Abstract
Mutations in the DSL (Delta, Serrate, Lag2) Notch (N) ligand Delta-like (Dll) 3 cause skeletal abnormalities in spondylocostal dysostosis, which is consistent with a critical role for N signaling during somitogenesis. Understanding how Dll3 functions is complicated by reports that DSL ligands both activate and inhibit N signaling. In contrast to other DSL ligands, we show that Dll3 does not activate N signaling in multiple assays. Consistent with these findings, Dll3 does not bind to cells expressing any of the four N receptors, and N1 does not bind Dll3-expressing cells. However, in a cell-autonomous manner, Dll3 suppressed N signaling, as was found for other DSL ligands. Therefore, Dll3 functions not as an activator as previously reported but rather as a dedicated inhibitor of N signaling. As an N antagonist, Dll3 promoted Xenopus laevis neurogenesis and inhibited glial differentiation of mouse neural progenitors. Finally, together with the modulator lunatic fringe, Dll3 altered N signaling levels that were induced by other DSL ligands.
Introduction
Functional studies of Notch (N) pathway genes have implicated this signaling system in the development of almost all structures within the vertebrate body plan. In particular, losses in core components (N1, Delta-like [Dll] 1, Dll3, presenilin-1, kuzbanian, and RBP-J) as well as in targets and modulators (Hes7, Mesp2, and lunatic fringe [LFng]) of the N signaling pathway all perturb the formation and patterning of somites (for review see Weinmaster and Kintner, 2003; Giudicelli and Lewis, 2004). Correct segmentation and patterning of somites is essential for proper axial skeletal formation, and mutations in Dll3 produce vertebral segmentation and rib defects in both spondylocostal dysostosis patients (Bulman et al., 2000; Turnpenny et al., 2003) and the pudgy mouse (Kusumi et al., 1998, 2004). Although it is clear that N signaling regulates somitogenesis, it is not clear which DSL (Delta, Serrate, Lag2) ligand activates N during this process. Of the DSL ligands that are expressed in the presomitic mesoderm (PSM), only Dll3 and Dll1 mutant mice display somitic defects; however, Dll3 and Dll1 mutant phenotypes differ with respect to the expression of somite markers and genes whose rhythmic expression is regulated by N (Dunwoodie et al., 2002; Zhang et al., 2002; Kusumi et al., 2004). Although it is difficult to discern from phenotypes and gene expression patterns alone, these different mutant phenotypes may reflect distinct roles for Dll1 and Dll3 in regulating N signaling during somitogenesis. In fact, the somite defects that are seen in Dll3 mutant mice are more similar to those reported in modulators of N signaling (LFng, Hes7, or Mesp2) rather than in mice lacking the well-characterized activating N ligand Dll1.
Activation of N signaling relies on contact between cells to allow the transmembrane DSL ligand on one cell to bind its receptor on an apposing cell. During its trafficking to the cell surface, N is constitutively processed by a furin-type protease producing a heterodimer that is composed of noncovalently associated extracellular and transmembrane subunits (Logeat et al., 1998). In response to ligand binding, the N heterodimer dissociates to release the extracellular domain from its membrane-bound portion (Sanchez-Irizarry et al., 2004; Weng et al., 2004). Removal of the extracellular domain is necessary for receptor activation that is mediated by proteolysis, first by a disintegrin and metalloprotease cleavage within the extracellular domain followed by a presenilin/γ-secretase intramembrane cleavage (for review see Mumm and Kopan, 2000; Weinmaster, 2000). These ligand-dependent cleavages allow the biologically active N intracellular domain (NICD) to be released from the plasma membrane and move to the nucleus, where it directly binds to the transcription factor CSL (CBF1, SuH, LAG-1). Through interactions with NICD, CSL is converted from a repressor into an activator of transcription to regulate N target gene expression. In addition to this well-characterized role for activation of N signaling through cell–cell interactions, DSL ligands have also been reported to cell autonomously antagonize N signaling in both vertebrate and invertebrate systems (Heitzler and Simpson, 1993; Henrique et al., 1997; Jacobsen et al., 1998; de Celis and Bray, 2000; Sakamoto et al., 2002; Itoh et al., 2003).
In this study, we show that Dll3 does not induce N signaling in multiple assay systems that measure the activation of N in response to DSL ligands. Our findings that Dll3 does not activate any of the known mammalian N receptors is in conflict with a previous study that found Dll3 activates N signaling (Dunwoodie et al., 1997). We find that, unlike other activating DSL ligands, Dll3 does not bind to cells expressing N receptors, and, conversely, N1 does not bind to Dll3-expressing cells. Although Dll3 did not bind or activate N when presented in trans, it cell autonomously inhibited N signaling that was induced by other DSL ligands in CSL gene reporter, Xenopus laevis primary neurogenesis, and mouse embryonic neural progenitor differentiation assays. Dll3 also cell autonomously attenuated the enhancement of Dll1-induced N signaling that was mediated by the modulator LFng, and Dll3 inhibition was reversed by LFng. This demonstrated that, together, Dll3 and LFng can modulate the levels of N signaling. Altogether, our analyses indicate that unlike other DSL ligands that either activate or inhibit N signaling depending on the cellular context, the primary function of Dll3 is to inhibit ligand-induced N signaling and, thereby, serve to attenuate the level of N signaling that is required to direct specific cell fates.
Results
Dll3 does not activate N signaling
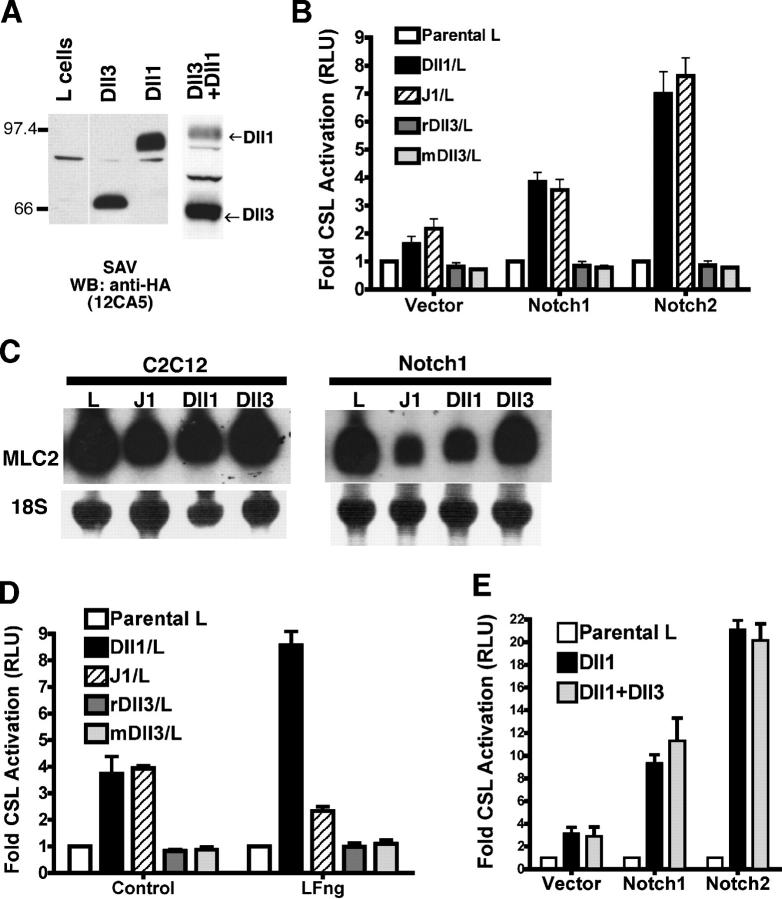
We isolated cDNA clones encoding rat Dll3 (rDll3) and engineered a full-length COOH terminally HA-tagged rDll3 for expression in L cells. An analysis of biotinylated cell surface proteins indicated that rDll3 and Dll1 are expressed to similar levels on the surface of expressing cells (Fig. 1 A). To determine which N receptors are activated by rDll3, we used a coculture assay to measure activation of the N downstream effector CSL (Nofziger et al., 1999; Hicks et al., 2000, 2002; Bush et al., 2001). In brief, NIH 3T3 cells were cotransfected with each of the known N receptors (N1–4) and with a CSL reporter construct containing multiple CSL-binding sites that were upstream of a luciferase gene (Hsieh et al., 1996). After coculture with Dll1, Jagged-1 (J1), rDll3, or parental L cell lines, quantitation of luciferase activity indicated the level of ligand-induced CSL-dependent N signaling. Dll1 and J1 activated CSL in N1-, N2- (Fig. 1 B), N3-, and N4-expressing cells (not depicted). However, rDll3 did not activate signaling from any of the four known N receptors (Fig. 1 B and not depicted) despite equivalent cell surface levels. This lack of activity was not limited to the HA-tagged rDll3 cDNA clone because cells expressing untagged mouse Dll3 (mDll3; Dunwoodie et al., 1997) were also inactive in this assay (Fig. 1 B). Moreover, Dll3 cells did not activate the CSL reporter that was expressed in C2C12 myoblasts, COS, or N2A neuroblastoma cells (unpublished data), indicating that Dll3 cannot activate N signaling in multiple cell types.
Figure 1.
Dll3 does not activate N signaling in trans. (A) Cell surface expression of HA-tagged Dll1, Dll3, and Dll1 + Dll3 L cell lines was determined by biotinylation, streptavidin (SAV) pull-down, and blotting with anti-HA mAb (12CA5). (B) NIH 3T3 cells transfected with N and CSL reporter were cocultured with J1, Dll1, Dll3, mDll3, or parental (L) cell lines and were assayed for luciferase activity. Error bars reflect the SD of the mean from three experiments. RLU, relative luciferase units. (C) Parental C2C12 and cell lines expressing N1 were cocultured with L, J1, Dll1, or Dll3 cells, and myogenesis was monitored by myosin light chain 2 (MLC2) mRNA expression. Loading and transfer of RNA was monitored by methylene blue staining of 18S rRNA. (D) NIH 3T3 cells transfected with N1, CSL reporter, and control or LFng were cocultured with Dll1, J1, rDll3, mDll3, or L cells and were assayed for luciferase activity (n = 3). (E) Parental L, Dll1, or Dll1 + Dll3 cells were cocultured with NIH 3T3 cells transfected with CSL reporter and N or vector. There was no statistically significant difference between Dll1 and Dll1 + Dll3 (n = 3).
Another measure of ligand-induced N signaling is the inhibition of myogenic differentiation of C2C12 myoblasts stably expressing N1 after coculture with cells expressing DSL ligands. In this assay, suppressed expression of the muscle structural gene myosin light chain 2 (MLC2) provides a readout of N signaling that is induced by ligands (Fig. 1 C; Nofziger et al., 1999; Bush et al., 2001). In contrast to Dll1 and J1 cells that strongly suppress the expression of MLC2, neither N1 nor N2 C2C12 myoblasts showed diminished MLC2 expression relative to parental L cells when cocultured with Dll3 cells (Fig. 1 C and not depicted). Therefore, Dll3 cells do not suppress C2C12 myogenic differentiation, which is consistent with our findings that Dll3 does not activate an N-responsive reporter construct (Fig. 1 B), providing an additional measure of Dll3's inability to activate N signaling.
LFng does not enable Dll3 to activate N1 signaling
We have previously reported that the glycosyltransferase LFng enhances Dll1-induced N signaling by using CSL reporter assays (Hicks et al., 2000; Yang et al., 2004). To determine whether LFng modification of N enables Dll3 to function as an activating ligand, NIH 3T3 cells were cotransfected with either alkaline phosphatase–tagged LFng or secreted alkaline phosphatase with N1 and a CSL reporter. Although LFng enhanced Dll1 activation of N1 and suppressed J1 activation of N1 as previously reported, neither rDll3 nor mDll3 activated N1 in the presence or absence of LFng (Fig. 1 D). Moreover, the other fringe family members radical and manic did not facilitate Dll3 activation of N1 or N2 (unpublished data). Together, these data suggest that fringe glycosylation of N does not enable Dll3 to function as an activating ligand.
Dll3 coexpressed with Dll1 does not perturb Dll1-induced N signaling
Given the inability of Dll3 to activate N signaling and the fact that Dll1 and Dll3 are coexpressed during development (Dunwoodie et al., 2002; Zhang et al., 2002; Takahashi et al., 2003), we asked whether Dll3 could alter Dll1-induced N signaling. Cells stably expressing both Dll1 and Dll3 were derived from the Dll1 line and were tested for CSL activation by either N1 or N2. Despite high levels of Dll3 expression on the surface of Dll1 + Dll3 cells relative to Dll1 cells (Fig. 1 A), they also activated the CSL reporter as found for Dll1 cells (Fig. 1 E), indicating that Dll3 does not antagonize Dll1-induced signaling.
Dll3 does not bind to N1 in trans
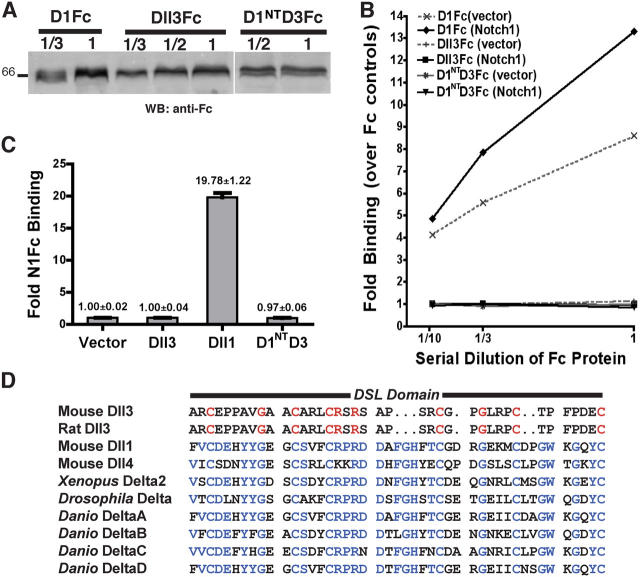
Because neither rDll3- nor mDll3-expressing cells activated N in either CSL reporter or myogenesis coculture assays, we determined whether Dll3 binds to N1. Based on the structure of a soluble D1Fc fusion protein that binds N1 and activates signaling, a Dll3Fc protein was generated by fusing the extracellular domain of Dll3 to Fc to allow clustering by anti-Fc antibodies, which is required for binding and activation (Hicks et al., 2000, 2002; Yang et al., 2004). When comparable amounts of Dll3Fc and D1Fc (Fig. 2 A) were assayed for binding to N1 cells, only DlFc binding was detected (Fig. 2 B). Furthermore, although a low level of D1Fc binding was detected with vector-transfected cells, which is presumably a result of endogenous N, Dll3Fc did not bind to either vector or N1-transfected 293T cells (Fig. 2 B). Using fluorescent microscopy to monitor binding, Dll3Fc did not bind to any of the known N receptors even though D1Fc binding was readily imaged (not depicted). Moreover, the coexpression of LFng with N1 or N2 did not enable Dll3Fc binding (not depicted).
Figure 2.
Dll3 does not bind to N. (A) Western Blot (WB) analysis using anti-Fc quantitated Dll3Fc, D1Fc, and D1NTD3Fc for binding assays in B. Numbers represent the dilution of condition media used in the binding assays. (B) 293T cells transfected with vector or N1 were assayed for binding of Fc, Dll3Fc, D1NTD3Fc, and D1Fc by flow cytometry. Fold binding over Fc control is plotted against serial dilutions. (C) Binding of N1Fc to 293T cells transfected with vector or HA-tagged Dll1, Dll3, or D1NTD3 is shown as fold binding over vector control (n = 3). Error bars represent SD. (D) Alignment of the DSL domains of rat and mDll3 with other Dll family members identifies Dll3 as highly divergent. Amino acid sequences conserved in Dll3 are red, whereas amino acid sequences conserved amongst the other Dll-related proteins are blue.
Although in agreement with our coculture data, the lack of detected Dll3Fc binding could result from low expression or misfolding of soluble Dll3Fc. Therefore, we determined whether a soluble N1Fc could bind Dll3-expressing cells. Although N1Fc binds to cells expressing either Dll1 (Fig. 2 C) or J1 (not depicted), N1Fc did not bind to Dll3 cells (Fig. 2 C). The lack of detected Dll3–N1 interactions in these binding assays is consistent with the inability of Dll3 L cells to activate N signaling in CSL reporter and myogenesis assays (Fig. 1, B and C).
When compared with other Dll ligands, it is obvious that the Dll3 DSL domain has not been conserved (Fig. 2 D). Given that the DSL domain is required for ligand binding and signaling (Henderson et al., 1997; Shimizu et al., 1999), our data suggested that the divergent Dll3 DSL module does not support binding to N when presented either on the surface of interacting cells or as a soluble protein. Because D1Fc binds to N-expressing cells and Dll1 activates N signaling, we reasoned that replacement of the Dll3 NH2-terminal and DSL domains (NT-DSL) with those of Dll1 (D1NT) would allow Dll3 to interact with N. To test this idea, we replaced the Dll3 NT-DSL in Dll3Fc with D1NT to produce a soluble D1NTD3Fc. Although comparable amounts of D1NTD3Fc, D1Fc, and Dll3Fc were used (Fig. 2 A), we were unable to detect the binding of D1NTD3Fc to N1-expressing cells. Moreover, when the D3NT sequences were replaced with D1NT in the full-length Dll3 HA-tagged protein (D1NTD3), cells expressing D1NTD3 did not bind N1Fc (Fig. 2 C). Together, our findings suggest that the divergent Dll3 DSL domain is not solely responsible for the lack of detected trans interactions for Dll3 and N receptor proteins.
Dll3 inhibits N signaling cell autonomously
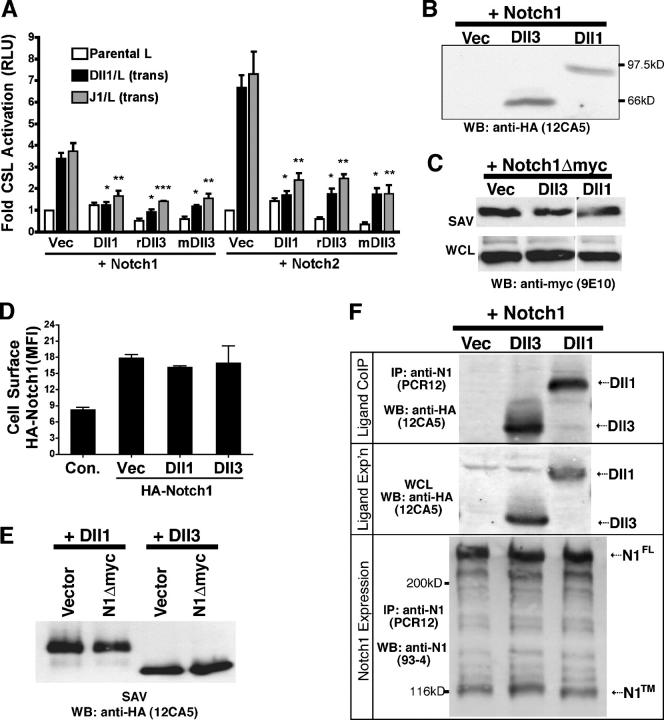
Because Dll3 did not bind or activate any of the known N receptors when presented in trans, we determined whether Dll3 inhibits N signaling when expressed with N in the same cell (cis or cell autonomously), as previously reported for other DSL family proteins (Henrique et al., 1997; Sakamoto et al., 2002; Itoh et al., 2003). To determine whether Dll3 could cell autonomously inhibit N signaling that is induced by other DSL ligands, NIH 3T3 cells transiently expressing either N1 or N2, CSL reporter, and either vector, Dll1, or Dll3 plasmids were cocultured with Dll1, J1, or L cells. In these assays, CSL reporter activity was decreased >60% when either Dll1 or Dll3 were coexpressed with either N1 or N2 (Fig. 3 A). Serrate has also been reported to cell autonomously inhibit N (de Celis and Bray, 2000; Kiyota and Kinoshita, 2004), and J1 that was coexpressed with N1 or N2 also suppressed CSL activation (unpublished data). Therefore, unlike other DSL ligands that both activate and inhibit N signaling, Dll3 cannot activate signaling in trans but effectively inhibits ligand-induced N signaling when coexpressed with N.
Figure 3.
Dll3 cell autonomously inhibits N signaling. (A) NIH 3T3 cells cotransfected with N1 or N2 and either HA-tagged Dll1, rat Dll3 (rDll3), mDll3, or vector along with a CSL reporter cocultured with Dll1, J1, or L cells. Error bars reflect the SD of the mean from four experiments (*, P < 0.01; **, P < 0.001; ***, P < 0.0001). (B) Western blot of HA-tagged Dll1 and Dll3 from NIH 3T3 cells indicates expression in CSL reporter assays in A. (C) 293T cells cotransfected with N1Δmyc and either vector, Dll1, or Dll3 plasmids and cell surface expression of N1 were analyzed after biotinylation/SAV pull-down and anti-myc (9E10) Western blotting. (D) 293T cells transfected with vector or HA-N1 and either vector, Dll1, or Dll3 plasmids were stained live, and mean fluorescence intensity was determined by FACS. Error bars represent SD. (E) 293T cells cotransfected with Dll1 or Dll3 and N1Δmyc or vector plasmids and cell surface expression of Dll1 or Dll3 were determined by biotinylation/SAV pull-down and anti-HA antibody (12CA5) Western blotting. (F) Lysates from 293T cells transfected with N1 and either HA-tagged Dll1, Dll3, or vector were incubated with N1 antibodies and HA antibody Western blot to detect Dll1 or Dll3 (12CA5; top) or with N1 antibody (93–4; bottom). Middle panel is a HA Western blot of Dll1 and Dll3 from WCL.
Dll3 cell-autonomous expression does not decrease cell surface N1
To ensure that the loss in N signaling, which is detected when either Dll3 or Dll1 were coexpressed with N, was not caused by decreased N1 cell surface expression, N1 cell surface levels in the presence of either Dll1 or Dll3 were determined. Biotinylation of cell surface proteins indicated that the coexpression of either Dll1 or Dll3 with N1 did not decrease the amount of N1 that was detected in whole cell lysates (WCLs; Fig. 3) or the level of N1 that was detected at the cell surface (streptavidin [SAV]; Fig. 3 C). These findings are in agreement with a study on N cell surface expression in cells coexpressing chick Delta1 and mouse N1 (Sakamoto et al., 2002). To more accurately quantitate the level of N1 cell surface expression, cells coexpressing either Dll1 or Dll3 with an NH2-terminal HA-tagged N1 (HA-N1) were stained with AlexaFluor488-conjugated HA antibody and were analyzed by flow cytometry. In agreement with our biotinylation data, the expression of Dll3 with N1 did not significantly alter cell surface N1 (Fig. 3 D), suggesting that losses in cell surface N1 cannot account for losses in signaling (Fig. 3 A). Furthermore, biotinylation analysis of Dll1 and Dll3 indicate that neither Dll1 nor Dll3 surface expression was altered when coexpressed with N1 (Fig. 3 E). This suggests that the overexpression of ligand and receptor in the same cell does not alter trafficking to the cell surface.
Dll3 directly interacts with N1 in coexpressing cells
Our binding studies did not detect interactions between Dll3 and N1 (Fig. 2, B and C); however, these experiments measured trans interactions rather than interactions between Dll3 and N1 within the same cell. Therefore, to detect cis interactions between Dll3 and N1, we determined whether Dll3 coimmunoprecipitated with N1. 293T cell lysates from cells coexpressing N1 and either vector, HA-tagged Dll3, or Dll1 were immunoprecipitated with N1 antibodies and immunoblotted with an HA antibody to detect Dll3 interaction with N1. Both Dll1 and Dll3 coimmunoprecipitated with N1 (Fig. 3 F). To control for postlysis interactions, lysates from Dll1- or Dll3-expressing cells were mixed with equal amounts of N1 lysate and were coimmunoprecipitated. Neither Dll3 nor Dll1 immunoprecipitated with N1 from mixed lysate controls (unpublished data). Therefore, in contrast to trans conditions, Dll3 stably interacts with N1, but the association requires the expression of both proteins in the same cell (cis conditions).
Dll3 promotes primary neurogenesis in X. laevis embryos, indicating a block in N signaling
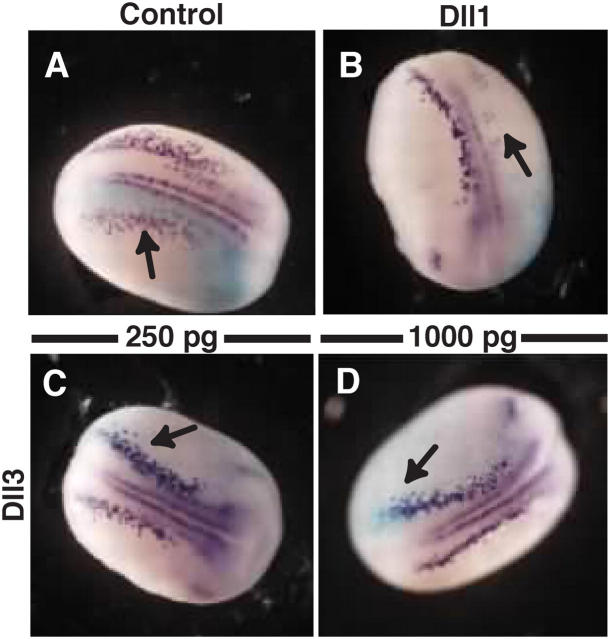
Our findings that Dll3 does not activate N signaling in cultured cells are at odds with a previous report in which Dll3 was shown to activate N signaling when tested in X. laevis embryos (Dunwoodie et al., 1997). In X. laevis, the formation of primary neurons can be used as a reliable readout of N signaling. In embryos in which N signaling is increased, the generation of primary neurons is markedly reduced, whereas the expression of N antagonists causes a reciprocal increase in primary neurons (Wettstein et al., 1997). Because Dll3 was previously reported to inhibit neurogenesis in this assay, we reexamined its activity in relation to that of Dll1. As previously demonstrated, injecting 250 pg Dll1 mRNA at the two-cell stage causes a marked decrease in the number of cells that express the neuronal marker β-tubulin at neural plate stages (Fig. 4, compare A with B; Chitnis et al., 1995; Dunwoodie et al., 1997). This decrease is indicative of activated N signaling (Chitnis et al., 1995) and is similar in nature to that observed when embryos are injected with NICD, XDelta1, or mDll4 (Shutter et al., 2000). In contrast, injecting either 250 pg or 1 ng mDll3 mRNA not only failed to inhibit neurogenesis but, in some cases, produced an increase in the number of β-tubulin–positive neurons (Fig. 4, C and D; and Table I). Thus, in our hands, Dll3 does not activate N signaling when ectopically expressed during neurogenesis but, instead, behaves as an N inhibitor. We cannot account for the difference between these results and those obtained with equivalent amounts of injected Dll3 mRNA from the same Dll3 clone, which was reported previously (Dunwoodie et al., 1997). One possibility is that a suppression or delay in neuronal differentiation is often an artifact that occurs in RNA injection experiments. Nonetheless, our results indicate that Dll3, over a large concentration range, primarily acts as an inhibitory ligand in the X. laevis assay, which is in line with our findings in mammalian cell culture assays. Importantly, Dll1 prevents neurogenesis at the same concentration (250 pg), whereas Dll3 promotes neurogenesis, highlighting the different activities of these DSL ligands.
Figure 4.
Injection of mDll3 mRNA promotes neurogenesis in X. laevis . Representatives of embryos injected with either synthetic lacZ mRNA alone (A) or in combination with 250 pg Dll1 mRNA (B), 250 pg mDll3 mRNA (C), or 1,000 pg mDll3 mRNA (D) and stained for X-gal (light blue) and β-tubulin (purple) expression. Arrows indicate injection site.
Table I. Quantitation of β-tubulin–positive neurons in D1- or mDll3-injected embryos.
| Injected mRNA | Increase in neurons |
Reduction or complete loss of neurons |
No change | Total embryos scored |
|---|---|---|---|---|
| Control (β-galactosidase) |
4 | 3 | 48 | 55 |
| Dll1 (250 pg) | 1 | 23 | 9 | 33 |
| Dll3 (250 pg) | 15 | 0 | 5 | 20 |
| Dll3 (1,000 pg) | 15 | 3 | 7 | 25 |
Embryos were injected and processed (Chitnis et al., 1995) to determine the relative amounts of β-tubulin that were scored either as an increase, reduction, or complete loss of neurons or as no change.
Dll3 antagonism of N signaling regulates neuronal and glial differentiation
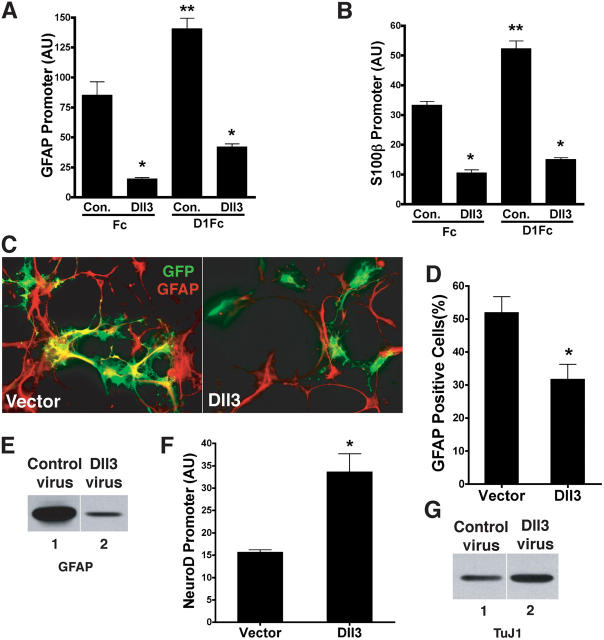
Both Dll1 and Dll3 are expressed in the developing brain (Dunwoodie et al., 1997; Campos et al., 2001), where N signaling is known to regulate the differentiation of progenitors into neurons and glia. We have previously reported that D1Fc inhibits neurogenesis and promotes gliogenesis in mammalian neural stem cells (NSCs; Morrison et al., 2000; Ge et al., 2002). To further demonstrate Dll3 antagonism of N signaling, we transfected cortical NSCs with Dll3 or vector and induced astrogliogenesis with D1Fc as previously described (Ge et al., 2002). In this system, Dll3 reduced expression of the astrocyte markers glial fibrillary acidic protein (GFAP) and S100β, which were measured by the activation of GFAP and S100β reporters (Fig. 5, A and B). Because both GFAP and S100β regulatory regions contain functional CSL-binding sites, they are direct targets of N activation and, thus, serve as readouts of N signaling as well as gliogenesis (Ge et al., 2002; Hermanson et al., 2002). Dll3 not only suppressed D1Fc-induced N signaling that was required to drive the transcription of GFAP and S100β, but it also antagonized signaling that was induced by endogenous ligands (Fig. 5, A and B; Fc treated). Even in the absence of D1Fc, NSCs ectopically expressing Dll3 did not express GFAP, and the number of GFAP-positive cells as well as the level of GFAP expression was decreased by Dll3 transfection (Fig. 5, C and D) or infection with Dll3 adenovirus (Fig. 5 E). In contrast to Dll1 that is known to block neurogenesis, Dll3 promoted neurogenesis of early (neurogenic) stage NSCs, as measured by increased NeuroD promoter activity (Fig. 5 F) and expression of the neuronal-specific marker β-tubulin (TuJ1; Fig. 5 G). Because N signaling prevents neurogenesis (Lewis, 1996), neural induction in the presence of Dll3 in cortical progenitors is in agreement with Dll3 functioning as a signaling antagonist of N in both our cell coculture (Fig. 3 A) and X. laevis injection assays (Fig. 4, C and D). Together, our findings suggest that Dll3 promotes neurogenesis and inhibits gliogenesis through antagonizing N and uncover a biological role for Dll3 as a negative regulator of cell fate decisions that are influenced by N signaling.
Figure 5.
Dll3 suppresses D1Fc-induced astrocytic differentiation. Embryonic day 11.5 mouse cortical NSCs cotransfected with reporters for GFAP (A) or S100β (B) and either vector or Dll3 were treated with control Fc or D1Fc to induce astrogliogenesis. (C) NSCs cotransfected with GFP and either vector or Dll3 were cultured with D1Fc for 4 d and stained for GFAP. (D) Quantitation of transfected cells expressing GFAP from six independent experiments. (E) GFAP expression in NSCs infected with control (lane 1) or Dll3 adenovirus (lane 2) as determined by Western blotting. (F) Neurogenic stage mouse cortical NSCs cotransfected with NeuroD promoter luciferase construct and vector or Dll3. Promoter activation is plotted as relative luciferase units for six experiments. P < 0.05. *, significant differences between Dll3 and vector; **, significant increase between Fc and D1Fc. Error bars represent SD. AU, arbitrary units. (G) Tubulin (TuJ1) expression in NSCs infected with control (lane 1) or Dll3 adenovirus (lane 2) as determined by Western blotting.
LFng and Dll3 function cell autonomously to modulate N signaling levels
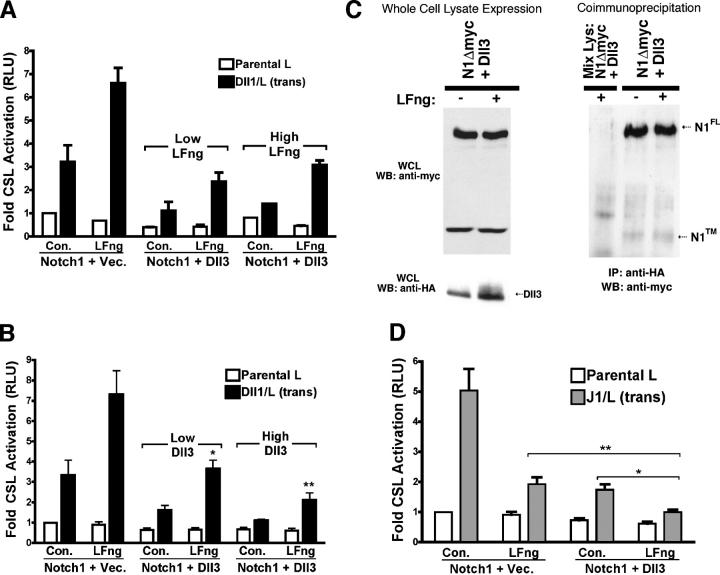
Although the loss of either Dll1 or Dll3 leads to defects in somitogenesis, Dll1 mutants display a complete loss of somite polarity markers, which is similar to RBP-J knockout mice, highlighting that Dll1 and RBP-J are core components of the N pathway (for review see Giudicelli and Lewis, 2004). In contrast, losses in Dll3 produce only disorganization in somite patterning, which is a phenotype that is strikingly similar to gains or losses in the N modulator LFng. These findings suggest that although Dll1 is an activating ligand that is absolutely required for N signaling, Dll3 and LFng may serve to regulate levels of N signaling during somitogenesis. To explore the relationship between LFng and Dll3 in modulating N signaling that is induced by other DSL ligands, we used the CSL reporter assay. When cells transiently expressing N1, CSL reporter, and either Dll3 or vector as well as low (100 ng) or high (500 ng) amounts of LFng DNA were cocultured with Dll1 cells, LFng appeared to counteract the inhibitory effects of Dll3 in a dose-dependent manner (Fig. 6 A). Specifically, increases in transfected LFng increased signaling even in the presence of Dll3, suggesting a dynamic interplay between LFng and Dll3 in modulating the level of N1 signaling. Conversely, LFng enhancement of Dll1-induced N signaling was suppressed by Dll3 in a dose-dependent manner (Fig. 6 B). Together, these experiments illustrate the dynamic nature of Dll1-induced N signaling in response to Dll3 and suggest that, like LFng, Dll3 also modulates N signaling.
Figure 6.
LFng and Dll3 dynamically modulate N signaling. (A) NIH 3T3 cells cotransfected with N1 and CSL reporter as well as vector or Dll3 with increasing amounts of LFng or control DNA were cocultured with L or Dll1 cells and assayed for luciferase activity. (B) NIH 3T3 cells cotransfected with N1 and CSL reporter as well as control or LFng, and either vector or increasing amounts of Dll3 DNA were cocultured with L or Dll1 cells. P < 0.01. *, a significant difference between vector and Dll3 (low); **, a significant difference between Dll3 (low) and Dll3 (high). n = 5. (C) Lysates from 293T cells cotransfected with N1Δmyc, HA-tagged Dll3, and LFng or control DNA incubated with anti-HA antibody to capture Dll3 immunoprecipitates, which were identified by anti-myc Western blotting to detect N1 (right). Lysate from N1 cells was mixed with equal amounts of D3 lysate and was analyzed alongside other samples (Mix Lys). WCLs were analyzed by anti-myc (top left) and anti-HA Western blotting (bottom left). (D) Parental L or J1 cells cocultured with NIH 3T3 cells that were transfected with N1, CSL reporter, vector, or Dll3 and control or LFng. (*, P < 0.005; **, P < 0.001; n = 7). Error bars represent SD.
Fringe proteins have been reported to override the cis-inhibitory effects of other DSL ligands in flies and chicks (Hukriede et al., 1997; de Celis and Bray, 2000; Sakamoto et al., 2002). It has been proposed that LFng modification of N and/or DSL ligand disrupts cis-inhibitory complexes to allow N activation by adjacent ligand cells. In this regard, two Dll3 EGF-like repeats have broad O-fucosylation consensus sequences, which is a prerequisite for LFng glycosylation (Panin et al., 2002). To determine whether LFng glycosylation of N1 or Dll3 disrupts interactions between Dll3 and N1, coimmunoprecipitation of N1 with Dll3 was measured in the presence of LFng. Irrespective of LFng coexpression, N1 coimmunoprecipitated with Dll3 (Fig. 6 C), and, conversely, Dll3 coimmunoprecipitated with N1 (not depicted), indicating that LFng does not alter cis interactions between Dll3 and N1. Even increasing amounts of LFng did not disrupt Dll3–N1 interactions (unpublished data).
Because LFng did not prevent interactions between Dll3 and N1, it seemed that LFng enhancement of Dll1 signaling in combination with Dll3-inhibitory effects could account for LFng reversing Dll3 cis inhibition. LFng both potentiates Dll1-induced N1 signaling and inhibits signaling by J1 (Hicks et al., 2000; Yang et al., 2004). Therefore, we reasoned that if LFng exclusively modulates trans signaling and has no effect on Dll3–N1 cis interactions, the coexpression of Dll3 and LFng with N1 should result in a greater inhibition of J1-induced N signaling than with either modulator alone. Conversely, if LFng functions directly to prevent cis inhibition by Dll3, then Dll3 coexpression should not further inhibit signaling by J1. The coexpression of either LFng or Dll3 suppressed J1-induced N1 signaling, whereas the coexpression of both LFng and Dll3 produced a stronger block in signaling (Fig. 6 D). This indicates that LFng does not disrupt Dll3 cis inhibition but rather functions to modulate the productivity of N signaling that was induced by trans ligand. LFng coexpression with Dll3 further inhibited N1 signaling that was induced by J1, yet reversed the Dll3 inhibition of signaling in response to Dll1. This suggests that, together, LFng and Dll3 could finely tune the levels of N signaling that are induced by different DSL ligands.
Discussion
In contrast to other DSL ligands, the reported activating ligand Dll3 (Dunwoodie et al., 1997) did not bind or activate N when tested in a number of different assays. Despite the inability of Dll3 to induce signaling from any of the known N receptors, we find that Dll3 is a potent antagonist of ligand-induced N signaling when coexpressed with N. Indicative and supportive of Dll3 antagonism of N signaling, we show that Dll3 promotes primary neurogenesis in X. laevis embryos and enhances neuronal differentiation of mouse cortical neural progenitors in vitro, whereas glial differentiation is reduced. In combination with LFng, which is a well-known modulator of ligand-induced N signaling, Dll3 regulates the level of signaling, suggesting that it may contribute to the dynamic changes in N signaling that are required in development.
Dll3 is a highly divergent DSL family member
Dll3 has 36, 41, 31, and 29% overall amino acid homology to mDll1, X. laevis Delta2, mDll4, and Drosophila melanogaster Delta, respectively, identifying Dll3 as the most divergent DSL member (Dunwoodie et al., 1997). Dll3 is also the shortest of the mammalian Dll ligands, with only six EGF-like repeats compared with eight repeats identified for Dll1 and Dll4. A comparison of the Dll3 DSL domain with mDll1, mDll4, X. laevis Dll2, D. melanogaster Delta, and zebrafish DeltaA-D highlights its divergence (Fig. 2 D). Interestingly, although a large number of Delta homologues have been identified in zebrafish, none appear to have the degenerate DSL domain that is characteristic of Dll3. The DSL domain is thought to be important in receptor binding and activation (Henderson et al., 1997; Shimizu et al., 1999). However, a soluble form of J1 containing only the NT-DSL fused to Fc binds poorly to N, and the addition of the first two J1 EGF-like repeats is required to enhance binding (Shimizu et al., 1999). Therefore, it was surprising that replacement of the Dll3 NT-DSL sequences with those of Dll1 in D1NTD3Fc did not promote binding to N1. In fact, if one assumes that Dll1 NT-DSL facilitates interactions, it seems that Dll3 EGF-like repeats antagonize Dll1 NT-DSL binding to N, suggesting that additional differences in Dll3 perturb Dll3–N interactions. In support of this idea, the replacement of NT-DSL sequences in full-length Dll3 with those of Dll1 did not promote N1Fc binding to D1NTD3 cells. These findings suggest that Dll3 EGF-like repeats do not function as reported for J1 EGF-like repeats, identifying additional differences between Dll3 and other DSL ligands. Interestingly, the second Dll3 EGF-like repeat is incomplete, and missense mutations map to this and other repeats in spondylocostal dysostosis patients (Bulman et al., 2000; Turnpenny et al., 2003), suggesting that these repeats are important for Dll3 function. An analysis of different Dll1–Dll3 chimeric proteins will be required to further investigate the structural differences between Dll1 and Dll3.
In addition to differences in Dll3 extracellular sequences, the intracellular domain is significantly smaller than that of Dll1 and other DSL family members (Dunwoodie et al., 1997). The Delta intracellular domain is required for the activation of N signaling, perhaps reflecting its role in endocytosis that is regulated through ubiquitination (for review see Le Borgne et al., 2005). Although the exact function of Delta ubiquitination in N signaling is not well understood, it is interesting to note that although the Dll1 intracellular domain contains 17 lysine residues, which are potential sites for ubiquitination, there are no lysines in the Dll3 intracellular domain. Given the importance of ubiquitination in Delta-induced N signaling, the lack of lysines in Dll3 is consistent with our findings that Dll3 is unable to activate N signaling.
Finally, the COOH terminus of Dll3 lacks a PDZ-binding motif that is present in other DSL proteins and directs different cellular responses through interactions with PDZ domain–containing proteins (Ascano et al., 2003; Pfister et al., 2003; Six et al., 2003; Wright et al., 2004), identifying additional functional differences for Dll3. Thus, in addition to extracellular changes that prevent N binding in trans, Dll3 appears to have undergone other alterations that do not promote signaling. Because our data suggest that Dll3 functions to inhibit rather than activate N signaling, associations of Dll3 with N pathway genes likely reflect a role for Dll3 as a signaling antagonist rather than as an activating ligand.
Dll3 as a modulator of cellular differentiation induced by N signaling
Dll1 and Dll3 have overlapping as well as distinct expression patterns in the developing cortex and spinal cord (Dunwoodie et al., 1997; Kusumi et al., 1998; Campos et al., 2001; Sparrow et al., 2002). Specifically, Dll1 is expressed within the ventricular zone, whereas Dll3 is located more laterally in a population of cells that is thought to be fated for terminal neuronal differentiation. It has been proposed that Dll1 ventricular cells express Dll3 after migrating away from the ventricular zone, and that this sequential expression of Dll1 and Dll3 is linked to progression toward a neuronal fate. Consistent with this idea, we find that ectopic expression of Dll3 in either X. laevis oocytes or neural progenitors blocks N signaling and promotes terminal neuronal differentiation. However, pudgy and Dll3 knockout mice display only subtle neuroepithelial defects in the lateral ventricles with a low penetrance of ∼50%, suggesting that if Dll3 does regulate neural differentiation, its effects must be transient. Nonetheless, our findings that Dll3 regulates neuronal cell fate are consistent with a role for Dll3 as a modulator of N signaling rather than as a core component of the pathway and support a role for Dll3 in altering progenitor cell fate through attenuating N signaling.
LFng and Dll3 function together to cell autonomously modulate N signaling
Loss of Dll3 causes severe defects in somite patterning and axial elongation, identifying a role for Dll3 in somitogenesis (Kusumi et al., 2004). Similar somite defects have been observed for both gains and losses in LFng (Serth et al., 2003), suggesting that Dll3 may also modulate the levels of N signaling during somite development, as proposed for LFng. Based on the published roles for fringe in modulation of N–ligand interactions, LFng and Dll3 were tested for their ability to coordinately regulate N signaling. We found that LFng modification of N did not enable Dll3 to function in trans as an activating ligand. Instead, LFng enhanced N signaling to override Dll3 cis-inhibitory effects on activation by Dll1. The effects of LFng did not prevent interactions between Dll3 and N1 even though both Dll3 and N1 contain multiple potential sites for LFng glycosylation. Our findings that the coexpression of LFng and Dll3 with N1 additively blocked J1-induced CSL activation suggest that LFng regulates trans ligand–N interactions, whereas Dll3 inhibits N activation through cis interactions. Altogether, our results suggest that Dll3 and LFng coordinately function in a cell-autonomous manner to modulate ligand-induced N signaling.
Spatial and temporal changes in N signaling within the PSM appear to be critical for proper segmental patterning (for review see Giudicelli and Lewis, 2004), and it has been proposed that N signaling in the PSM must fall below a certain threshold during each segmental cycle to ensure proper somite patterning (Serth et al., 2003). Given our observations that Dll3 and LFng function together to dynamically regulate ligand-induced N signaling, it is tempting to speculate that Dll3 may participate in the negative feedback loops that regulate cyclic activation of N signaling during somitogenesis.
Dll3 is an inhibitor of N signaling
We have found that Dll3 is not an activating ligand for N but rather functions to cell autonomously inhibit signaling. In support of Dll3 as an N antagonist, findings in mice have also proposed that Dll3 counteracts the activity of Dll1 in regulating N signaling during somite patterning (Takahashi et al., 2003). Consistent with Dll3 functioning as a negative regulator, the expression of an N target gene, N-regulated ankyrin repeat protein (NRARP), is extinguished in nascent somites in mice lacking either N1 or Dll1 but is increased in pudgy mice (Krebs et al., 2001). Perhaps, Dll1-induced N signaling activates NRARP expression, whereas Dll3 antagonizes this signaling, and, thus, a loss of Dll3 leads to an increase in N signaling and a consequential increase in NRARP expression. In contrast, expression of the N target gene Hes5 is either absent or reduced in the PSM of Dll3 mutant embryos (Dunwoodie et al., 2002), which is supportive of Dll3 as an activator of N signaling. However, it is important to note that Dll1 mutant embryos that are defective in N signaling show a reduction or loss of Hes5 in both the PSM and neural tube, whereas Dll3 mutants maintain strong Hes5 expression in the neural tube (Barrantes et al., 1999; Dunwoodie et al., 2002). Because somitogenesis, unlike the developing nervous system, requires cyclic N signaling, losses in Dll3 may adversely affect negative feedback loops that are required to maintain proper levels of N signaling in the PSM in order to affect Hes5 expression. Nonetheless, Dll3 and Dll1 mutant somite phenotypes are clearly different, suggesting that Dll3 and Dll1 have distinct functions in regulating N signaling during somitogenesis (Kusumi et al., 2004). Our findings support this idea, and we suggest that the different activities identified in this study for Dll1 and Dll3 may account for the distinct mutant phenotypes. Finally, our finding that Dll3 has diverged to function solely as an inhibitor of N signaling that is induced by other DSL ligands is reminiscent of reports for other signaling antagonists that are structurally related to their activating ligands but function to inhibit rather than activate signaling (Vinos and Freeman, 2000; Daluiski et al., 2001).
Materials and methods
Cell lines and mammalian expression constructs
Parental cell lines were obtained from the American Type Culture Collection and were propagated as suggested. Stable C2C12 cell lines expressing N1 and L cell lines expressed Dll1 or J1 have been described previously (Lindsell et al., 1995; Hicks et al., 2000). Stable Dll3-expressing cells were generated by using hypoxanthine and thymine selection as previously described for J1-expressing L cells (Lindsell et al., 1995).
rDll3 was isolated from an embryonic day 13 rat brain cDNA library (GenBank/EMBL/DDBJ accession no. AF084576) based on homology to mDll3 (cDNA obtained from S. Dunwoodie, University of New South Wales, Sydney, Australia; Dunwoodie et al., 1997) and was tagged with triple tandem repeat of the influenza virus HA epitope. HA-tagged rDll3 and the previously described Dll1 were subcloned into pCS2 expression vectors (Turner and Weintraub, 1994). Dll3Fc was generated by fusing the extracellular domain of Dll3 (1–1476 bp) to Fc and subcloning into the pcDNA3 expression vector (Invitrogen). D1NTD3Fc and D1NTD3 were constructed by replacing the first 651 bp of Dll3Fc or HA-tagged Dll3, respectively, with the NH2 terminus and DSL domain of Dll1 (1–725 bp). N1Δmyc replaced the COOH-terminal 436 amino acids of full-length rat N1 with six myc epitopes in the pCS2+mt vector (Yang et al., 2004). NH2-terminal HA-tagged N1 (HA-N1) was generated by inserting the triple HA epitope immediately downstream of the signal peptide by using a PCR overlap strategy and subcloning into pBOS.
Biotinylation, immunoprecipitation, and Western blot analysis
Cell surface proteins were labeled with biotin and isolated as previously described (Bush et al., 2001). In brief, 293T cells were cotransfected with 0.5 g N1Δmyc and 0.5 g of either Dll3, Dll1, or vector per 60-mm dish using a standard Hepes-based calcium phosphate precipitation method. After 48 h, the cells were washed with cold PBS and were incubated in PBS containing 0.5 mg/ml Sulfo-NHS-Biotin (Pierce Chemical Co.) for 1 h at 4°C. Cells were washed with glycine buffer (20 mM Tris, pH 7.4, 300 mM NaCl, 0.1% BSA, and 100 mM glycine) and were incubated in glycine buffer for another 15 min. Cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, and 0.1% SDS) containing protease inhibitors (1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin). WCLs were incubated with SAV-immobilized beads (Pierce Chemical Co.) at 4°C for 10–12 h.
Coimmunoprecipitation experiments (for Fig. 3 F) also used 293T cells that were transfected with calcium phosphate and were washed with cold PBS but lysed in Triton X-100 buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, and 1% Triton X-100). WCLs were incubated on ice for 1 h with a mixture of anti-N1 intracellular domain pAbs (PCR12 and 93–4) at 1:200 each. For pull-downs between LFng modified N1 and Dll3 (Fig. 6 C), the immunoprecipitation protocols were based on methods published by Sakamoto et al. (2002). In brief, 293T cells were plated at a density of 1.5 × 106 cells per 60-mm dish and were transfected with LipofectAMINE (Invitrogen) according to the manufacturer's instructions using 0.3 μg N1, Dll3, or vector and 1 μg of secreted alkaline phosphatase or LFng DNA. Cells were lysed in 1% Triton X-100 lysis buffer (100 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, and 1% Triton X-100; Sakamoto et al., 2002). Lysate was incubated with a mAb anti-HA (3F10; Roche Biosciences) and rabbit anti–rat Ig for 30 min–1 h on ice. Immunoprecipitates were collected on protein A agarose beads (Invitrogen). Specific proteins were identified after SDS/PAGE, transferred to NitroBind membrane (Osmonics), probed with 93–4 (1:1,000), 12CA5 (1:2,000), or 9E10 (1:1,000; Santa Cruz Biotechnology, Inc.) antibodies, and were detected using ECL Plus Western blotting detection system (GE Healthcare) and a scanner (Typhoon 9410; GE Healthcare).
CSL gene reporter assay
CSL reporter was provided by D. Hayward (Johns Hopkins School of Medicine, Baltimore, MD). NIH 3T3 cells were cotransfected at 70% confluence according to the manufacturer's suggested protocols for LipofectAMINE (Invitrogen) with 0.25 μg CSL reporter, 0.005 μg Renilla luciferase (RLCMV), 0.1μg N DNA, and 0.1 μg vector or cis ligand DNA. 24 h posttransfection, cells were cocultured with ligand-expressing L cell lines or the parental L cell line control, and lysates were collected 48 h posttransfection and assayed using the Dual Luciferase Reagent system (Promega).
Cell surface labeling and ligand-binding assays
HEK 293T cells were calcium phosphate transfected with vector or HA-N1 and either vector, Dll1, or Dll3 plasmids. To detect N1 cell surface expression, cells were also transfected with pHcRed (BD Biosciences and CLONTECH Laboratories, Inc.), and 48 h posttransfection, cells were stained live with an AlexaFluor488-conjugated HA mouse monoclonal (16B12; Molecular Probes). The pHcRed-expressing cells emitting fluorescence at 660 nm were assayed for mean fluorescence intensity at 530 nm to detect the AlexaFluor488 signal using FACSCalibur (Becton Dickinson) as previously described (Hicks et al., 2002; Yang et al., 2004). For binding studies, D1Fc, Dll3Fc, D1NTD3Fc, or Fc conditioned medium was generated as previously described (Hicks et al., 2000, 2002) and was diluted to similar levels of expression. N1Fc containing the first 12 EGF-like repeats of rat N1 fused to Fc was purchased from R&D Systems. 293T cells were plated in 24-well plates and transfected with Transfast (Promega) using 60 ng of either vector or N1 and Dll1, Dll3, or D1NTDll3 DNA per well according to the manufacturer's recommended protocols. 48 h posttransfection, cells were incubated for 45 min at 37°C in blocking media (DME containing 10% goat serum and 1% BSA). To determine the level of binding, Fc fusion proteins were preclustered with FITC-conjugated goat anti–human Fc antibody (1:200; Jackson ImmunoResearch Laboratories) at 4°C for 1 h. Serial dilutions of the clustered ligand (Fig. 2, A and B) or 1 μg/ml N1Fc were added to cells for 40–60 min at 37°C. After binding, the cells were resuspended and washed in wash buffer (PBS, 0.2% BSA, and 0.1% NaN3) and were analyzed by FACScan (BD Biosciences).
X. laevis embryo injections
Single blastomeres of two- or four-cell stage X. laevis embryos were injected with 50 pg synthetic lacZ mRNA alone or in combination with Dll1 or Dll3 mRNA. Embryos were stained for X-gal and β-tubulin expression at the neural plate stage as previously described (Chitnis et al., 1995; Wettstein et al., 1997). X. laevis embryos were imaged at RT in ethanol using a photomicroscope (Wild M420; Leica) with an Apozoom lens at 32× and a camera (model DP11; Olympus). Images were acquired using Olympus software and were processed using Adobe Photoshop to adjust brightness and contrast, which was applied to the entire image.
Mouse cortical NSC culture and differentiation assays
NSCs were isolated and cultured from embryonic day 11.5 BALB/c mouse cortex as previously described (Ge et al., 2002). Progenitors were cultured in concentrated and clustered Fc or D1Fc conditioned medium as previously described (Morrison et al., 2000). For reporter assays, NSCs were plated onto polyornithine/fibronectin-coated 96-well plates at a density of 2–4 × 106 cells/plate. Using Fugene 6 transfection method (Roche Biosciences), cells were cotransfected with vector or Dll3 as well as with promoters of interest driving the firefly luciferase (pGL3) and a constitutively active thymidine kinase promoter driving Renilla luciferase (TK-pRL) as internal controls for transfection efficiency. 24 h posttransfection, cells were lysed, and promoter activities were assayed using the Promega Dual Luciferase Assay kit. NSC cultures were infected with control or Dll3 adenovirus as previously described (Sun et al., 2001). GFAP or TuJ1 expression was determined by immunoblotting using standard protocols (Ge et al., 2002). NSC cultures were stained with Cy3- and Cy2-conjugated antibodies and were imaged at RT using a microscope (model BX51; Olympus) with a UplanApo 20×/0.7 objective (Olympus) and a camera (model IKH027779; Olympus). Images were acquired using Magna FIRE software (Optronics) and were processed using Adobe Photoshop for cropping, resizing, and merging.
Acknowledgments
We thank Drs. Eddy DeRobertis, Ellen Robey, Larry Zipursky, and Yumiko Saga for helpful comments, S. Dunwoodie for the mDll3 cDNA, Che Hutson for help with HA-N1 construction, and D. Hayward for the CBF1 reporter. Flow cytometry was performed in the UCLA/JCCC Flow Cytometry Core Facility.
This work was supported by the National Institutes of Health (grant NS31885), Stop Cancer (grant to G. Weinmaster), the Ruth L. Kirschstein National Research Service Award (grant GM-07185 to J.T. Nichols), JCCC, and the Chemistry-Biology Interphase Training Program (grant GM-08946 to E. Ladi).
E. Ladi's present address is Department of Molecular and Cellular Biology, University of California, Berkeley, Berkeley, CA 94720.
Abbreviations used in this paper: Dll, Delta-like; GFAP, glial fibrillary acidic protein; LFng, lunatic fringe; mDll, mouse Dll; MLC2, myosin light chain 2; NICD, Notch intracellular domain; NRARP, Notch-regulated ankyrin repeat protein; NSC, neural stem cell; NT-DSL, NH2-terminal and DSL domains; PSM, presomitic mesoderm; rDll, rat Dll; SAV, streptavidin; WCL, whole cell lysate.
References
- Ascano, J.M., L.J. Beverly, and A.J. Capobianco. 2003. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J. Biol. Chem. 278:8771–8779. [DOI] [PubMed] [Google Scholar]
- Barrantes, I.B., A.J. Elia, K. Wunsch, M.H. De Angelis, T.W. Mak, J. Rossant, R.A. Conlon, A. Gossler, and J.L. de la Pompa. 1999. Interaction between N signalling and Lunatic fringe during somite boundary formation in the mouse. Curr. Biol. 9:470–480. [DOI] [PubMed] [Google Scholar]
- Bulman, M.P., K. Kusumi, T.M. Frayling, C. McKeown, C. Garrett, E.S. Lander, R. Krumlauf, A.T. Hattersley, S. Ellard, and P.D. Turnpenny. 2000. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet. 24:438–441. [DOI] [PubMed] [Google Scholar]
- Bush, G., G. diSibio, A. Miyamoto, J.B. Denault, R. Leduc, and G. Weinmaster. 2001. Ligand-induced signaling in the absence of furin processing of N1. Dev. Biol. 229:494–502. [DOI] [PubMed] [Google Scholar]
- Campos, L.S., A.J. Duarte, T. Branco, and D. Henrique. 2001. mDll1 and mDll3 expression in the developing mouse brain: role in the establishment of the early cortex. J. Neurosci. Res. 64:590–598. [DOI] [PubMed] [Google Scholar]
- Chitnis, A., D. Henrique, J. Lewis, D. Ish-Horowicz, and C. Kintner. 1995. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 375:761–766. [DOI] [PubMed] [Google Scholar]
- Daluiski, A., T. Engstrand, M.E. Bahamonde, L.W. Gamer, E. Agius, S.L. Stevenson, K. Cox, V. Rosen, and K.M. Lyons. 2001. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat. Genet. 27:84–88. [DOI] [PubMed] [Google Scholar]
- de Celis, J.F., and S.J. Bray. 2000. The Abruptex domain of N regulates negative interactions between N, its ligands and Fringe. Development. 127:1291–1302 [DOI] [PubMed] [Google Scholar]
- Dunwoodie, S.L., D. Henrique, S.M. Harrison, and R.S. Beddington. 1997. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 124:3065–3076. [DOI] [PubMed] [Google Scholar]
- Dunwoodie, S.L., M. Clements, D.B. Sparrow, X. Sa, R.A. Conlon, and R.S. Beddington. 2002. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development. 129:1795–1806. [DOI] [PubMed] [Google Scholar]
- Ge, W., K. Martinowich, X. Wu, F. He, A. Miyamoto, G. Fan, G. Weinmaster, and Y.E. Sun. 2002. N signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci. Res. 69:848–860. [DOI] [PubMed] [Google Scholar]
- Giudicelli, F., and J. Lewis. 2004. The vertebrate segmentation clock. Curr. Opin. Genet. Dev. 14:407–414. [DOI] [PubMed] [Google Scholar]
- Heitzler, P., and P. Simpson. 1993. Altered epidermal growth factor-like sequences provide evidence for a role of N as a receptor in cell fate decisions. Development. 117:1113–1123. [DOI] [PubMed] [Google Scholar]
- Henderson, S.T., D. Gao, S. Christensen, and J. Kimble. 1997. Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol. Biol. Cell. 8:1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique, D., E. Hirsinger, J. Adam, I. Le Roux, O. Pourquie, D. Ish-Horowicz, and J. Lewis. 1997. Maintenance of neuroepithelial progenitor cells by Delta-N signalling in the embryonic chick retina. Curr. Biol. 7:661–670. [DOI] [PubMed] [Google Scholar]
- Hermanson, O., K. Jepsen, and M.G. Rosenfeld. 2002. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 419:934–939. [DOI] [PubMed] [Google Scholar]
- Hicks, C., S.H. Johnston, G. diSibio, A. Collazo, T.F. Vogt, and G. Weinmaster. 2000. Fringe differentially modulates Jagged1 and Delta1 signalling through N1 and N2. Nat. Cell Biol. 2:515–520. [DOI] [PubMed] [Google Scholar]
- Hicks, C., E. Ladi, C. Lindsell, J. Hsieh, S. Hayward, A. Collazo, and G. Weinmaster. 2002. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. J. Neurosci. Res. 68:655–667. [DOI] [PubMed] [Google Scholar]
- Hsieh, J.J., T. Henkel, P. Salmon, E. Robey, M.G. Peterson, and S.D. Hayward. 1996. Truncated mammalian N1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hukriede, N.A., Y. Gu, and R.J. Fleming. 1997. A dominant-negative form of Serrate acts as a general antagonist of N activation. Development. 124:3427–3437. [DOI] [PubMed] [Google Scholar]
- Itoh, M., C.H. Kim, G. Palardy, T. Oda, Y.J. Jiang, D. Maust, S.Y. Yeo, K. Lorick, G.J. Wright, L. Ariza-McNaughton, et al. 2003. Mind bomb is a ubiquitin ligase that is essential for efficient activation of N signaling by Delta. Dev. Cell. 4:67–82. [DOI] [PubMed] [Google Scholar]
- Jacobsen, T.L., K. Brennan, A.M. Arias, and M.A. Muskavitch. 1998. Cis-interactions between Delta and N modulate neurogenic signalling in Drosophila. Development. 125:4531–4540. [DOI] [PubMed] [Google Scholar]
- Kiyota, T., and T. Kinoshita. 2004. The intracellular domain of X-Serrate-1 is cleaved and suppresses primary neurogenesis in Xenopus laevis. Mech. Dev. 121:573–585. [DOI] [PubMed] [Google Scholar]
- Krebs, L.T., M.L. Deftos, M.J. Bevan, and T. Gridley. 2001. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the N signaling pathway. Dev. Biol. 238:110–119. [DOI] [PubMed] [Google Scholar]
- Kusumi, K., E.S. Sun, A.W. Kerrebrock, R.T. Bronson, D.C. Chi, M.S. Bulotsky, J.B. Spencer, B.W. Birren, W.N. Frankel, and E.S. Lander. 1998. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat. Genet. 19:274–278. [DOI] [PubMed] [Google Scholar]
- Kusumi, K., M.S. Mimoto, K.L. Covello, R.S. Beddington, R. Krumlauf, and S.L. Dunwoodie. 2004. Dll3 pudgy mutation differentially disrupts dynamic expression of somite genes. Genesis. 39:115–121. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., A. Bardin, and F. Schweisguth. 2005. The roles of receptor and ligand endocytosis in regulating N signaling. Development. 132:1751–1762. [DOI] [PubMed] [Google Scholar]
- Lewis, J. 1996. Neurogenic genes and vertebrate neurogenesis. Curr. Opin. Neurobiol. 6:3–10. [DOI] [PubMed] [Google Scholar]
- Lindsell, C.E., C.J. Shawber, J. Boulter, and G. Weinmaster. 1995. Jagged: a mammalian ligand that activates N1. Cell. 80:909–917. [DOI] [PubMed] [Google Scholar]
- Logeat, F., C. Bessia, C. Brou, O. LeBail, S. Jarriault, N. Seiday, and A. Israel. 1998. The N1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. 95:8108–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., S.E. Perez, Z. Qiao, J.M. Verdi, C. Hicks, G. Weinmaster, and D.J. Anderson. 2000. Transient N activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 101:499–510. [DOI] [PubMed] [Google Scholar]
- Mumm, J.S., and R. Kopan. 2000. N signaling: from the outside in. Dev. Biol. 228:151–165. [DOI] [PubMed] [Google Scholar]
- Nofziger, D., A. Miyamoto, K.M. Lyons, and G. Weinmaster. 1999. N signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 126:1689–1702. [DOI] [PubMed] [Google Scholar]
- Panin, V.M., L. Shao, L. Lei, D.J. Moloney, K.D. Irvine, and R.S. Haltiwanger. 2002. Notch ligands are substrates for protein O-fucosyltransferase-1 and Fringe. J. Biol. Chem. 277:29945–29952. [DOI] [PubMed] [Google Scholar]
- Pfister, S., G.K. Przemeck, J.K. Gerber, J. Beckers, J. Adamski, and M. Hrabe de Angelis. 2003. Interaction of the MAGUK family member Acvrinp1 and the cytoplasmic domain of the N ligand Delta1. J. Mol. Biol. 333:229–235. [DOI] [PubMed] [Google Scholar]
- Sakamoto, K., O. Ohara, M. Takagi, S. Takeda, and K. Katsube. 2002. Intracellular cell-autonomous association of N and its ligands: a novel mechanism of N signal modification. Dev. Biol. 241:313–326. [DOI] [PubMed] [Google Scholar]
- Sanchez-Irizarry, C., A.C. Carpenter, A.P. Weng, W.S. Pear, J.C. Aster, and S.C. Blacklow. 2004. N subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol. Cell. Biol. 24:9265–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serth, K., K. Schuster-Gossler, R. Cordes, and A. Gossler. 2003. Transcriptional oscillation of Lunatic fringe is essential for somitogenesis. Genes Dev. 17:912–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K., S. Chiba, K. Kumano, N. Hosoya, T. Takahashi, Y. Kanda, Y. Hamada, Y. Yazaki, and H. Hirai. 1999. Mouse Jagged1 physically interacts with N2 and other N receptors. Assessment by quantitative methods. J. Biol. Chem. 274:32961–32969. [DOI] [PubMed] [Google Scholar]
- Shutter, J.R., S. Scully, W. Fan, W.G. Richards, J. Kitajewski, G.A. Deblandre, C.R. Kintner, and K.L. Stark. 2000. Dll4, a novel N ligand expressed in arterial endothelium. Genes Dev. 14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- Six, E., D. Ndiaye, Y. Laabi, C. Brou, N. Gupta-Rossi, A. Israel, and F. Logeat. 2003. The N ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc. Natl. Acad. Sci. USA. 100:7638–7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrow, D.B., M. Clements, S.L. Withington, A.N. Scott, J. Novotny, D. Sillence, K. Kusumi, R.S. Beddington, and S.L. Dunwoodie. 2002. Diverse requirements for N signalling in mammals. Int. J. Dev. Biol. 46:365–374. [PubMed] [Google Scholar]
- Sun, Y., M. Nadal-Vicens, S. Misono, M.Z. Lin, A. Zubiaga, X. Hua, G. Fan, and M.E. Greenberg. 2001. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 104:365–376. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., T. Inoue, A. Gossler, and Y. Saga. 2003. Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development. 130:4259–4268. [DOI] [PubMed] [Google Scholar]
- Turner, D.L., and H. Weintraub. 1994. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8:1434–1447. [DOI] [PubMed] [Google Scholar]
- Turnpenny, P.D., N. Whittock, J. Duncan, S. Dunwoodie, K. Kusumi, and S. Ellard. 2003. Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the N signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J. Med. Genet. 40:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinos, J., and M. Freeman. 2000. Evidence that Argos is an antagonistic ligand of the EGF receptor. Oncogene. 19:3560–3562. [DOI] [PubMed] [Google Scholar]
- Weinmaster, G. 2000. N signal transduction: a real rip and more. Curr. Opin. Genet. Dev. 10:363–369. [DOI] [PubMed] [Google Scholar]
- Weinmaster, G., and C. Kintner. 2003. Modulation of notch signaling during somitogenesis. Annu. Rev. Cell Dev. Biol. 19:367–395. [DOI] [PubMed] [Google Scholar]
- Weng, A.P., A.A. Ferrando, W. Lee, J.P. Morris IV, L.B. Silverman, C. Sanchez-Irizarry, S.C. Blacklow, A.T. Look, and J.C. Aster. 2004. Activating mutations of N1 in human T cell acute lymphoblastic leukemia. Science. 306:269-271. [DOI] [PubMed] [Google Scholar]
- Wettstein, D.A., D.L. Turner, and C. Kintner. 1997. The Xenopus homolog of Drosophila Suppressor of Hairless mediates N signaling during primary neurogenesis. Development. 124:693–702. [DOI] [PubMed] [Google Scholar]
- Wright, G.J., J.D. Leslie, L. Ariza-McNaughton, and J. Lewis. 2004. Delta proteins and MAGI proteins: an interaction of N ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development. 131:5659–5669. [DOI] [PubMed] [Google Scholar]
- Yang, L.T., J.T. Nichols, C. Yao, J.O. Manilay, E.A. Robey, and G. Weinmaster. 2004. Fringe glycosyltransferases differentially modulate N1 proteolysis induced by Delta1 and Jagged1. Mol. Biol. Cell. 16:927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., C.R. Norton, and T. Gridley. 2002. Segmentation defects of N pathway mutants and absence of a synergistic phenotype in lunatic fringe/radical fringe double mutant mice. Genesis. 33:21–28. [DOI] [PubMed] [Google Scholar]