Abstract
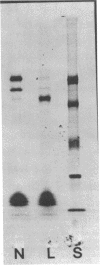
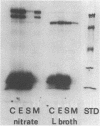
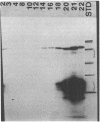
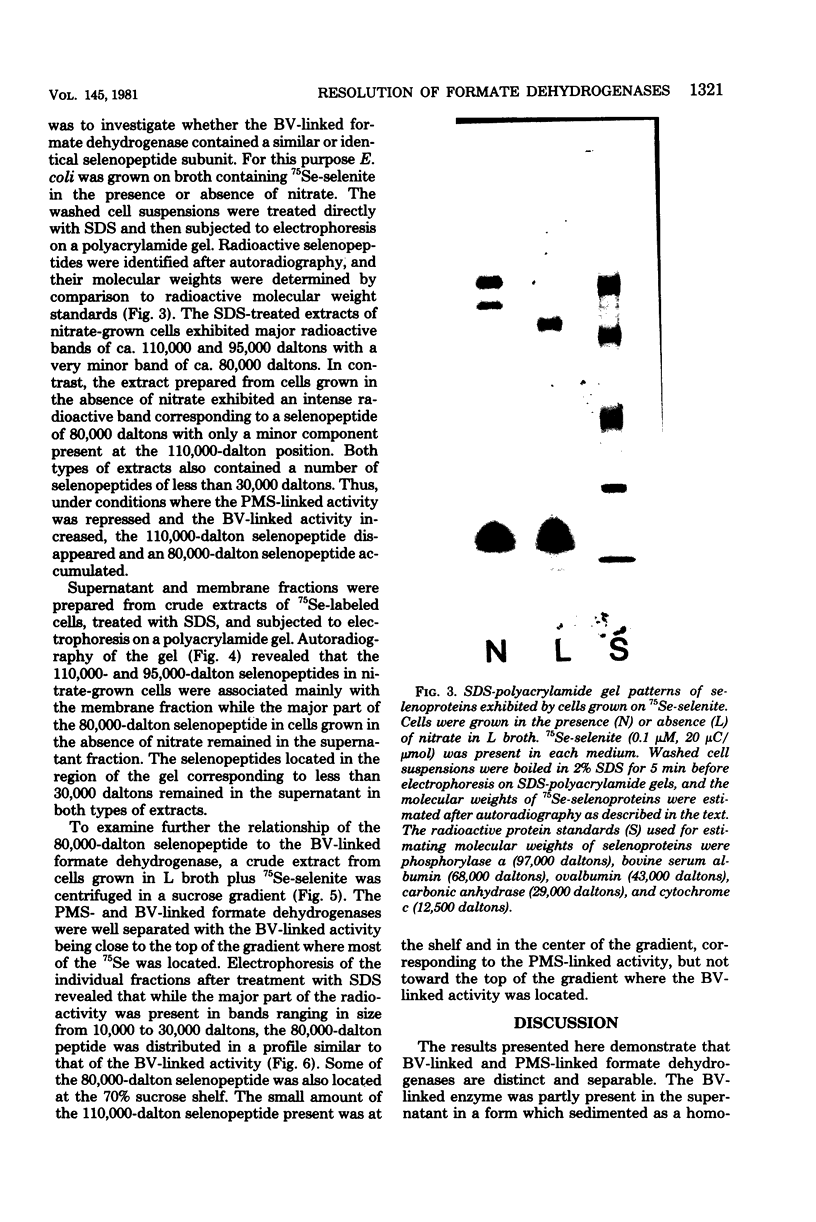
Formate dehydrogenase, a component activity of two alternative electron transport pathways in anaerobic Escherichia coli, has been resolved as two distinguishable enzymes. One, which was induced with nitrate reductase as a component of the formate-nitrate reductase pathway, utilized phenazine methosulfate (PMS) in preference to benzyl viologen (BV) as an artificial electron acceptor and appeared to be exclusively membrane-bound. A second formate dehydrogenase, which was induced as a component of the formate hydrogenlyase pathway, appeared to exist both as a membrane-bound form and as a cytoplasmic enzyme; the cytoplasmic activity was resolved completely from the PMS-linked activity on a sucrose gradient. When E. coli was grown in the presence of 75Se-selenite, a 110,000-dalton selenopeptide, previously shown to be a component of the PMS-linked enzyme, was induced and repressed with this activity. In contrast, an 80,000-dalton selenopeptide was induced and repressed with the BV-linked activity and exhibited a distribution similar to the BV-linked formate dehydrogenase in cell fractions and in sucrose gradients. The results indicate that the two formate dehydrogenases are distinguishable on the basis of their artificial electron acceptor specificity, their cellular localization, and the size of their respective selenoprotein components.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. W., Baum P. R., Gesteland R. F. Processing of adenovirus 2-induced proteins. J Virol. 1973 Aug;12(2):241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Jackson C. E., Fukumoto H. T., Chang G. W. Formate dehydrogenase mutants of Salmonella typhimurium: a new medium for their isolation and new mutant classes. Mol Gen Genet. 1979;177(1):95–101. doi: 10.1007/BF00267258. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Casse F., Pascal M. C. Isolation and phenotypes of mutants from Salmonella typhimurium defective in formate hydrogenlyase activity. J Bacteriol. 1972 May;110(2):766–768. doi: 10.1128/jb.110.2.766-768.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippaux M., Pascal M. C., Casse F. Formate hydrogenlyase system in Salmonella typhimurium LT2. Eur J Biochem. 1977 Jan 3;72(1):149–155. doi: 10.1111/j.1432-1033.1977.tb11234.x. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- GEST H., PECK H. D., Jr A study of the hydrogenlyase reaction with systems derived from normal and anaerogenic coli-aerogenes bacteria. J Bacteriol. 1955 Sep;70(3):326–334. doi: 10.1128/jb.70.3.326-334.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Keesey J. K., Jr, Bigelis R., Fink G. R. The product of the his4 gene cluster in Saccharomyces cerevisiae. A trifunctional polypeptide. J Biol Chem. 1979 Aug 10;254(15):7427–7433. [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957 Jun;73(6):706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PINSENT J. The need for selenite and molybdate in the formation of formic dehydrogenase by members of the coli-aerogenes group of bacteria. Biochem J. 1954 May;57(1):10–16. doi: 10.1042/bj0570010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: participation of specific formate dehydrogenase and cytochrome b1 components in nitrate reduction. J Bacteriol. 1969 Sep;99(3):720–729. doi: 10.1128/jb.99.3.720-729.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Showe M. K., DeMoss J. A. Nitrate reductase complex of Escherichia coli K-12: isolation and characterization of mutants unable to reduce nitrate. J Bacteriol. 1969 Mar;97(3):1291–1297. doi: 10.1128/jb.97.3.1291-1297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruíz-Herrera J., Alvarez A., Figueroa I. Solubilization and properties of formate dehydrogenases from the membrane of Escherichia coli. Biochim Biophys Acta. 1972 Dec 7;289(2):254–261. doi: 10.1016/0005-2744(72)90075-7. [DOI] [PubMed] [Google Scholar]
- Showe M. K., DeMoss J. A. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol. 1968 Apr;95(4):1305–1313. doi: 10.1128/jb.95.4.1305-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyhach R. J., Hawrot E., Satre M., Kennedy E. P. Increased synthesis of phosphatidylserine decarboxylase in a strain of Escherichia coli bearing a hybrid plasmid. Altered association of enzyme with the membrane. J Biol Chem. 1979 Feb 10;254(3):627–633. [PubMed] [Google Scholar]
- Wimpenny J. W., Cole J. A. The regulation of metabolism in facultative bacteria. 3. The effect of nitrate. Biochim Biophys Acta. 1967 Oct 9;148(1):233–242. doi: 10.1016/0304-4165(67)90298-x. [DOI] [PubMed] [Google Scholar]