Abstract
Receptor-stimulated Ca2+ signals come in several flavors. The Ca2+ signals can be decoded linearly or by integration of the response. How the duration of the signal conveyed by cytosolic Ca2+ concentration ([Ca2+]i) changes is regulated is not well understood. Liu et al. (Liu, Q., S.A. Walker, D. Gao, J.A. Taylor, Y.-F. Dai, R.S. Arkell, M.D. Bootman, H.L. Roderick, P.J. Cullen, and P.J. Lockyer. 2005. J. Cell Biol. 170:183–190) now report an example of decoding based on the differential regulation of Ras function by two Ca2+-sensitive Ras inhibitors: Ca2+-promoted Ras activator (CAPRI), which extends the duration of the effect of Ca2+ on Ras activity, and Ras GTPase activating-like protein (RASAL), which functions as a linear decoder of the Ca2+ signal.
The Ca2+ signal generated by physiological agonist concentrations is most often in the form of Ca2+ oscillations that initiate at discrete cellular sites and can either remain confined to a particular cellular microdomain (such as the apical pole of secretory cells) or propagate to all cellular domains in the form of repetitive Ca2+ waves (Petersen, 2004). The amplitude and frequency of the Ca2+ oscillations and speed of the Ca2+ waves are regulated by the intensity of the stimulus (Berridge et al., 2000). Furthermore, the pattern of the Ca2+ oscillations and waves are receptor specific in the same cell (Kiselyov et al., 2003). These intricate Ca2+ signals regulate cellular activities as diverse as vision and neurotransmitter release on a micro- or millisecond time scale and gene regulation on the scale of hours or days (Berridge et al., 2003).
How is the same Ca2+ signal decoded to regulate multiple functions with different spatial and temporal characteristics? A simple form of decoding is an exact translation of the Ca2+ signal to a physiological response. An example is direct binding of Ca2+ to effector proteins, such as the Ca2+-activated Cl− and K+ channels in epithelia (Melvin et al., 2005). Another example is the uptake by the mitochondria of Ca2+ released from the ER to regulate enzymes involved in mitochondrial energy metabolism (Hajnoczky et al., 1995). A more complex form of decoding is achieved by differential Ca2+ sensitivity of the sensors. An example is the family of synaptotagmins, which display a range of apparent affinities for Ca2+ to confer specific Ca2+ dependence to exocytotic events (Sudhof, 2004). The most intricate form of decoding requires translating the information in the frequency and amplitude of Ca2+ oscillations into a physiological response, such as the activation of the NFAT, OAP, and NF-κB transcription factors (Dolmetsch et al., 1998; Li et al., 1998; Tomida et al., 2003). The molecular mechanism responsible for this form of decoding is not known.
Although we intuitively assume that decoding of complex Ca2+ signals involves activation of multiple Ca2+ sensors with different spatial and temporal characteristics that converge on the same pathway, to date there has been no clear example of this. The study by Liu et al. (2005) shows the selective response of two Ras GTPase-activating proteins (GAPs), Ca2+-promoted Ras activator (CAPRI), and Ras GTPase activating-like protein (RASAL) to complex Ca2+ signals.
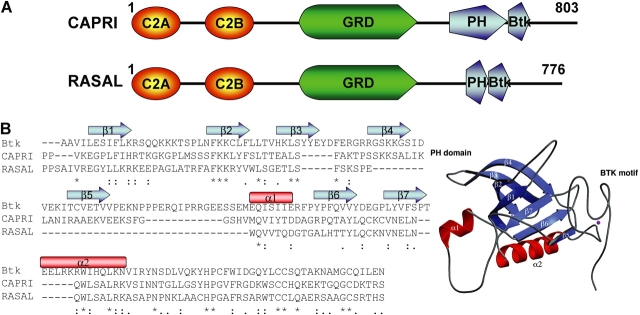
Ras proteins are binary switches that cycle between the GTP-active and GDP-inactive forms and regulate many signaling pathways, including the MAPK cascade, to regulate cell proliferation and differentiation (Downward, 2003). The intensity and duration of Ras activation is determined by the relative activity of the guanine nucleotide exchange factors (GEFs) that catalyze the exchange of GDP for GTP to activate Ras and the GAPs that catalyze the intrinsic Ras GTPase and inactivate Ras. Ras is activated by tyrosine kinase-linked receptors through recruitment and activation of Ca2+-independent GEFs and GAPs (Takai et al., 2001), and by Ca2+-mobilizing receptors, including G protein–coupled receptors, that recruit and activate Ca2+-dependent GEFs and GAPs (Cullen and Lockyer, 2002). Previous work identified CAPRI and RASAL as Ca2+-dependent Ras GAPs (Lockyer et al., 2001). CAPRI and RASAL are members of the GAP1 family that also includes GAP1IP4BP and GAP1m (Cullen, 1998). As illustrated in Fig. 1 A for RASAL and CAPRI, the family is typified by four conserved structural domains; tandem C2 domains; a GAP-related domain (GRD); a pleckstrin homology (PH) domain, and a Bruton's tyrosine kinase motif (Btk). The C2 domains of GAP1IP4BP and GAP1m do not mediate Ca2+-dependent plasma membrane (PM) translocation (Lockyer et al., 1997); however, the C2 domains are essential for the Ca2+-dependent translocation of CAPRI and RASAL.
Figure 1.
Structural motif prediction and alignment of PH domains. Structural motif prediction and alignment of PH domains was done using the 3D-PSSM Web Server V 2.6.0 at http://www.sbg.bio.ic.ac.uk/~3dpssm. The structure of BTK PH domain was taken from Hyvonen and Saraste (1997).
In an earlier work, Walker et al. (2004) showed that RASAL faithfully translates the changes in [Ca2+]i to PM translocation events and inactivation of Ras. Intense receptor stimulation resulted in a single [Ca2+]i transient and a matching transient translocation of RASAL to the PM, whereas weak receptor stimulation generated Ca2+ oscillations and a synchronized oscillatory translocation of RASAL. Translocation required RASAL's tandem C2 domains—a finding confirmed in Liu et al. (2005). Hence, the response of RASAL to [Ca2+]i indicates that RASAL is a linear decoder of [Ca2+]i signals.
Although the translocation of CAPRI to the PM is also dependent on its tandem C2 domains, Liu et al. (2005) report that CAPRI does not behave as a linear decoder of [Ca2+]i changes. The initial [Ca2+]i increase triggered by intense or weak stimulation caused C2 domain–dependent translocation of CAPRI to the PM. CAPRI remained at the PM despite the reduction of [Ca2+]i to a plateau or maintained Ca2+ oscillations. Switching between RASAL and CAPRI C2 domains converted CAPRI to a linear decoder of Ca2+ oscillations and conferred persistent retention of RASAL at the PM. Importantly, Liu et al. (2005) found that the removal of external Ca2+ to prevent Ca2+ influx by store-operated channels converted CAPRI translocation from persistent to transient, and removal of the stimulus rapidly retrieved CAPRI from the PM. The dependence of CAPRI's retention at the PM on Ca2+ influx indicates that CAPRI is a sensor of Ca2+ influx. Ca2+ influx may maintain high Ca2+ levels next to the PM. The requirement for persistent agonist stimulation may reflect dependence on receptor-mediated lipid metabolism and binding of the PH domain of CAPRI to phosphatidylinositides because mutation of the conserved tryptophan of the PH domain of CAPRI (W664A) resulted in a transient, rather than persistent, translocation of CAPRI to the PM. Sequence analysis suggests that the PH domains of CAPRI and RASAL are most similar to that of Bruton's tyrosine kinase (Hyvonen and Saraste, 1997). The predicted CAPRI PH domain is more intact than the RASAL PH domain (Fig. 1 B) and it is functional, as evident from the effect of the W664A mutation. The RASAL PH domain lacks important structural motifs, which may render it nonfunctional. Indeed, Liu et al. (2005) show that PM retention segregates with CAPRI's PH domain.
The cooperation between the tandem C2 domains and the PH domain makes CAPRI an integrator of the Ca2+ signal. As a consequence, the duration of CAPRI's retention at the PM, and therefore its action as a GAP, is markedly extended. Hence, the study of Liu et al. (2005) provides an example for a mechanism by which the timing and duration of the Ca2+ signal can be differentially decoded. CAPRI and RASAL are both deactivators of Ras, and their combined action can be used to decode the complex forms of the receptor-evoked Ca2+ oscillations and waves.
The findings of Liu et al. (2005) raise several questions. For example, do the cells use the properties of the two GAPs to regulate physiological functions of different duration? Recent work showed that activation of Ras and the ERK/MAPK cascade is sensitive to the frequency of Ca2+ oscillations (Kupzig et al., 2005). What is the role of CAPRI and/or RASAL in this form of regulation? How do the tandem C2 domains of CAPRI mediate [Ca2+]i-dependent translocation while the PH domain mediates Ca2+ influx-dependent retention? Decoding of Ca2+ signals by CAPRI and RASAL is only one side of the equation. Two families of Ca2+-dependent GEFs have been identified: Ras-GRF1 and Ras-GRF2, and the Ras-GRP/CalDAG GEFs (Cullen and Lockyer, 2002). At present, however, it is not known how these GEFs decode Ca2+ signals. Further insight into this process is needed before we can solve the puzzle of how a complex pathway like the Ras/MAPK pathway decodes agonist-evoked Ca2+ signaling.
Acknowledgments
Note added in proof. An additional mechanism to regulate the duration of Ras signaling is through transcriptional regulation of RASAL and CAPRI expression levels. A recent study, searching for tumor suppressors that affect the activity of wild-type Ras, reported that the transcription factor PITX1 increased the expression of RASAL (Kolfschoten et al., 2005). The down-regulation of PITX1, resulting in a reduced level of RASAL and an increased level of active Ras, was found in several prostate, bladder, and colon cancers. Restoring PITX1 expression to colon cancer cell lines inhibited tumorigenesis in a Ras-dependent manner (Kolfschoten et al., 2005). These studies establish RASAL as a tumor suppressor whose long-term activity can be controlled by expression, and whose acute activity can be controlled by [Ca2+]i.
Abbreviations used in this paper: CAPRI, Ca2+-promoted Ras activator; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; PH, pleckstrin homology; PM, plasma membrane; RASAL, Ras GTPase activating-like protein.
References
- Berridge, M.J., P. Lipp, and M.D. Bootman. 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1:11–21. [DOI] [PubMed] [Google Scholar]
- Berridge, M.J., M.D. Bootman, and H.L. Roderick. 2003. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4:517–529. [DOI] [PubMed] [Google Scholar]
- Cullen, P.J. 1998. Bridging the GAP in inositol 1,3,4,5-tetrakisphosphate signalling. Biochim. Biophys. Acta. 1436:35–47. [DOI] [PubMed] [Google Scholar]
- Cullen, P.J., and P.J. Lockyer. 2002. Integration of calcium and Ras signalling. Nat. Rev. Mol. Cell Biol. 3:339–348. [DOI] [PubMed] [Google Scholar]
- Dolmetsch, R.E., K. Xu, and R.S. Lewis. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 392:933–936. [DOI] [PubMed] [Google Scholar]
- Downward, J. 2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 3:11–22. [DOI] [PubMed] [Google Scholar]
- Hajnoczky, G., L.D. Robb-Gaspers, M.B. Seitz, and A.P. Thomas. 1995. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 82:415–424. [DOI] [PubMed] [Google Scholar]
- Hyvonen, M., and M. Saraste. 1997. Structure of the PH domain and Btk motif from Bruton's tyrosine kinase: molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16:3396–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov, K., D.M. Shin, and S. Muallem. 2003. Signalling specificity in GPCR-dependent Ca2+ signalling. Cell. Signal. 15:243–253. [DOI] [PubMed] [Google Scholar]
- Kolfschoten, I.G., B. van Leeuwen, K. Berns, J. Mullenders, R.L. Beijersbergen, R. Bernards, P.M. Voorhoeve, and R. Agami. 2005. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 121:849–858. [DOI] [PubMed] [Google Scholar]
- Kupzig, S., S.A. Walker, and P.J. Cullen. 2005. The frequencies of calcium oscillations are optimized for efficient calcium-mediated activation of Ras and the ERK/MAPK cascade. Proc. Natl. Acad. Sci. USA. 102:7577–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., J. Llopis, M. Whitney, G. Zlokarnik, and R.Y. Tsien. 1998. Cell-permeant caged InsP3 ester shows that Ca2+ spike frequency can optimize gene expression. Nature. 392:936–941. [DOI] [PubMed] [Google Scholar]
- Liu, Q., S.A. Walker, D. Gao, J.A. Taylor, Y.-F. Dai, R.S. Arkell, M.D. Bootman, H.L. Roderick, P.J. Cullen, and P.J. Lockyer. 2005. CAPRI and RASAL impose different modes of information processing on Ras due to contrasting temporal filtering of Ca2+. J. Cell Biol. 170:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer, P.J., J.R. Bottomley, J.S. Reynolds, T.J. McNulty, K. Venkateswarlu, B.V. Potter, C.E. Dempsey, and P.J. Cullen. 1997. Distinct subcellular localisations of the putative inositol 1,3,4,5-tetrakisphosphate receptors GAP1IP4BP and GAP1m result from the GAP1IP4BP PH domain directing plasma membrane targeting. Curr. Biol. 7:1007–1010. [DOI] [PubMed] [Google Scholar]
- Lockyer, P.J., S. Kupzig, and P.J. Cullen. 2001. CAPRI regulates Ca2+-dependent inactivation of the Ras-MAPK pathway. Curr. Biol. 11:981–986. [DOI] [PubMed] [Google Scholar]
- Melvin, J.E., D. Yule, T. Shuttleworth, and T. Begenisich. 2005. Regulation of fluid and electrolyte secretion in salivary gland acinar cells. Annu. Rev. Physiol. 67:445–469. [DOI] [PubMed] [Google Scholar]
- Petersen, O.H. 2004. Local and global Ca2+ signals: physiology and pathophysiology. Biol. Res. 37:661–664. [DOI] [PubMed] [Google Scholar]
- Sudhof, T.C. 2004. The synaptic vesicle cycle. Annu. Rev. Neurosci. 27:509–547. [DOI] [PubMed] [Google Scholar]
- Takai, Y., T. Sasaki, and T. Matozaki. 2001. Small GTP-binding proteins. Physiol. Rev. 81:153–208. [DOI] [PubMed] [Google Scholar]
- Tomida, T., K. Hirose, A. Takizawa, F. Shibasaki, and M. Iino. 2003. NFAT functions as a working memory of Ca2+ signals in decoding Ca2+ oscillation. EMBO J. 22:3825–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S.A., S. Kupzig, D. Bouyoucef, L.C. Davies, T. Tsuboi, T.G. Bivona, G.E. Cozier, P.J. Lockyer, A. Buckler, G.A. Rutter, et al. 2004. Identification of a Ras GTPase-activating protein regulated by receptor-mediated Ca2+ oscillations. EMBO J. 23:1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]