Abstract
In diploid organisms, meiosis reduces the chromosome number by half during the formation of haploid gametes. During meiotic prophase, telomeres transiently cluster at a limited sector of the nuclear envelope (bouquet stage) near the spindle pole body (SPB). Cohesin is a multisubunit complex that contributes to chromosome segregation in meiosis I and II divisions. In yeast meiosis, deficiency for Rec8 cohesin subunit induces telomere clustering to persist, whereas telomere cluster–SPB colocalization is defective. These defects are rescued by expressing the mitotic cohesin Scc1 in rec8Δ meiosis, whereas bouquet-stage exit is independent of Cdc5 pololike kinase. An analysis of living Saccharomyces cerevisiae meiocytes revealed highly mobile telomeres from leptotene up to pachytene, with telomeres experiencing an actin- but not microtubule-dependent constraint of mobility during the bouquet stage. Our results suggest that cohesin is required for exit from actin polymerization–dependent telomere clustering and for linking the SPB to the telomere cluster in synaptic meiosis.
Introduction
The formation of haploid cells from diploid progenitors requires a tightly regulated series of differentiation steps. In meiosis, homologous chromosomes (homologues) undergo pairing and crossing over, which are prerequisites for the reductional segregation that compensates for genome doubling at fertilization. Specialized meiotic cohesin complexes are assembled along meiotic chromosomes and underlie the axial elements (AEs) of the synaptonemal complex (SC; for reviews see Jessberger, 2002; Page and Hawley, 2003). AEs consist of Rec8-containing meiotic cohesin complexes (Rec8 replaces mitotic Scc1/Mcd1/Rad21), STAG3 (instead of mitotic Scc3/SA1 or SA2; Klein et al., 1999; Watanabe and Nurse, 1999; Pezzi et al., 2000; Prieto et al., 2001, 2002; Siomos et al., 2001; James et al., 2002; Eijpe et al., 2003; Molnar et al., 2003; Pasierbek et al., 2003; Couteau et al., 2004), condensin (Yu and Koshland, 2003), and specific AE proteins such as mammalian SCP3 and SCP2 (Lammers et al., 1994; Offenberg et al., 1998; Yuan et al., 2000), or Caenorhabditis elegans Him3 (Zetka et al., 1999), Arabidopsis thaliana and Brassica oleracea Asy1 (Armstrong et al., 2002), or Saccharomyces cerevisiae Red1 (Smith and Roeder, 1997).
The induction of meiosis causes telomeres to attach to the inner nuclear membrane. Once attached, they move along the inner nuclear envelope and transiently cluster in a minimal time window at the leptotene–zygotene transition to form a so-called chromosomal bouquet, which is highly conserved among eukaryotes (for reviews see Zickler and Kleckner, 1998; Scherthan, 2001). In leptotene, the transesterase Spo11 generates DNA double strand breaks (DSBs; Bergerat et al., 1997; Keeney et al., 1997; Mahadevaiah et al., 2001) that are instrumental in homology searching and in initiating crossing over (for review see Keeney, 2001; Lichten, 2001). The extent of homologue pairing increases during zygotene and is fortified by the SC, a ribbon-like structure that forms between homologues and regulates crossing over (von Wettstein et al., 1984; Bishop and Zickler, 2004; Page and Hawley, 2004). During diplotene, the nuclear envelope disintegrates, chromosomes condense, and sister kinetochores attach as a single unit to microtubules (MTs) from the same spindle pole in metaphase I (monopolar attachment). The subsequent release of arm cohesion mediates reductional segregation in the first meiotic division (for reviews see Page and Hawley, 2003; Petronczki et al., 2003).
The existence of a temporal overlap between homologue pairing and telomere clustering has led to the suggestion that telomere clustering supports homologue pairing (Loidl, 1990; Dernburg et al., 1995). Indeed, in the absence of telomere clustering in budding and fission yeast, homologue pairing is significantly delayed or perturbed (Cooper et al., 1998; Nimmo et al., 1998; Trelles-Sticken et al., 2000; Ding et al., 2004).
In vegetative cells of budding yeast, telomeres localize to a few clusters at the periphery of the nucleus (Klein et al., 1992; Gotta et al., 1996). After induction of meiosis, yeast telomeres disperse over the nuclear periphery and subsequently cluster at the spindle pole body (SPB; the centrosome equivalent of yeast; Trelles-Sticken et al., 1999). The meiotic telomere protein Ndj1/Tam1 (Chua and Roeder, 1997; Conrad et al., 1997), but not Spo11, Kar3, or Sir3, is required for telomere attachment and clustering (Trelles-Sticken et al., 2000, 2003). In asynaptic prophase I fission yeast, telomere clustering is regulated by the mating pheromone signaling pathway (Chikashige et al., 1997; Yamamoto et al., 2004). Telomere proteins Taz1 and Rap1 contribute to attachment and clustering (Cooper et al., 1998; Nimmo et al., 1998; Chikashige and Hiraoka, 2001; Kanoh and Ishikawa, 2001), which is maintained during much of prophase I when the nucleus undergoes sweeping movements (Chikashige et al., 1994). In the maize pam1 mutant and in rye meiocytes treated with the MT drug colchicine, telomere clustering fails (Cowan and Cande, 2002; Golubovskaya et al., 2002). However, telomere clustering in plants and fission yeast does not require cytoplasmic MTs (Chikashige et al., 1994; Ding et al., 1998; Cowan and Cande, 2002).
Because little is known about the requirements for and dynamics of telomere movements in species with synaptic meiosis, we probed the role of the cytoskeleton and cohesin in this process and investigated meiotic telomere clustering in live and fixed budding yeast meiocytes.
Results
Meiotic telomere mobility is constrained at the bouquet stage
To investigate telomere dynamics in living meiocytes, we created S. cerevisiae SK1 strains in which the endogenous Rap1 protein was replaced with a GFP-tagged version of the protein (see Materials and methods and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). Rap1 is telomere associated in vegetative and meiotic cells (Longtine et al., 1989; Klein et al., 1992). This allowed us to track the general behavior of telomeres in live cells by using an approach similar to that of Hayashi et al. (1998). Rap1-GFP strains underwent meiotic divisions (as measured by the appearance of bi- and tetranucleate cells) with wild-type kinetics (unpublished data). We obtained a series of time-lapse images from individual live cells (see Materials and methods) and recorded Rap1-GFP signal dynamics over time periods of 7 or 44 min in prophase I. Nuclei were identified by diffuse GFP fluorescence and were staged by the alignment of observed telomeric Rap1-GFP signal distribution patterns with substage-specific patterns that were previously established by telomere FISH and Zip1 immunostaining. For instance, wild-type meiocyte nuclei with a single telomere cluster were identified as being zygotene nuclei, as bouquet formation occurs during this substage of yeast meiosis (Trelles-Sticken et al., 1999). Because small Rap1-GFP signals faded rapidly, we recorded a series of time-lapse images (at 0.5- and 5-s intervals) at a fixed focal plane and limited the analysis to the time period during which signals could be traced. This approach was also used successfully in a previous study (Heun et al., 2001). Taking images that were spaced apart by minutes or hours was not feasible for recording meiotic telomere dynamics/clustering in wild-type yeast meiosis (Hayashi et al., 1998).
Telomeres in living vegetative (premeiotic) cells were grossly immobile, as seen in videos lasting 7 (not depicted) and 44 min (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1), which is consistent with previous observations (Heun et al., 2001). The few vegetative perinuclear telomere signals underwent locally restricted movements (Fig. 1 A) with a mean velocity of 9.5 ± 3.07 μm/min (n = 29 trajectories), which was determined by trajectory analysis (i.e., particle tracking; see Materials and methods) that revealed the velocity of telomere spot movement (Fig. 1 B). Meiocytes arrested in premeiotic S-phase by hydroxyurea treatment also exhibited only a few peripheral telomere signal clusters that were similar to those of premeiotic cells (unpublished data), which, in agreement with previous FISH analysis (Trelles-Sticken et al., 1999, 2000), suggests that the resolution of peripheral vegetative telomere clusters occurs at the end of or shortly after premeiotic S-phase.
Figure 1.
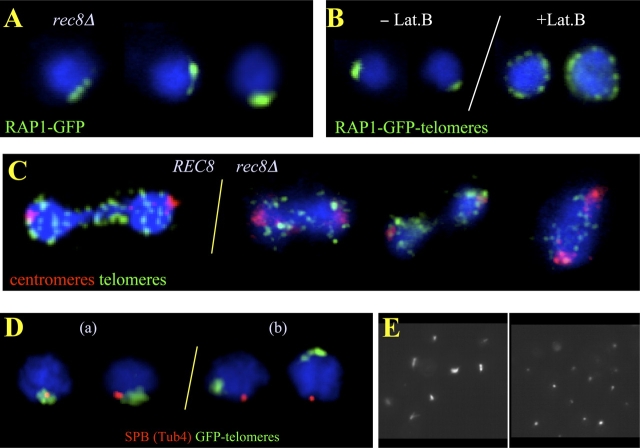
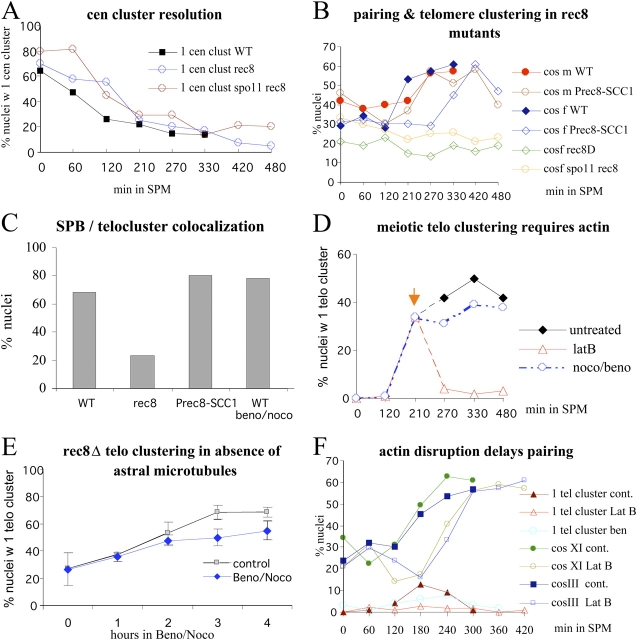
Meiotic telomere dynamics and exit from the bouquet stage require functional cohesin. (A) Projections of trajectories (red threads) of tracked Rap1-GFP telomere spots from a time-lapse image series displayed over an image frame of the respective nucleus. Vegetative telomere clusters (veg.) displaying locally accumulated trajectory projections as a result of constrained motility range. Leptotene telomeres (L) move across the nuclear periphery, which leads to a web of trajectories around the nucleus. Bouquet nucleus (B) with trajectories confined to a limited sector of the nuclear periphery, indicative of telomere clustering. Pachytene nucleus (P) with trajectories around the nucleus, consistent with unconstrained, peripheral telomere movements around the nucleus. Bar, 1 μm. (B) Cumulative plot displaying the velocities of single telomere signals derived from movements of tracked GFP-Rap1 signals in an image series captured every 0.5 s during 7 min. Each trajectory represents a traceable telomere signal through an image series (focal plane fixed at nuclear equator). The trajectories are ranked according to speed. It is evident that in the wild type, vegetative telomere movements are slower than leptotene/bouquet movements, whereas pachytene telomere movements are the most rapid ones. In rec8Δ cells, vegetative telomere velocities match those of wild type, whereas telomere velocities in bouquet-like rec8Δ meiocytes are variable. (C) Analysis of bouquet frequencies by telomere FISH to meiotic time courses. The wild-type frequency of nuclei with a single telomere cluster reaches a maximum after 210 min in sporulation medium (SPM). Wild-type time courses were shorter because meiotic divisions appeared after 270 min and rendered FISH patterns difficult to interpret. rec8Δ and rec8Δ spo11Δ nuclei maintain a telomere cluster up to meiosis I division (compare C with E). Telomere cluster frequency drops after 420 min in rec8Δ spo11Δ meiosis when meiotic divisions become abundant (E). Meiotic expression of the mitotic cohesin SCC1 (Prec8-SCC1) restores bouquet-stage exit in a rec8Δ background, leading to low frequencies of nuclei with a single telomere cluster. Reduction of meiotic CDC5 expression by SCC1 promoter control in REC8 meiosis (Pscc1-CDC5) does not increase telomere clustering. A delay in bouquet formation is evident in red1Δ, Prec8-SCC1, and Pscc1-CDC5 meiosis. (D) Development of FACS profiles in wild-type (WT), rec8Δ, and rec8Δ spo11Δ sporulation indicates that all strains pass through S-phase. G1/2, cell-cycle stage gap1/2. (E) Timing of meiotic divisions in wild-type, rec8Δ, and rec8Δ spo11Δ strains derived by scoring bi- and tetranucleate cells (100–200 per time point). Divisions were significantly delayed in rec8Δ meiosis and occurred at a maximum of 20% (even in longer time courses; not depicted), whereas the spo11Δ mutation eliminated the DSB-dependent prophase I arrest.
Entry into prophase I leads to a rimlike telomere distribution (Trelles-Sticken et al., 1999, 2000). Leptotene meiocytes with a rimlike Rap1-GFP telomere distribution were also detected by live cell imaging and were characterized by rapidly moving, usually dim, and small perinuclear GFP telomere signals in videos that spanned 7 and 44 min (Fig. 1 A; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). Leptotene Rap1-GFP signals moved with a mean velocity of 12.6 ± 3.89 μm/min (n = 19 trajectories; Fig. 1 B). The low intensity of individual Rap1-GFP telomere signals is likely to be a bleaching effect (Hayashi et al., 1998), which prevented the recording of videos with >500 images. The few, occasionally seen strong leptotene Rap1-GFP signals suggest that telomeres form miniclusters before congregating into the large cluster of the bouquet stage, which seems to be the case in plants (Carlton et al., 2003).
In the subsequent bouquet stage, telomere movements were restricted to a limited area of the nuclear periphery (Fig. 1 A and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1), which, in fixed cells, is represented by a single telomere signal at the SPB (Trelles-Sticken et al., 1999). In videos with frame rates of 0.5 s, telomeres moved in a restricted area of the nuclear periphery (Video 3). The meiotic telomere cluster repeatedly split and reformed, which indicates that telomeres are motile during this stage as well. The mean velocity of telomere GFP signals was 12.5 ± 4.48 μm/min (n = 20 trajectories), which is similar to the mean velocity of leptotene telomeres (Fig. 1 B). It was not possible to record the formation of a tight telomere cluster in videos with 5-s exposure intervals, which underlines the transient nature of the SK1 bouquet stage and is consistent with earlier observations (Hayashi et al., 1998). A single, large telomere cluster in a favorable SK1 meiocyte was observed to persist for ∼30 s before it resolved and formed again (Video 3), suggesting that telomeres in the bouquet stage are highly mobile and that several telomeres move together. During a longer time period (i.e., in the range of minutes), a single telomere cluster may repeatedly dissolve and reform, making a more detailed analysis of telomere dynamics during this time period impossible. To overcome this drawback of the fast-sporulating SK1 strain, we searched for a mutant with enduring telomere clustering (see next section).
In postbouquet pachytene cells, GFP telomeres underwent rapidly oscillating movements over the entire nuclear periphery (Fig. 1 A) and had a mean velocity of 16.3 ± 5.66 μm/min (n = 31 trajectories). Rapid telomere movements (Fig. 1 B) were accompanied by oscillating nuclear deformations (Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). Peripherally dispersed telomere FISH signals have also been noted in fixed pachytene meiocytes (Trelles-Sticken et al., 1999).
Rec8 deletion induces persistent clustering
To analyze the molecular requirements of meiotic telomere dynamics in more detail, we used FISH with pantelomeric and pancentromeric probes (telomere/centromere FISH; Trelles-Sticken et al., 2000) and screened for mutants with enduring telomere clustering. FISH analysis was first performed in mutants that lack proteins implied in perinuclear telomere location in vegetative budding yeast cells, such as Mlp1 and -2 nuclear pore extensions (Strambio-de-Castillia et al., 1999; Galy et al., 2000) and the Hdf1 (Ku70 in mammals) nonhomologous end-joining repair protein that is required for perinuclear tethering of vegetative telomeres (Laroche et al., 1998).
Telomere FISH revealed transient telomere clustering in mlp1Δ, mlp2Δ, and hdf1Δ meioses, which occurred with a delay in the latter mutant (unpublished data). In contrast, telomere clustering persisted in most prophase I cells lacking the meiotic cohesin Rec8 (Fig. 1 C and see Fig. 2 A), whereas the wild-type SK1 strain showed a maximum of 11% bouquet nuclei (n = 100) 270 min after the induction of meiosis (Fig. 1 C).
Figure 2.
Telomere clustering in rec8Δ meiosis is displaced from the SPB and requires actin polymerization. (A) Rap1-GFP–tagged telomeres (green) in rec8Δ meiocytes display one telomere cluster. (B) rec8Δ meiocyte nuclei before (−Lat B) and after (+Lat B) inhibition of actin polymerization by Lat B treatment, which leads to peripherally dispersed telomeres. (C) Anaphase I figures of wild-type (left) and rec8Δ meiocytes (right) show absence of a telomere cluster. Centromeres (red) lead the anaphase movements, whereas telomeres (green) are seen as numerous spots trailing behind. (D) Spatial relationships of the telomere cluster (green, Rap1-GFP) and SPB (red, Tub4) in rec8Δ meiocytes. Class a nuclei depict abundant patterns of telomere cluster–SPB association in wild-type meiosis. In rec8Δ meiosis, the telomere cluster is often separated from the SPB (class b; Table I). (E) Image field showing tubulin staining (FITC channel, gray scale) of cells from meiotic cultures without (left) and with (right) benomyl and nocodazole treatment. The treated cells display spots as a result of residual SPB staining that is resistant to MT drugs (Hasek et al., 1987; Lillie and Brown, 1998).
Rec8 is required for sister chromatid cohesion, recombination, AE/SC formation, and homologue segregation (Klein et al., 1999). Flow cytometry of our rec8Δ strain showed that it passed through premeiotic S-phase (Fig. 1 D). Centromere clustering was resolved with wild-type kinetics after induction of rec8Δ meiosis (see Fig. 3 A). In agreement with previous data (Klein et al., 1999), our rec8Δ strains underwent meiotic divisions, but these were greatly delayed (Fig. 1 E), and bi- and tetranucleate cells never exceeded 23% in prolonged time courses. After 24 h, only 22% of cells sporulated (vs. 94% in wild-type cells), as assayed by the formation of asci with one or more spores (>100 cells analyzed for each experiment). Altogether, these observations suggest that most rec8Δ meiocytes arrest in prophase I, most likely in an aberrant zygotene-like stage as DSBs are formed (Klein et al., 1999) and telomere clustering persists.
Figure 3.
Cohesin and actin polymerization are required for normal telomere and chromosome dynamics in SK1 meiosis. (A) Centromere cluster resolution as measured by the frequency of nuclei with a single centromere FISH cluster (Trelles-Sticken et al. 1999). Centromere cluster resolution is similar in wild-type (WT), rec8Δ (rec8), and rec8Δ spo11Δ (spo11 rec8) time courses. (B) Cosmid FISH pairing (cos f, interstitial to XI; cos m, localizing to HML; Trelles-Sticken et al., 2000) as scored by the presence of one large FISH cos-signal cluster in rec8Δ strains. In rec8Δ and rec8Δ spo11Δ cells, this was only seen at a low level (cos f), and meiosis-specific pairing levels never installed with progression through sporulation. Similar values were noted for cos m (not depicted for clarity of display). It should be noted that up to four FISH signals were seen in rec8Δ meiosis, consistent with a lack of sister chromatid cohesion. A single FISH signal may relate to persistent premeiotic association and to a few rec8Δ cells not entering meiosis. Pairing is restored by meiotic expression of the mitotic cohesin SCC1 (Prec8-SCC1). The interstitial chromosome XI region shows a delay in homologue pairing, whereas the telomere of III does not (n > 100 in all time points). (C) SPB colocalization with the telomere cluster in wild-type (WT; n = 60) cells as measured by FISH and Tub4 costaining. In rec8Δ meiocytes (rec8; n = 100), this colocalization is significantly reduced, whereas it is restored in cells expressing the mitotic cohesin Scc1 (Prec8-SCC1; n = 100). Telomere–SPB colocalization is not affected by the presence of benomyl/nocodazole MT drugs (beno/noco; n = 60; anti–Rap1-GFP and Tub4 costaining). (D) Time course experiments with addition of MT and actin poisons 210 min after meiosis induction (orange arrow) in rec8Δ SK1 meiosis. Combined nocodazole/benomyl treatment induced only a slight reduction in the frequency of bouquet-like meiocytes, consistent with results in live cells (C). However, Lat B treatment (latB) dissociated the telomere cluster (n ≥ 100 nuclei were evaluated in all time points and strains). (E) Benomyl/nocodazole treatment of live rec8Δ Rap1-GFP meiocytes fails to eliminate telomere clustering. MT drugs were added 4 h after meiosis induction, and samples were taken before addition (0). After each hour, cells were evaluated for the presence of a single telomere cluster. Error bars represent SD of three experiments; n = 100. (F) Pairing of cos f and m (B) in MT-depleted (ben), and wild-type REC8 (closed symbols, cont.) time courses as measured by FISH in actin-depleted (Lat B) meiosis. The frequency of cells with one telomere cluster is marked by red, open triangles. Inhibition of actin polymerization throughout sporulation prevents telomere clustering and delays homologous pairing for 1–2 h.
Most live rec8Δ Rap1-GFP meiocytes displayed persistent telomere clustering after 7 h in sporulation medium. In these bouquet-like meiocytes, telomeres formed a few miniclusters that constantly underwent restricted movements within a limited region of the nuclear periphery (Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). Recurrent reformation of a single, tight telomere cluster in bouquet-like rec8Δ meiocytes mirrored observations in the short-lived bouquet stage of wild-type SK1 meiocytes (Fig. 1 A and Video 3) and in meiosis of a W303 strain (not depicted). Most rec8Δ cells still displayed dynamic telomere clustering even 24 h after the induction of meiosis (unpublished data).
Trajectory analysis revealed that telomeres in premeiotic rec8Δ cells undergo locally restricted movements with a mean velocity of 9.2 ± 2.6 μm/min (n = 82 trajectories), which is similar to the velocities observed in wild-type cells (Fig. 1 B), thus suggesting that the absence of Rec8 has no consequences for premeiotic telomere motility. In bouquet-like rec8Δ meiocytes, telomeres underwent short-range movements at the cluster site and had a mean velocity of 14.4 μm/min and a maximum velocity of ≤36.6 μm/min (n = 123 trajectories), which is almost as rapid as wild-type pachytene telomeres (mean velocity of 16.3 μm/min and maximum velocity of 35.1 μm/min).
Telomere FISH time course analysis of rec8Δ meiosis revealed that 69% of nuclei (n = 100) had a single telomere cluster even 480 min after the induction of meiosis. After 10 h in sporulation medium, we observed a few rec8Δ meiocytes that had escaped prophase I arrest and were beginning to undergo divisions (Fig. 1 E). In such early anaphase I cells, telomere clustering had been dissolved (Fig. 2 C), which suggests that cells that escape the rec8Δ arrest resolve telomere clustering before entry into meiosis I. This makes it unlikely that the division itself disrupts the meiotic telomere cluster.
rec8Δ-dependent telomere clustering is maintained in the absence of DSBs
Defects in recombination elicit an arrest in prophase I (Roeder and Bailis, 2000) and can lead to an extended duration of telomere clustering (Scherthan, 2003; Storlazzi et al., 2003). To determine whether rec8Δ-dependent telomere cluster maintenance is altered when DSBs are absent, we investigated rec8Δspo11Δ meiosis, which lacks DSBs as a result of the absence of the Spo11 trans-esterase (Bergerat et al., 1997; Keeney et al., 1997). In meiotic time courses, the frequency of rec8Δ spo11Δ bouquet-like cells was similar to rec8Δ meiosis but was reduced at late time points (Fig. 1 C). The frequency of telomere clustering in rec8Δ spo11Δ meiosis (41% at 330 min) was about twice as high as in the spo11Δ single mutant (Trelles-Sticken et al., 1999), suggesting that Rec8 is epistatic over Spo11 on telomere cluster maintenance. The rec8Δ spo11Δ strain underwent meiotic divisions with wild-type kinetics (Fig. 1 E). As DSBs are not repaired in rec8Δ meiosis (Klein et al., 1999), our data suggest that the prophase I arrest in rec8Δ meiosis is DSB dependent but that rec8Δ-promoted telomere cluster maintenance does not require DSBs.
Cohesin is required for telomere cluster–SPB colocalization
To assess whether the compromised assembly of AEs underlies persistent telomere clustering in rec8Δ meiosis, we investigated red1Δ meiosis, which lacks AEs (Smith and Roeder, 1997) and has defective recombination and significantly reduced homologue pairing (Rockmill and Roeder, 1990; Nag et al., 1995). red1Δ meiosis displayed delayed, but wild-type, telomere clustering frequencies with a maximum of 10% red1Δ bouquets at 330 min, whereas 10% wild-type bouquets were seen at 210 min (n ≥ 100 in both strains; Fig. 1 C). Thus, defective AE formation in yeast does not lead to increased levels of telomere clustering.
Homology searching and pairing in yeast appears to be DSB dependent (Loidl et al., 1994; Weiner and Kleckner, 1994; Lichten, 2001). DSBs form but are hyperresected in rec8Δ meiosis (Klein et al., 1999). When we evaluated homologue pairing in rec8Δ meiocytes by FISH with cosmids homologous to the chromosome III left telomere and an interstitial site on chromosome XI (Trelles-Sticken et al., 2000), we found that meiosis-specific pairing never installed in rec8Δ or in rec8Δ spo11Δ meioses (see Fig. 3 B and not depicted), which is consistent with synaptic defects in both spo11Δ and rec8Δ strains (Loidl et al., 1994; Klein et al., 1999). This indicates that even extensive telomere clustering cannot ameliorate the pairing defects that are caused by the absence of cohesin and axial cores.
Telomere cluster–SPB colocalization is considered a hallmark of the bouquet stage in budding yeast (Trelles-Sticken et al., 1999) and other organisms (Zickler and Kleckner, 1998; Bass, 2003). To determine whether this colocalization is affected by the absence of cohesin, the relative positioning of the telomere cluster and SPB was investigated by Rap1-GFP and tubulin/SPB immunostaining in rec8Δ meiocytes. In the wild type, 68% of the cells displayed telomere cluster–SPB colocalization (Fig. 2 D, Table I, and see Fig. 3 C) 210 min after the induction of meiosis. This was significantly different in rec8Δ meiosis, in which only 23% of meiocyte nuclei displayed this colocalization (Fig. 2 D, Table I, and see Fig. 3 C). As telomere cluster–SPB association was absent in 32% of wild-type meiocytes, it could be that the telomere cluster first forms away from the SPB and becomes positioned at this location at a later time point. Alternatively, telomere cluster–SPB colocalization may be released before telomeres disperse from the cluster site. Because the single telomere cluster continuously splits into a few closely spaced miniclusters and reforms, it may be that Rec8, cohesin, and some yet unidentified cofactors may help to tether mobile telomeres to the SPB region.
Table I. Telomere cluster–SPB colocalization.
| n | Overlap/Touching (A) | Separated (B) | |
|---|---|---|---|
| rec8Δ 210′ | 100 | 23 | 77 |
| rec8Δ 330′ | 100 | 30 | 70 |
| rec8Δ 480′ | 100 | 11 | 89 |
| WT | 60 | 68 | 32 |
Categories A and B as depicted in Fig. 2 D.
Telomere cluster resolution depends on cohesin
To test whether cohesin or the Rec8 protein, by itself, is crucial for the maintenance of telomere clustering in the rec8Δ mutant, we studied telomere behavior in a strain that expressed the mitotic cohesin component Scc1 from the meiotic Rec8 promoter (hereafter designated Prec8-SCC1; Toth et al., 2000). We found that the expression of Scc1 in Prec8-SCC1 meiocytes (confirmed by Scc1-HA immunofluorescence; not depicted) restored both telomere cluster–SPB association (Fig. 3 C) and the timely resolution of the telomere cluster (Fig. 1 C). Moreover, the pairing defect in rec8Δ meiosis was restored by meiotic SCC1 expression, albeit with a delay for the interstitial region of chromosome XI (Fig. 3 B). Restoration of pairing in Prec8-SCC1 meiosis suggests that the DSBs formed can now promote homologous strand interactions.
Meiotic telomere clustering is not controlled by Cdc5-dependent Rec8 phosphorylation
It has been shown that late aspects of Rec8 function are regulated by Cdc5 pololike kinase (Clyne et al., 2003). Thus, the influence of Rec8 phosphorylation on telomere clustering was analyzed by telomere FISH in a strain deficient for meiotic CDC5 expression as a result of SCC1 promoter control of CDC5 (Pscc1-CDC5; Clyne et al., 2003). It was found that Pscc1-CDC5 meiosis displayed the same dynamics of telomere cluster formation and resolution as the Prec8-SCC1 mutant (Fig. 1 C). The wild-type–like telomere dynamics that were observed suggest that the resolution of meiotic telomere clustering is not under the control of Cdc5-dependent Rec8 phosphorylation.
Meiotic telomere clustering in yeast occurs in the presence of MT-disrupting drugs
Next, we searched for other components that are involved in meiotic telomere clustering. Wild-type and rec8Δ meiocytes were treated with MT-disrupting drugs because, in plants, telomere clustering is sensitive to the MT drug colchicine (Cowan and Cande, 2002). The administration of a combination of the MT-disrupting drugs nocodazole and benomyl abolished astral MTs that were found in about one third of meiotic cells and left only residual fluorescence at the SPB (Fig. 2 E), which was noted previously (Hasek et al., 1987; Jacobs et al., 1988; Lillie and Brown, 1998). The absence of radial MTs was seen over the course of 3 h after administration of the drugs, the time frame in which all live cell recordings were taken. However, the administration of MT drugs at different time points never eliminated telomere clustering (unpublished data). Although unlikely, the minimal MT fluorescence at the SPB could mean that some residual MTs drive telomere clustering.
Telomere FISH to MT drug–treated rec8Δ meiocytes revealed only a modest reduction of the high rec8Δ-induced bouquet frequency (Fig. 3 D). When MT drugs were added 4 h after meiosis induction to live rec8Δ meiocytes (before most rec8Δ cells engage in telomere clustering), telomere clustering increased with time but remained below untreated control levels during the next 4 h (Fig. 3 E). A recording of live benomyl/nocodazole-treated rec8Δ meiocytes revealed that nuclear deformations and clustering was present, and telomeres were motile during 7-min (Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1) and 44-min (not depicted) recordings. Analysis of live MT drug–treated rec8Δ meiocytes (n = 43) showed that telomere minicluster velocity was slightly reduced as compared with a DMSO-treated control (n = 51; i.e., 9.9 ± 3.8 μm/min vs. 11.5 ± 5.3 μm/min, respectively, with the differences being statistically not significant; t test, P = 0.078). Because drug addition before bouquet formation did not inhibit telomere clustering (Fig. 3, E and F) or telomere cluster–SPB colocalization (Fig. 3 C), it is likely that MTs stabilize telomere clustering but are not absolutely necessary for telomere cluster formation and maintenance in budding yeast meiosis.
Meiotic telomere clustering in S. cerevisiae requires actin
Because recent studies suggest vital roles for actin in nuclear processes (Hofmann et al., 2004; Holaska et al., 2004; Forest et al., 2005), we tested the role of actin in telomere clustering by the administration of latrunculin B (Lat B), which binds to and reversibly prevents the polymerization of G-actin (Coue et al., 1987). Much to our surprise, Lat B addition to living rec8Δ meiocytes induced the rapid dispersion of clustered telomeres over the nuclear periphery (Fig. 3 D and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). This dispersion was detected as a rimlike telomere FISH signal pattern in meiocytes that were fixed after Lat B treatment (Fig. 2 B). The continuous presence of Lat B in meiotic cultures inhibited telomere cluster formation in the wild type (Fig. 3 F) and prevented sporulation, which was accompanied by the presence of arrested cells with deformed nuclei in meiotic time courses (unpublished data). When we incubated rec8Δ meiocytes with cytochalasin D, which also inhibits actin polymerization (Peterson and Mitchison, 2002), there was a 1-h delay of telomere dispersion relative to the addition of the drug (the same effect as with Lat B), possibly owing to the reduced cell permeability of cytochalasin D (Greer and Schekman, 1982). Because vegetative telomere movements were not affected by the inhibition of actin polymerization (unpublished data), our observations suggest a novel role for actin in bringing about and maintaining meiotic telomere clustering, which is distinct from its known role in meiosis I division (Forer and Pickett-Heaps, 1998; LaFountain et al., 1999).
Nuclei of wild-type pachytene cells undergo deformations that are associated with rapid telomere movements (Video 4). When we investigated whether actin also plays a role in telomere movements at postbouquet stages, we found that inhibition of actin polymerization stopped rapid telomere movements and nuclear deformations and that Lat B–treated nuclei obtained a round shape (Video 8, available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1). Telomere spots were still motile and had a mean velocity of 12.6 ± 2.7 μm/min (n = 23 trajectories), which is similar to leptotene telomere velocities and is significantly reduced as compared with wild-type pachytene telomere velocities (t test, P < 0.05). These observations suggest that actin not only mediates telomere clustering but also drives nuclear deformations at pachytene.
Discussion
Prophase I telomeres are highly dynamic
Using a GFP-tagged version of the S. cerevisiae telomere-binding protein Rap1, we recorded, for the first time, dynamic telomere redistribution in synaptic meiosis. The strains that were used underwent meiotic divisions with wild-type kinetics. We recorded videos, which were, because of the bleaching of GFP signals, restricted to <500 frames. Live cell imaging showed that telomeres of vegetative cells formed a few peripheral clusters that underwent locally constrained movements, which coincides with earlier analysis (Heun et al., 2001). Leptotene telomeres rapidly moved around the periphery of nuclei (round-shaped nuclei discriminated leptotene from deformed pachytene nuclei). In bouquet nuclei, the motile telomeres accumulated in a limited region of the nuclear envelope. This conformation is indicative of the leptotene/zygotene transition and was only maintained for ∼30 s in rapid SK1 wild-type meiosis. The short duration of this stage explains why telomere clustering could not be detected by time-lapse videos with large intervals between frames (Hayashi et al., 1998). In postbouquet pachytene cells, telomeres continuously underwent rapid perinuclear movements that were accompanied by nuclear deformation. Overall, the telomere dynamics observed in live cells confirm the sequence of peripheral telomere redistribution that was deduced from the FISH analysis of fixed yeast cells from meiotic time courses (Trelles-Sticken et al., 1999, 2000).
MTs are dispensable for telomere clustering
Previously, it was noted in the meiosis of plants and mammals that a colchicine-sensitive mechanism seems to control meiotic telomere clustering and/or nuclear movements (Salonen et al., 1982; Cowan and Cande, 2002). In the asynaptic meiosis of fission yeast, MT disruption inhibited nuclear movements, whereas telomeres remain clustered in dhc − cells that lack nuclear mobility (Yamamoto et al. 1999). The application of a combination of MT-disrupting drugs to S. cerevisiae meiocytes failed to inhibit or dissolve telomere clustering, which suggests that microtubules are not absolutely required either for telomere cluster formation or for its maintenance in this synaptic yeast. Nevertheless, the disruption of astral MTs led to a moderate reduction in telomere clustering in drug-treated wild-type and rec8Δ meiocytes, as measured by FISH or in live cells. Altogether, it may be that the polarized microtubule cytoskeleton provides a framework in which telomeres can undergo ordered motility to and from the SPB, which is a possibility that requires further experiments in live cells.
Actin is required for telomere clustering and nuclear motility
We were surprised to observe that the inhibition of actin polymerization by Lat B treatment prevented meiotic telomere clustering in the wild type and disintegrated the persistent telomere cluster in rec8Δ meiocytes. The latter was also seen, with a delay, in meiocytes treated with the actin drug cytochalasin D. However, actin disruption did not delocalize telomeres toward the nuclear interior, suggesting that actin somehow constrains telomere movements to a limited nuclear envelope sector but is dispensable for attachment. Bouquet disruption by actin depolymerization considerably delayed homologue pairing, which is reminiscent of delayed homologue pairing in yeast meiocytes lacking the meiotic telomere protein Ndj1 (Chua and Roeder, 1997; Conrad et al., 1997). However, unlike Lat B treatment, the ndj1Δ mutation delocalizes meiotic telomeres toward the nuclear interior (Trelles-Sticken et al., 2000).
Although our study could not determine whether cytoplasmic or nuclear actin is responsible for telomere clustering, we favor the hypothesis that nuclear actin (for reviews see Pederson and Aebi, 2002; Franke, 2004) might displace telomeres toward the SPB-bearing sector of the nuclear envelope. In this proposed mechanism, the actin-perpetuated gathering of motile telomeres would compress chromosome ends into a limited nuclear region. Interestingly, actin-related proteins localize to telomeric but not to centromeric heterochromatin in Schizosaccharomyces pombe (Ueno et al., 2004). The highly dynamic telomere movements and deformation of S. cerevisiae pachytene nuclei could involve cytoplasmic and nuclear actin. In other systems, nuclear actin has been localized to emerin and nuclear pore extensions at the nucleoplasmic face of the nuclear envelope (Hofmann et al., 2001; Holaska et al., 2004) and is implied in nuclear envelope assembly (Krauss et al., 2003) and in intranuclear transport (Forest et al., 2005). On the other hand, the actin cytoskeleton is important for control of the nuclear shape (Guilak, 1995; Abe et al., 2004).
Cohesin is required for exit from meiotic telomere clustering
Enforcement of telomere clustering seems to occur when neither the homologue nor the sister chromatid is available for repair or when defects in recombination are present (Trelles-Sticken et al., 1999, 2003; Scherthan, 2003; Storlazzi et al., 2003). Telomere clustering in the bouquet stage likely facilitates homology testing and pairing (Rockmill and Roeder, 1998; Niwa et al., 2000; Trelles-Sticken et al., 2000; Ding et al., 2004).
Telomere clustering was greatly increased in rec8Δ meiocytes that were arrested in prophase I. This arrest was likely the consequence of unrepaired DSBs, because a spo11Δ mutation removed DSBs and restored meiotic divisions and the resolution of telomere clustering before meiotic divisions. Nevertheless, extended telomere clustering was maintained in rec8Δ spo11Δ meiosis, suggesting that Rec8 imparts a DSB-independent role in telomere clustering. Given that bouquet formation plays a role in the instigation of homologue interactions, extended telomere clustering in rec8Δ meiosis may be a response to the absence of sister chromatid cohesion. Initiation of the recombinational repair of DSBs would be impeded by spatially separate homologous chromosomes and chromatids. Alternatively, the processing of telomere repeats, which requires Ndj1, may occur during the bouquet stage (Joseph et al., 2005) and may also require cohesin function. Bouquet exit is regulated by cohesion because meiotic expression of the Scc1 cohesin in rec8Δ cells reestablished exit from telomere clustering. Moreover, meiotic Scc1 expression also restored defective telomere cluster–SPB colocalization in rec8Δ meiocytes.
Cohesin-dependent telomere cluster regulation does not require Cdc5 pololike kinase (Clyne et al., 2003) because shutting off Cdc5 activity in meiosis did not increase bouquet frequencies. It may be speculated that telomeric cohesin may recruit other factors contributing to the regulation of telomere cluster duration, which may involve the meiotic telomere protein Ndj1 and other factors influencing the proposed actin-dependent mechanism for the telomere clustering. Interestingly, Ndj1 is required for telomere clustering and contributes to sister chromatid cohesion at meiotic telomeres (Chua and Roeder, 1997; Conrad et al., 1997; Trelles-Sticken et al., 2000). Moreover, Ndj1 has been found to interact with the SPB component Msp3 (Uetz et al., 2000), which raises the possibility that Ndj1 could be involved in telomere/SPB tethering.
Recently, mice mutant for Rec8, unlike budding yeast mutants, assemble AEs (Bannister et al., 2004), and mice deficient for the meiotic cohesin Smc1β are defective in recombination, telomere attachment, and clustering (Revenkova et al., 2004). Thus, further analysis is required to determine whether the cohesin- and actin-dependent telomere cluster maintenance in budding yeast meiosis applies to other species.
Materials and methods
Strains
Genotypes of SK1 strains that were used are listed in Table S1. Standard genetic procedures (Elble, 1992) were applied to generate the rec8Δ rap1::RAP1-GFP-LEU2 strain. In brief, RAP1 gene replacement was performed in REC8-deleted haploid strains by transformation (Elble, 1992) with a Pst1-digested plasmid coding for the GFP-tagged version of RAP1 (plasmid provided by A. Ehrenhofer-Murray, Max-Planck-Institute for Molecular Genetics, Berlin, Germany). Haploid clones of opposing mating types were mated, and zygotes were picked to obtain homozygous diploid rec8Δ rap1::RAP1-GFP-LEU2 strains. Strains expressing Scc1 under the Rec8 promoter (Toth et al., 2000) and CDC5 under the Scc1 promoter (Clyne et al., 2003) were provided by K. Nasmyth (Institute of Molecular Pathology, Vienna, Austria).
Cell culture and drug treatment
Culture conditions, sporulation of strains, and meiotic cell preparation were performed as described previously (Scherthan and Trelles-Sticken, 2002). Nocodazole and benomyl (Calbiochem) were simultaneously used at 15 and 20 μg/ml in culture medium, respectively. Lat B (Sigma-Aldrich) was used at 30 μM, and cytochalasin D (Sigma-Aldrich) was used at 100 μg/ml. Drugs were added 210 and 90 min after the induction of meiosis in rec8Δ and wild-type sporulation, respectively. Control cultures received a drug solvent (DMSO) at the same concentration.
For meiotic cell preparation, cultures were grown in presporulation medium to a density of 2 × 107 cells/ml and were transferred to sporulation medium (2% KAc) at a density of 4 × 107 cells/ml (Roth and Halvorson, 1969). Aliquots from the sporulating cultures were transferred at different time points during the meiotic time course to Eppendorf tubes at 0°C containing 0.1 vol of acid-free 37% formaldehyde (Merck). After 30 min, cells were removed from the fixative, washed with 1× SSC, and spheroplasted with 100 μg/ml Zymolyase 100T (Seikagaku) in 0.8 M sorbitol, 2% KAc, and 10 mM DTT. Spheroplasting was terminated by adding 10 vol of ice-cold 1 M sorbitol. Nuclear spreads were prepared as described previously (Trelles-Sticken et al., 2000). Sporulation was assayed by the formation of asci with one or more spores after 24 h in sporulation medium.
Flow cytometry
Cells were induced to sporulate, and 500 μl of meiotic cells from each time point were harvested by centrifugation and fixed in 70% ethanol. After one wash in 50 mM citrate buffer, pH 7.4, cells were RNase A–digested for 2 h at 50°C (0.25 mg RNase A/ml citrate buffer) followed by the addition of 50 μl proteinase K (from a 20 mg/ml-H2O stock) and were further incubated at 50°C for 2.5 h. Finally, 1 ml of 16 μg/ml propidium iodide and 50 mM sodium citrate solution were added to the tube, and DNA content was determined by flow cytometry using FACS (Becton Dickinson).
Fluorescent in situ hybridization, DNA probes, and immunofluorescence
FISH to yeast cells, DNA probes to all telomeres and centromeres, and their labeling were performed as described previously (Scherthan and Trelles-Sticken, 2002). Pairing of an internal locus on the left arm of chromosome XI and of HML near the left telomere of chromosome III was assessed as described previously (Trelles-Sticken et al., 2000). Antibodies to Tub4 (gift of L. Marschall, California State University, East Bay, Hayward, CA) and Zip1 (gift of G.S. Roeder, Yale University, New Haven, CT) were used in immunofluorescence as described previously (Trelles-Sticken et al., 2000).
Imaging
FISH preparations were evaluated by using the Isis fluorescence image analysis system (MetaSystems) fitted to an epifluorescence microscope (Axioskop; Carl Zeiss MicroImaging, Inc.) equipped with appropriate fluorescence filters (Chroma Technology Corp.).
Live cells were investigated by using a temperature-controlled microscope room equipped with an epifluorescence microscope (Axiovert; Carl Zeiss MicroImaging, Inc.). Cells were recorded with a 100× plan-neofluar oil-immersion lens (NA 1.35; Carl Zeiss MicroImaging, Inc.) attached to a PIFOC z-SCAN (Physik Instrumente) and a 12-bit black-and-white CCD digital camera (PCO; SensiCam) that were controlled by TILLvisION v4.0 software (TILL Phototonics). GFP or fluorochromes were excited by using a monochromator (Polychrome IV; TILL Photonics) in combination with a GFP filter set or a quadruple band-pass beam splitter and barrier filter (Chroma Technology Corp.). The digital images displayed were processed by using Photoshop 6.0 (Adobe) and were adjusted in contrast and brightness to match the fluorescence image seen in the microscope.
Live cells were sporulated and attached to Con A–coated coverslips (Chikashige et al., 1994). Cells were immediately transferred to the microscope setting in a temperature-controlled room. Images of live cells were recorded every 0.5 s for 7 min or every 5 s for 44 min, with an exposure time of 300 ms for each frame, by using a preset focal plane at the nuclear equator. We chose to record two-dimensional images to limit bleaching and to be able to observe the cell for a longer time period. Telomere spot movements were analyzed with DiaTrack 2.3 software (Semasopht) in the time-lapse series by interactively tracking each visible spot in subsequent images until it left the focal plane. The same software was used to create histograms of velocity, which were created along trajectories and show the distribution of particle speed (the length of a trajectory, computed as the sum of the lengths of displacement vectors composing it divided by the lifetime of the particle). All trajectories that were created over the investigated time interval within one nucleus were superimposed to reveal a measure of the area of the focal plane over which telomere movements occurred. Generally, we were able to track spots over 13 consecutive images, whereas in a few cases, spots could be tracked for up to 69 frames.
Online supplemental material
The strains used in this study are listed in Table S1. Video 1 shows a vegetative wild-type nucleus with Rap1-GFP telomere clusters. Video 2 shows wild-type leptotene meiocyte nuclei displaying small, whitish Rap1-GFP telomere dots that move rapidly over the nuclear periphery. Video 3 shows a wild-type meiocyte nucleus displaying the transient formation of a telomere cluster after 1.4 min at the lower-right sector of the nuclear periphery. Video 4 shows pachytene wild-type nuclei displaying vigorous telomere movements and nuclear deformation. Video 5 shows three rec8Δ bouquet nuclei, which display moving telomeres that are restricted to a limited region of the nuclear periphery. Video 6 shows the poisoning of telomere clustering in rec8Δ meiocytes by Lat B. Video 7 shows telomere cluster movements in the presence of nocodazole/benomyl. Video 8 shows telomere and nuclear movements before and at the addition of Lat B in a wild-type pachytene cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200501042/DC1.
Acknowledgments
We thank L. Marschall for anti-Tub4 antibodies, G.S. Roeder for anti-ZIP1 antibodies, A. Ehrenhofer-Murray for the Rap1-GFP-LEU2 plasmid, and K. Nasmyth for the SCC1 strains. We thank M. Siomos and anonymous reviewers for their constructive comments on the manuscript.
H. Scherthan thanks the Deutsche Forschungsgemeinschaft (grant Sche350/8-4) and H.-H. Ropers for support. J. Loidl acknowledges support from the Austrian Science Fund (grant P16282). There are no conflicts of interest.
E. Trelles-Sticken and C. Adelfalk contributed equally to this paper.
Edgar Trelles-Sticken's present address is Schuetzenstrasse 31, D-12105 Berlin, Germany.
Abbreviations used in this paper: AE, axial element; DSB, DNA double strand break; Lat B, latrunculin B; MT, microtubule; SC, synaptonemal complex; SPB, spindle pole body.
References
- Abe, T., K. Takano, A. Suzuki, Y. Shimada, M. Inagaki, N. Sato, T. Obinata, and T. Endo. 2004. Myocyte differentiation generates nuclear invaginations traversed by myofibrils associating with sarcomeric protein mRNAs. J. Cell Sci. 117:6523–6534. [DOI] [PubMed] [Google Scholar]
- Armstrong, S.J., A.P. Caryl, G.H. Jones, and F.C. Franklin. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115:3645–3655. [DOI] [PubMed] [Google Scholar]
- Bannister, L.A., L.G. Reinholdt, R.J. Munroe, and J.C. Schimenti. 2004. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 40:184–194. [DOI] [PubMed] [Google Scholar]
- Bass, H.W. 2003. Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cell. Mol. Life Sci. 60:2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerat, A., B. de Massy, D. Gadelle, P.C. Varoutas, A. Nicolas, and P. Forterre. 1997. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 386:414–417. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., and D. Zickler. 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 117:9–15. [DOI] [PubMed] [Google Scholar]
- Carlton, P.M., C.R. Cowan, and W.Z. Cande. 2003. Directed motion of telomeres in the formation of the meiotic bouquet revealed by time course and simulation analysis. Mol. Biol. Cell. 14:2832–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige, Y., and Y. Hiraoka. 2001. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr. Biol. 11:1618–1623. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D.Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko, M. Yanagida, and Y. Hiraoka. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science. 264:270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige, Y., D.Q. Ding, Y. Imai, M. Yamamoto, T. Haraguchi, and Y. Hiraoka. 1997. Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J. 16:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, P.R., and G.S. Roeder. 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11:1786–1800. [DOI] [PubMed] [Google Scholar]
- Clyne, R.K., V.L. Katis, L. Jessop, K.R. Benjamin, I. Herskowitz, M. Lichten, and K. Nasmyth. 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 5:480–485. [DOI] [PubMed] [Google Scholar]
- Conrad, M.N., A.M. Dominguez, and M.E. Dresser. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 276:1252–1255. [DOI] [PubMed] [Google Scholar]
- Cooper, J.P., Y. Watanabe, and P. Nurse. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 392:828–831. [DOI] [PubMed] [Google Scholar]
- Coue, M., S.L. Brenner, I. Spector, and E.D. Korn. 1987. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213:316–318. [DOI] [PubMed] [Google Scholar]
- Couteau, F., K. Nabeshima, A. Villeneuve, and M. Zetka. 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14:585–592. [DOI] [PubMed] [Google Scholar]
- Cowan, C.R., and W.Z. Cande. 2002. Meiotic telomere clustering is inhibited by colchicine but does not require cytoplasmic microtubules. J. Cell Sci. 115:3747–3756. [DOI] [PubMed] [Google Scholar]
- Dernburg, A.F., J.W. Sedat, W.Z. Cande, and H.W. Bass. 1995. The cytology of telomeres. Telomeres. E.H. Blackburn and C.W. Greider, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 295–338.
- Ding, D.Q., Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 1998. Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci. 111:701–712. [DOI] [PubMed] [Google Scholar]
- Ding, D.Q., A. Yamamoto, T. Haraguchi, and Y. Hiraoka. 2004. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 6:329–341. [DOI] [PubMed] [Google Scholar]
- Eijpe, M., H. Offenberg, R. Jessberger, E. Revenkova, and C. Heyting. 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 160:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble, R. 1992. A simple and efficient procedure for transformation of yeasts. Biotechniques. 13:18–20. [PubMed] [Google Scholar]
- Forer, A., and J.D. Pickett-Heaps. 1998. Cytochalasin D and latrunculin affect chromosome behaviour during meiosis in crane-fly spermatocytes. Chromosome Res. 6:533–549. [DOI] [PubMed] [Google Scholar]
- Forest, T., S. Barnard, and J.D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429–431. [DOI] [PubMed] [Google Scholar]
- Franke, W.W. 2004. Actin's many actions start at the genes. Nat. Cell Biol. 6:1013–1014. [DOI] [PubMed] [Google Scholar]
- Galy, V., J.C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. [DOI] [PubMed] [Google Scholar]
- Golubovskaya, I.N., L.C. Harper, W.P. Pawlowski, D. Schichnes, and W.Z. Cande. 2002. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics. 162:1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S.M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer, C., and R. Schekman. 1982. Actin from Saccharomyces cerevisiae. Mol. Cell. Biol. 2:1270–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilak, F. 1995. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J. Biomech. 28:1529–1541. [DOI] [PubMed] [Google Scholar]
- Hasek, J., I. Rupes, J. Svobodova, and E. Streiblova. 1987. Tubulin and actin topology during zygote formation of Saccharomyces cerevisiae. J. Gen. Microbiol. 133:3355–3363. [DOI] [PubMed] [Google Scholar]
- Hayashi, A., H. Ogawa, K. Kohno, S.M. Gasser, and Y. Hiraoka. 1998. Meiotic behaviours of chromosomes and microtubules in budding yeast: relocalization of centromeres and telomeres during meiotic prophase. Genes Cells. 3:587–601. [DOI] [PubMed] [Google Scholar]
- Heun, P., T. Laroche, K. Shimada, P. Furrer, and S.M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science. 294:2181–2186. [DOI] [PubMed] [Google Scholar]
- Hofmann, W., B. Reichart, A. Ewald, E. Muller, I. Schmitt, R.H. Stauber, F. Lottspeich, B.M. Jockusch, U. Scheer, J. Hauber, and M.C. Dabauvalle. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)– and constitutive transport element (CTE)–containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, W.A., L. Stojiljkovic, B. Fuchsova, G.M. Vargas, E. Mavrommatis, V. Philimonenko, K. Kysela, J.A. Goodrich, J.L. Lessard, T.J. Hope, et al. 2004. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat. Cell Biol. 6:1094–1101. [DOI] [PubMed] [Google Scholar]
- Holaska, J.M., A.K. Kowalski, and K.L. Wilson. 2004. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed]
- Jacobs, C.W., A.E. Adams, P.J. Szaniszlo, and J.R. Pringle. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, R.D., J.A. Schmiesing, A.H. Peters, K. Yokomori, and C.M. Disteche. 2002. Differential association of SMC1alpha and SMC3 proteins with meiotic chromosomes in wild-type and SPO11-deficient male mice. Chromosome Res. 10:549–560. [DOI] [PubMed] [Google Scholar]
- Jessberger, R. 2002. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 3:767–778. [DOI] [PubMed] [Google Scholar]
- Joseph, I., D. Jia, and A.J. Lustig. 2005. Ndj1p-dependent epigenetic resetting of telomere size in yeast meiosis. Curr. Biol. 15:231–237. [DOI] [PubMed] [Google Scholar]
- Kanoh, J., and F. Ishikawa. 2001. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr. Biol. 11:1624–1630. [DOI] [PubMed] [Google Scholar]
- Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1–53. [DOI] [PubMed] [Google Scholar]
- Keeney, S., C.N. Giroux, and N. Kleckner. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 88:375–384. [DOI] [PubMed] [Google Scholar]
- Klein, F., T. Laroche, M.E. Cardenas, J.F. Hofmann, D. Schweizer, and S.M. Gasser. 1992. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J. Cell Biol. 117:935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, F., P. Mahr, M. Galova, S.B. Buonomo, C. Michaelis, K. Nairz, and K. Nasmyth. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 98:91–103. [DOI] [PubMed] [Google Scholar]
- Krauss, S.W., C. Chen, S. Penman, and R. Heald. 2003. Nuclear actin and protein 4.1: Essential interactions during nuclear assembly in vitro. Proc. Natl. Acad. Sci. USA. 100:10752–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain, J.R., Jr., A.J. Siegel, and G.K. Rickards. 1999. Chromosome movement during meiotic prophase in crane-fly spermatocytes: IV. Actin and the effects of cytochalasin D. Cell Motil. Cytoskeleton. 43:199–212. [DOI] [PubMed] [Google Scholar]
- Lammers, J.H., H.H. Offenberg, M. van Aalderen, A.C. Vink, A.J. Dietrich, and C. Heyting. 1994. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 14:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche, T., S.G. Martin, M. Gotta, H.C. Gorham, F.E. Pryde, E.J. Louis, and S.M. Gasser. 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8:653–656. [DOI] [PubMed] [Google Scholar]
- Lichten, M. 2001. Meiotic recombination: breaking the genome to save it. Curr. Biol. 11:R253–R256. [DOI] [PubMed] [Google Scholar]
- Lillie, S.H., and S.S. Brown. 1998. Smy1p, a kinesin-related protein that does not require microtubules. J. Cell Biol. 140:873–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl, J. 1990. The initiation of meiotic chromosome pairing: the cytological view. Genome. 33:759–778. [DOI] [PubMed] [Google Scholar]
- Loidl, J., F. Klein, and H. Scherthan. 1994. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J. Cell Biol. 125:1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., N.M. Wilson, M.E. Petracek, and J. Berman. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16:225–239. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah, S.K., J.M. Turner, F. Baudat, E.P. Rogakou, P. de Boer, J. Blanco-Rodriguez, M. Jasin, S. Keeney, W.M. Bonner, and P.S. Burgoyne. 2001. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27:271–276. [DOI] [PubMed] [Google Scholar]
- Molnar, M., E. Doll, A. Yamamoto, Y. Hiraoka, and J. Kohli. 2003. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116:1719–1731. [DOI] [PubMed] [Google Scholar]
- Nag, D.K., H. Scherthan, B. Rockmill, J. Bhargava, and G.S. Roeder. 1995. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 141:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo, E.R., A.L. Pidoux, P.E. Perry, and R.C. Allshire. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 392:825–828. [DOI] [PubMed] [Google Scholar]
- Niwa, O., M. Shimanuki, and F. Miki. 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 19:3831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenberg, H.H., J.A. Schalk, R.L. Meuwissen, M. van Aalderen, H.A. Kester, A.J. Dietrich, and C. Heyting. 1998. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 26:2572–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S.L., and R.S. Hawley. 2003. Chromosome choreography: the meiotic ballet. Science. 301:785–789. [DOI] [PubMed] [Google Scholar]
- Page, S.L., and R.S. Hawley. 2004. The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20:525–558. [DOI] [PubMed] [Google Scholar]
- Pasierbek, P., M. Födermayr, V. Jantsch, M. Jantsch, D. Schweizer, and J. Loidl. 2003. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp. Cell Res. 289:245–255. [DOI] [PubMed] [Google Scholar]
- Pederson, T., and U. Aebi. 2002. Actin in the nucleus: what form and what for? J. Struct. Biol. 140:3–9. [DOI] [PubMed] [Google Scholar]
- Peterson, J.R., and T.J. Mitchison. 2002. Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton. Chem. Biol. 9:1275–1285. [DOI] [PubMed] [Google Scholar]
- Petronczki, M., M.F. Siomos, and K. Nasmyth. 2003. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 112:423–440. [DOI] [PubMed] [Google Scholar]
- Pezzi, N., I. Prieto, L. Kremer, L.A. Perez Jurado, C. Valero, J. Del Mazo, A.C. Martinez, and J.L. Barbero. 2000. STAG3, a novel gene encoding a protein involved in meiotic chromosome pairing and location of STAG3-related genes flanking the Williams-Beuren syndrome deletion. FASEB J. 14:581–592. [DOI] [PubMed] [Google Scholar]
- Prieto, I., J.A. Suja, N. Pezzi, L. Kremer, A.C. Martinez, J.S. Rufas, and J.L. Barbero. 2001. Mammalian STAG3 is a cohesin specific to sister chromatid arms in meiosis I. Nat. Cell Biol. 3:761–766. [DOI] [PubMed] [Google Scholar]
- Prieto, I., N. Pezzi, J.M. Buesa, L. Kremer, I. Barthelemy, C. Carreiro, F. Roncal, A. Martinez, L. Gomez, R. Fernandez, et al. 2002. STAG2 and Rad21 mammalian mitotic cohesins are implicated in meiosis. EMBO Rep. 3:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova, E., M. Eijpe, C. Heyting, C.A. Hodges, P.A. Hunt, B. Liebe, H. Scherthan, and R. Jessberger. 2004. Cohesin SMC1beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6:555–562. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G.S. Roeder. 1990. Meiosis in asynaptic yeast. Genetics. 126:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and G.S. Roeder. 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 12:2574–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, G.S., and J.M. Bailis. 2000. The pachytene checkpoint. Trends Genet. 16:395–403. [DOI] [PubMed] [Google Scholar]
- Roth, R., and H.O. Halvorson. 1969. Sporulation of yeast harvested during logarithmic growth. J. Bacteriol. 98:831–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen, K., J. Paranko, and M. Parvinen. 1982. A colcemid-sensitive mechanism involved in regulation of chromosome movements during meiotic pairing. Chromosoma. 85:611–618. [DOI] [PubMed] [Google Scholar]
- Scherthan, H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2:621–627. [DOI] [PubMed] [Google Scholar]
- Scherthan, H. 2003. Knockout mice provide novel insights into meiotic chromosome and telomere dynamics. Cytogenet. Genome Res. 103:235–244. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., and E. Trelles-Sticken. 2002. Yeast FISH: delineation of chromosomal targets in vegetative and meiotic yeast cells. Springer Lab Manual on FISH Technology. T. Liehr, editor. Springer-Verlag New York Inc., Berlin/New York. 329–345.
- Siomos, M.F., A. Badrinath, P. Pasierbek, D. Livingstone, J. White, M. Glotzer, and K. Nasmyth. 2001. Separase is required for chromosome segregation during meiosis I in Caenorhabditis elegans. Curr. Biol. 11:1825–1835. [DOI] [PubMed] [Google Scholar]
- Smith, A.V., and G.S. Roeder. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136:957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi, A., S. Tesse, S. Gargano, F. James, N. Kleckner, and D. Zickler. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes Dev. 17:2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia, C., G. Blobel, and M.P. Rout. 1999. Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol. 144:839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, A., K.P. Rabitsch, M. Galova, A. Schleiffer, S.B. Buonomo, and K. Nasmyth. 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 103:1155–1168. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., J. Loidl, and H. Scherthan. 1999. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112:651–658. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken, E., M.E. Dresser, and H. Scherthan. 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken, E., J. Loidl, and H. Scherthan. 2003. Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J. Cell Sci. 116:2431–2442. [DOI] [PubMed] [Google Scholar]
- Ueno, M., T. Murase, T. Kibe, N. Ohashi, K. Tomita, Y. Murakami, M. Uritani, T. Ushimaru, and M. Harata. 2004. Fission yeast Arp6 is required for telomere silencing, but functions independently of Swi6. Nucleic Acids Res. 32:736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- von Wettstein, D., S.W. Rasmussen, and P.B. Holm. 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18:331–413. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and P. Nurse. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464. [DOI] [PubMed] [Google Scholar]
- Weiner, B.M., and N. Kleckner. 1994. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 77:977–991. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., R.R. West, J.R. McIntosh, and Y. Hiraoka. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol. 145:1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T.G., Y. Chikashige, F. Ozoe, M. Kawamukai, and Y. Hiraoka. 2004. Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J. Cell Sci. 117:3875–3886. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., and D.E. Koshland. 2003. Meiotic condensin is required for proper chromosome compaction, SC assembly, and resolution of recombination-dependent chromosome linkages. J. Cell Biol. 163:937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., J.G. Liu, J. Zhao, E. Brundell, B. Daneholt, and C. Hoog. 2000. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 5:73–83. [DOI] [PubMed] [Google Scholar]
- Zetka, M.C., I. Kawasaki, S. Strome, and F. Muller. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13:2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner. 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32:619–697. [DOI] [PubMed] [Google Scholar]