotype (x and y > 1).
Using this assay, we analyzed strains lacking Nup2p, Nup60p, Htz1p, and Gsp2p. Δhtz1 and Δnup60 cells showed significantly increased and decreased rates in the initial OFF state, respectively (Fig. 6 c). These data support the proposed function of Htz1p in establishing the active chromatin state (Galy et al., 2000; Feuerbach et al., 2002; Meneghini et al., 2003; Mizuguchi et al., 2004). In contrast, cells lacking Nup2p or Gsp2p exhibited near wild-type activity in establishing silent chromatin.
Considering the intimate relationship between establishing silent chromatin and BA, it is also plausible that Gsp2p and Nup2p affect both activation and repression to similar extents, in which case deletion of either gene might not change the steady-state distribution of the silenced and active states in a population. To get at this point, we asked whether, once established, these different strains could maintain the silenced or OFF state. Interestingly, Δnup2, Δnup60, and Δgsp2 cells were twice as likely as WT cells to switch back to the ON state, as determined by their ability to reinitiate cell division (Fig. 6 c). By comparison, Δhtz1 cells exhibited a 50% increase in the ability to maintain the OFF state, as expected. In summary, these experiments enabled the detection of the subtle differences in the activities of these factors. Nup2p and Gsp2p were required for the stable maintenance, but not the establishment, of a silenced domain. In contrast, Nup60p and Htz1p are involved in both processes, but while Nup60p promotes silencing, Htz1p promotes activation. Given that Nup2p and Gsp2p harbor BA, but Htz1p and Nup60p do not, these results suggest that a dynamic balance between repression and activation is required for BA. Together with the expression array data, these results support a role for Nup2p in controlling the expression mediated through Nup2p-dependent BA.
Discussion
Convergence of nuclear functions at the NPC
Given that the NPC is one of a limited number of positional markers in the nucleus, it is no surprise that cells have taken advantage of this structure to organize nuclear events. Indeed, yeast nups have been linked to Mlp-dependent gene silencing (Galy et al., 2000; Feuerbach et al., 2002), BA (Ishii et al., 2002), ribosome maturation (Ho et al., 2000), unspliced mRNA retention (Galy et al., 2004), and de novo formation of NPCs and the NE (Marelli et al., 2001). Lying at the crossroads between the nuclear interior and the nuclear periphery, the NPC has long been suggested, but only recently shown, to interact with actively transcribed genes, enabling ready access of transcription factors to specific sites of the genome and, likewise, newly transcribed mRNA to the cytoplasm—a function termed “gene gating” (Blobel, 1985; Casolari et al., 2004). Beyond these interpretations, we further propose that the NPC plays an active role in chromatin organization by facilitating the transition of chromatin between activity states, which as previously proposed, are partially imposed by three-dimensional position of a gene within the nucleus (Galy et al., 2000; Gartenberg et al., 2004).
Dynamic model of endogenous NPC-mediated boundary function
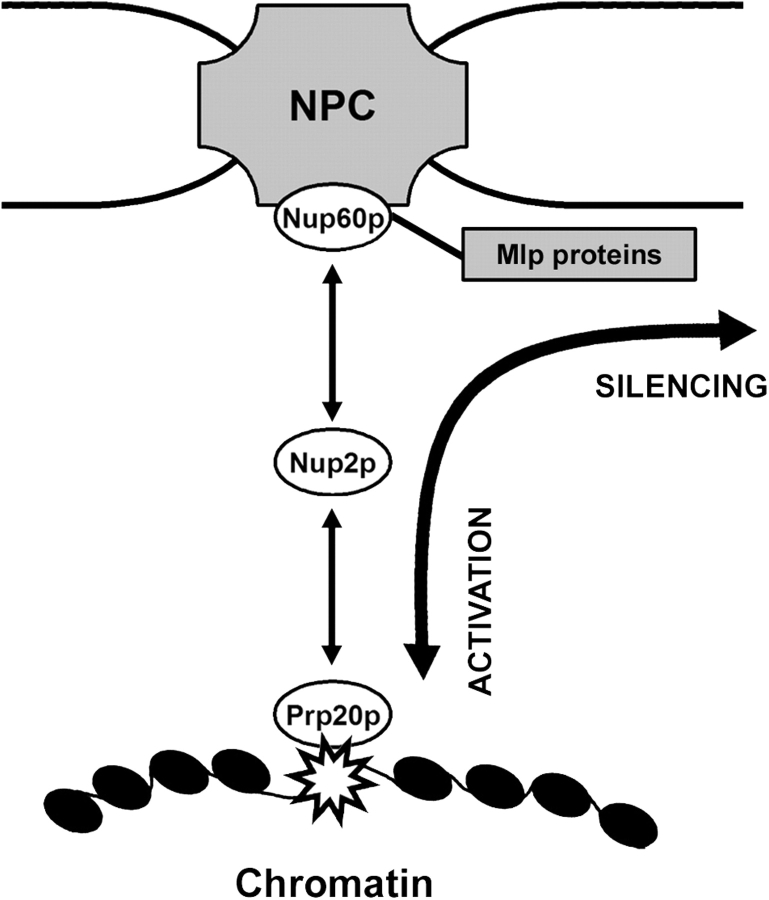
In developing a model of endogenous NPC-mediated BA, we must consider the high molecular burden placed on NPCs. Our previous work on Nup2p led us to propose that it provides a soluble scaffold to decrease the residence time of transport complexes at the NPC, facilitating transport and relieving congestion at NPCs. Extending this paradigm to Nup2p-mediated BA, we propose that DNA boundaries utilize NPCs as waypoints, not stable anchors, en route to subnuclear compartments that promote different transcriptional states. Once associated with the nuclear face of the NPC, chromatin can enter the peripheral silencing apparatus (a process proposed to involve the Mlp proteins [Galy et al., 2000; Feuerbach et al., 2002]), be selectively activated at the periphery (Casolari et al., 2004), or be released to freely diffuse to the nuclear interior. We propose that the mobility of Nup2p is the key to this BA, as no other nups harbor this activity and all the transport factors that exhibit BA transiently associate with NPCs (Ishii et al., 2002). Thus, we envision that Nup2p promotes transcriptional plasticity by effectively “greasing the wheels” of DNA movement to and from NPCs.
Previous models of NPC-mediated BA incorporated a static mechanism involving the stable anchoring of chromatin to the nuclear basket of the NPC, but several observations argue against this. For example, the static model predicts that nucleoporins associate with both active and silenced ORFs (on either side of a chromosomal boundary); yet, nups are found to enrich specifically with active ORFs (Casolari et al., 2004). A dynamic model explains why Nup2p is unique—it is not stably associated with the NPC—and might also provide an explanation of the lack of correlation between our ChIP-CHIP datasets and those put forth by Casolari et al. (2004). That is, differences in the rates of association, or residence times, of coding and noncoding regions of chromatin with NPCs could give rise to unique association patterns when using ORFs or intergenic microarrays. In other words, if actively transcribed chromatin associates more avidly with NPCs than inactive chromatin, then highly transcribed ORFs would be more likely to cross-link to this structure during the ChIP-CHIP procedure.
Endogenous NPC-mediated BA also requires a mechanism for chromatin to associate with the NPC, and we have presented several lines of evidence indicating that the Prp20p–Nup2p interaction provides this critical link. In vitro binding data reveal a robust interaction between Prp20p and Nup2p, but immunopurification and localization data suggest that it is transient in vivo. Thus, we suggest that chromosomal regions interacting with Prp20p attain an equilibrium distribution between subnuclear regions that promote active and silent states, and that these transitions are facilitated through Nup2p-dependent associations with the NPC (Fig. 7). An alternative but not mutually exclusive interpretation of the data is that rather than recruit chromatin to the NPC, Nup2p targets factors important for boundary function from the NPC to specific chromatin sites, thus contributing to boundary maintenance without the requirement of NPC association.
Figure 7.
Dynamic model of NPC-mediated BA. Boundaries (star), marked by Prp20p, are proposed to be mobile but spatially restricted within the nucleus due to their transient Nup2p-dependent association with NPCs. The complexation of DNA with the NPC represents an unstable reaction intermediate from which the DNA can either enter the perinuclear silencing region through Nup60p or detach from the NPC, free to enter the nuclear interior.
By linking the function of NPCs in BA with their role in Mlp-dependent silencing, this model also explains the defect in BA that we observed in cells lacking the Mlp proteins, as without them, regions marked for silencing are unable to efficiently enter the peripheral silencing matrix (Galy et al., 2000; Feuerbach et al., 2002). It also agrees with the observation by Laemmli's group that simply tethering DNA to the nuclear periphery is not sufficient to establish boundaries in the boundary trap assay (Ishii et al., 2002). This dynamic aspect of chromatin also agrees with observations by Heun et al. (2001) that telomeric chromatin is predominantly constrained to perinuclear regions, yet is still subject to occasional large movements into the nuclear interior.
Interestingly, Prp20p was originally identified as SRM1, a suppressor of the mating defect in MATα cells that lack STE3, which encodes the a-factor receptor (Clark and Sprague, 1989). A temperature-sensitive allele of Prp20p (srm1-1) partially restored the ability of MATα Δste3 cells to mate with cells of the opposite mating type, indicating that these mutants underwent the mating response despite their insensitivity to α-factor. In light of the data implicating Prp20p in chromatin organization, it is possible that the inability of strains harboring srm1-1 to efficiently maintain chromosome boundaries results in the aberrant expression of normally silenced genes involved in the mating response, thereby allowing cells to mate in the absence of the pheromone receptor.
The chromatin connection—links to nucleocytoplasmic transport
A major unanswered question is how transport complexes are specifically targeted to regions of DNA and how this might be related to chromatin organization. Perhaps the guanylyl-nucleotide exchange factor activity of Prp20p or its association with nucleosomes, which has been shown to be transient in higher eukaryotes (Cushman et al., 2004), is modulated dependent on its heterochromatic or euchromatic localization, thereby providing a function in both gene activation and silencing. Although speculative, our data provide preliminary evidence for the involvement of Htz1p and Gsp2p with Prp20p in these processes. Eukaryotic cells use a variety of means to epigenetically regulate gene expression, including the exploitation of the three-dimensional architecture of the nucleus. Understanding the mechanisms by which the NPC, its associated factors, and nuclear transport contribute to these functions is fundamental to a comprehensive view of how cells express their genome.
Materials and methods
Strains and plasmids
For immunopurification studies, strains encoding genes of interest tagged COOH terminally with Staphylococcus aureus protein A (prA) were generated in the DF5 background (ura3-52 trp1-901 leu2-3,112 his3-200) by previously described techniques (Aitchison et al., 1995; Rout et al., 2000). For in vivo formaldehyde cross-linked immunopurifications, a strain expressing Nup2p-prA and Prp20p-myc from their respective endogenous promoters was created using the Nup2p-prA parental strain by homologous recombination of a Prp20p-specific PCR product generated with the plasmid pFA6a-13Myc-kanMX6 and Prp20p-specific primers as previously described (Longtine et al., 1998). Genetic interaction studies were performed with G418 (Fisher Scientific) selectable Δnup2, Δnup60, Δnup53, and Δhtz1 null mutant strains (MATα geneX::kanMX4) contained within the S. cerevisiae Deletion Project library (Invitrogen), the temperature-sensitive M316/1A strain (MATa prp20-7) (Amberg et al., 1993), and a nourseothricin (Werner Bioagents) selectable Δhtz1 strain (MATa htz1::natMX4), derived by replacing the KAN R cassette in the S. cerevisiae Deletion Project HTZ1/htz1::kanMX4 diploid strain with a NAT R cassette followed by sporulation and tetrad dissection to isolate a MATa htz1::natMX4 haploid. Boundary trap strains were generated in the KIY54 background (MATa leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 hml-EΔi-UASg-ADE2-UASg-URA3), which, along with a nup2::kanMX4 derivative (YGA2), were gifts of Ulrich Laemmli (University of Geneva, Geneva, Switzerland; Ishii et al., 2002). Other single-deletion mutants of KIY54 were created by homologous recombination using G418 or nourseothricin selectable deletion cassette PCR products generated from genomic DNA of corresponding mutants in the S. cerevisiae Deletion Project library or nourseothricin-switched derivatives thereof. In a like manner, two independent Δmlp1/Δmlp2 double mutants were generated by sequential integration of mlp1::kanMX4 followed by mlp2::natMX4 or mlp1::natMX4 then mlp2::kanMX. All deletions were confirmed by gene-specific PCR. Expression arrays were performed using the wild-type strain, BY4742 ( MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and its nup2::kanMX4 derivative. The parental strain used for subtelomeric gene expression variegation studies, YTI448 (MATa bar1Δ::hisG ade2Δ::hisG can1-100 his3-11,15 leu2-3 trp1-1 ura3-1 VR:: α2ADE2-TEL), was a gift of Hiroyuki Araki (National Institute of Genetics, Mishima, Japan; Ishii et al., 2002), and null mutants were obtained by homologous recombination as described above. For chromatin localization studies, myc-tagged versions of Prp20p, Nup2p, and Oaf1p were created in the BY4742 background using the plasmid pFA6a-13Myc-kanMX6 (Longtine et al., 1998).
Plasmids used to genomically tag genes allowing expression of in-frame COOH-terminal fusions to S. aureus prA were described previously (Aitchison et al., 1995; Rout et al., 2000). The GST fusion plasmid pGST-PRP20 was a gift of Michael Rexach (University of California, Santa Cruz, Santa Cruz, CA). A PCR product encoding NUP2 generated from yeast genomic DNA using oligonucleotides containing flanking BamHI and EcoRI restriction sites was ligated in-frame with GST into the bacterial expression vector pGEX-2TK (GE Healthcare) to create pGST-NUP2. The pGST-CFP plasmid was created by ligating the SalI-NotI fragment containing the ECFP gene from pECFP (BD Biosciences) into pGEX-4T1 (GE Healthcare) to create the parent vector pGEX-4T1-ECFP in which the ECFP gene was frame-shifted −1 relative to the GST reading frame. This initial clone was then linearized and blunted at the XbaI site to generate in-frame clones, which were screened for GST-CFP expression using an Axiophot fluorescence microscope (Carl Zeiss MicroImaging, Inc.). To generate pGST-NUP2-CFP, the NUP2 gene was PCR amplified from genomic DNA using oligonucleotides containing EcoRI sites and a +1 frame-shift encoded in the reverse primer such that, after ligation into pGEX-4T1-ECFP, a GST-NUP2-CFP is expressed. When required, G418-selectable null mutants were switched to nourseothricin selection using the plasmid p4339 (pCRII-TOPO::MX4-natR), a gift of Charles Boone (University of Toronto, Toronto, Ontario, Canada; Tong et al., 2001). The plasmids pGBC11, pGBC11-BEAF-C, pGBC11-Cse1[474–960]-GFP, and pGBC12-NUP2[1–720] used in the boundary trap assays were provided by Ulrich Laemmli and pGBC12-PRP20-GFP, pGBC12-NUP60-GFP, and pGBC12-HTZ1-GFP plasmids were constructed in a like manner (Ishii et al., 2002).
Purification of prA-tagged chimeras from yeast whole-cell extracts and identification of copurifying proteins
Immunopurifications of Nup2p, Nup60p, Nup49p, Prp20p, and Kap95p were performed as described previously (Dilworth et al., 2001) with the following modifications. For cell disruption, samples were passed seven times through a microfluidizer (model M-110S; Microfluidics). In place of IgG sepharose, we used M-280 tosylactivated Dynabeads (Dynal) conjugated to rabbit affinity-purified antibody to mouse IgG (Cappel) by manufacturer's suggested protocols. Rather than elute bound proteins over a magnesium gradient, beads were washed 10 times with 4 ml of wash buffer containing 50 mM MgCl2 eluted with 0.1% SDS prewarmed to 42°C and TCA precipitated. Protein pellets were resolved by SDS-PAGE and Coomassie blue visible bands were excised and identified by LC-MS/MS using standard techniques (Eng et al., 1994). Alternatively, protein pellets were resuspended in 100 μl 50 mM ammonium bicarbonate, pH 9.0, containing 2 ng/μL porcine sequencing grade trypsin (Promega), incubated at 37°C overnight, and dried in a speed-vac at which point peptides were identified by LC-MS/MS. Immunopurifications from formaldehyde cross-linked cells were performed as described above, except that log-phase cultures were treated with formaldehyde for 15 min (1% final concentration) before harvesting and lysis. Histone acetylation and methylation levels were determined as described previously (van Leeuwen et al., 2002; Smith et al., 2003; Tackett et al., 2005).
GST binding experiments
Expression and purification of GST fusion proteins and in vitro binding experiments were performed as described previously (Lee and Aitchison, 1999). The ability of bacterially expressed and purified fusions to associate with the Prp20p–nucleosome complex was determined by two complementary methods. First, the Prp20p-prA immunopurification procedure was repeated as above, but the complex was not eluted from the dynabeads; rather, the beads were washed twice with transport buffer (Lee and Aitchison, 1999), divided into equal fractions, and incubated for 30 min at room temperature with the indicated GST fusion proteins in transport buffer. The unbound fraction was collected and the beads were washed four times with transport buffer, and were then eluted with SDS-PAGE sample buffer to obtain the bound fraction. Samples were resolved by SDS-PAGE, transferred to nitrocellulose, and GST-containing proteins and Prp20p-prA were identified by immunoblotting using a monoclonal mouse antibody directed against GST (Sigma-Aldrich). In the second experiment, GST-CFP and GST-Nup2p-CFP were immobilized on glutathione-Sepharose beads and incubated with yeast lysates prepared from strains expressing Prp20p-prA, which we detected in bound and unbound fractions by immunoblotting with rabbit affinity-purified antibody to mouse IgG (Cappel).
Boundary trap assay
These experiments were performed as described previously (Ishii et al., 2002).
mRNA expression arrays and ChIP-CHIPs
Expression microarray analyses were performed as described previously (Smith et al., 2002) using arrays composed of spotted oligonucleotides (70 bases) representing each yeast ORF, except that spot finding and quantitation were performed using AnalyzerDG (MolecularWare, Inc.). ChIP-CHIP experiments were performed as described previously (Ren et al., 2000) up to and including microarray hybridization and washing. Protocols can be found at http://jura.wi.mit.edu/young_public/regulatory_network/Location_analysis_protocol.pdf. We used in-house generated arrays composed of spotted PCR products between 60 and 1,500 bp in length. Each PCR product covers a single intergenic region in entirety, except in instances where an intergenic region is >1,500 bp in length, in which case multiple PCR products were obtained in order to cover the entire intergenic region. The remaining stages of ChIP-CHIP analyses were performed as for mRNA expression microarrays (Smith et al., 2002). Significant data from at least three independent microarrays were analyzed using Excel (Microsoft) to assess telomeric biases (see supplemental data). The statistical significance of observed differences in histogram plots were assessed using the two-sample Kolmogorov-Smirnov test, which, with a large enough sample, is able to detect any difference between the two population distributions from which the samples were chosen, based on the maximum vertical distance between the two sample cumulative distribution functions (Hollander and Wolfe, 1973).
Genetic interactions
Double-mutant strains were created by crossing G418-selectable MATα null strains from the S. cerevisiae Deletion Project library either to a MATa strain harboring the temperature-sensitive allele of PRP20, prp20-7, or to a nourseothricin-selectable MATa Δhtz1 strain. In crosses involving prp20-7, diploid cells were selected by growth at 37°C on media containing G418, while crosses involving Δhtz1 were selected by growth at 30°C on media containing G418 and nourseothricin. Diploid strains were sporulated and tetrads were dissected by standard techniques. For each combination, four double-mutant spores were isolated and analyzed along with parental single mutants by serially spotting overnight cultures grown at 23°C in rich (YPD) media onto YPD plates in triplicate and incubating for 72 h at 23°C, 60 h at 30°C or 48 h at 37°C to allow colony formation.
Single-cell TPE assays
Log-phase cultures pregrown at 30°C in liquid YPD supplemented with 40 mg/l adenine (YPDA), were treated with 3 μg/ml α-factor for 4 h to allow shmoo formation. The initial proportion of shmooing cells was determined by visual scoring of at least 100 cells and then cultures were diluted 1:100 in sterile water, sonicated briefly in a water bath sonicator and spotted onto YPDA containing 3 μg/ml α-factor (YPDAα) for OFF maintenance studies, which involved arraying grids containing 16–30 shmooing cells for each genotype on YPDAα using a dissection microscope (Eclipse model E-400; Nikon) and periodic visualization over 20 h. For both initial OFF and OFF maintenance experiments, the data were normalized to wild-type controls to account for run to run variability.
Online supplemental material
Fig. S1 shows results from gradient fractionation experiments, which revealed that Nup2p is competent to bind to nucleosome associated Prp20p Fig. S2 shows evidence of an in vivo interaction between Nup2p and Prp20p obtained from immunopurification studies using cross-linked cell lysates. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200509061/DC1.
Acknowledgments
We thank U.K. Laemmli, H. Araki, G. Sprague, C. Boone, C. Cole, M. Rexach and J. Young for strains and plasmids used in this study and M. Rout and T. Galitski for thoughtful discussion. We also wish to acknowledge the expert technical assistance of B. Marzolf and M. Johnson of the ISB array facility
This research was supported by operating and salary support from Canadian Institutes of Health Research and Alberta Heritage Foundation for Medical Research to J.D. Aitchison, D.J. Dilworth, J.J. Smith, and R.W. Wozniak; and from the National Institutes of Health to B.T. Chait (RR00862), J.D. Aitchison, and B.T. Chait (RR022220) and A.J. Tackett (GM066496).
Abbreviations used in this paper: BA, boundary activity; ChIP-CHIP, chromatin immunopurification microarray; Gbd, Gal4p DNA binding domain; NE, nuclear envelope; NPC, nuclear pore complex; Nup, nucleoporin; prA, protein A.
References
- Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi, M., M.W. Clark, U. Vijayraghavan, and J. Abelson. 1990. A yeast mutant, PRP20, altered in mRNA metabolism and maintenance of the nuclear structure, is defective in a gene homologous to the human gene RCC1 which is involved in the control of chromosome condensation. Mol. Gen. Genet. 224:72–80. [DOI] [PubMed] [Google Scholar]
- Aitchison, J.D., M.P. Rout, M. Marelli, G. Blobel, and R.W. Wozniak. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J. Cell Biol. 131:1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, N., H. Hagan, J.E. Lopilato, and A.H. Corbett. 2001. Functional analysis of the yeast Ran exchange factor Prp20p: in vivo evidence for the RanGTP gradient model. Mol. Genet. Genomics. 265:851–864. [DOI] [PubMed] [Google Scholar]
- Amberg, D.C., M. Fleischmann, I. Stagljar, C.N. Cole, and M. Aebi. 1993. Nuclear PRP20 protein is required for mRNA export. EMBO J. 12:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E.D., A.M. Neiman, D.C. Zappulla, and R. Sternglanz. 1998. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 394:592–595. [DOI] [PubMed] [Google Scholar]
- Blobel, G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, J.W., K.D. Belanger, M.I. Sannella, and L.I. Davis. 1999. The yeast nucleoporin Nup2p is involved in nuclear export of importin alpha/Srp1p. J. Biol. Chem. 274:32360–32367. [DOI] [PubMed] [Google Scholar]
- Casolari, J.M., C.R. Brown, S. Komili, J. West, H. Hieronymus, and P.A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. [DOI] [PubMed] [Google Scholar]
- Casolari, J.M., C.R. Brown, D.A. Drubin, O.J. Rando, and P.A. Silver. 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K.L., and G.F. Sprague Jr. 1989. Yeast pheromone response pathway: characterization of a suppressor that restores mating to receptorless mutants. Mol. Cell. Biol. 9:2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell, M., and S.M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9:199–205. [DOI] [PubMed] [Google Scholar]
- Cushman, I., D. Stenoien, and M.S. Moore. 2004. The dynamic association of RCC1 with chromatin is modulated by Ran-dependent nuclear transport. Mol. Biol. Cell. 15:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning, D., B. Mykytka, N.P. Allen, L. Huang, B. Al, and M. Rexach. 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol. 154:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth, D.J., A. Suprapto, J.C. Padovan, B.T. Chait, R.W. Wozniak, M.P. Rout, and J.D. Aitchison. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol. 153:1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall, C., S. Kandels-Lewis, and B. Seraphin. 1995. A family of Ran binding proteins that includes nucleoporins. Proc. Natl. Acad. Sci. USA. 92:7525–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., and R.T. Kamakaka. 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., C.R. Adams, J. Rine, and R.T. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, J., A. McCormack, and J.R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5:976–989. [DOI] [PubMed] [Google Scholar]
- Fawcett, D.W., W. Bloom, and E. Raviola. 1994. A Textbook of Histology. Chapman & Hall, New York. 964 pp.
- Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier, E. Gilson, J.C. Olivo-Marin, H. Scherthan, and U. Nehrbass. 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4:214–221. [DOI] [PubMed] [Google Scholar]
- Floer, M., G. Blobel, and M. Rexach. 1997. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J. Biol. Chem. 272:19538–19546. [DOI] [PubMed] [Google Scholar]
- Fourel, G., E. Revardel, C.E. Koering, and E. Gilson. 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18:2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., F. Magdinier, and E. Gilson. 2004. Insulator dynamics and the setting of chromatin domains. Bioessays. 26:523–532. [DOI] [PubMed] [Google Scholar]
- Galy, V., J.C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. [DOI] [PubMed] [Google Scholar]
- Galy, V., O. Gadal, M. Fromont-Racine, A. Romano, A. Jacquier, and U. Nehrbass. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 116:63–73. [DOI] [PubMed] [Google Scholar]
- Gartenberg, M.R., F.R. Neumann, T. Laroche, M. Blaszczyk, and S.M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell. 119:955–967. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T.I., and V.G. Corces. 2001. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 35:193–208. [DOI] [PubMed] [Google Scholar]
- Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S.M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling, D.E., O.M. Aparicio, B.L. Billington, and V.A. Zakian. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 63:751–762. [DOI] [PubMed] [Google Scholar]
- Hanlon, S.E., and J.D. Lieb. 2004. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr. Opin. Genet. Dev. 14:697–705. [DOI] [PubMed] [Google Scholar]
- Hediger, F., K. Dubrana, and S.M. Gasser. 2002. a. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J. Struct. Biol. 140:79–91. [DOI] [PubMed] [Google Scholar]
- Hediger, F., F.R. Neumann, G. Van Houwe, K. Dubrana, and S.M. Gasser. 2002. b. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12:2076–2089. [DOI] [PubMed] [Google Scholar]
- Heun, P., T. Laroche, K. Shimada, P. Furrer, and S.M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science. 294:2181–2186. [DOI] [PubMed] [Google Scholar]
- Ho, J.H., G. Kallstrom, and A.W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander, M., and D.A. Wolfe. 1973. Nonparametric Statistical Methods. John Wiley & Sons, Inc., New York. 528 pp.
- Ideker, T., V. Thorsson, J.A. Ranish, R. Christmas, J. Buhler, J.K. Eng, R. Bumgarner, D.R. Goodlett, R. Aebersold, and L. Hood. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 292:929–934. [DOI] [PubMed] [Google Scholar]
- Iida, T., and H. Araki. 2004. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U.K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 109:551–562. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., M.C. Keogh, N. Datta, C. Sawa, O.W. Ryan, H. Ding, R.A. Haw, J. Pootoolal, A. Tong, V. Canadien, et al. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 12:1565–1576. [DOI] [PubMed] [Google Scholar]
- Kurdistani, S.K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell. 117:721–733. [DOI] [PubMed] [Google Scholar]
- Lee, D.C., and J.D. Aitchison. 1999. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna. J. Biol. Chem. 274:29031–29037. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Loo, S., and J. Rine. 1994. Silencers and domains of generalized repression. Science. 264:1768–1771. [DOI] [PubMed] [Google Scholar]
- Maillet, L., F. Gaden, V. Brevet, G. Fourel, S.G. Martin, K. Dubrana, S.M. Gasser, and E. Gilson. 2001. Ku-deficient yeast strains exhibit alternative states of silencing competence. EMBO Rep. 2:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., J.J. Smith, S. Jung, E. Yi, A.I. Nesvizhskii, R.H. Christmas, R.A. Saleem, Y.Y. Tam, A. Fagarasanu, D.R. Goodlett, et al. 2004. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 167:1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini, M.D., M. Wu, and H.D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 112:725–736. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W.H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 303:343–348. [DOI] [PubMed] [Google Scholar]
- Noguchi, E., N. Hayashi, N. Nakashima, and T. Nishimoto. 1997. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol. Cell. Biol. 17:2235–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai, C.Y., and V.G. Corces. 2002. The nuclear pore complex and chromatin boundaries. Trends Cell Biol. 12:452–455. [DOI] [PubMed] [Google Scholar]
- Ren, B., F. Robert, J.J. Wyrick, O. Aparicio, E.G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, et al. 2000. Genome-wide location and function of DNA binding proteins. Science. 290:2306–2309. [DOI] [PubMed] [Google Scholar]
- Rexach, M., and G. Blobel. 1995. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 83:683–692. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., and J.D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593–16596. [DOI] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.M., P.R. Gafken, Z. Zhang, D.E. Gottschling, J.B. Smith, and D.L. Smith. 2003. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal. Biochem. 316:23–33. [DOI] [PubMed] [Google Scholar]
- Smith, J.J., M. Marelli, R.H. Christmas, F.J. Vizeacoumar, D.J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R.A. Rachubinski, and J.D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.S., and J.D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241–254. [DOI] [PubMed] [Google Scholar]
- Solsbacher, J., P. Maurer, F. Vogel, and G. Schlenstedt. 2000. Nup2p, a yeast nucleoporin, functions in bidirectional transport of importin alpha. Mol. Cell. Biol. 20:8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett, A.J., D.J. Dilworth, M.J. Davey, M. O'Donnell, J.D. Aitchison, M.P. Rout, and B.T. Chait. 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H., M. Evangelista, A.B. Parsons, H. Xu, G.D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C.W. Hogue, H. Bussey, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368. [DOI] [PubMed] [Google Scholar]
- van Leeuwen, F., P.R. Gafken, and D.E. Gottschling. 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 109:745–756. [DOI] [PubMed] [Google Scholar]
- Wente, S.R. 2000. Gatekeepers of the nucleus. Science. 288:1374–1377. [DOI] [PubMed] [Google Scholar]