Abstract
4E-transporter (4E-T) is one of several proteins that bind the mRNA 5′cap-binding protein, eukaryotic initiation factor 4E (eIF4E), through a conserved binding motif. We previously showed that 4E-T is a nucleocytoplasmic shuttling protein, which mediates the import of eIF4E into the nucleus. At steady state, 4E-T is predominantly cytoplasmic and is concentrated in bodies that conspicuously resemble the recently described processing bodies (P-bodies), which are believed to be sites of mRNA decay. In this paper, we demonstrate that 4E-T colocalizes with mRNA decapping factors in bona fide P-bodies. Moreover, 4E-T controls mRNA half-life, because its depletion from cells using short interfering RNA increases mRNA stability. The 4E-T binding partner, eIF4E, also is localized in P-bodies. 4E-T interaction with eIF4E represses translation, which is believed to be a prerequisite for targeting of mRNAs to P-bodies. Collectively, these data suggest that 4E-T interaction with eIF4E is a priming event in inducing messenger ribonucleoprotein rearrangement and transition from translation to decay.
Introduction
The regulation of mRNA biogenesis and decay plays an important role in the control of gene expression. mRNA decay regulates protein levels, eliminates aberrant mRNAs, and is an essential host defense mechanism during viral infection (Coller and Parker, 2004). There are three major mRNA decay pathways (Coller and Parker, 2004; Parker and Song, 2004). The general pathway involves deadenylation followed by decapping of the mRNA. In yeast, transcripts are primarily degraded in a 5′ to 3′ direction by the Xrn1 exonuclease (Muhlrad et al., 1995; Schwartz and Parker, 1999; Coller and Parker, 2004; Parker and Song, 2004), whereas in higher eukaryotes, mRNAs mostly are eliminated in a 3′ to 5′ fashion by the exosome after deadenylation (Chen et al., 2001; Hilleren et al., 2001; Wang and Kiledjian, 2001; Wilusz et al., 2001; Mukherjee et al., 2002; Tourriere et al., 2002). mRNA degradation also can occur through a second pathway that involves endonucleolytic cleavage of mRNAs (Schoenberg and Chernokalskaya, 1997), and in addition, through specialized mechanisms, including nonsense-mediated decay and nonstop decay (for reviews see Wilusz et al., 2001; Tourriere et al., 2002; Coller and Parker, 2004; Parker and Song, 2004).
mRNA decapping is a crucial step in general and specialized mRNA decay (Parker and Song, 2004). Decapping is mediated by a heterodimeric complex composed of Dcp1 and Dcp2 (Beelman et al., 1996; Dunckley and Parker, 1999; Lykke-Andersen, 2002; van Dijk et al., 2002; Wang et al., 2002). Numerous factors that regulate decapping activity have been identified in yeast and mammals. The Lsm 1–7 complex, Pat1p, Dhh1p, Edc1p, Edc2, and Edc3 are positive regulators of decapping, characterized in yeast (Coller and Parker, 2004; Parker and Song, 2004).
Most mammalian mRNAs studied are the target of degradation by the 3′-5′exosome (Chen et al., 2001; Mukherjee et al., 2002; Wang and Kiledjian, 2001). Nonetheless, decapping activity similar in function to the yeast Dcp1p has been detected in HeLa cells (Gao et al., 2001); mammalian homologues of Dcp1/2 (Lykke-Andersen, 2002; Wang et al., 2002), the Lsm proteins (Achsel et al., 1999), and Dhh1p (Smillie and Sommerville, 2002) also exist. Furthermore, Xrn1 homologues have been identified in mammals (Bashkirov et al., 1997; Ingelfinger et al., 2002). However, it has not been demonstrated unequivocally whether deadenylated-decapped mRNAs are subject to 5′ or 3′ exonucleolytic decay in mammalian cells in vivo (discussed in Wang and Kiledjian, 2001; Mukherjee et al., 2002; Wilusz and Wilusz, 2004).
The mRNA 5′ cap structure and the 3′ polyA tail play important roles in the decapping process (Wilusz et al., 2001; Coller and Parker, 2004). The polyA inhibits decapping, likely through the polyA binding protein (PABP) interaction with eIF4G, a component of the eIF4F cap-binding complex (Coller and Parker, 2004). eIF4F also contains the cap binding protein, eukaryotic initiation factor 4E (eIF4E), and the RNA helicase, eIF4A (Gingras et al., 1999). The interaction of PABP with eIF4G enhances the binding of eIF4F to the mRNA (Kahvejian et al., 2005), and thus, hinders the access of the decapping complex to the cap. Consequently, translation and decapping are antagonistic. Consistent with this idea, mutations in eIF4E and eIF4G can lead to an increase in decapping in yeast (Schwartz and Parker, 1999). Importantly, decapping requires the removal of eIF4F from the 5′ cap structure, which, in turn, necessitates a transition from a translationally active messenger RNP (mRNP) to one destined for decay. This transition entails the replacement of initiation factors on the mRNA with decapping factors (Tharun and Parker, 2001; Coller and Parker, 2004; Parker and Song, 2004; Teixeira et al., 2005). The switch from translation to decay occurs concomitantly with aggregation of mRNPs into distinct cytoplasmic foci, termed processing bodies (P-bodies), in yeast (Sheth and Parker, 2003), or GW bodies (Eystathioy et al., 2003) or Dcp1a bodies in mammals (Cougot et al., 2004); these are referred to as P-bodies throughout this article. The decapping enzyme, Dcp1/2, and decapping associated factors, Lsm1-7, Dhh1p/p54, and Xrn1, are concentrated in these foci (Ingelfinger et al., 2002; van Dijk et al., 2002; Sheth and Parker, 2003; Cougot et al., 2004; Liu et al., 2004). However, little is known about the recruitment mechanism of mRNAs to P-bodies, or how initiation factors are exchanged with decapping enzymes.
We reported earlier the cloning of 4E-transporter (4E-T), which interacts with eIF4E through a conserved eIF4E recognition motif (YXXXXLΦ, where Φ is a hydrophobic amino acid) that is also found in eIF4G and 4E-BP. 4E-T is a nucleocytoplasmic shuttling protein that mediates the nuclear import of eIF4E through the importin α/β pathway. At steady state, 4E-T is predominantly cytoplasmic and is concentrated in bodies that conspicuously resemble the recently described P-bodies (Dostie et al., 2000a; Sheth and Parker, 2003). Here, we report that 4E-T colocalizes with eIF4E and decapping factors in bona fide P-bodies and controls mRNA turnover. Significantly, depletion of 4E-T from cells enhances mRNA stability.
Results
4E-T colocalizes with decapping factors
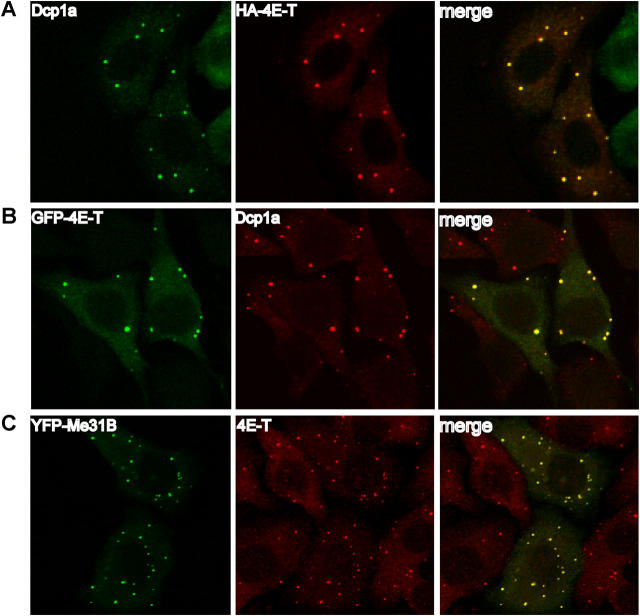
To determine whether 4E-T associates with P-bodies, the localization of 4E-T and P-body components was analyzed by immunofluorescence in HeLa cells. Cells transfected with HA–4E-T were analyzed by double-label immunofluorescence microscopy using anti-HA antibody and an affinity-purified anti-Dcp1a antibody that recognizes the endogenous protein (see Materials and methods). Transfected HA–4E-T and endogenous Dcp1a colocalized in the cytoplasm in discrete foci that strongly resemble P-bodies (Fig. 1 A). Similar results were obtained with a GFP-tagged 4E-T (Fig. 1 B). To show that endogenous 4E-T also localizes to P-bodies, its colocalization with Me31B was studied (Me31B is the Drosophila homologue of the human decapping factor Dhh1p/p54, and similar to Dcp1a, is an established marker of P-bodies [Sheth and Parker, 2003; Tseng-Rogenski et al., 2003]; Me31B shares 71% identity with Dhh1p/p54 and was shown to form cytoplasmic granules in germline cells [Nakamura et al., 2001]). HeLa cells transfected with myc-EYFP-Me31B were stained with an affinity-purified anti–4E-T antibody, and analyzed by microscopy. Endogenous 4E-T colocalizes with myc-EYFP-Me31B in discrete cytoplasmic foci (Fig. 1 C). Thus, endogenous and transfected 4E-T colocalize with bona fide decapping factors in cytoplasmic foci, which fulfill the criteria of P-bodies.
Figure 1.
4E-T colocalizes with decapping factors in P-bodies. (A and B) Transfected 4E-T colocalizes with endogenous Dcp1a. HeLa cells transfected with HA–4E-T (A) or GFP–4E-T (B) were stained with anti-Dcp1a to visualize endogenous Dcp1a. The localization of HA–4E-T was determined by indirect immunofluorescence with anti-HA (Covance) and Alexa Fluor 594 anti–mouse IgG. Dcp1a was revealed with anti-Dcp1a and either Alexa Fluor 488 anti–rabbit IgG (A) or Alexa Fluor 594 anti–mouse IgG (B). The colocalization of HA–4E-T or GFP–4E-T with Dcp1a appears yellow (right). (C) Endogenous 4E-T colocalizes with transfected myc-EYFP-Me31B. HeLa cells were transfected with myc-EYFP-Me31B and colocalization with endogenous 4E-T was performed with anti–4E-T and Texas Red–conjugated anti–rabbit IgG. Colocalization appears yellow (right).
4E-T down-regulates cap-dependent translation
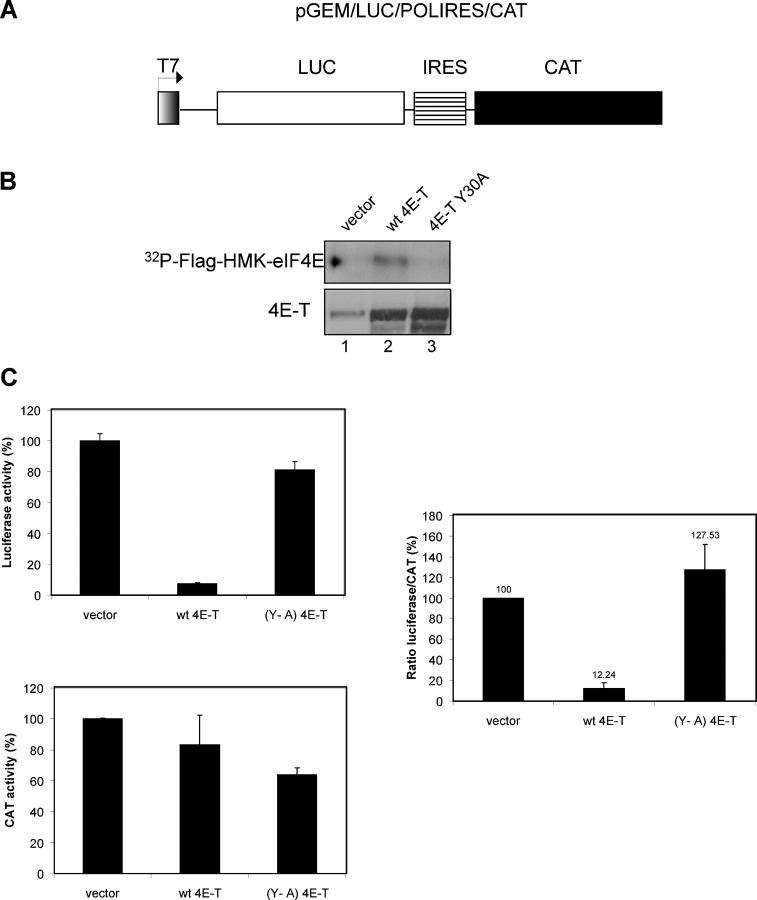
In the yeast Saccharomyces cerevisiae, targeting of mRNAs to P-bodies is believed to occur as a consequence of cessation of translation (Schwartz and Parker, 1999; Tharun and Parker, 2001; Teixeira et al., 2005). Thus, it is conceivable that changes in mRNP composition that lead to translational arrest result in targeting the mRNA to P-bodies in mammals. 4E-T interacts with eIF4E through a conserved binding site, which also is found in eIF4G (Mader et al., 1995), and thus, might promote targeting of mRNAs to P-bodies by inhibiting translation initiation. To address this possibility, the inhibitory effect of 4E-T on translation was examined in vivo. HeLa cells infected with recombinant vaccinia virus vTF7-3 (to synthesize the T7 RNA polymerase in the cytoplasm) were cotransfected transiently with plasmid DNA containing a 4E-T cDNA and a bicistronic reporter construct expressed from a T7 promoter. The bicistronic reporter construct consists of the luciferase (LUC) and chloramphenicol acetyl transferase (CAT) cistrons, separated by the poliovirus internal ribosome entry site (IRES; Pause et al., 1994) (Fig. 2 A). Translation of the LUC cistron proceeds in a cap-dependent manner, whereas translation of the CAT cistron is cap-independent and serves as a surrogate control for mRNA levels. (Note that quantification of the RNA by Northern blotting could not be performed because the vaccinia virus T7 system produces heterogenous RNA transcripts, generating smeared bands [Fuerst and Moss, 1989]. This is attributed to the lack of an authentic T7 RNA polymerase termination sequence. However, this has no deleterious effect on mRNA turnover or translation [Fuerst and Moss, 1989].) Only a fraction (5–10%) of T7-transcribed RNAs are capped by the vaccinia-capping enzyme (Fuerst and Moss, 1989). However, uncapped RNA is translated inefficiently in vivo and in vitro (Both et al., 1975; Shatkin, 1976; Sonenberg et al., 1980; Fuerst and Moss, 1989; Peng and Schoenberg, 2005; and unpublished data). Thus, even if only a small fraction of the T7-transcribed RNAs is capped in the T7 vaccinia virus system, the uncapped products largely are untranslated (∼0.4–1% of uncapped compared with capped RNA; unpublished data). All constructs were expressed to similar levels (Fig. 2 B, bottom panel, compare lanes 2–4). The binding of 4E-T to eIF4E was demonstrated by Far Western blotting with [32P]HMK-eIF4E (Fig. 2 B, lane 2, top panel). The 4E-T mutant, Y30A, which is mutated in the YXXXXLΦ motif, did not bind to eIF4E, as expected (Fig. 2 B, lane 3, top panel). LUC expression was decreased significantly by overexpression of wild-type 4E-T (Fig. 2 C, left, top panel), whereas CAT activity remained relatively unchanged (Fig. 2 C, left, bottom panel). In contrast, the Y30A mutant of 4E-T did not affect LUC or CAT expression. Therefore, overexpression of wild-type 4E-T, but not the mutant Y30A, strongly inhibited (88%) translation (Fig. 2 C). In conclusion, overexpression of 4E-T in HeLa cells results in the repression of translation only when bound to eIF4E.
Figure 2.
4E-T inhibits cap-dependent translation in vivo. (A) Schematic presentation of the bicistronic reporter plasmid (pGEM-LUC-POLIRES-CAT). HeLa cells were infected with the vaccinia virus vTF7-3, and transfected transiently with the reporter plasmid and pcDNA3-4E-T constructs. (B, bottom panel) Western blot analysis of the transfected cells was performed with anti-4E-T antibody. Lane 1, pcDNA3; lane 2, pcDNA3-4E-T; lane 3, pcDNA3–4E-T–Y30A. The interaction of the wt and mutant 4E-T protein with eIF4E was examined by Far Western analysis (top panel) using a [32P]-labeled eIF4E probe. (C) LUC and CAT activity were measured 16 h after transfection. The LUC/CAT activity ratio is expressed as a percentage of the control (pcDNA3) set at 100%. The experiments were performed four times in duplicate.
eIF4E localizes to P-bodies
Because 4E-T localizes to P-bodies, and because it interacts with eIF4E, it was pertinent to ask whether eIF4E also localizes to P-bodies, and whether this localization requires 4E-T. To this end, we performed immunofluorescence analysis of endogenous eIF4E in HeLa cells using a monoclonal antibody, which recognizes eIF4E in the cytoplasm (Kimball et al., 2003). Although eIF4E is distributed throughout the cytoplasm, it is concentrated in foci reminiscent of P-bodies (Fig. 3 A). This staining pattern is observed in two different HeLa cell sublines: HeLa CCL2 and HeLa S3 (Carter et al., 1995), but with qualitative differences (Fig. 3, A and B–E, respectively), and in mouse embryo fibroblasts (not depicted). Furthermore, transiently transfected HA-eIF4E also displays a similar cytoplasmic distribution pattern (see below, Fig. 4 A).
Figure 3.
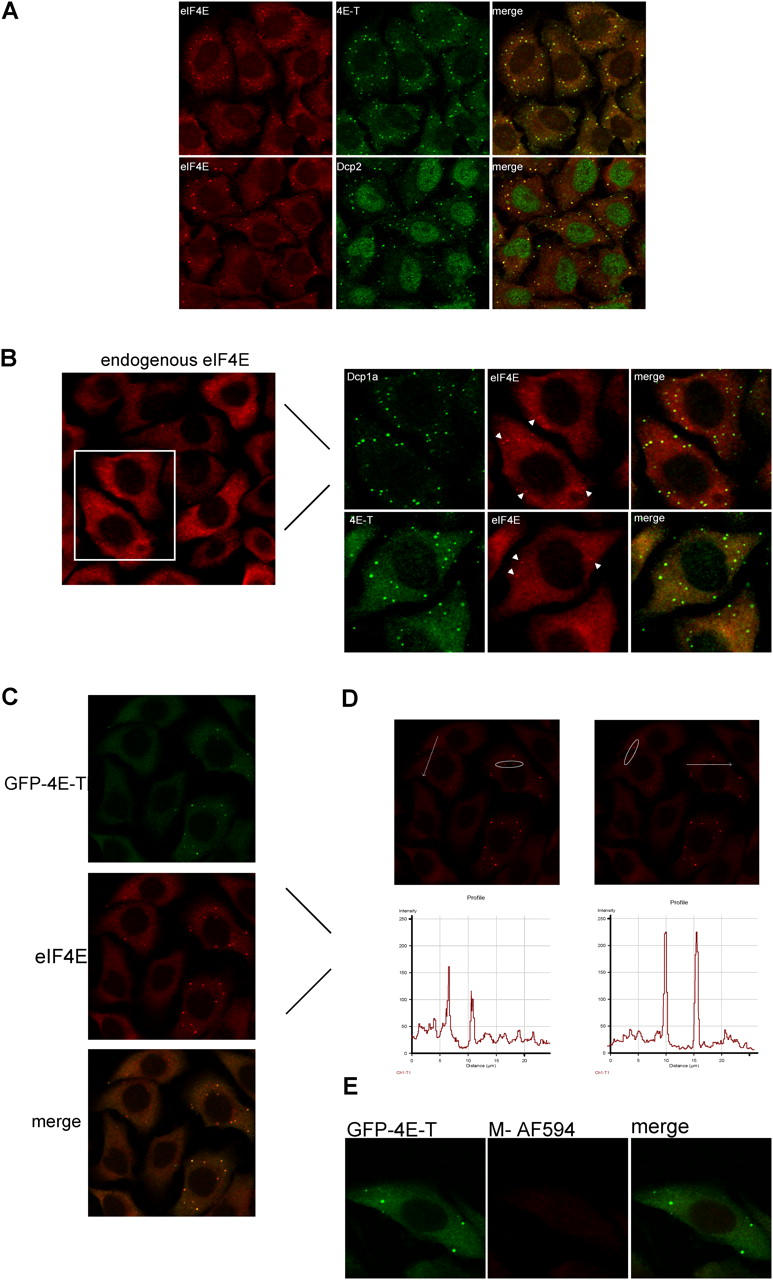
eIF4E colocalizes with 4E-T, Dcp1a, and Dcp2 in P-bodies. (A) HeLa cells (CCL2) were fixed and the localization of eIF4E was determined by indirect immunofluorescence with mouse anti-eIF4E monoclonal antibody and Alexa Fluor 594 anti–mouse IgG. The localization of 4E-T was examined with anti–4E-T and that of Dcp2 with anti-Dcp2 and Alexa Fluor 488 anti–rabbit IgG. The colocalization of these factors appears yellow in the merged image. (B) The colocalization of endogenous eIF4E with 4E-T and Dcp1a was examined with anti–4E-T and anti-Dcp1a in HeLa S3 as above. The left panel demonstrates endogenous eIF4E localization in HeLa S3. Zoomed images (right panel) display colocalization of endogenous eIF4E and Dcp1a in P-bodies. Some of the more distinct P-bodies are indicated by arrowheads. (C) HeLa cells were transfected with GFP-4E-T and the staining of endogenous eIF4E was performed with anti-eIF4E antibody as described above. (D) The intensity of fluorescence of endogenous eIF4E in P-bodies from GFP–4E-T–transfected cells (from panel C) was compared against the intensity of fluorescence of eIF4E from nontransfected cells along the path, which is indicated by the arrow. The P-bodies along the path of the arrow are circled in the adjacent panel for comparison for better visibility. The signal was allowed to bleach before quantification to avoid a saturated signal. The graphs plot the intensity of fluorescence against the distance (μm) traversed by the arrow. (E) HeLa cells transfected with GFP-4E-T were incubated with Alexa Fluor 594 anti–mouse IgG (M-AF594) without previous incubation with anti-eIF4E antibody to demonstrate the integrity of the green filter.
Figure 4.
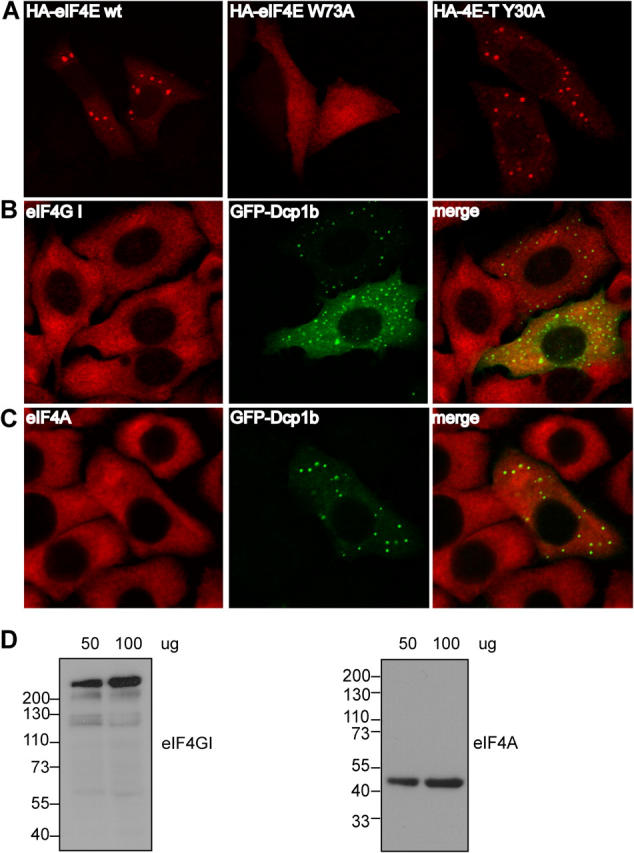
eIF4E requires interaction with 4E-T for localization to P-bodies, whereas the other components of the eIF4F complex are excluded from P-bodies. (A) HeLa cells were transfected with HA-eIF4E wt, HA-eIF4E W73A, or HA-4E-T Y30A. Localization of proteins was studied by indirect immunofluorescence with anti-HA (Covance) and Alexa Fluor 594 anti–mouse IgG. (B and C) HeLa cells were transfected with GFP-Dcp1b and staining of endogenous eIF4GI (B) or eIF4A (C) was detected with anti-eIF4GI and Texas Red conjugated anti–rabbit IgG or with anti-eIF4A and Alexa Fluor 594 anti–mouse IgG. (D) HeLa extract (50 and 100 μg) was resolved by SDS-8% PAGE and a Western blot was performed with rabbit anti–eIF4GI polyclonal antibody. For eIF4A detection, proteins were resolved by SDS-10% PAGE and Western blot was performed with a mouse anti-eIF4A monoclonal antibody.
Double-label immunofluorescence analysis shows that eIF4E colocalized with 4E-T and Dcp2 in P-bodies (Fig. 3 A). eIF4E exhibited a more pronounced association with P-bodies in CCL2 (Fig. 3 A) as compared with HeLa S3 (Fig. 3 B). eIF4E colocalized with Dcp1a and 4E-T in P-bodies, albeit staining of eIF4E in these structures is not as apparent as the staining of 4E-T and Dcp1a in HeLa S3 (Fig. 3 B). Strikingly, overexpression of GFP–4E-T resulted in enhanced staining of eIF4E in P-bodies (Fig. 3 C). Because localization of eIF4E in P-bodies was more prominent, and the foci appeared larger and brighter in GFP–4E-T overexpressing cells than in parental cells, these data suggest that 4E-T might tether eIF4E to P-body structures. This effect is illustrated in a graph plotting the intensity of the eIF4E signal (red channel) in two P-bodies marked by an arrow in cells overexpressing 4E-T levels (Fig. 3 D, right panel) and in cells expressing endogenous 4E-T levels (Fig. 3 D, left panel; the respective P-bodies traversed by the arrow are circled in the adjacent panel for comparison). The signal intensity of eIF4E was approximately twofold higher in cells overexpressing 4E-T as compared with endogenous 4E-T levels, when P-bodies of weaker intensity were selected from cells overexpressing 4E-T to avoid signal saturation of the plot. Similar results were obtained with a monoclonal antibody against eIF4E from Transduction Laboratories (610270), and with a monoclonal antibody (10C6), which recognizes eIF4E predominantly in the nucleus (Lejbkowicz et al., 1992; Dostie et al., 2000b; unpublished data). The enhanced eIF4E signal is not due to leakage of the FITC signal, because no significant signal was detected in the red channel when cells transfected with GFP–4E-T were stained with the secondary antibody alone (Fig. 3 E).
To determine whether the association of eIF4E with P-bodies requires its interaction with 4E-T, we performed immunofluorescence analysis using wild-type HA-eIF4E and a mutant lacking the 4E-T binding site (eIF4E W73A) (Dostie et al., 2000a). Low eIF4E cDNA amounts were transfected transiently into HeLa cells to obtain a predominantly cytoplasmic (Dostie et al., 2000a) and P-body localization (Fig. 4 A). Cytoplasmic eIF4E levels in HA-eIF4E–expressing cells were similar to endogenous levels. However, a more pronounced cytoplasmic foci staining was observed in ∼40% of the transfected cells, presumably because more eIF4E is expressed in these cells. In contrast, HA-eIF4E W73A did not localize to P-bodies, and was distributed uniformly between the cytoplasm and the nucleus (Fig. 4 A). Therefore, loss of 4E-T interaction (and also interaction with eIF4G and 4E-BPs) (Mader et al., 1995) leads to deregulated cellular localization, which results in diffuse staining throughout the cell. In addition, because the HA–4E-T (Y30A) mutant, which is defective in eIF4E binding (Dostie et al., 2000a), still localizes to P-bodies, these results suggest that 4E-T is required for the association of eIF4E with P-bodies, but not vice versa (see also Fig. 5 C).
Figure 5.
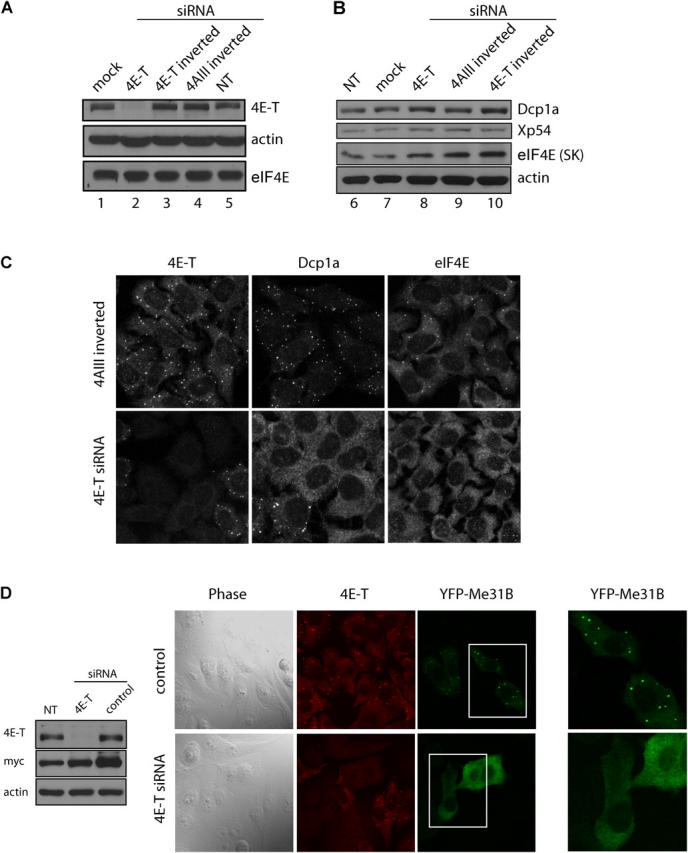
Depletion of 4E-T from HeLa cells results in disappearance of decapping factors from P-bodies. (A) HeLa cells were transfected with siRNA against 4E-T or control siRNAs (4AIII inverted or 4E-T inverted). 48 h after transfection, cells were split into chamber slides for immunofluorescence analysis [see (C)] and into a 6-cm dish for Western blot analysis. 60 h after transfection, protein cell extracts were prepared and 20 μg of extract was resolved by SDS-10% PAGE. Western blotting was performed with rabbit anti–4E-T and anti-eIF4E antibodies, or mouse anti–β-actin monoclonal antibody (Sigma-Aldrich). 4E-T levels were quantified against β-actin, which served as a loading control, and the level of protein in the negative control (4AIII inverted) transfected cells was set as 100. (B) HeLa extract (20 μg) as in (A) was resolved by 10% SDS-PAGE and Western blotting was performed with rabbit anti-Dcp1a and anti-Xenopus p54 (Xp54) antibodies, or monoclonal mouse anti-eIF4E (SK) and anti–β-actin antibodies. (C) HeLa (CCL2) cells were transfected with control siRNA (4AIII inverted) or 4E-T siRNA and indirect immunofluorescence was performed 60 h after transfection with rabbit anti–4E-T and anti-Dcp1a antibodies, or monoclonal mouse anti-eIF4E antibody. (D) HeLa S3 cells were transfected with 4E-T siRNA or control siRNA (4AIII inverted). 24 h after transfection, cells were transfected with myc-EYFP-Me31B; 36 h later, extracts were collected for SDS-PAGE analysis or fixed for immunofluorescence. Cell extracts were resolved by 10% SDS-PAGE and Western blot analysis with rabbit anti–4E-T, or monoclonal anti-myc (9E10) and anti–β-actin was performed. Indirect immunofluorescence was performed to assess the localization of 4E-T, and the localization of myc-EYFP-Me31B was assessed by direct immunofluorescence. The right most panels show the enlarged bordered image.
Because eIF4E is part of the heterotrimeric eIF4F complex, it was important to determine whether eIF4A and eIF4G also localize to P-bodies. Because 4E-T and eIF4G are expected to compete for binding to eIF4E, no other eIF4F complex components—with the exception of eIF4E—were expected to localize to P-bodies. To investigate this, the localization of endogenous eIF4GI and eIF4A was studied in HeLa cells transfected with GFP-Dcp1b. Western blot analysis with anti-eIF4A, which recognizes eIF4AI and eIF4AII (Edery et al., 1983), and anti-eIF4GI revealed protein bands corresponding to the expected size of the proteins (Fig. 4 D). Immunofluorescence analysis with these antibodies shows that eIF4GI and eIF4A do not localize to P-bodies because neither protein colocalizes with the P-body marker, GFP-Dcp1b (Fig. 4, B and C). Similar results were obtained using two other anti-eIF4GI antibodies (unpublished data). Thus, eIF4E, but neither eIF4GI nor eIF4A, localized to P-bodies in vivo. It is highly likely that eIF4GII behaves in a similar manner, because it exhibits very similar properties to eIF4GI (Gradi et al., 1998).
Short interfering RNA against 4E-T results in decreased localization of decapping factors to mammalian P-bodies
To investigate whether 4E-T plays a role in the formation of P-bodies, cellular 4E-T protein levels were reduced by RNA interference (RNAi). Western blotting (Fig. 5 A) and immunofluorescence analysis (Fig. 5 C) demonstrate that 4E-T was reduced strongly (85%) after treatment with a short interfering RNA (siRNA) against 4E-T, but not by the corresponding inverted sequence (4E-T inverted) (Fig. 5 A). Another nonspecific siRNA that was used as a control, 4AIII inverted (Ferraiuolo et al., 2004), failed to affect 4E-T protein levels (Fig. 5, A and C). 4E-T depletion caused Dcp1a and eIF4E to disperse throughout the cytoplasm and no longer concentrate in foci (Fig. 5 C; cells that were unaffected in their staining in P-bodies are those that had not taken up the siRNA against 4E-T, as confirmed by immunostaining with anti-4E-T antibody: not depicted). The disappearance of P-bodies was not observed in cells that were transfected with the nonspecific control siRNA. No changes in Dhh1p/p54, Dcp1a, and eIF4E protein levels occurred in 4E-T–depleted cells as compared with nonspecific siRNA controls (Fig. 5, A and B).
The effect of 4E-T depletion on Me31B localization also was examined (Fig. 5 D). The tagged version of Drosophila Me31B was used because the antibody against endogenous p54, used in Fig. 5 B for Western analysis, is not suitable for immunofluorescence. HeLa cells transfected with 4E-T or control siRNAs were transfected with the myc-EYFP-Me31B plasmid, and analyzed by Western blotting and double label immunofluorescence (Fig. 5 D). 4E-T levels were reduced efficiently in 4E-T siRNA-treated cells, and the myc-EYFP-Me31B protein was expressed in cells treated with control or 4E-T siRNAs (left panel). In control cells, myc-EYFP-Me31B was predominantly cytoplasmic and concentrated in P-bodies (right panel). However, when 4E-T levels were reduced, Me31B exhibited a diffuse staining pattern throughout the cytoplasm and reduced or no staining in P-bodies (right panel). Thus, reduction of 4E-T protein levels causes mislocalization of eIF4E and decapping factors away from P-bodies.
Cycloheximide inhibits localization of 4E-T to P-bodies, whereas leptomycin B treatment does not affect its localization
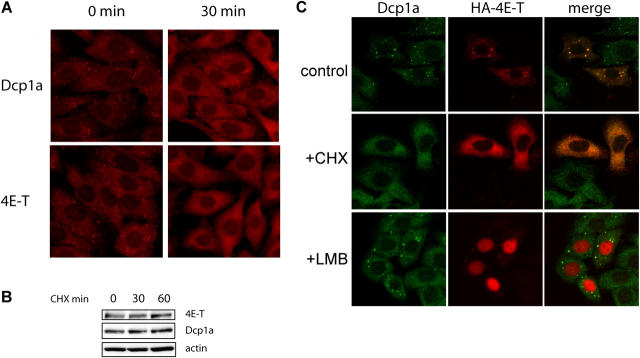
In yeast and mammalian cells, cycloheximide treatment releases decapping factors from P-bodies (Sheth and Parker, 2003; Cougot et al., 2004). Cycloheximide inhibits mRNA translation at the elongation step, which prevents mRNA degradation because of its protection by ribosomes (Ross, 1995). To study whether 4E-T localization to P-bodies is dependent on mRNA availability, the effect of cycloheximide on the localization of 4E-T was examined. HeLa cells were treated with cycloheximide (10 μg/ml), and analyzed by immunofluorescence with anti-Dcp1a and anti–4E-T antibodies. A near complete loss of 4E-T and Dcp1a from P-bodies was observed after 30 min of cycloheximide treatment (Fig. 6 A); this suggested that the accumulation of 4E-T in P-bodies is dependent on mRNA availability. This effect is not due to a reduction in 4E-T protein levels. Western blot analysis demonstrates that the levels of 4E-T and Dcp1a remain unchanged throughout the cycloheximide treatment (Fig. 6 B). Similar results were obtained in HeLa cells that were transfected transiently with HA-4E-T (Fig. 6 C, middle panel).
Figure 6.
Effect of cycloheximide and LMB on 4E-T localization in P-bodies. (A) HeLa cells were treated with cycloheximide (CHX; 10 μg/ml) for 30 min, and immunofluorescence staining of 4E-T and Dcp1a were examined as described above. (B) HeLa cell extract (20 μg) treated with cycloheximide (CHX; 10 μg/ml) for 30 and 60 min was resolved by 10% SDS-PAGE and Western blot analysis with rabbit anti–4E-T and anti-Dcp1a, or monoclonal mouse β-actin was performed. (C) HeLa cells were transfected with HA–4E-T. 36 h after transfection, the medium was replaced with fresh medium (control) or medium containing CHX (100 μg/ml), and cells were incubated for 3 h before fixation. Alternatively, media containing LMB (5 ng/ml) was added to cells and incubated for 5 h before fixation. The colocalization of HA–4E-T was determined by indirect immunofluorescence with anti-HA (Covance) and that of Dcp1a with anti-Dcp1a antibody as described above. The colocalization of HA–4E-T and Dcp1a appears yellow.
Leptomycin B (LMB) is an inhibitor of chromosome region maintenance-1–dependent protein nuclear export. We previously reported that 4E-T is a shuttling protein that exits the nucleus through the chromosome region maintenance-1 pathway (Dostie et al., 2000a). Because Dhh1p/p54 also shuttles between the nucleus and the cytoplasm (Smillie and Sommerville, 2002), it is possible that nuclear export is required for the localization of 4E-T and decapping factors to P-bodies. To test this hypothesis, the effect of LMB on the localization of 4E-T and Dcp1a to P-bodies was studied. LMB treatment resulted in the accumulation of 4E-T in the nucleus, but did not affect its association with P-bodies (Fig. 6 C, bottom panel). The distribution of Dcp1a also was unaffected by the drug (bottom panel). Thus, the presence of 4E-T in P-bodies is dependent on mRNA, but is independent of its nucleocytoplasmic shuttling.
4E-T knockdown increases mRNA half-life
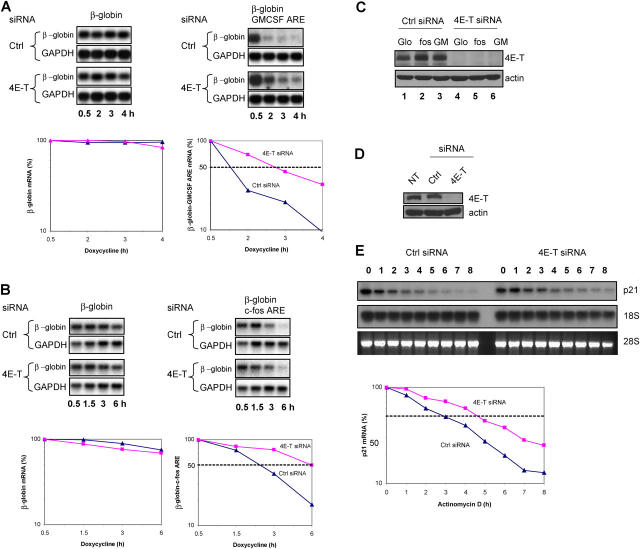
The results described above suggest that 4E-T might play a role in promoting mRNA degradation. To test this hypothesis, we analyzed the turnover of several mRNAs in 4E-T–depleted and control cells. A β-globin reporter that is under the control of a tetracycline-responsive promoter was fused at its 3′ end to one of two distinct adenine/uridine rich elements (AREs) (Stoecklin et al., 2004), GM-CSF or c-fos. The reporter gene—either lacking or containing the ARE—was cotransfected with a plasmid encoding the activator of transcription (pTet-Off) into HeLa cells that were transfected previously with 4E-T or control siRNA. Doxycycline was added to block transcription and RNA was collected at the indicated times and analyzed by Northern blotting. As expected, β-globin lacking an ARE remained stable throughout the doxycycline treatment, whereas β-globin containing the GM-CSF or c-fos AREs were degraded rapidly, as was reported previously (Chen et al., 1995) (Fig. 7, A and B). However, 4E-T diminution in HeLa cells resulted in significant stabilization (>2-fold) of β-globin/GM-CSF and β-globin/c-fos ARE-containing mRNA, with no effect on β-globin reporter mRNA lacking an ARE (Fig. 7, A and B). Western blot analysis demonstrates that 4E-T protein levels were diminished efficiently in HeLa cells that were transfected with the reporter plasmids and in the presence of doxycycline (Fig. 7 C). The effect of 4E-T depletion on endogenous c-fos mRNA turnover also was analyzed. Similar to transfected β-globin containing the destabilizing element of c-fos, endogenous c-fos was stabilized in the absence of 4E-T (unpublished data).
Figure 7.
siRNA-mediated depletion of 4E-T affects mRNA turnover. HeLa cells were transfected with siRNA against 4E-T or a control (Ctrl) siRNA (4AIII inverted). Protein and RNA were harvested for use in Western (C and D) and Northern (A, B, and E) analysis, respectively. (A and B) HeLa cells were transfected with siRNA against 4E-T or with control siRNA and later cotransfected with pTet-β-globin or pTet-β-globin c-fos or GM-CSF ARE and pTet Off. Cells were treated with doxycycline (1 μg/ml) ∼72 h after transfection to block transcription. Total RNA was isolated at 0.5, 2, 3, and 4 h for β-globin/GM-CSF ARE assays (A) and 0.5, 1.5, 3, and 6 h for β-globin/c-fos ARE assays (B) and analyzed by Northern blot. GAPDH served as a loading control. mRNA half-lives were calculated from Northern blots and normalized against GAPDH levels. (C) Protein extracts were collected at 6 h after treatment with doxycycline (1 μg/ml) from cells that were transfected with the reporter plasmid and pTetOff and resolved by SDS-PAGE. Lanes 1 and 4: glo = β-globin + pTetOff; lanes 2 and 5: fos = β-globin/c-fos ARE + pTetOff; lanes 3 and 6: GM = β-globin/GMCSF ARE + pTetOff. (D) Protein extracts were collected at the latest time point of actinomycin D treatment and resolved by 10% SDS-PAGE. Western blot analysis was done with anti–4E-T and anti–β-actin antibodies. NT = nontransfected. (E) At 48 h after transfection, the half-lives of p21 mRNA were assessed by using actinomycin D (5 μg/ml) for the indicated amount of time. Total RNA (5 μg) was resolved on 1.3% formaldehyde gel and analyzed by Northern blotting. 28S levels and 18S levels served as a loading marker. mRNA half-lives were calculated from Northern blots and normalized against 32P-labeled 18S levels and plotted on a graph with the zero time point set at 100.
The stability of the labile p21 mRNA, which contains an ARE (Lal et al., 2004), also was examined in 4E-T–depleted cells. Western blotting analysis demonstrates that 4E-T RNAi strongly reduced the amount of 4E-T (>80%, Fig. 7 D). p21 mRNA stability was determined by Northern blot analysis after actinomycin D treatment to inhibit de novo transcription. p21 mRNA levels were quantified in cells that were treated with 4E-T siRNAs and in nondepleted cells, and normalized to 18S rRNA levels (Fig. 7 E). 4E-T depletion resulted in a modest (∼1.5 fold), but significant, increase in the mRNA half-life (Fig. 7 E). The difference in p21 mRNA stability achieved with 4E-T depletion is related inversely to that obtained when the mRNA stabilizer, HuR, is depleted from cells (∼2-fold decrease; Lal et al., 2004). Thus, 4E-T promotes degradation of mRNAs that contain an ARE in their 3′ untranslated region.
Discussion
Here we report a new role for 4E-T in mammalian mRNA degradation. We demonstrate that 4E-T inhibits translation, colocalizes with decapping factors in cytoplasmic P-bodies, and decreases mRNA stability. 4E-T interacts with eIF4E through a shared recognition motif (YXXXXLΦ) also found in eIF4G (Dostie et al., 2000a). Similar to other cap-dependent translation inhibitors, 4E-T is likely to inhibit translation by competing with eIF4G for binding to eIF4E and preventing formation of the eIF4F complex (Pause et al., 1994; Poulin et al., 1998). Accordingly, overexpression of 4E-T wt, but not of a mutant defective in eIF4E binding, strongly inhibited cap-dependent translation of a reporter mRNA in vivo (Fig. 2).
The transition of an mRNA from a translationally active mRNP to one destined for decay is believed to be a consequence of translational inhibition, especially in yeast (Tharun and Parker, 2001; Teixeira et al., 2005). Remarkably, 4E-T is present in P-bodies, which are cytoplasmic foci containing factors involved in mRNA decay (Ingelfinger et al., 2002; van Dijk et al., 2002; Sheth and Parker, 2003; Cougot et al., 2004; Liu et al., 2004) (Fig. 1). In this study, we also show that eIF4E is localized to mammalian P-bodies (Fig. 3). Overexpression of 4E-T in HeLa cells caused a marked increase of eIF4E in P-bodies (Fig. 3). This observation suggests that translation inhibition and targeting of eIF4E to P-bodies are causally related. The accumulation of eIF4E in P-bodies requires interaction with 4E-T, because an eIF4E mutant that fails to bind to 4E-T cannot localize to P-bodies (Fig. 4), and depletion of 4E-T resulted in loss of eIF4E from P-bodies (Fig. 5). However, the localization of 4E-T to P-bodies does not require interaction with eIF4E, because a 4E-T mutant defective in binding to eIF4E also localizes to P-bodies (Fig. 4).
eIF4GI and eIF4A did not localize to mammalian processing bodies (Fig. 4), which is in agreement with studies that demonstrated that mRNA pools from P-bodies are distinct from translating pools (Schwartz and Parker, 1999; Tharun and Parker, 2001; Teixeira et al., 2005). These observations favor a model whereby translation inhibition that results from disruption of the translation initiation complex occurs outside P-bodies, and precedes P-body formation.
As was shown for Dcp1a (Sheth and Parker, 2003; Cougot et al., 2004), cycloheximide treatment of HeLa cells reduced the amount of 4E-T associated with P-bodies (Fig. 6). This result indicates that ongoing mRNA translation regulates the association of 4E-T with P-bodies, and suggests that 4E-T is not a permanent P-body constituent, but rather localizes to these structures in an mRNA-dependent manner. Importantly, 4E-T RNAi treatment results in decreased decapping activity in HeLa cells as evidenced by diminished localization of Dcp1a and p54 to P-bodies. Taken together, these data support a model whereby interaction of 4E-T with eIF4E acts as a priming event that leads to mRNP remodeling and mRNA decay.
In addition to their role in mRNA decay, P-bodies were suggested to function as mRNA storage sites (Sheth and Parker, 2003; Coller and Parker, 2004). For example, the Drosophila decapping factor Me31B, is concentrated in cytoplasmic granules in germline cells where bicoid and oskar mRNAs are translationally masked (Nakamura et al., 2001). In addition, the Xenopus equivalent, Xp54, is a major constituent of maternal mRNA storage particles where translation is repressed (Ladomery et al., 1997; Minshall and Standart, 2004). Thus, it is possible that under certain conditions, the presence of eIF4E within P-bodies might permit the transition of a translationally repressed/stored mRNA to a translationally competent state.
Homology searches using Blast algorithms failed to identify 4E-T yeast homologues (Dostie et al., 2000a), and eIF4E was shown not to localize to P-bodies in yeast (unpublished data). Therefore, 4E-T might represent a more evolutionarily complex mRNA decay/storage regulation pathway in higher eukaryotes. The Drosophila 4E-T homologue, Cup, was reported to mediate translational repression of nanos and oskar by interacting with eIF4E and 3′ trans-acting factors (Wilhelm and Smibert, 2005). Interestingly, the RNA-binding protein Smaug, which interacts with Cup, recently was shown to recruit the CCR4 deadenylase complex in Drosophila embryos (Semotok et al., 2005).
An important finding in this paper is that reduction in 4E-T results in an increase in mRNA stability (Fig. 7). All of the mRNAs tested here are ARE-containing mRNAs, which are believed to be subject to 3′-5′ degradation by the exosome, based on in vitro decay assays (Chen et al., 2001; Mukherjee et al., 2002). The major deadenylase of AU-containing mRNAs in mammalian cells is believed to be poly (A) ribonuclease (Gao et al., 2000; Lai et al., 2003). Therefore, 3′-5′degradation would require destabilization of the interaction between PABP and eIF4G to allow entry and association of poly (A) ribonuclease with the cap (Wilusz et al., 2001). Therefore, 4E-T may instigate the dissociation of PABP from eIF4G by binding to eIF4E and displacing eIF4G. The decay machinery, which processes mRNA through the 5′-3′ exonucleolytic pathway, is present in P-bodies. Recent studies have implied that the 5′-3′ and 3′-5′ pathways converge at these sites. For instance, mRNA decay enzymes involved in 5′-3′ and 3′-5′ decay are recruited by the ARE binding proteins, tristetraprolin and butyrate response factor (Lykke-Andersen and Wagner, 2005). Moreover, tristetraprolin, which mediates ARE degradation by recruiting the exosome (Chen et al., 2001), also is found localized in P-bodies (Kedersha et al., 2005). Because the disappearance of P-bodies with 4E-T RNAi was not synonymous with complete stabilization of p21 mRNA, it is possible that this process occurs outside of P-bodies or that its deadenylation/decay occurs by way of a distinct pathway from c-fos and GM-CSF mRNAs.
In conclusion, the interaction of 4E-T with eIF4E in cells has two consequences: import of eIF4E to the nucleus (Dostie et al., 2000a) and the targeting of eIF4E to sites of mRNA decay. The function of eIF4E in the nucleus is under investigation; a proposed nuclear function of eIF4E is to export a subset of mRNAs from the nucleus (Lai and Borden, 2000; Rousseau et al., 1996). The dual role of 4E-T also might serve to sequester the limiting factor in translation initiation, eIF4E, as a means of maintaining translational homeostasis. It will be important to examine how 4E-T is regulated. It is reported that 4E-T is a phosphoprotein (Pyronnet et al., 2001); therefore, it would be interesting to know what pathways regulate its phosphorylation and activity.
While this paper was under review, two reports were published which demonstrated that eIF4E is found in P-bodies (Andrei et al., 2005; Kedersha et al., 2005), and that 4E-T is also concentrated in P-bodies (Andrei et al., 2005).
Materials and methods
Plasmids and cell culture
HA–4E-T, HA–4E-T (Y30A), HA-eIF4E, HA-eIF4E (W73A) (Dostie et al., 2000a), and GFP-Dcp1b (Cougot et al., 2004) were described previously. GFP–4E-T was generated by subcloning the coding region of 4E-T from pcDNA3-4E-T (Dostie et al., 2000a) into pFRED143 (provided by G. Pavlakis, National Cancer Institute, Frederick, MD). Myc-EYFP-Me31B plasmid was a gift from A. Nakamura (University of Tsukuba, Tsukuba, Japan).
Construction of tetracycline-regulated β-globin expression plasmids
The starting plasmid, CMV-glo-SPA, which contains the human β-globin gene with the 3′ untranslated region and polyadenylation site replaced with a multiple cloning site and a strong synthetic polyadenylation element (SPA), was described previously (Das Gupta et al., 1998). The first generation tetracycline-regulated globin construct pTet-O-βAc-glo-SPA was prepared by digesting CMV-glo-SPA with HindIII, end-filling with Klenow fragment DNA polymerase, then digesting with NcoI. The globin-SPA fragment was gel purified and cloned into pGTetOβAcLuc3 (provided by J. Garcia-Sanz, Universidad Autãnoma, Madrid, Spain) in which the LUC insert was removed by digesting with BamHI, followed by end-filling with Klenow fragment DNA polymerase plus digestion with NcoI. The c-fos ARE was recovered from the plasmid pBBB+AREcfos (provided by A.-B. Shyu, University of Texas Medical School, Houston, TX) by PCR amplification with primers BBBARE1 (5′-CGCTCTAGACAGAAGGTGGTGGCTGGTGTG-3′) and BBBARE2 (5′-GCGTCTAGACTCAAGGGGCTTCATGATGTC-3′). The GM-CSF ARE was recovered similarly by PCR from pBBB+AREGM-CSF using primers BBBARE1 and BBBARE3 (5′-CCCTCTAGAGCTGGTTATTGTGCTGTCTCA-3′). The PCR products were digested with XbaI and inserted into XbaI-digested Tet-O-βAc-glo-SPA to create plasmids pTet-O-βAc-glo-ARE[cfos]-SPA and pTet-O-βAc-glo-ARE[GM-CSF]-SPA. These were used to prepare the second-generation plasmids that were used in this study. The region spanning the tetracycline operator and β-actin promoter elements of each of these plasmids was removed by digestion with Nhe1 and Nco1, and replaced with the region spanning the tetracycline operator elements and minimal CMV promoter regions of pTREmyc. The latter was prepared by PCR amplification of pTREmyc using primers BM127 (5′-ATGCCATGGTGTCTAGCACGCG-3′) and BM128 (5′-CCGCTAGCCACGAGGCCCTTTCGTCTCG-3′). The final plasmids are designated pTet-CMVmin-glo-SPA, pTet-CMVmin-glo-ARE[cfos]-SPA, and pTet-CMVmin-glo-ARE [GM-CSF]-SPA.
HeLa S3 cells from M. Wilkinson (M.D. Anderson Cancer Center, Houston, TX) (Carter et al., 1995) and HeLa CCL2 (American Type Culture Collection) were cultured in DMEM supplemented with 10% FBS. HeLa S3 cells usually were used unless specified otherwise.
siRNA transfections
siRNA annealed duplexes were purchased from Dharmacon and sequences are listed below. HeLa cells were transfected at 30% confluency in 6-well plates with siRNA (300 pmole final) using oligofectamine according to the manufacturer's instructions. siRNA complexes were left overnight, and cells were processed 48 to 72 h later. 4E-T siRNA 5′-GAACAAGAUUAUCGACCUA dTdT-3′; 4AIII inverted 5′-CGACCAGAG-CTTAAGGTGA dTdT-3′; 4E-T inverted 5′-CGUACCGUGGAAUAGUUCC dTdT-3′.
Transient transfections and immunofluorescence
Transfections of DNA plasmids were performed by the calcium phosphate method as described previously (Dostie et al., 2000a). HeLa cells were transfected at 50% confluency with 5 μg (or 8 μg for GFP-4E-T) of plasmid DNA, and were processed 36 h after transfection. For transfection of myc-EYFP-Me31B (Fig. 5 D), siRNA transfections were performed in 6-well plates as described above; after 24 h, 2 μg of myc-EYFP-Me31B was transfected by the calcium phosphate method. 48 h after siRNA transfection, cells were trypsinized into chamber slides and processed for immunofluorescence 12 h later. Immunofluorescence was performed as described previously (Dostie et al., 2000a). In brief, cells were plated onto Lab-Tek chamber slides (Nunc) 16 h before fixation. Cells were fixed with formaldehyde, permeabilized, blocked, and incubated with affinity-purified rabbit anti–4E-T (1:100) (Dostie et al., 2000a), affinity-purified rabbit anti-Dcp1a (1:200) (a gift from J. Lykke-Andersen, University of Colorado, Boulder, CO), and anti-Dcp2 (1:20) (Liu et al., 2004) antibodies, or monoclonal anti-eIF4A (1:100) (Edery et al., 1983), anti-eIF4E (1:10) (Kimball et al., 2003), and anti-HA (1:1,000) (Covance) antibodies for 2 h at RT. Cells were washed extensively and incubated with Texas red or AlexaFluor 488 or 594 conjugated secondary antibody (1:200) (Molecular Probes) for 30 min. Nuclei were stained with Hoechst dye 33258 (Sigma-Aldrich). Images were taken from a 63× objective of a Zeiss LSM 510 confocal microscope. Immunofluorescence analysis for each experiment was performed three to five times, and the most representative results are displayed.
Western blotting
Cells were washed twice with ice cold PBS and pelleted at maximum speed for 10 min in a microcentrifuge. Cell pellets were frozen on dry ice and thawed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.5% SDS, and a mixture of protease inhibitors [Roche Diagnostics]). Extracts were incubated on ice for 10 min and centrifuged for 5 min at maximum speed to recover the supernatant. Proteins were resolved by SDS/PAGE, transferred to nitrocellulose membrane, blocked, and incubated with rabbit anti–4E-T (1:1,000) (Dostie et al., 2000a), anti-Dcp1a (1:1,000), anti-Xp54 (1:2,000) (Smillie and Sommerville, 2002), and anti-eIF4E 5851 (1:1,000) (Frederickson et al., 1991) polyclonal antibodies or monoclonal anti-eIF4A (1:1,000) (Edery et al., 1983), anti-eIF4E (1:100) (Kimball et al., 2003), anti-HA (1:2,000) (Covance), and anti-β-actin (1:5,000) (Sigma-Aldrich) antibodies.
Vaccinia virus infection, Far Western analysis, and translation assay
HeLa cells (80% confluency) were infected with recombinant vaccinia virus for 1 h at 37°C (Fuerst et al., 1986). Cells were rinsed once with serum-free media and transfected with 5 μg each of the reporter plasmid (pGEM-LUC-POLIRES-CAT) and pcDNA3–4E-T wt or mutant constructs using lipofectin (15 μg; Invitrogen) as previously reported (Pause et al., 1994). After 16 h, cell extracts were collected and analyzed for LUC activity using a LUC assay system (Promega), and activity was measured with a luminometer; CAT expression was analyzed using ELISA (Boehringer). Far Western analysis was performed on transfected cell extracts as described previously (Dostie et al., 2000a)
Northern blotting
Total RNA was isolated using a QIAGEN RNAeasy kit according to the manufacturer's instructions. 5 μg of total RNA was separated on a 1.3% agarose/formaldehyde gel, transferred to a Hybond-N membrane (GE Healthcare), and probed with 32P-labeled, random-primed DNA probe to detect p21 (Lal et al., 2004) or 32P-end labeled oligonucleotide, using terminal deoxynucleotidyl transferase (Invitrogen), to detect 18S rRNA (Lal et al., 2004). For β-globin reporter assays, 4E-T or control siRNA was transfected into HeLa cells as described above. 36 h later, pTet-β-globin reporter plasmids and pTet Off (encoding for tTA, transactivator) were cotransfected. 12 h later, cells were split 1:6 and treated 16 to 20 h later with doxycycline (1 μg/ml). RNA was resolved on a formaldehyde gel as above and probed with 32P-labeled, random-primed DNA probe for detecting β-globin (Stoecklin et al., 2004) or GAPDH.
Acknowledgments
We thank H. Imataka for providing unpublished data. We thank J. Lykke-Andersen for anti-Dcp1a, M. Kiledjian (Rutgers University, Piscataway, NJ) for anti-Dcp2, S. Kimball (The Pennsylvania State University College of Medicine, Hershey, PA) for monoclonal anti-eIF4E, H. Trachsel (University of Berne, Berne, Switzerland) for anti-eIF4A, and J. Sommerville (University of St. Andrews, St. Andrews, Scotland) and N. Standart (University of Cambridge, Cambridge, England) for anti-Xp54 antibodies. We are grateful to B. Seraphin (Centre Nationale de la Recherche Scientifique, Gif-sur-Yvette, France) for GFP-Dcp1b plasmid, A. Nakamura for myc-EYFP-Me31B plasmid, and M. Gorospe (National Institutes of Health, Bethesda, MD) for p21 and 18S oligomers. We thank J. Garcia-Sanz for pGTetOβAcLuc3, and A.B. Shyu for pBBB+AREc-fos plasmids, respectively. We thank I. Gallouzi, F. Tremblay, and P. Cho for critical reading of the manuscript, and J. Laliberte for technical support with the confocal microscopy. We are indebted to C. Lister and S. Perreault for assistance.
This research was supported by a grant from the Canadian Institute of Health Research (CIHR) and U.S. Public Health Service (PHS) grants GM38277 and GM55407 to D.R. Schoenberg. E.L. Murray was supported by PHS grant T32 CA09338. N. Sonenberg is a CIHR distinguished scientist and a Howard Hughes Medical Institute International Scholar.
J. Dostie's present address is Department of Biochemistry and Molecular Pharmacology and Program in Gene Function and Expression, University of Massachusetts Medical School, Worcester, MA 01605.
Abbreviations used in this paper: 4E-T, 4E-transporter; ARE, adenine/uridine rich element; CAT, chloramphenicol acetyl transferase; eIF4E, eukaryotic initiation factor 4E; LMB, leptomycin B; LUC, luciferase; mRNP, messenger RNP; PABP, polyA binding protein; P-body, processing body; RNAi, RNA interference; siRNA, short interfering RNA; SPA, synthetic polyadenylation element.
References
- Achsel, T., H. Brahms, B. Kastner, A. Bachi, M. Wilm, and R. Luhrmann. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei, M.A., D. Ingelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov, V.I., H. Scherthan, J.A. Solinger, J.M. Buerstedde, and W.D. Heyer. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136:761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C.A., A. Stevens, G. Caponigro, T.E. LaGrandeur, L. Hatfield, D.M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 382:642–646. [DOI] [PubMed] [Google Scholar]
- Both, G.W., A.K. Banerjee, and A.J. Shatkin. 1975. Methylation-dependent translation of viral messenger RNAs in vitro. Proc. Natl. Acad. Sci. USA. 72:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, M.S., J. Doskow, P. Morris, S. Li, R.P. Nhim, S. Sandstedt, and M.F. Wilkinson. 1995. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 270:28995–29003. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., R. Gherzi, S.E. Ong, E.L. Chan, R. Raijmakers, G.J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 107:451–464. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., N. Xu, and A.B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 15:5777–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861–890. [DOI] [PubMed] [Google Scholar]
- Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Gupta, J., H. Gu, E. Chernokalskaya, X. Gao, and D.R. Schoenberg. 1998. Identification of two cis-acting elements that independently regulate the length of poly(A) on Xenopus albumin pre-mRNA. RNA. 4:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., M. Ferraiuolo, A. Pause, S.A. Adam, and N. Sonenberg. 2000. a. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., F. Lejbkowicz, and N. Sonenberg. 2000. b. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 148:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T., and R. Parker. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery, I., M. Humbelin, A. Darveau, K.A. Lee, S. Milburn, J.W. Hershey, H. Trachsel, and N. Sonenberg. 1983. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J. Biol. Chem. 258:11398–11403. [PubMed] [Google Scholar]
- Eystathioy, T., A. Jakymiw, E.K. Chan, B. Seraphin, N. Cougot, and M.J. Fritzler. 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 9:1171–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraiuolo, M.A., C.S. Lee, L.W. Ler, J.L. Hsu, M. Costa-Mattioli, M.J. Luo, R. Reed, and N. Sonenberg. 2004. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc. Natl. Acad. Sci. USA. 101:4118–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson, R.M., K.S. Montine, and N. Sonenberg. 1991. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol. Cell. Biol. 11:2896–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst, T.R., and B. Moss. 1989. Structure and stability of mRNA synthesized by vaccinia virus-encoded bacteriophage T7 RNA polymerase in mammalian cells. Importance of the 5′ untranslated leader. J. Mol. Biol. 206:333–348. [DOI] [PubMed] [Google Scholar]
- Fuerst, T.R., E.G. Niles, F.W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA. 83:8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., D.T. Fritz, L.P. Ford, and J. Wilusz. 2000. Interaction between a poly(A)-specific ribonuclease and the 5′ cap influences mRNA deadenylation rates in vitro. Mol. Cell. 5:479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, M., C.J. Wilusz, S.W. Peltz, and J. Wilusz. 2001. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 20:1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913–963. [DOI] [PubMed] [Google Scholar]
- Gradi, A., H. Imataka, Y.V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T.H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 413:538–542. [DOI] [PubMed] [Google Scholar]
- Ingelfinger, D., D.J. Arndt-Jovin, R. Luhrmann, and T. Achsel. 2002. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Kahvejian, A., Y.V. Svitkin, R. Sukarieh, M.N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M.J. Fitzler, D. Scheuner, R.J. Kaufman, D.E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball, S.R., R.L. Horetsky, D. Ron, L.S. Jefferson, and H.P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273–C284. [DOI] [PubMed] [Google Scholar]
- Ladomery, M., E. Wade, and J. Sommerville. 1997. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 25:965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, H.K., and K.L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene. 19:1623–1634. [DOI] [PubMed] [Google Scholar]
- Lai, W.S., E.A. Kennington, and P.J. Blackshear. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23:3798–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, A., K. Mazan-Mamczarz, T. Kawai, X. Yang, J.L. Martindale, and M. Gorospe. 2004. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23:3092–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejbkowicz, F., C. Goyer, A. Darveau, S. Neron, R. Lemieux, and N. Sonenberg. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA. 89:9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.W., X. Jiao, H. Liu, M. Gu, C.D. Lima, and M. Kiledjian. 2004. Functional analysis of mRNA scavenger decapping enzymes. RNA. 10:1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., and E. Wagner. 2005. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 19:351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader, S., H. Lee, A. Pause, and N. Sonenberg. 1995. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 15:4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall, N., and N. Standart. 2004. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 32:1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., C.J. Decker, and R. Parker. 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15:2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, D., M. Gao, J.P. O'Connor, R. Raijmakers, G. Pruijn, C.S. Lutz, and J. Wilusz. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., R. Amikura, K. Hanyu, and S. Kobayashi. 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 128:3233–3242. [DOI] [PubMed] [Google Scholar]
- Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121–127. [DOI] [PubMed] [Google Scholar]
- Pause, A., G.J. Belsham, A.C. Gingras, O. Donze, T.A. Lin, J.C. Lawrence Jr., and N. Sonenberg. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 371:762–767. [DOI] [PubMed] [Google Scholar]
- Peng, J., and D.R. Schoenberg. 2005. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA. 11:1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin, F., A.C. Gingras, H. Olsen, S. Chevalier, and N. Sonenberg. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002–14007. [DOI] [PubMed] [Google Scholar]
- Pyronnet, S., J. Dostie, and N. Sonenberg. 2001. Suppression of cap-dependent translation in mitosis. Genes Dev. 15:2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA. 93:1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg, D.R., and E. Chernokalskaya. 1997. Ribonucleases involved in eukaryotic mRNA turnover. mRNA Metabolism and Post-Transcriptional Gene Regulation. J.B. Harford and D.R. Morris, editors. Wiley-Liss, Inc., New York. 217–240.
- Schwartz, D.C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semotok, J.L., R.L. Cooperstock, B.D. Pinder, H.K. Vari, H.D. Lipshitz, and C.A. Smibert. 2005. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 15:284–294. [DOI] [PubMed] [Google Scholar]
- Shatkin, A.J. 1976. Capping of eucaryotic mRNAs. Cell. 9:645–653. [DOI] [PubMed] [Google Scholar]
- Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie, D.A., and J. Sommerville. 2002. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 115:395–407. [DOI] [PubMed] [Google Scholar]
- Sonenberg, N., H. Trachsel, S. Hecht, and A.J. Shatkin. 1980. Differential stimulation of capped mRNA translation in vitro by cap binding protein. Nature. 285:331–333. [DOI] [PubMed] [Google Scholar]
- Stoecklin, G., T. Stubbs, N. Kedersha, S. Wax, W.F. Rigby, T.K. Blackwell, and P. Anderson. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23:1313–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., U. Sheth, M.A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 11:371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., and R. Parker. 2001. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 8:1075–1083. [DOI] [PubMed] [Google Scholar]
- Tourriere, H., K. Chebli, and J. Tazi. 2002. mRNA degradation machines in eukaryotic cells. Biochimie. 84:821–837. [DOI] [PubMed] [Google Scholar]
- Tseng-Rogenski, S.S., J.L. Chong, C.B. Thomas, S. Enomoto, J. Berman, and T.H. Chang. 2003. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 31:4995–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 99:12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., and M. Kiledjian. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell. 107:751–762. [DOI] [PubMed] [Google Scholar]
- Wilhelm, J.E., and C.A. Smibert. 2005. Mechanisms of translational regulation in Drosophila. Biol. Cell. 97:235–252. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20:491–497. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., M. Wormington, and S.W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237–246. [DOI] [PubMed] [Google Scholar]