Abstract
The tumor suppressor lethal giant larvae (Lgl) plays a critical role in epithelial cell polarization. However, the molecular mechanism by which Lgl carries out its functions is unclear. In this study, we report that the yeast Lgl proteins Sro7p and Sro77p directly interact with Exo84p, which is a component of the exocyst complex that is essential for targeting vesicles to specific sites of the plasma membrane for exocytosis, and that this interaction is important for post-Golgi secretion. Genetic analyses demonstrate a molecular pathway from Rab and Rho GTPases through the exocyst and Lgl to SNAREs, which mediate membrane fusion. We also found that overexpression of Lgl and t-SNARE proteins not only improves exocytosis but also rescues polarity defects in exocyst mutants. We propose that, although Lgl is broadly distributed in the cells, its localized interaction with the exocyst and kinetic activation are important for the establishment and reenforcement of cell polarity.
Introduction
Cell polarity is a basic feature of eukaryotic cells from budding yeast to mammalian epithelia. Studies in a wide variety of systems have begun to reveal evolutionally conserved mechanisms that regulate cytoskeleton and membrane traffic for the generation and maintenance of cell polarity (Mostov et al., 2003; for reviews see Wodarz, 2002; Nelson, 2003). Lethal (2) giant larvae (Lgl) was first identified in Drosophila melanogaster as a tumor suppressor (Gateff, 1978). Its homologues were later identified in yeast and mammalian cells (Kagami et al., 1998; Larsson et al., 1998; Lehman et al., 1999; Musch et al., 2002; Widberg et al., 2003). Research in flies and mammals demonstrated that Lgl proteins play important roles in the establishment of epithelial cell polarity and in the asymmetric distribution of cell fate determinants in neuroblasts during development (Manfruelli et al., 1996; Bilder et al., 2000; Ohshiro et al., 2000; Peng et al., 2000; Musch et al., 2002). Genetic and morphological analyses suggest that Lgl proteins function in concert with two other tumor suppressors: Scrib and Dlg (Bilder et al., 2000). Recently, it was shown that Lgl interacts with the apical domain Par6–Par3–aPKC complex and is phosphorylated by aPKC (Betschinger et al., 2003; Justice and Jan, 2003; Plant et al., 2003). In mammalian fibroblasts, it was shown that Lgl phosphorylation is important for cell migration (Plant et al., 2003). In D. melanogaster, Lgl phosphorylation may help to exclude Lgl function from the apical membrane (Betschinger et al., 2003).
The yeast homologues of Lgl—Sro7p and Sro77p—were originally identified as high-copy suppressors of Rho3p mutants, which is important for actin organization and polarized exocytosis (Kagami et al., 1998). Sro7p and Sro77p are homologous in sequence and share overlapping functions (Kagami et al., 1998; Larsson et al., 1998). Recent studies suggest that Lgl proteins regulate the late stage of exocytosis (Lehman et al., 1999; Musch et al., 2002; Widberg et al., 2003). For example, in epithelial cells, Lgl associates with syntaxin 4, the t-SNARE protein that mediates vesicle fusion at the basolateral domain of the plasma membrane (Musch et al., 2002). In yeast, Sro7p and Sro77p directly interact with the t-SNARE protein Sec9p, and double deletion of SRO7 and SRO77 genes results in exocytosis defects (Lehman et al., 1999). Based on the findings above, it is speculated that the role of Lgl in cell polarization may be attributed to its function in exocytosis, and it may promote targeted vesicle fusion at specific areas of the plasma membrane (Bilder et al., 2000; Musch et al., 2002). However, Lgl proteins themselves may not be able to confer the targeting information, as its distribution in the cell is not very restrictive. For example, in fly cells, Lgl is localized to both the cytosol and cell cortex (Bilder et al., 2000; Ohshiro et al., 2000; Peng et al., 2000). In epithelial cells, Lgl is distributed in the cytosol and is recruited to the lateral membrane after cell–cell contact–initiated polarization (Musch et al., 2002). Although aPKC phosphorylation may help to limit the function of Lgl to the basolateral domain (Betschinger et al., 2003), there is evidence that vesicle targeting may be restricted to the region of the lateral membrane close to the junctional complex. (Louvard, 1980; Kreitzer et al., 2003). In yeast, Sro7p is distributed in the cytosol and along the entire plasma membrane (Larsson et al., 1998; Lehman et al., 1999). However, exocytosis is restricted to the bud tips in growing daughter cells. Therefore, there must be additional mechanisms that spatially restrict and/or kinetically promote the action of Lgl in exocytosis at specific areas of the plasma membrane.
The exocyst is an evolutionally conserved octameric protein complex consisting of Sec3p, Sec5p, Sec6p, Sec8p, Sec10p, Sec15p, Exo70p, and Exo84p. This complex is localized to specific sites of the plasma membrane, where it tethers secretory vesicles for subsequent SNARE assembly and exocytosis (Lipschutz and Mostov, 2002; Novick and Guo, 2002; Hsu et al., 2004). How the exocyst, as a tethering complex (Whyte and Munro, 2002), communicates with and activates SNAREs is unknown. The exocyst is polarized to sites of active exocytosis and cell surface expansion. In epithelial cells, the exocyst localizes to trans-Golgi and recycling endosomes (Yeaman et al., 2001; Fölsch et al., 2003) and is recruited to the vicinity of adherens junctions upon the generation of apical–basolateral asymmetry, where it mediates exocytosis exclusively at the basolateral membrane (Grindstaff et al., 1998). In yeast, exocyst proteins localize to the bud tips and mother–daughter junctions, which are sites of active exocytosis and cell growth (TerBush and Novick, 1995; Finger et al., 1998, Guo et al., 1999a). This pattern of localization is in contrast to that of t-SNAREs (Sso1p and Sso2p), which are distributed along the entire cell membrane (Brennwald et al., 1994). The yeast exocyst is a direct downstream effector of the Rab protein Sec4p (Guo et al., 1999b). In addition, the yeast exocyst is under the control of the Rho family of small GTPases, which coordinates actin organization and the membrane traffic for polarized cell growth (Adamo et al., 1999, 2001; Guo et al., 1999b; Robinson et al., 1999; Zhang et al., 2001).
In this study, we report that the yeast Lgl proteins Sro7/77p directly interact with a component of the exocyst, Exo84p, and disruption of this interaction leads to defects in exocytosis. Genetic analyses using an array of yeast mutants demonstrate a pathway in which the exocyst and Sro7 mediate signaling from the Rho3p and Rab protein Sec4p to control SNARE assembly. We also demonstrate that overexpression of Lgl and t-SNAREs not only helps improve exocytosis kinetically in exocyst mutants but also rescues the polarity defects in these cells. Our results suggest that the interaction of Lgl with the exocyst and upstream small GTPases at specific domains of the plasma membrane is important for localized SNARE assembly and polarized exocytosis. Conversely, up-regulation of Lgl and downstream t-SNARE functions may reenforce cell polarity. Altogether, our studies shed light on the molecular function of Lgl in exocytosis and cell polarization.
Results
Sro7p and Sro77p interact with the exocyst component Exo84p
In a search for proteins that interact with the exocyst, we identified Sro7p as a binding protein for Exo84p in a yeast two-hybrid screen. Further analyses indicate that Sro7p and its homologue Sro77p interact with Exo84p but not with any other component of the exocyst complex (Fig. 1 A). Domain mapping showed that the NH2 terminus of Exo84p (aa 1–327) binds Sro7p and Sro77p (Fig. 1 B). Sro7p and Sro77p were expressed at similar levels in the yeast host cells that were used in the two-hybrid assay (Fig. 1 C).
Figure 1.
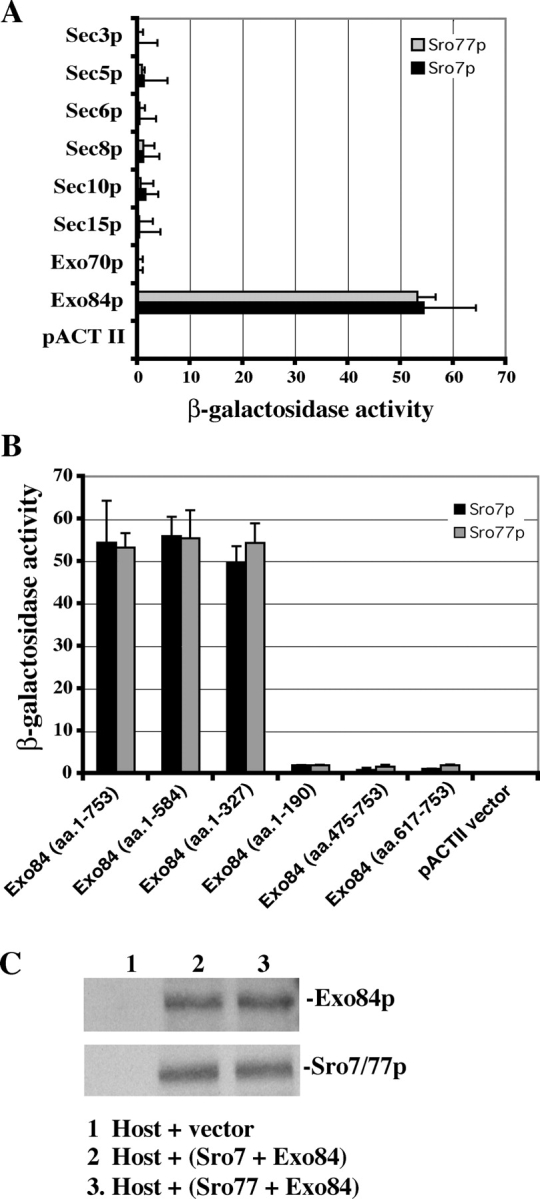
The yeast two-hybrid assay of the interaction between Sro7p/Sro77p and Exo84p. (A) Sro7p and Sro77p interact with Exo84p but not with other members of the exocyst complex. SRO7 and SRO77 were subcloned into the pOBD8 vector. The construct was used to test the interactions of Sro7p and Sro77p with eight members of the exocyst. The average and SD of β-galactosidase activities were determined from three independent experiments. (B) The interaction of different fragments of Exo84p with both Sro7p and Sro77p were tested by the yeast two-hybrid assay. The region of Exo84p that binds Sro7p and Sro77p was mapped to the NH2-terminal 327 amino acids of Exo84p. Error bars represent SD. (C) Sro7p and Sro77p were expressed at the same level in the two-hybrid host cells (Y190). Lysates from yeast cells used in the two-hybrid assay coexpressing Exo84p and Sro7p or Sro77p were subjected to 8% SDS-PAGE. Amounts of Sro7p, Sro77p, and Exo84p were detected by Western blotting using anti-HA mAb against the HA tag engineered at the NH2 termini of expressed proteins in two-hybrid vectors. The fusion proteins of Sro7p and Sro77p migrated at almost the same position on SDS-PAGE. (lane 1) Host cells transformed with the pACTII vector; (lane 2) host cells expressing Sro7p and Exo84p; (lane 3) host cells expressing Sro77p and Exo84p.
The two-hybrid results were confirmed by in vitro binding assays (Fig. 2). The full-length Sro7p was synthesized by in vitro transcription and translation in the presence of [35S]methionine. Then, the radiolabeled Sro7p was incubated with Exo84p NH2-terminal domain (aa 1–320) GST fusion protein (GST-Exo84N) that was purified from Escherichia coli. As shown in Fig. 2 A, >70% of Sro7p (the double bands) binds to GST-Exo84N Sepharose beads. The binding was also performed in the opposite direction, in which Exo84p was in vitro translated, and Sro7p was expressed in yeast as a protein A fusion that was purified by using IgG–Sepharose beads (Fig. 2 B). More than 75% of input Exo84p bound to the protein A–Sro7p Sepharose beads. To obtain the affinity of the interaction between Exo84p and Sro7p, we purified the protein A–tagged Sro7p from yeast lysates and purified GST-Exo84N from E. coli. Various amounts of purified GST-Exo84N were incubated with 10 nM protein A–Sro7p that was conjugated to IgG–Sepharose beads for the binding reaction. The protein samples were subjected to SDS-PAGE, and the gels were stained by Coomassie blue to visualize the proteins. The bound and free GST-Exo84N were quantified and plotted with a single rectangular hyperbola equation: B = BmaxC/(Kd + C), where Kd is/represents the dissociation constant, B is bound, and C is free GST-exo84N (Fig. 2 C). The dissociation constant was calculated from each plot by nonlinear regression; average Kd = 46 ± 6 nM (n = 4).
Figure 2.
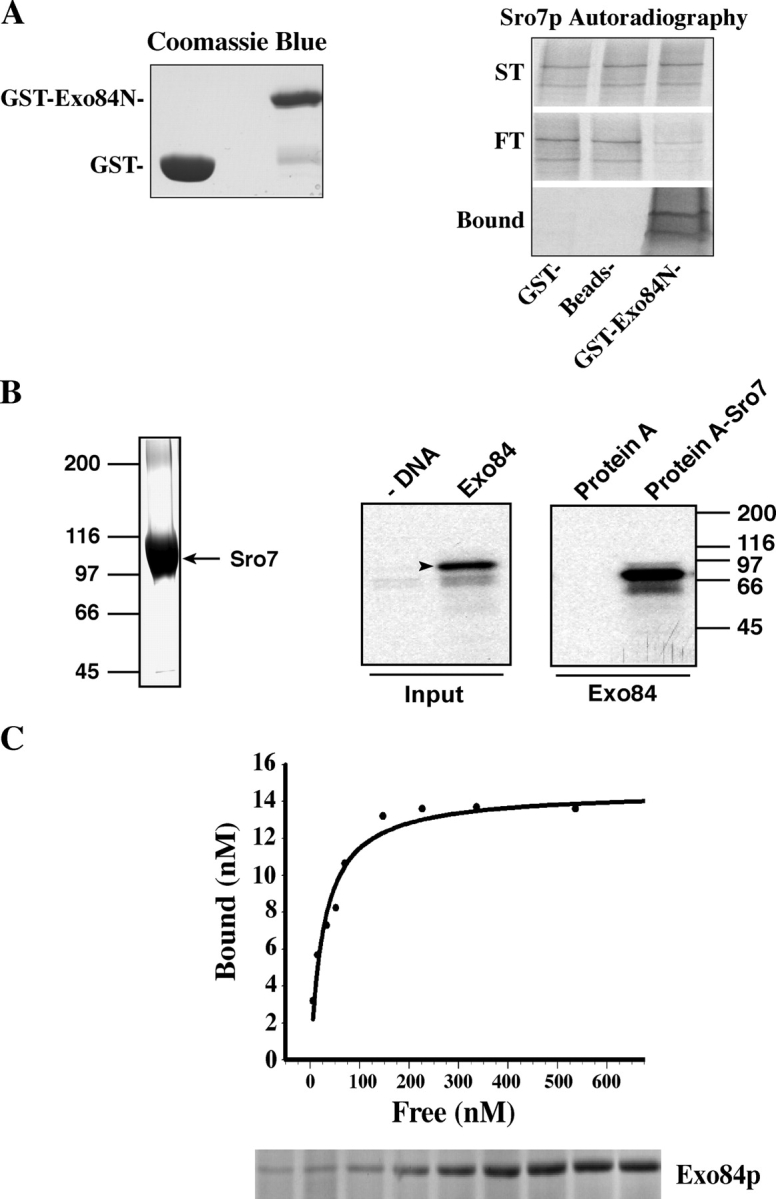
Sro7p interacts with Exo84p in vitro. (A) Sro7p specifically binds to the NH2 terminus of Exo84p in vitro. The NH2 terminus of Exo84p (aa 1–320) was expressed as a GST fusion protein and was immobilized to glutathione–Sepharose 4B; Sro7p was in vitro translated in the reticulocyte lysate system in the presence of [35S]methionine. The proteins as well as controls (GST–Sepharose beads and unconjugated Sepharose beads) were used in the binding reaction. The starting materials (ST), bound, and unbound (FT) Sro7p were analyzed by electrophoresis and autoradiography. (left) Coomassie blue staining of input GST and GST-Exo84N fusion protein. (B) Protein A–Sro7 recombinant protein was expressed in yeast and was purified by IgG–Sepharose beads. An aliquot of the fusion protein that bound to Sepharose beads was shown in the Coomassie blue–stained gel (left). Full-length Exo84p was in vitro transcribed and translated in the presence of [35S]methionine (arrowhead). Exo84p that bound to the Sepharose beads was shown in the autoradiograph. 50% of input Exo84p used in this binding assay is shown to the left. As a control, protein A–Sepharose beads were used in the binding reaction. (C) The dissociation constant of Sro7p–Exo84p interaction. Protein A–Sro7p was purified from yeast and was conjugated to IgG–Sepharose beads. Various amounts of purified GST-Exo84N were incubated with protein A–Sro7p Sepharose beads for the binding reaction. The protein samples were subjected to SDS-PAGE, and the gels were stained by Coomassie blue. Bound and free GST-Exo84N were quantified and plotted with a single rectangular hyperbola equation (B = BmaxC/[Kd + C]) by using SigmaPlot software. (bottom) Bound GST-Exo84N stained by Coomassie blue.
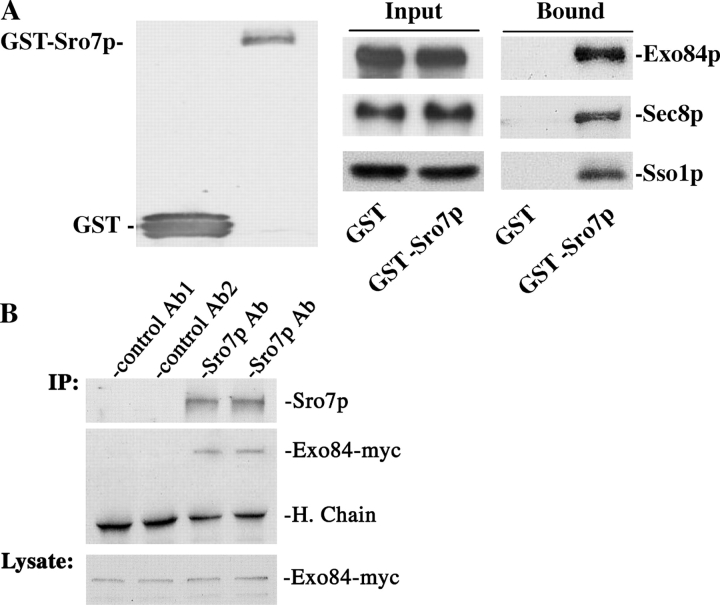
To test whether this interaction takes place in vivo, GST-Sro7p was expressed in a yeast strain with its endogenous Exo84 (the chromosomal copy) tagged by the myc epitope (Exo84p-myc). As shown in Fig. 3 A, Exo84p coprecipitated with GST-Sro7p that was bound to the glutathione–Sepharose beads. Sec8p also coprecipitated, suggesting that other members of the exocyst complex indirectly bind to Sro7p in the cell through Exo84p. As a control, GST does not bind to Exo84p or Sec8p. Using this assay, we were also able to demonstrate the interaction between Sro7p and the t-SNARE protein Sso1p, which is consistent with our previous observation (Lehman et al., 1999). In addition to testing in vivo binding by using GST-tagged Sro7p, we also performed immunoprecipitation experiments so that the binding of Sro7p and Exo84p at their endogenous levels could be examined. As shown in Fig. 3 B, the anti-Sro7p antibody was able to precipitate Sro7p, and Exo84p was coprecipitated. The amount of Exo84p that coimmunoprecipitated with Sro7p was ∼1.2% of the input. As controls, polyclonal antibodies against GFP or GST failed to immunoprecipitate these proteins.
Figure 3.
Sro7p interacts with Exo84p in vivo. (A) GST-Sro7p was coexpressed with myc-tagged Exo84p or Sec8p in yeast cells. Lysates from the GST vector control and GST-Sro7p strains were used for coprecipitation with glutathione–Sepharose 4B to test the interaction between Sro7p and exocyst proteins. The precipitates were separated by SDS-PAGE and were subjected to Western blot analyses using anti-GST antibody to detect GST and GST-Sro7p (left), anti-myc antibody to detect Exo84-myc and Sec8-myc (right, top and middle), and a rabbit anti-Sso1p pAb to detect Sso1p (right bottom). The same amount of Exo84p, Sec8p, and Sso1p were used (Input) in the assay. (B) Coimmunoprecipitation of Sro7p and Exo84p. Yeast cells with their chromosomal copy of myc-tagged Exo84 were grown to log phase. Cell lysates were used for immunoprecipitation with rabbit anti-Sro7p antibody. Anti-GFP (control Ab1) and anti-GST (control Ab2) pAbs were used as negative controls. The precipitates were separated by SDS-PAGE and were subjected to Western blot analyses to detect Exo84-myc and Sro7p. (bottom) The same amounts of Exo84-myc were used as starting materials (1% of the input). H. Chain, heavy chains of the antibodies.
Generation and characterization of exo84 mutants that are defective in Sro7/77p interaction
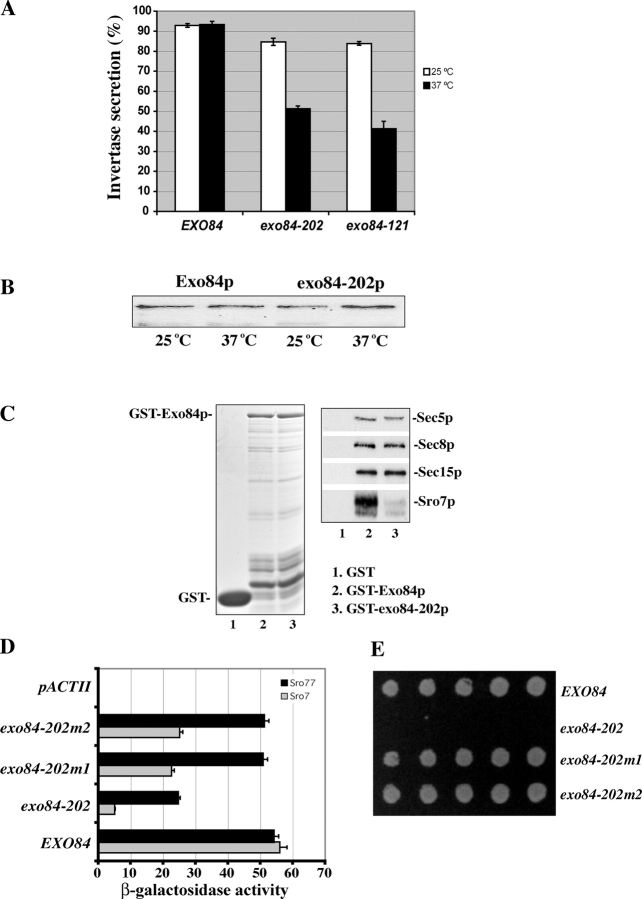
To elucidate the functional implication of the Exo84p–Sro7/77p interaction, we generated mutant yeast strains with a disruption of this interaction (Table I). Although the NH2 terminus of Exo84p interacts with Sro7/77p, our yeast two-hybrid assay and binding experiments suggest that the COOH-terminal portion of Exo84p mediates its association with other exocyst components (unpublished data). Because Exo84p interacts with Sro7/77p and other exocyst components through distinct regions, we performed “error-prone” PCR to generate exo84 mutants with mutations restricted to the NH2 terminus of Exo84p. The mutants were selected if the cells were inviable at 18 or 37°C and were defective in secretion of the invertase to the media. After random mutagenesis and plasmid shuffling, we screened ∼10,000 yeast colonies and identified one mutant allele, exo84-202, which survives at the permissive temperature of 25°C but not at 37°C. We also examined the secretion of invertase in the mutants. As shown in Fig. 4 A, ∼85% of the invertase produced in exo84-202 cells was secreted into the media at 25°C. After a 2-h shift to 37°C, only 51% of the invertase was secreted. As a positive control, we have included another exo84 mutant, exo84-121, in the assay. exo84-121 contains mutations at the COOH terminus of Exo84p and is defective in binding to other exocyst components (unpublished data). Only 42% of the invertase was secreted to the media in this mutant at 37°C. As a positive control, the wild-type cells were able to secret >90% of the invertase at 37°C. We found that the levels of the mutant exo84-202 protein remained unchanged after the cells were shifted to the restrictive temperature for 2 h (Fig. 4 B). In addition to the invertase assay, we also examined the mutant cells by thin-section EM. Several vesicles accumulated in exo84-202 cells after shifting to the restrictive temperature of 37°C (see Fig. 5 B). These vesicles were 80–100 nm in diameter, which is typical of post-Golgi secretory vesicles. In contrast, there were few such vesicles in the wild-type cells. These results indicate that disruption of the Sro7/77p–Exo84p interaction blocks post-Golgi secretion.
Table I. Yeast strains and genotypes.
| Strain | Genotype |
|---|---|
| NY768 | Matα ura3-52 leu2-3, 112 sec1-1 |
| NY770 | Matα ura3-52 leu2-3, 112 sec2-41 |
| NY772 | Matα ura3-52 leu2-3, 112 sec3-2 |
| NY774 | Matα ura3-52 leu2-3, 112 sec4-8 |
| NY776 | Matα ura3-52 leu2-3, 112 sec5-24 |
| NY778 | Matα ura3-52 leu2-3, 112 sec6-4 |
| NY780 | Matα ura3-52 leu2-3, 112 sec8-9 |
| NY782 | Matα ura3-52 leu2-3, 112 sec9-4 |
| NY784 | Matα ura3-52 leu2-3, 112 sec10-2 |
| NY786 | Matα ura3-52 leu2-3, 112 sec15-1 |
| NY2450 | Mata leu2-3, 112 ura3-52 his3Δ200 sec3::KanM |
| GY1201 | Matα ura3-52 leu2-3, 112 his3Δ200 trp1 exo70::HIS3 Gal+ (CEN exo70-38 LEU2) |
| GY1215 | Matα ura3-52 leu2-3, 112 his3Δ200 trp1 Gal+ |
| GY1236 | Mata ura3-52 leu2-3, 112 his3Δ200 trp1 exo84::HIS3 (CEN exo84-121 LEU2) LA− |
| GY1264 | Mata ura3-52 leu2-3, 112 his3Δ200 trp1 exo84::HIS3 (CEN EXO84 URA3) |
| GY1466 | Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sro77::KanM (BY4741, ATCC #4003134) sro7::URA3 |
| GY1617 | Matα ura3-52 leu2-3, 112 his3Δ200 trp1 sec8::(SEC8-12XMYC LEU2) |
| GY1717 | Matα ura3-52 leu2-3, 112 his3Δ200 trp1 exo84::(Exo84-12XMYC LEU2) |
| GY1853 | Mata, his3Δ1, leu-2Δ0, met15Δ0, ura3Δ0, sro77::KanM, exo84::HIS3 (TEF-Exo84 URA CEN) |
| GY1964 | Mata ura3-52 leu2-3, 112 his3Δ200 trp1 exo84::HIS3 (CEN exo84-202-12XMYC LEU2) |
| GY1965 | Mata, his3Δ1, leu-2Δ0, met15Δ0, ura3Δ0, sro77::KanM, exo84::HIS3 (TEF-exo84-202-12myc LEU2 CEN) |
| GY1967 | Mata ura3-52 leu2-3, 112 his3Δ200 trp1 exo84::HIS3 (CEN EXO84-12XMYC LEU2) |
| GY2180 | Mata ura3-52 leu2-3, 112 his3Δ200 trp1 exo70::HIS3 (CEN EXO70 URA3) |
| GY2239 | Matα ura3-52 leu2-3, 112 his3Δ200 trp1 sec15::(SEC15-12xMYC LEU2) |
| GY2245 | Mata, ura3-52, leu-2-3, 112, his3Δ200, trp1, LA−, sro7::URA3 exo84::HIS3 (TEF-exo84-202-12myc LEU2 CEN) |
Figure 4.
Characterization of the exo84-202 mutant that is defective in Exo84p–Sro7p interaction. (A) The exo84-202 mutant is defective in invertase secretion. The invertase secretion assay was performed at 25°C and, after a shift to 37°C for 2 h, a comparison of the secretion defect of exo84-202 with those of wild type and another exo84 mutant allele, exo84-121, was shown (n = 3). (B) The expression level of Exo84 protein is the same in the exo84-202 mutant and the wild-type cells. Wild-type and exo84-202 yeast cells were cultured to midlog phase and were shifted to 37°C for 2 h. 20 μg of the lysates from each sample was subjected to SDS-PAGE. The amount of Exo84p was detected by Western blotting. (C) Exo84-202p is defective in binding Sro7p but maintains its interaction with the exocyst components. Lysates from four yeast strains expressing Sro7-3Xmyc, Sec5-3XHA, Sec8-12Xmyc, or Sec15-12Xmyc were incubated with the GST fusion proteins of the wild-type (GST-Exo84p) or mutant form of Exo84p (GST–exo84-202p). Sro7p, Sec5p, Sec8p, and Sec15p that bound to the GST fusion proteins were analyzed by Western blot using mAbs against the tagged epitopes (right). (left) Coomassie blue–stained gel showing the inputs of GST fusion proteins used in the binding assay. Lane 1, GST; lane 2, GST-Exo84p; lane 3, GST–exo84-202p. (D) The interaction of Sro7p and Sro77p with exo84 mutant proteins tested by yeast two-hybrid assay. The exo84-202 mutant harbors point mutations, which result in changes of two amino acids (E217G and L243P). The exo84-202m1 (L243P) and exo84-202m2 (E217G) mutants were engineered by restoring one of the two mutations back to the wild-type sequence. The average and SD of the β-galactosidase activities were determined from three independent experiments. Error bars represent SD. (E) Growth of the wild type (EXO84), exo84-202, exo84-202m1, and exo84-202m2 on an SC plate at the restrictive temperature 37°C.
Figure 5.
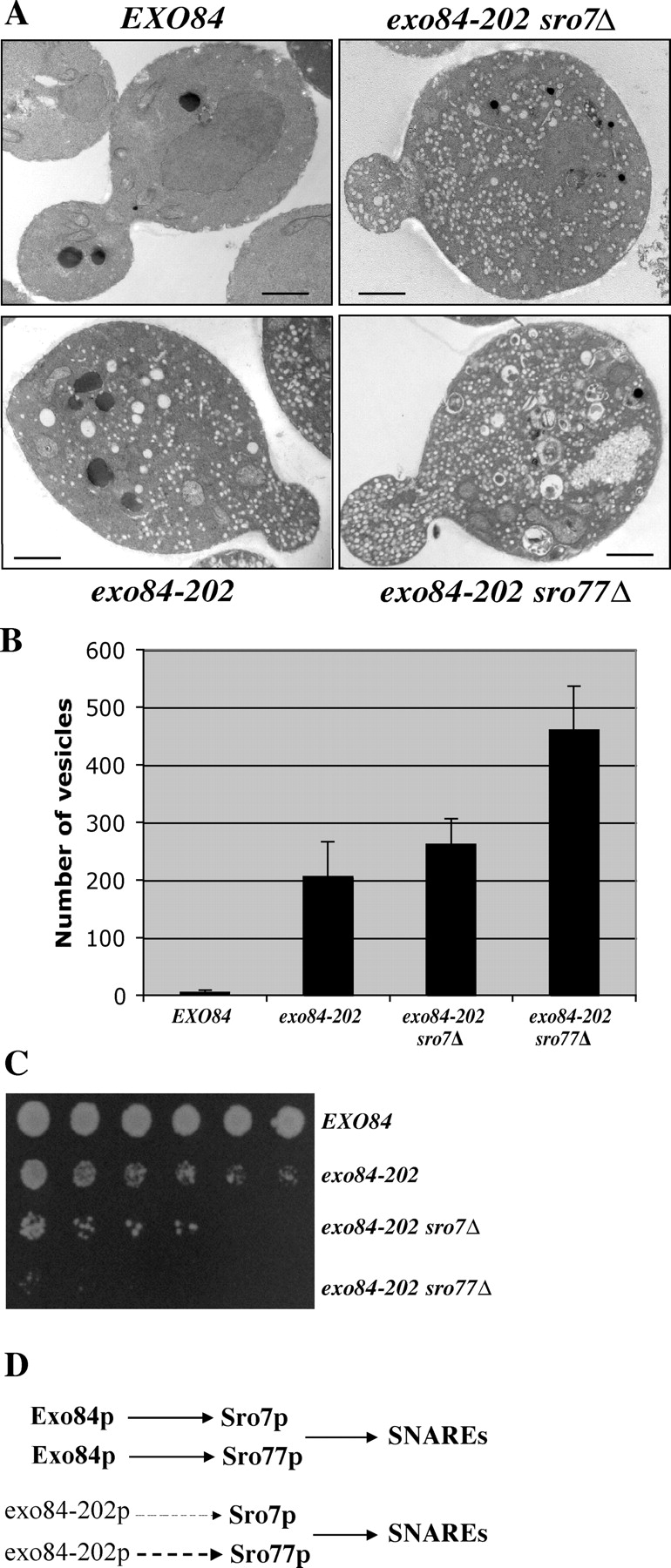
Growth defects and accumulation of post-Golgi secretory vesicles in exo84-202 and sro7/77 mutants. (A) Wild-type, exo84-202, exo84-202 sro7Δ, and exo84-202 sro77Δ cells were grown to early log phase at 25°C, shifted to 37°C for 2 h, and processed for thin-section EM. Bars, 1 μm. (B) Quantification of the number of secretory vesicles/section in the wild-type and mutant cells (n = 20). Error bars represent SD. (C) Growth of wild-type, exo84-202, exo84-202 sro7Δ, and exo84-202 sro77Δ cells on the yeast extract/peptone/glucose plate at 34°C. The yeast cells were plated in 1:10:20:40:80:160 dilutions. (D) Exo84p interacts with both Sro7p and Sro77p in the cells, forming parallel pathways that regulate SNARE assembly. The exo84-202 protein maintains a weak interaction with Sro77p (dashed line) and an even weaker interaction with Sro7p (thin dashed line). Deletion of SRO7 or SRO77 in the exo84-202 strain background further disrupts the pathway, leading to more severe growth and secretion defects.
The exo84-202 mutant harbors two point mutations (E217G and L243P) at the Sro7/77p-binding region. Binding assays indicate that the mutant Exo84p has a much weaker interaction with Sro7p, whereas it maintains its interaction with other exocyst proteins (Fig. 4 C). In fact, both of the point mutations in exo84-202 are needed to disrupt the binding between Exo84p and Sro7/77p, as recovering either one of them to the wild-type sequence greatly restored the binding of Exo84p to Sro7p and almost completely restored binding to Sro77p (Fig. 4 D). In addition, the recovery of binding in the mutants correlates with the recovery of their growth at 37°C (Fig. 4 E). This may explain why many mutants had to be screened to obtain the exo84-202 allele, as both mutations are needed for the phenotypes.
The two-hybrid analysis (Fig. 4 D) also suggests that the mutant exo84-202 protein still maintains a weak interaction with Sro7p. Moreover, its interaction with Sro77p was ∼43% of the wild-type level. These residual interactions may explain why the secretion defect in the exo84-202 mutant is not as significant as those observed for other sec genes. Sro7p and Sro77p are functionally redundant in yeast genome, as the deletion of only one of them does not cause any secretion defects (Lehman et al., 1999). We speculated that further disruption of the interaction between the exo84-202 and Sro proteins by the deletion of Sro7p (or especially Sro77p) in the exo84-202 strain may aggravate secretion defects in the cells. Therefore, we generated exo84-202 sro7Δ and exo84-202 sro77Δ strains and examined their secretion defects by EM. As shown in Fig. 5 A, exo84-202 cells accumulate secretory vesicles (average, 206 ± 60.6 vesicles/section) at 37°C. Further deletion of SRO7 in the exo84-202 background resulted in an increased accumulation of vesicles (262 ± 44.5 vesicles/section). In exo84-202 sro77Δ cells, there was a much larger increase in the number of secretory vesicles (460 ± 76.7 vesicles/section), indicating severe secretion defects. The severity of secretion defects in these mutants strongly correlated with the degree of binding disruption. We have also examined the growth of these cells on plates. As shown in Fig. 5 C, exo84-202 sro7Δ grows poorly compared with exo84-202 at 34°C (a semipermissive temperature for exo84-202). exo84-202 sro77Δ cells were barely able to grow under the same condition. These results suggest that further disruption of the Sro7/77p–Exo84p interaction in the exo84-202 background aggravates secretion and growth defects in the cells. The most likely explanation for the observed results is that Exo84p interacts with both Sro7p and Sro77p in the cells, forming parallel pathways for the regulation of exocytosis (Fig. 5 D). Deletion of SRO7 or SRO77 in the exo84-202 background removes one of the two parallel pathways, thus aggravating the exocytosis defects.
The sro7Δ sro77Δ–null mutant demonstrates cold sensitive growth defects (Lehman et al., 1999), whereas the exo84-202 mutant has growth defects at 37°C. It is likely that the functions of Sro7/77p are not limited to exocytosis (see the next section); therefore, mutations in EXO84 do not lead to the same phenotypes as sro7Δ sro77Δ null. Conversely, the functions of Exo84p may not be limited to its interaction with Sro7/77p. exo84-202 must also affect a process that is unrelated to Sro7/77p, although we currently cannot identify it. Therefore, the mutants generated in our study do not completely resemble the sro7Δ sro77Δ–null mutant. However, our detailed characterization of exo84-202, exo84-202 sro7Δ, and exo84-202 sro77Δ mutants clearly indicate the close relationship between Exo84p and Sro7/77p in exocytosis. Furthermore, exo84-202 proves to be a useful allele in the following genetic analyses of the Sro7/77p function in exocytosis.
Genetic analyses of the regulatory pathway involving the exocyst and Sro7/77p
Making use of our collection of yeast mutants, we next performed genetic experiments that were aimed at understanding the contexts of the Exo84p–Sro7/77p interaction. We found that SRO7 overexpression rescued exo84-202 growth at the restrictive temperature (Table II). SRO7 also suppressed several other exocyst mutants, including sec3-2, sec5-19, sec8-9, sec10-2, sec15-1, and exo70-38, which is similar to the results obtained by Lehman et al. (1999). We also found that overexpression of SRO7 can rescue SEC3 and EXO70 deletion (Table II and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200502055/DC1), suggesting that the regulatory function of SRO7 is very likely downstream of the exocyst. However, overexpression of SRO7 cannot bypass EXO84 deletion. This observation and the results reported by Wiederkehr et al. (2004) suggest that Exo84p and several other exocyst components (Sec6p, Sec8p, Sec10p, and Sec15p) form the core tethering machinery for exocytosis. Sro7p, as a regulator in this process, cannot bypass the core vesicle-tethering proteins.
Table II. High-dosage suppression of the exocyst mutants by SRO7 .
| Mutants | Suppression by 2μ SRO7 |
|---|---|
| sec1-1 | — |
| sec2-41 | — |
| *sec3-2 | +++ |
| *sec3Δ | ++ |
| sec4-8 | — |
| *sec5-19 | ++ |
| *sec6-4 | — |
| *sec8-9 | + |
| sec9-4 | — |
| *sec10-2 | + |
| *sec15-1 | ++ |
| *exo70-38 | +++ |
| *exo70Δ | ++ |
| *exo84-202 | ++ |
| *exo84Δ | — |
Growth of post-Golgi secretory mutants transformed with the SRO7 2μ plasmid at the restrictive temperatures.*, The exocyst mutants. Plus signs are used to score the suppression strength of the mutants by 2μ SRO7: ++++, the highest degree of rescuing activity (the same growth rate as the wild type cells); +, the lowest rescuing activity (slight growth at the restrictive temperature).
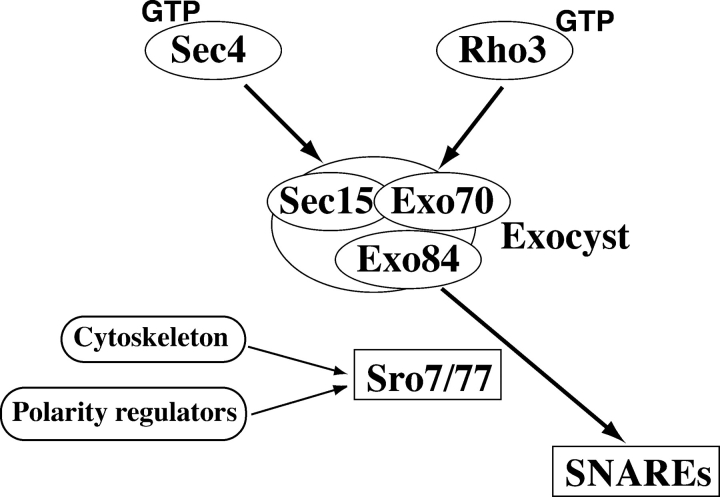
We have also tested the ability of all known yeast genes that were implicated in post-Golgi secretion to rescue exo84-202, exo70-38, and sro7Δ sro77Δ. As summarized in Table III, overexpression of SEC4, EXO70, SRO7, the downstream t-SNAREs SSO1/SSO2 and SEC9, and the t-SNARE–associated protein Sec1p rescued exo84-202. In addition, Rho3p, but not other Rho-family proteins, strongly rescued exo84-202. An almost identical set of proteins also showed rescuing activities toward exo70-38. In particular, RHO3, RHO4, SRO7, and EXO84 showed strong suppression activities toward exo70-38. It was previously shown that Exo70p interacts with activated Rho3p (Rho3-GTP; Adamo et al., 1999; Robinson et al., 1999). Our genetic analyses suggest that Exo70p and Exo84p are probably mediators for Rho3p signaling. It was also shown that Sec4p-GTP interacts with the exocyst component Sec15p and controls the efficient assembly of the exocyst complex (Guo et al., 1999b). Therefore, it is most likely that Sec4p functions upstream of the exocyst and Sro7p. Combining all of the genetic data, we propose a pathway in which the exocyst complex mediates the signaling from Rho3p and Sec4p to Sro7p, which controls SNARE assembly and the fusion of secretory vesicles at specific domains of the plasma membrane (Fig. 6). However, the genetic results do not totally exclude the possibility that Sec4p and Rho3p regulate Sro7p in parallel to the exocyst.
Table III.
High-dosage suppression tests on exo84, exo70, and sro7 Δ sro77Δ mutants
| 2μ plasmids | exo84-202 | exo70-38 | sro7Δ sro77Δ |
|---|---|---|---|
| SEC2 | — | — | — |
| SEC4 | ++ | + | — |
| SEC3 | — | — | — |
| SEC5 | — | — | — |
| SEC6 | — | — | — |
| SEC8 | — | — | — |
| SEC10 | — | — | — |
| SEC15 | — | — | — |
| EXO70 | +++ | ++++ | — |
| EXO84 | ++++ | +++ | — |
| SEC9 | ++ | ++ | — |
| SEC1 | + | + | — |
| SSO2 | +++ | ++ | — |
| SRO7 | ++ | +++ | +++ |
| CDC42 | — | — | — |
| RHO1 | — | — | — |
| RHO2 | — | — | — |
| RHO3 | ++ | +++ | — |
| RHO4 | — | ++ | — |
| pRS426 | — | — | — |
Growth of exo84 and exo70 mutants and the sro7Δ sro77Δ transformed with 2μ plasmids containing the yeast Rho family of small GTPases and genes involved in the late stage of exocytosis. The pRS426 vector was used as a negative control. Scoring for temperature-sensitive mutants is similar to that used in Table II: ++++, the highest degree of rescuing activity (the same growth rate as the wild type cells); +, the lowest rescuing activity (slight growth at the restrictive temperature).
Figure 6.
The exocyst and Sro7/77p mediate the signaling from Rho3p and Sec4p to the SNAREs. The GTP-bound form of Sec4p controls the exocyst through its direct interaction with Sec15p (Guo et al., 1999b), whereas the GTP-bound form of Rho3p controls the exocyst through its interaction with Exo70p (Adamo et al., 1999; Robinson et al., 1999). Exo84p directly interacts with Sro7/77p. Genetic analyses suggest that the exocyst complex mediates the signaling from Rho3p and Sec4p to Sro7/77p, which regulate SNARE assembly. Sro7p and Sro77p play an important regulatory role in the communication between the exocyst and t-SNAREs. The genetic analyses do not totally exclude the possibility that Sec4p and Rho3p regulate Sro7p in parallel to the exocyst. Besides interacting with the secretory machinery, Sro7p and Sro77p may also interact with cytoskeleton and polarity regulators. Other members of the exocyst complex are not shown in the diagram.
We found that none of the tested genes besides SRO7 and SRO77 themselves can rescue sro7Δ sro77Δ. It is possible that the role of Lgl proteins in exocytosis is not their sole function in cells. Therefore, loss of Lgl function cannot be rescued by compensation in the exocytosis pathway alone. It was shown that Sro7/77p interacts with Kin1/2p and myosins in yeast (Kagami et al., 1998; Elbert et al., 2005). In higher eukaryotes, Lgl proteins were shown to interact with myosins and the Par6–Par3–aPKC complex (Strand et al., 1994; Bilder et al., 2000; Ohshiro et al., 2000; Peng et al., 2000; Betschinger et al., 2003; Plant et al., 2003; Tanentzapf and Tepass, 2003). Lgl is involved in both epithelial polarization and the asymmetric distribution of cell fate determinants, especially in flies. It is possible that Lgl proteins interact with separate sets of molecules in the cell to carry out their different, but related, functions.
SRO7 and SSO2 rescue the polarity defects of exocyst mutants
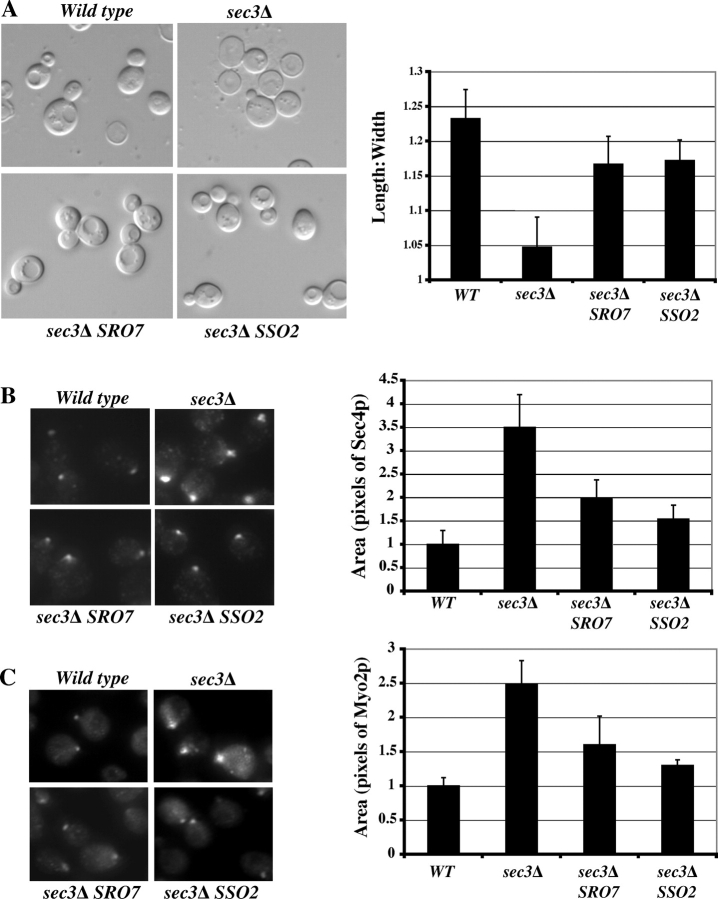
The above genetic analyses have shown that overexpression of SRO7 rescues several exocyst mutants. Then, we asked whether the Lgl function in kinetic regulation of exocytosis spatially contributes to cell polarization. The exocyst component Sec3p demarcates the subdomains of the plasma membrane for efficient and precise targeting of the secretory vesicles for exocytosis (Finger et al., 1998; Wiederkehr et al., 2003). Although vesicles can still be transported to daughter cells in the absence of Sec3p, they cannot find the appropriate sites within the buds. This results in isotropic membrane fusion along the entire daughter cell membrane rather than at the tip of the daughter membrane (the bud tip). After generations of growth, the sec3Δ cells are “rounder” in shape for both mother and daughter cells (lower cell length/width ratio), which is different from the normally ellipsoid shapes of wild-type yeast cells (Wiederkehr et al., 2003). Despite secretion and polarity defects, sec3Δ cells can survive at 25°C, allowing us to investigate the role of SRO7 by using genetics and cell biological methods. As shown in Fig. 7 A, sec3Δ cells grow poorly at 25°C and can barely survive at 34°C. However, cells that were transformed with 2μ plasmids containing SRO7 or SSO2 have much-improved growth at 25°C and become viable at 34°C. We have also compared the cell doubling times in liquid culture at 25°C. As shown in Fig. 7 B, doubling time of the sec3Δ cells (202 min) was more than two times longer than that of the wild-type cells (80 min). However, overexpression of SRO7 or SSO2 accelerated the growth rate of sec3Δ cells near the wild-type level. We measured invertase secretion in these cells at 25°C and 34°C. As shown in Fig. 7 C, sec3Δ cells accumulate 25% of the produced invertase. Overexpression of SRO7 or SSO2 greatly reduced the amounts of invertase in the cells, and growth and secretion properties correlated with each other.
Figure 7.
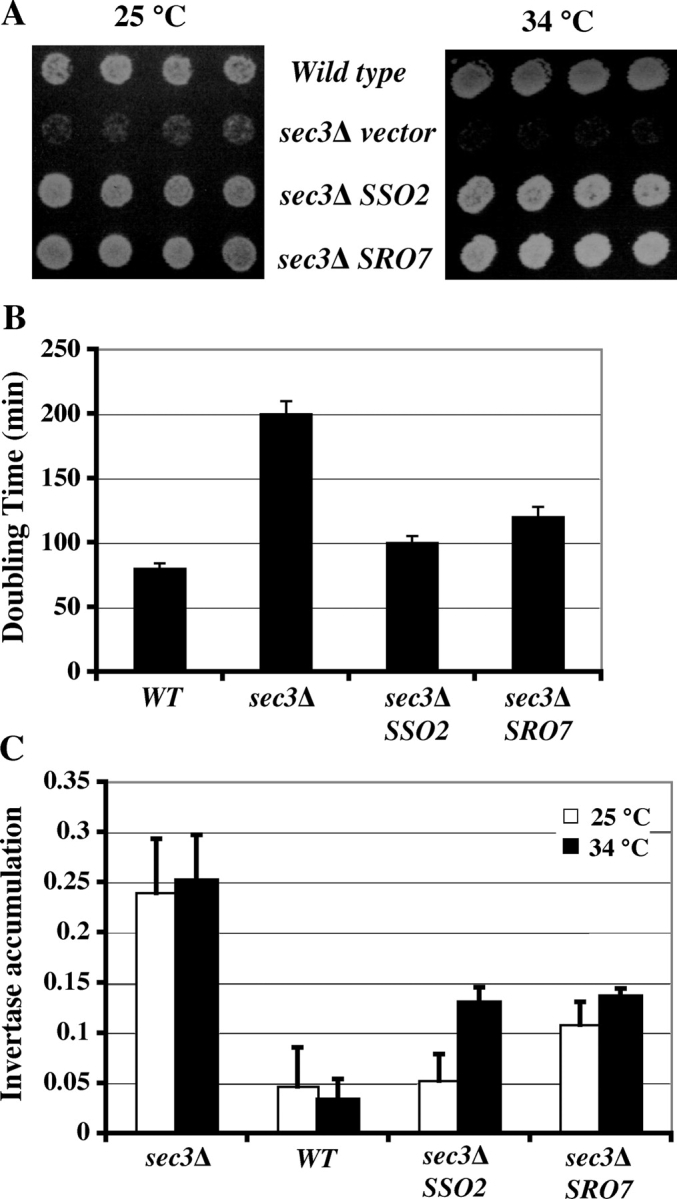
Rescue of the growth and secretion defects of sec3 Δ by overexpression of SRO7 and SSO2. (A) Wild-type and sec3Δ haploid strains carrying SSO2 or SRO7 2μ plasmids, or the empty vector, were grown on SC plates at 25°C (left) and 34°C (right). SSO2 or SRO7 high-copy plasmids partially rescue the growth of sec3Δ cells. (B) Growth of indicated strains at 25°C was assayed by measuring the OD (OD600) after dilution from overnight culture in SC medium. Doubling times of cells at the exponential growth phase were measured (n = 3). (C) Invertase secretion defects were measured in wild-type, sec3Δ, and sec3Δ strains transformed with 2μ SRO7 or SSO2 in SC medium at 25 or 34°C. Accumulation of intracellular invertase was expressed as internal invertase/(internal invertase + external invertase). n = 3. Error bars represent SD.
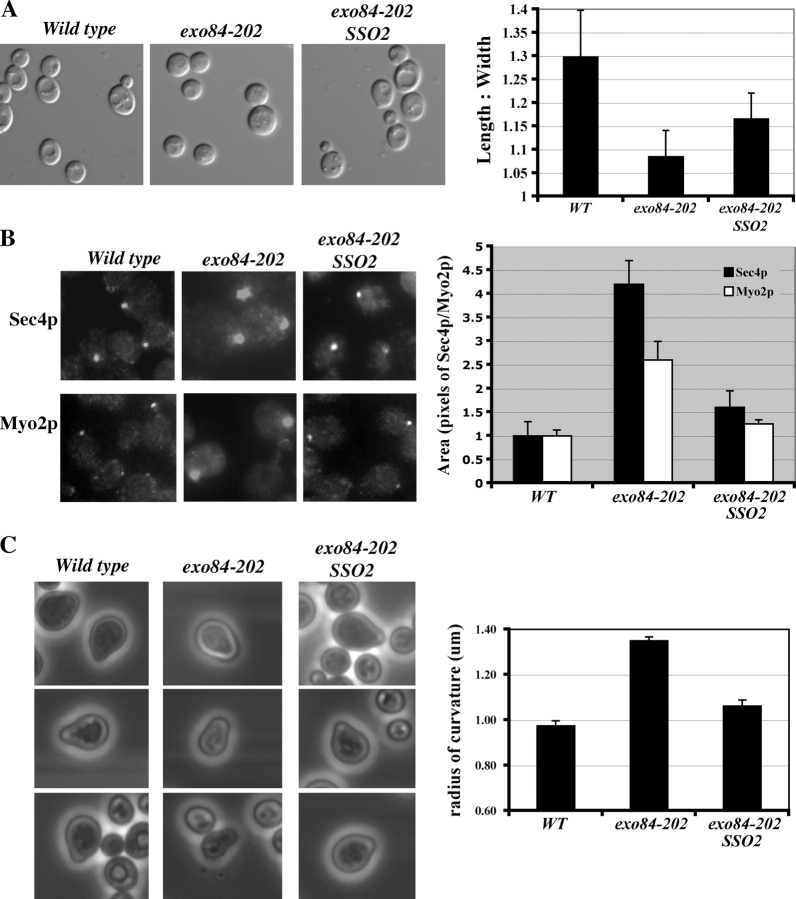
Next, we examined the morphologies of these cells. As shown in Fig. 8 A, differential interference contrast images show the rounder shapes of sec3Δ cells in contrast to the ellipsoidal shapes of wild-type cells, which is consistent with the previous observation (Wiederkehr et al., 2003). Transformation of SRO7 or SSO2 2μ plasmids partially rescued the round morphology of sec3Δ cells. The cell shapes were quantified as the axial ratio (length/width) as previously described (Sheu et al., 2000; Wiederkehr et al., 2003). Overexpression of SRO7 or SSO2 significantly increased the axial ratio of sec3Δ cells (Fig. 8 A, right).
Figure 8.
Rescue of the polarity defect of sec3 Δ by overexpression of SRO7 and SSO2. (A) Cell morphology of wild-type, sec3Δ, and sec3Δ strains transformed with 2μ SRO7 or SSO2 (left). The average length/width ratios of 100 individual mid-log-phase cells from each indicated strain were plotted (right). The immunofluorescence signals of Sec4p (B) and Myo2p (C) in the buds of various strains (left) are shown. Compared with wild-type cells, Sec4p and Myo2p were less concentrated in the buds of sec3Δ cells. In sec3Δ cells overexpressing SRO7 or SSO2, the immunofluorescence signals became concentrated. The areas of the immunofluorescence signals of Sec4p and Myo2p were quantified as pixels by Openlab software and were plotted (right; n = 100). The area in wild-type cells was designated as 1. Error bars represent SD.
Because the rounder morphology in sec3Δ cells is likely caused by less restricted vesicle targeting to the bud tip in sec3Δ cells, we further examined the distribution of Sec4p, which associates with secretory vesicles that are delivered to the daughter cells. As shown in Fig. 8 B, the immunofluorescence signals of Sec4p are diffused in the buds, whereas in wild-type cells, the signals are much more focused. The spread pattern in sec3Δ can be rescued by overexpressing SRO7 or SSO2. We have also examined the localization of Myo2p by immunofluorescence. Myo2p is a class V myosin that mediates the transport of post-Golgi secretory vesicles along the actin cables to the bud (Govindan et al., 1995; Pruyne et al., 1998; Schott et al., 1999). Myo2p spreads out in the buds of sec3Δ cells, which is similar to Sec4p, and this diffuse pattern was partially rescued by overexpression of SRO7 or SSO2 (Fig. 8 C). Overall, our results indicate that overexpression of SRO7 and the downstream t-SNARE SSO2 helps to rescue the polarity defects caused by the absence of the spatial landmark protein Sec3p.
Morphology of exo84-202 mutant cells
Although overexpression of SRO7 can rescue the cell polarity defects of sec3Δ, we examined whether the exo84-202 mutant, in which the physical interaction between the exocyst and Sro7p is disrupted, has polarity defects. Examination of the morphology of exo84-202 cells indicated that this mutant, like sec3Δ, is indeed rounder in shape (Fig. 9 A). This rounder shape can be observed even at 25°C and becomes more obvious at elevated temperatures. Also, like sec3Δ, Sec4p and Myo2p are less restricted in daughter cells (Fig. 9 B). Actin, although slightly diffused, remains polarized to the daughter cells (unpublished data), which is similar to observations made by Lehman et al. (1999) in sro7Δ sro77Δ cells. We have also examined whether exo84-202 cells form normal mating projections (“shmoos”) in response to α-factor treatment. As shown in Fig. 9 C (right), in contrast to the “sharper” shmoos formed in wild-type cells, exo84-202 cells project rounder shmoos. Because SSO2 is a strong suppressor for exo84-202 (Table III), we examined whether SSO2 can rescue the polarity defects of exo84-202. As shown in Fig. 9, SSO2 overexpression greatly improved the polarity of exo84-202 cells.
Figure 9.
Polarity defects in exo84-202 cells. (A) Cell morphology of wild-type, exo84-202, and exo84-202 cells transformed with 2μ SSO2 (exo84-202 SSO2) at 25°C. The average length/width ratios of 100 midlog-phase cells from each sample were plotted on the right. The exo84-202 cells are rounder in shape. Overexpression of SSO2 partially rescued this polarity defect. (B) Immunofluorescence staining of Sec4p and Myo2p in the buds. The areas of the immunofluorescence signals of Sec4p and Myo2p were quantified as pixels by Openlab software, and the average of 100 cells were plotted (right). The area of wild-type cells was designated as 1. The immunofluorescence signals in exo84-202 cells are more spread out. Overexpression of SSO2 partially restored the signals to that of the wild-type cells. (C) Shmoo morphologies of wild-type, exo84-202, and exo84-202 SSO2 cells after α-factor treatment. Images of representative examples of shmooing yeast cells are shown. The curvature of the shmoo tips was measured and plotted (right). Error bars represent SD
Discussion
We report that the yeast Lgl proteins Sro7p and Sro77p interact with Exo84p, which is a component of the exocyst complex. The Sro7/77p–exocyst interaction is important for exocytosis, as disruption of this binding leads to secretion defects. Genetic analyses demonstrate a pathway from upstream small GTPases to SNARE-mediated membrane fusion. It was speculated that the exocyst, a vesicle-tethering complex, communicates with and activates downstream SNAREs for subsequent fusion of the vesicles with the plasma membrane (Whyte and Munro, 2002; Hsu et al., 2004). However, the physical association between the exocyst and SNARE proteins has not been identified. Our finding, for the first time, reveals a molecular link between the exocyst and t-SNAREs. Because overexpression of SRO7/77 rescues several secretory mutants and the deletion of SRO7 and SRO77 causes severe secretion defects, Lgl proteins in yeast are probably positive regulators in the secretory pathway and may help to promote coupling between the tethering complex (the exocyst) and the membrane-fusion machinery (SNAREs) for exocytosis at the plasma membrane.
The Sro7/77p–SNARE interaction is a critical positive regulatory event for SNARE assembly and membrane fusion, and our biochemical and genetic analyses raise the possibility that the exocyst and upstream small GTPases provide the activation signal or spatial restriction for Sro7/77p function. It was shown that t-SNARE proteins and Sro7/77p are evenly distributed along the entire plasma membrane (Brennwald et al., 1994; Larsson et al., 1998; Lehman et al., 1999), whereas the exocyst is concentrated to the bud tip (TerBush and Novick, 1995; Finger et al., 1998; Guo et al., 1999a). Sro7/77p–exocyst interaction may be important for restricting exocytosis to specific sites of the plasma membrane for polarized cell growth. Although the in vitro binding affinity for Exo84p–Sro7p interaction is high (Kd = 46 ± 6 nM), their in vivo interaction may be restricted to the bud tip, and this interaction may be modulated by several regulators such as Kin1/2p (Elbert et al., 2005). Overall, our data suggest a model in which Exo84p interacts with Sro7/77p, which then promotes SNARE-mediated membrane fusion at specific regions of the plasma membrane. Although the yeast Lgl proteins themselves are not polarized in distribution, they may achieve their function in polarized exocytosis kinetically through their localized interaction with the exocyst.
Does the function of Lgl in exocytosis contribute to cell polarization? Taking advantage of the simple morphology of budding yeast, we examined the effects of SRO7 and t-SNARE SSO2 overexpression on the exocyst mutant with polarity defects. The sec3Δ and exo84-202 cells are rounder in shape. These phenotypes resemble those of the polarisome mutants that play important regulatory roles during cell polarization in yeast (Sheu et al., 2000). We found that overexpression of SRO7 and t-SNARE SSO2 not only helped to rescue the secretion defects but also significantly improved the polarization of these cells. Neither Sro7p nor Sso2p has a polarized distribution pattern in the cells. Particularly, Sso2p, which is a yeast t-SNARE protein responsible for the fusion reaction of lipid bilayers, does not contain any targeting information in its sequence. The rescuing effects on cell polarity from Sro7p and Sso2p, therefore, probably contributed through their positive roles in promoting secretion kinetically in the cells. Consistent with this observation, it was recently shown that overexpression of Sec1p, a protein that interacts with t-SNARE, also rescues the polarity defect of sec3Δ cells (Wiederkehr et al., 2004). Our data suggest that, although the localized activation of Lgl may be controlled by the exocyst and small GTPases, increased Lgl and SNARE functions may conversely contribute to cell polarization. This reciprocal regulation may “fine tune” or reenforce cell polarity, leading to tightly restricted secretion at the tip of the bud. In this regard, it is interesting to note that a previous study demonstrated that delivery of the polarisome component Bud6p/Aip3p to the bud relies on the functional secretory pathway (Jin and Amberg, 2000). We do not exclude the possibility that other yet unidentified functions of Lgl may also contribute to this rescuing effect. Future identification and characterization of Lgl-interacting proteins may help to elucidate the role(s) of Lgl in cell polarization.
The core mechanisms for the generation and maintenance of cell polarity are evolutionally conserved. The sequences and many functions of the exocyst and Lgl proteins are also conserved. Previous analysis shows that both fly and mammalian Lgl can partially rescue the yeast sro7Δ sro77Δ mutant (Kagami et al., 1998; Larsson et al., 1998). It will be interesting to extend this study to higher eukaryotes to examine whether Lgl and the exocyst carry out their functions in targeting regulators of cell morphogenesis for the establishment and reenforcement of cell polarity.
Materials and methods
Yeast strains and procedures
Standard methods were used for yeast media, growth, and genetic manipulations. The yeast strains used in the project are listed in Table I.
Yeast two-hybrid assays
The cDNAs encoding exocyst components and Sro7p and Sro77p were subcloned in pACTII and pOBD8 vectors, respectively, and were expressed in Y190 cells. Interactions were detected by measuring whole cell β-galactosidase activities as previously described (Guo et al., 1999a).
Recombinant proteins
For purification of recombinant Sro7p, a protein A sequence was engineered at the NH2 terminus of Sro7p, and the fusion protein was expressed under the control of the ADH1 promoter in yeast. Yeast lysates containing the recombinant protein were incubated with IgG–Sepharose beads. For in vitro synthesis of radiolabeled Sro7p, Sro7 was subcloned into pcDNA3, the purified plasmid was added to the rabbit reticulocyte lysate–coupled in vitro transcription–translation system under the T7 promoter (Promega) in the presence of [35S]methionine, and the plasmid was processed as described by the manufacturer. The in vitro–synthesized Sro7 is shown in Fig. 2 A as doublets, which either resulted from the degradation or downstream initiation of translation by the translation machinery in the reticulocyte lysate system. For in vitro synthesis of radiolabeled Exo84p, Exo84 cDNA was amplified by PCR and was directly used in the in vitro transcription–translation reaction.
In vitro binding assays
For binding of the radiolabeled Exo84p to protein A–Sro7p Sepharose beads, 5 μl [35S]methionine-labeled in vitro transcription–translation reaction mixture was diluted to 85 μl in binding buffer (10 mM Hepes/KOH, pH 7.4, 140 mM KCl, 2 mM MgCl2, 0.5% Triton X-100, and protease inhibitors) and was preincubated with Sepharose beads for 15 min on ice followed by centrifugation. The resulting supernatant was used for a 100-μl binding reaction using 2 μM immobilized protein A–Sro7p. The reaction mixture was incubated for 90 min at 4°C, and the supernatants were separated from the pellets by centrifugation. A similar procedure was used to test the binding between radiolabeled Sro7p and GST-Exo84N conjugated to glutathione–Sepharose beads, except that the binding reaction took place in 500 μl of binding buffer (20 mM Hepes-NaOH, 140 mM KCl, 2 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, and 1 mM PMSF) for 4 h at 4°C. The samples were subjected to SDS-PAGE and autoradiography. Quantitation was performed using a phosphoimager (model Storm; Molecular Dynamics) with ImageQuant software (Molecular Dynamics).
To obtain the dissociation constant of the Sro7p–Exo84p interaction, various amounts of purified GST-Exo84N were incubated with 10 nM protein A–Sro7p conjugated to IgG–Sepharose beads in 1,000 μl of binding buffer. The reaction mixtures were incubated at RT for 2 h followed by washing. The protein samples were subjected to SDS-PAGE, and the gels were stained by Coomassie blue. The bound and free GST-Exo84N were quantified with ImageQuant and were plotted with a single rectangular hyperbola equation (B = BmaxC/[Kd + C]) using the SigmaPlot software (Systat Software, Inc.). The dissociation constant was calculated from each plot by nonlinear regression.
Coprecipitation assays
Yeast cells expressing GST or GST-Sro7p in a CEN plasmid under the TEF promoter were grown to early log phase, and cell lysates were prepared by disrupting the cell wall using glass beads in buffer A (20 mM Hepes, 150 mM KCl, 1 mM EDTA, 1 mM DTT, 0.5% NP-40, and protease inhibitors). The lysates were incubated with glutathione–Sepharose 4B for 1 h at 4°C. The beads were washed, and proteins that were precipitated with the GST fusion proteins from lysates were analyzed by Western blot. For immunoprecipitation of Sro7p and Exo84p at their endogenous levels, Exo84p was tagged with the myc epitope by chromosomal integration, and detergent extracts were prepared by using buffer A. A rabbit anti-Sro7p antibody was used at a 1:300 dilution in the lysates for precipitation, and precipitated Sro7p and Exo84p were detected by Western blot analysis. As controls, rabbit anti-GFP and anti-GST pAbs (Convance, Inc.) were used in the same procedure.
Generation of exo84 mutants
For generation of the exo84-202 mutant, mutagenesis focusing on the NH2 terminus of Exo84p was performed by using error-prone PCR as previously described (Zhang et al., 2001). The forward primer covers 300 bp upstream of the starting codon, and the reverse primer covers the Exo84 ORF bp 1–900 region. Mutagenized PCR products were mixed with linearized CEN LEU2 plasmid containing the Exo84 promoter and bp 900–2260 tagged with a 12Xmyc sequence at the 3′ end. Then, this mixture was transformed into GY1264, in which the endogenous EXO84 gene was disrupted by HIS3 and supplemented with a CEN URA3 EXO84 plasmid to allow gap repair between the PCR products and the linearized plasmid. Transformants were selected on synthetic complete media (SC) Leu–His–Ura plates at 25°C and were replicated onto SC Leu–His plates containing 1 mg/ml 5-fluoroorotic acid to select for the loss of the CEN URA3 EXO84 plasmid. The selected colonies were replicated onto three sets of SC Leu–His plates and were incubated at 18°C, 25°C, and 37°C, respectively, to allow the identification of temperature-sensitive mutants. Possible mutants were confirmed by the retransformation of isolated plasmids into the GY1264 host strain. In addition to the exo84-202 strain, we also generated strains with the SRO7 or SRO77 gene deleted in the exo84-202 background (exo84-202 sro7Δ and exo84-202 sro77Δ, respectively) by replacement of the chromosomal copy of SRO genes with KanM cassette. The desired strains were selected by 0.2 mg/ml G418 on SC plates and were confirmed by PCR. Besides exo84-202, we have also generated other exo84 mutants by random mutagenesis of the whole Exo84 ORF by using a similar error-prone PCR strategy (Zhang et al., 2005). One such mutant, exo84-121, was used in this study.
Light microscopy
Immunofluorescence staining of yeast cells was performed as previously described (Walch-Solimena et al., 1997). Rabbit anti-Sec4p and anti-Myo2p pAbs were used at a 1:1,000 dilution followed by a secondary AlexaFluor594-conjugated goat anti–rabbit IgG antibody (Molecular Probes). The digital images were captured by a fluorescence microscope (model DM IRB; Leica) using a 100× objective and a high resolution CCD camera (model ORCA-ER; Hamamatsu Photonics). Immunofluorescence signals were quantified as pixels by using OpenLab 3.1.4 software (Improvision). For morphological analysis, the length and width of yeast cells were measured on differential interference contrast images using OpenLab software, which is basically similar to that used in the Saccharomyces cerevisiae Morphological Database (http://scmd.gi.k.u-tokyo.ac.jp/). The roundness of the cells was calculated as the axial ratio (length/width) in the mother cells. To observe shmoo formation, Mat a yeast cells were cultured to OD600 = 0.2 followed by 1 mg/ml α-factor treatment. Light microscopy images were taken 6 h after α-factor addition. The curvature radius of the shmoo tips was analyzed by using the OpenLab 3.1.4 program.
EM
Cells were collected by vacuum filtration using a 0.45-μm nitrocellulose membrane and were fixed for 1 h at RT in 0.1 M cacodylate, pH 7.4, 3% formaldehyde, 1 mM MgCl2, and 1 mM CaCl2. The cells were spheroplasted and fixed with 1% glutaraldehyde (in PBS, pH 7.4) at 4°C overnight. The spheroplasts were washed in 0.1 M cacodylate buffer and were postfixed twice with ice-cold 0.5% OsO4 and 0.8% potassium for 10 min each. After dehydration and embedding in Spurr's epoxy resin (Polysciences, Inc.), thin sections were cut and transferred onto 600 mesh uncoated copper grids (Ernest Fullam, Inc) and were poststained with uranyl acetate and lead citrate. Cells were observed on a transmission electron microscope (model 1010; JEOL) at 100,000×.
Online supplemental material
Fig. S1 shows that SRO7 overexpression rescues exo70Δ. The supplemental materials and methods paragraph explains the methods used for the rescuing experiment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200502055/DC1.
Acknowledgments
We thank Allison Zajac for her technical help and Dr. John Murray for his advice on the EM experiments. We also thank Dr. Peter Novick for many helpful discussions and Dr. Lennart Adler for reagents.
This work is supported by grants from the National Institutes of Health (RO1-GM64690), American Cancer Society, and the Pew Scholar Program in Biomedical Sciences (to W. Guo).
X. Zhang and P. Wang contributed equally to this work.
Abbreviations used in this paper: Lgl, lethal giant larvae; SC, synthetic complete media.
References
- Adamo, J., G. Rossi, and P. Brennwald. 1999. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell. 10:4121–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamo, J., J. Moskow, A. Gladfelter, D. Viterbo, D. Lew, and P. Brennwald. 2001. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 155:581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger, J., K. Mechtler, and J. Knoblich. 2003. The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature. 422:326–330. [DOI] [PubMed] [Google Scholar]
- Bilder, D., M. Li, and N. Perrimon. 2000. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 289:113–116. [DOI] [PubMed] [Google Scholar]
- Brennwald, P., B. Kearns, K. Champion, S. Keranen, V. Bankaitis, and P. Novick. 1994. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 79:245–258. [DOI] [PubMed] [Google Scholar]
- Elbert, M., G. Rossi, and P. Brennwald. 2005. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol. Biol. Cell. 16:532–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger, F., T. Hughes, and P. Novick. 1998. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 92:559–571. [DOI] [PubMed] [Google Scholar]
- Fölsch, H., M. Pypaert, S. Maday, L. Pelletier, and I. Mellman. 2003. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J. Cell Biol. 163:351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateff, E. 1978. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 200:1448–1459. [DOI] [PubMed] [Google Scholar]
- Grindstaff, K., C. Yeaman, N. Anandasabapathy, S. Hsu, E. Rodriguez-Boulan, R. Scheller, and W. Nelson. 1998. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 93:731–740. [DOI] [PubMed] [Google Scholar]
- Govindan, B., R. Bowser, and P. Novick. 1995. The role of Myo2, a yeast class V myosin, in vesicular transport. J. Cell Biol. 128:1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., A. Grant, and P. Novick. 1999. a. Exo84p is an exocyst protein essential for secretion. J. Biol. Chem. 274:23558–23564. [DOI] [PubMed] [Google Scholar]
- Guo, W., D. Roth, C. Walch-Solimena, and P. Novick. 1999. b. The exocyst is an effecter for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18:1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, S., D. TerBush, M. Abraham, and W. Guo. 2004. The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 233:243–265. [DOI] [PubMed] [Google Scholar]
- Jin, H., and D.C. Amberg. 2000. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol. Biol. Cell. 11:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice, N.J., and Y.N. Jan. 2003. A lethal giant kinase in cell polarity. Nat. Cell Biol. 5:273–274. [DOI] [PubMed] [Google Scholar]
- Kagami, M., A. Toh-e, and Y. Matsui. 1998. Sro7p, a Saccharomyces cerevisiae counterpart of the tumor suppressor l(2)gl protein, is related to myosins in function. Genetics. 149:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer, G., J. Schmoranzer, S.H. Low, X. Li, Y. Gan, T. Weimbs, S.M. Simon, and E. Rodriguez-Boulan. 2003. Three-dimensional analysis of post-Golgi carrier exocytosis in epithelial cells. Nat. Cell Biol. 5:126–136. [DOI] [PubMed] [Google Scholar]
- Larsson, K., F. Bohl, I. Sjostrom, N. Akhtar, D. Strand, B. Mechler, R. Grabowski, and L. Adler. 1998. The Saccharomyces cerevisiae SOP1 and SOP2 genes, which act in cation homeostasis, can be functionally substituted by the Drosophila lethal(2)giant larvae tumor suppressor gene. J. Biol. Chem. 273:33610–33618. [DOI] [PubMed] [Google Scholar]
- Lehman, K., G. Rossi, J. Adamo, and P. Brennwald. 1999. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146:125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipschutz, J., and K. Mostov. 2002. Exocytosis: the many masters of the exocyst. Curr. Biol. 12:R212–R214. [DOI] [PubMed] [Google Scholar]
- Louvard, D. 1980. Apical membrane aminopeptidase appears at site of cell-cell contact in cultured kidney epithelial cells. Proc. Natl. Acad. Sci. USA. 77:4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfruelli, P., N. Arquier, W. Hanratty, and M. Semeriva. 1996. The tumor suppressor gene, lethal(2)giant larvae (1(2)g1), is required for cell shape change of epithelial cells during Drosophila development. Development. 122:2283–2294. [DOI] [PubMed] [Google Scholar]
- Mostov, K., T. Su, and M. ter Beest. 2003. Polarized epithelial membrane traffic: conservation and plasticity. Nat. Cell Biol. 5:287–293. [DOI] [PubMed] [Google Scholar]
- Musch, A., D. Cohen, C. Yeaman, W.J. Nelson, E. Rodriguez-Boulan, and P.J. Brennwald. 2002. Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in Madin-Darby canine kidney cells. Mol. Biol. Cell. 13:158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, W.J. 2003. Adaptation of core mechanisms to generate cell polarity. Nature. 422:766–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick, P., and W. Guo. 2002. Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol. 12:247–249. [DOI] [PubMed] [Google Scholar]
- Ohshiro, T., T. Yagami, C. Zhang, and F. Matsuzaki. 2000. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 408:593–596. [DOI] [PubMed] [Google Scholar]
- Peng, C., L. Manning, R. Albertson, and C. Doe. 2000. The tumor-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 408:596–600. [DOI] [PubMed] [Google Scholar]
- Plant, P., J. Fawcett, D. Lin, A. Holdorf, K. Binns, S. Kulkarni, and T. Pawson. 2003. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat. Cell Biol. 5:301–308. [DOI] [PubMed] [Google Scholar]
- Pruyne, D.W., D.H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931–1945. [DOI] [PubMed] [Google Scholar]
- Robinson, N.G., L. Guo, J. Imai, A. Toh-E, Y. Matsui, and F. Tamanoi. 1999. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol. Cell Biol. 19: 3580–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, D., J. Ho, D. Pruyne, and A. Bretscher. 1999. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 147:791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., Y. Barral, and M. Snyder. 2000. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell Biol. 20:5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand, D., R. Jakobs, G. Merdes, B. Neumann, A. Kalmes, H. Heid, I. Husmann, and B. Mechler. 1994. The Drosophila lethal(2)giant larvae tumor suppressor protein forms homo-oligomers and is associated with nonmuscle myosin II heavy chain. J. Cell Biol. 127:1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanentzapf, G., and U. Tepass. 2003. Interactions between the crumbs, lethal giant larvae and bazooka pathways in epithelial polarization. Nat. Cell Biol. 5:46–52. [DOI] [PubMed] [Google Scholar]
- TerBush, D., and P. Novick. 1995. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 130:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena, C., R.N. Collins, and P.J. Novick. 1997. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 137:1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte, J.R., and S. Munro. 2002. Vesicle tethering complexes in membrane traffic. J. Cell Sci. 115:2627–2637. [DOI] [PubMed] [Google Scholar]
- Widberg, C., N. Bryant, M. Girotti, S. Rea, and D. James. 2003. Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J. Biol. Chem. 278:35093–35101. [DOI] [PubMed] [Google Scholar]
- Wiederkehr, A., Y. Du, M. Pypaert, S. Ferro-Novick, and P. Novick. 2003. Sec3p is needed for the spatial regulation of secretion and for the inheritance of the cortical endoplasmic reticulum. Mol. Biol. Cell. 14:4770–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr, A., J.O. De Craene, S. Ferro-Novick, and P. Novick. 2004. Functional specialization within a vesicle tethering complex: bypass of a subset of exocyst deletion mutants by Sec1p or Sec4p. J. Cell Biol. 167:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz, A. 2002. Establishing cell polarity in development. Nat. Cell Biol. 4:E39–E44. [DOI] [PubMed] [Google Scholar]
- Yeaman, C., K.K. Grindstaff, J.R. Wright, and W.J. Nelson. 2001. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J. Cell Biol. 155:593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., E. Bi, P. Novick, L. Du, K. Kozminski, J. Lipschutz, and W. Guo. 2001. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 276:46745–46750. [DOI] [PubMed] [Google Scholar]
- Zhang X., A. Zajac, J. Zhang, P. Wang, M. Li, J. Murray, D. TerBush, and W. Guo. 2005. The critical role of Exo84p in the organization and polarized localization of the exocyst complex. J. Biol. Chem. 280:20356–20364. [DOI] [PubMed] [Google Scholar]