Abstract
Centrosomes act as sites of microtubule growth, but little is known about how the number and stability of microtubules emanating from a centrosome are controlled during the cell cycle. We studied the role of the TACC3–XMAP215 complex in this process by using purified proteins and Xenopus laevis egg extracts. We show that TACC3 forms a one-to-one complex with and enhances the microtubule-stabilizing activity of XMAP215 in vitro. TACC3 enhances the number of microtubules emanating from mitotic centrosomes, and its targeting to centrosomes is regulated by Aurora A–dependent phosphorylation. We propose that Aurora A regulation of TACC3 activity defines a centrosome-specific mechanism for regulation of microtubule polymerization in mitosis.
Introduction
In animal cells, microtubules are nucleated from centrosomes and grow out with their plus ends that lead into the cytoplasm (for review see Howard and Hyman, 2003). The number and stability of microtubule plus ends growing from centrosomes changes through the cell cycle, which is concomitant with assembly of the mitotic spindle (Hannak et al., 2001; Kline-Smith and Walczak, 2004; Piehl et al., 2004). One obvious mechanism controlling microtubule assembly at centrosomes is modulation of the microtubule nucleation rate (for review see Job et al., 2003). Indeed, the amount of γ-tubulin increases at centrosomes as cells enter mitosis (Khodjakov and Rieder, 1999; Hannak et al., 2002). However, another equally plausible mechanism is modulation of the stability of nucleated plus ends. The stability of microtubule plus ends is dependent on the rate at which microtubules interconvert between growing and shrinking (called the catastrophe rate) and between shrinking and growing (called the rescue rate; Walker et al., 1988; Howard and Hyman, 2003).
Xenopus laevis egg extracts have been particularly useful for studying the regulation of microtubule polymerization from centrosomes. Because meiotic spindles in Xenopus egg extracts can form without centrosomes by nucleation and stabilization of microtubules around chromosomes (Heald et al., 1996; Karsenti and Vernos, 2001; Sampath et al., 2004), it is possible to study separately the regulation of microtubule polymerization around mitotic centrosomes or chromosomes. In Xenopus egg extracts, the dynamic behavior of microtubules in the cytoplasm is regulated, in large part, by the activity of XMAP215, which is a member of a conserved family of microtubule-associated proteins (Kinoshita et al., 2002; Gard et al., 2004). XMAP215 both stimulates the growth rate and opposes catastrophe activity of XKCM1/XlMCAK, which is member of the Kin I/kinesin-13 family (Walczak et al., 1996; Tournebize et al., 2000; Noetzel et al., 2005). Physiological microtubule dynamics can be reconstituted in vitro by a mixture of XMAP215 and mitotic centromere-associated kinesin (MCAK) together with tubulin (Kinoshita et al., 2001). Although XMAP215 and MCAK clearly have a central role in determining microtubule dynamics in Xenopus egg extracts, we know little about how these proteins are regulated during cell cycle progression. However, these proteins are localized to centrosomes, suggesting that they could be involved in the regulation of microtubule growth from centrosomes (Kinoshita et al., 2002; Gard et al., 2004).
A clue as to the possible mechanisms regulating XMAP215 family proteins at centrosomes came from the discovery that the Drosophila melanogaster member of the XMAP215 family is associated with D-TACC (D. melanogaster transforming acidic coiled coil; Cullen and Ohkura, 2001; Lee et al., 2001). Mutants in TACC family members reduce microtubule number and prevent the localization of XMAP215 family proteins to centrosomes or spindles in D. melanogaster (Cullen and Ohkura, 2001; Lee et al., 2001), Caenorhabditis elegans (Bellanger and Gönczy, 2003; Le Bot et al., 2003; Srayko et al., 2003), yeasts (Usui et al., 2003; Sato et al., 2004), and human cells (Gergely et al., 2003). Beyond the fact that TACC family members are required to localize XMAP215 family members to centrosomes, we know little about the role of TACC in the modulation of microtubule dynamics and in regulating the activity of XMAP215. In this study, we investigated these issues by using purified proteins and Xenopus egg extracts. Maskin is a Xenopus TACC homologue (Stebbins-Boaz et al., 1999); in this paper, we call it Xenopus TACC3 because of its sequence similarity to the mammalian TACC3 subfamily (Still et al., 2004). We demonstrate that a Xenopus TACC3 is a cell cycle–dependent regulator of XMAP215.
Results
TACC3 forms a complex with and stimulates the activity of XMAP215
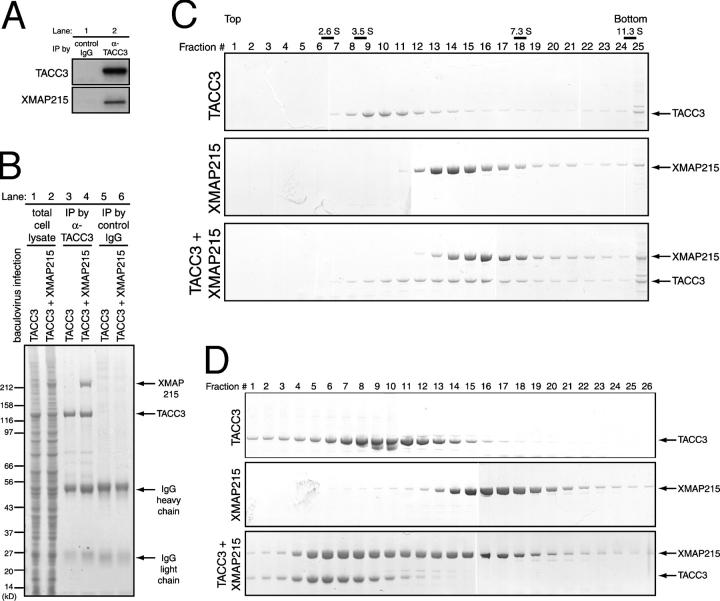
To look at the role of the TACC3–XMAP215 complex in microtubule assembly, we studied the activity of the complex in vitro. We first confirmed via immunoprecipitation that native TACC3 and XMAP215 form a protein complex in Xenopus egg extracts (Fig. 1 A). When coexpressed in baculovirus-infected insect cells, recombinant TACC3 and XMAP215 proteins also form a complex (Fig. 1 B). By mixing purified recombinant proteins and performing sucrose gradient centrifugation as well as gel filtration, we showed that TACC3 and XMAP215 form a one-to-one complex with a native molecular mass of ∼366 kD (Fig. 1, C and D).
Figure 1.
TACC3 forms a complex with XMAP215 in Xenopus egg extracts and in vitro. (A) Coimmunoprecipitation in Xenopus egg extracts. Immunoprecipitation (IP) was performed using control IgG (lane 1) or anti-TACC3 antibody (lane 2). The blots were probed with anti-TACC3 (top) or anti-XMAP215 (bottom). (B) Coimmunoprecipitation in the extracts of baculovirus-infected insect cells. Total lysates of TACC3 baculovirus single infected cells and TACC3 (lane 1) and XMAP215 virus double infected cells (lane 2) were prepared for immunoprecipitation. In each cell lysate, immunoprecipitation were performed using anti-TACC3 (lanes 3 and 4) or control IgG antibody (lanes 5 and 6). Total cell lysate and immunoprecipitates that were dissolved in sample buffer were subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue. (C) Fractions from the top and bottom of a continuous sucrose density gradient centrifugation with purified proteins. Fractions collected after 3–15% continuous sucrose gradient centrifugation with TACC3 alone (top), XMAP215 alone (middle), or a mixture of TACC3 and XMAP215 (bottom) were subjected to SDS-PAGE. The gels were stained with Coomassie brilliant blue. (D) Gel filtration with purified proteins. TACC3 alone (top), XMAP215 alone (middle), or a mixture of TACC3 and XMAP215 (bottom) were analyzed by gel filtration. Fractions were collected and subjected to SDS-PAGE, and the gels were stained with Coomassie brilliant blue.
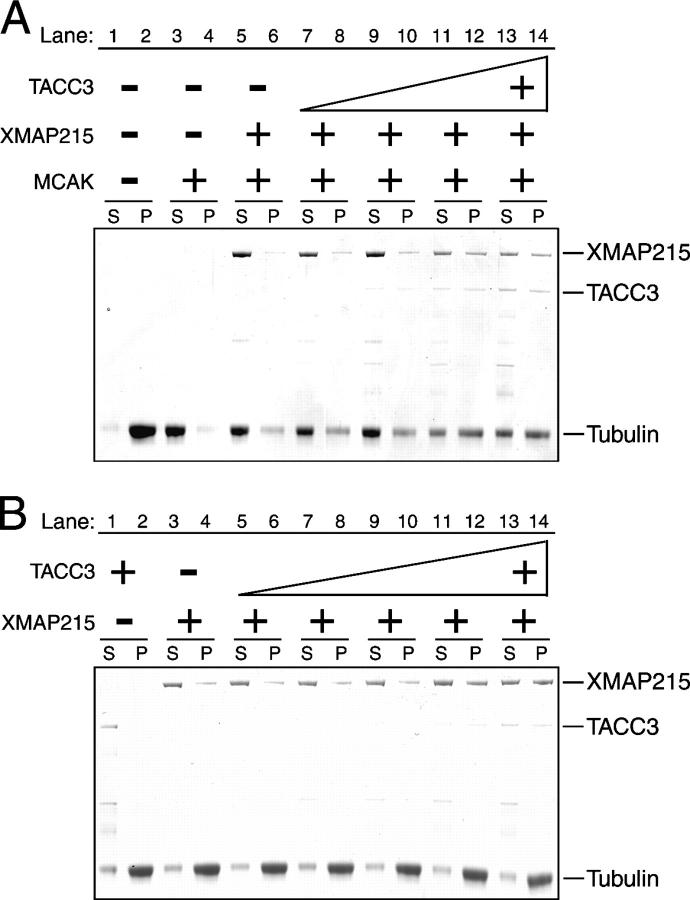
Using a mixture of these purified proteins, we monitored the ability of the TACC3–XMAP215 complex to oppose the microtubule-destabilizing activity of MCAK by using a microtubule sedimentation assay (Desai et al., 1999a). In the presence of MCAK, microtubules were completely destabilized (Fig. 2 A, lanes 1 and 2) as reported previously (Desai et al., 1999a). The addition of 500 nM XMAP215 resulted in a slight reduction of MCAK's microtubule-destabilizing activity (Fig. 2 A, lanes 3 and 4). However, XMAP215 that complexed with TACC3 more efficiently opposed the destabilizing activity of MCAK (Fig. 2 A, lanes 7–14). TACC3 alone had no detectable effect on MCAK activity (not depicted). One possibility is that TACC3 increases the microtubule-stabilizing activity of XMAP215 by enhancing the affinity of XMAP215 for microtubules. Indeed, microtubule sedimentation analyses that were performed in the absence of MCAK indicated that the TACC3–XMAP215 complex has a higher affinity for microtubules than XMAP215 alone (Fig. 2 B). We have previously shown that XMAP215 opposes MCAK in a dose-dependent manner (Kinoshita et al., 2001). Therefore, we conclude that TACC3 stabilizes microtubules in vitro by forming a complex with XMAP215, increasing the affinity of XMAP215 for microtubules, and, thus, enhancing the antagonizing activity against MCAK.
Figure 2.
TACC3 increases the antagonizing activity of XMAP215 against MCAK. (A) Sedimentation analysis to monitor MCAK-dependent microtubule destabilization activity. 2.5 μM GMPCPP microtubules were mixed with 125 nM MCAK (lanes 3–14) or control buffer (lanes 1 and 2) in presence of ATP. 500 nM XMAP215 (lanes 5–14) and increasing concentrations of TACC3 from 250 (lanes 7 and 8), 500 (9 and 10), and 1,000 (11 and 12) to 1,500 nM (13 and 14) or control buffer (5 and 6) were added in reactions. Reaction mixtures were sedimented after a 38-min incubation at 30°C, supernatants (S) and pellets (P) were subjected to SDS-PAGE, and the gel was stained with Coomassie brilliant blue. (B) Sedimentation assay to monitor XMAP215 affinity to microtubules. 2.5 μM GMPCPP microtubules were mixed with 1,250 nM TACC3 (lanes 1 and 2), 500 nM XMAP215 (3 and 4), or 500 nM XMAP215 in addition to increasing concentrations of TACC3 from 250 (5 and 6), 500 (7 and 8), 750 (9 and 10), and 1,000 (11 and 12) to 1,250 nM (13 and 14). Mixtures were incubated for 38 min at 30°C and sedimented, and supernatants (S) and pellets (P) were analyzed by SDS-PAGE.
TACC3 is required for mitotic microtubule assembly around centrosomes by opposing the activity of MCAK
To confirm that TACC3 regulates microtubule growth under physiological conditions, we analyzed meiotic spindle assembly in cycling extracts (cycled spindles; Murray, 1991; Desai et al., 1999b). Specifically, we used antibodies to immunodeplete TACC3 from Xenopus egg extracts and examined spindle assembly in the depleted extracts. To our surprise, meiotic spindles that assembled in the absence of TACC3 were indistinguishable from spindles that assembled in mock-depleted extracts (Fig. S1, available at http://jcb.org/cgi/content/full/jcb.200503023/DC1) despite the depletion of TACC3 to undetectable levels. Under similar depletion conditions for XMAP215, no spindles were observed. This was unexpected because TACC mutants have phenotypes in mitotic systems that were previously studied (see Introduction; Raff, 2002).
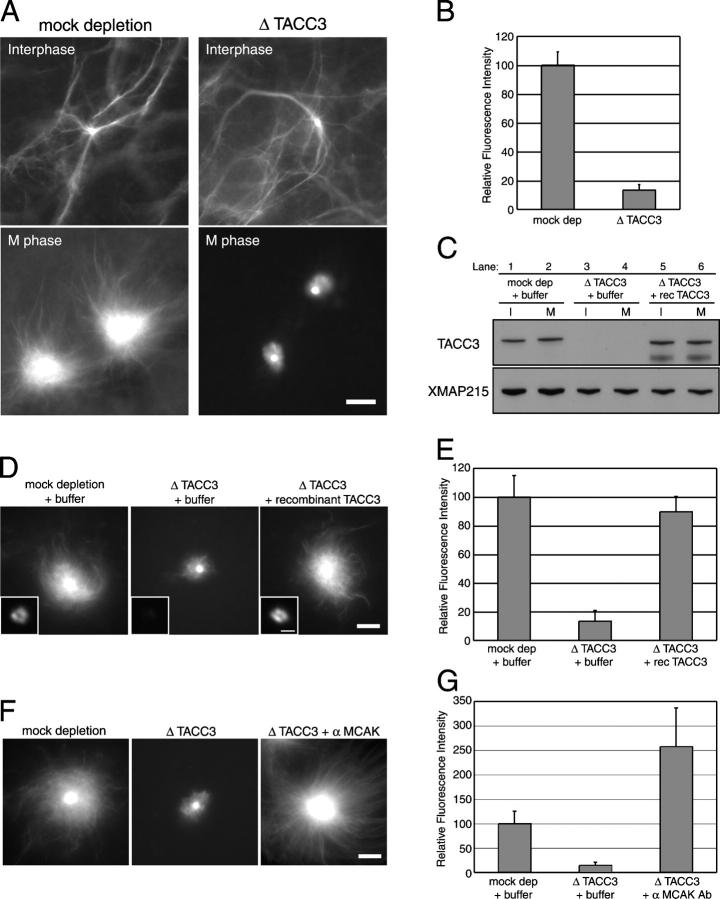
In all systems in which localization has been examined, TACC and XMAP215 are found at centrosomes, suggesting that TACC3 could have a centrosome-specific role in mitosis. Accordingly, we looked specifically at the role of TACC3 in microtubule growth from mitotic centrosomes. Traditionally, centrosomal microtubule assembly has been examined in Xenopus egg extracts by adding human centrosomes directly to mitotic extracts (Belmont et al., 1990). However, the formation of a mitotic centrosome is likely to be a complex process involving cell cycle transition. To create mitotic centrosomes, we added isolated centrosomes to interphase extracts and drove the extracts into mitotic (M) phase by the addition of a nondegradable cyclin B (cyclin B Δ90; Glotzer et al., 1991). In mock-depleted extracts, microtubule assembly around centrosomes was activated by the induction of M phase (Fig. 3 A, left). In TACC3-depleted extracts, the microtubule number around centrosomes was decreased to 13% of that in control extracts (Fig. 3 A, right; and B). Importantly, adding back recombinant TACC3 protein to depleted extracts increased microtubule assembly from mitotic centrosomes, returning it to the levels that were found in control extracts (Fig. 3, C–E). In contrast to M phase–induced extracts, no detectable effect of TACC3 depletion was observed on microtubule assembly in interphase extracts (Fig. 3 A, top). We confirmed that TACC3 was localized to these mitotic centrosomes in mock-depleted and TACC3 add-back extracts but not in TACC3-depleted extracts (Fig. 3 D, insets). We conclude that TACC3 is required specifically for microtubule assembly around mitotic centrosomes in Xenopus egg extracts.
Figure 3.
TACC3 is required for mitosis-specific microtubule assembly around centrosomes. (A) Microtubules were visualized by the addition of fluorescently labeled tubulin in control mock-depleted (left) and TACC3-depleted extracts (right). Centrosomes were added to interphase extracts (top), and then the extracts were driven into M phase by the addition of nondegradable cyclin B (bottom). Bar, 10 μm. (B) Quantification of tubulin fluorescence in A. (C) Immunoblots of depleted extracts. The extracts were immunodepleted with control IgG (lanes 1 and 2; mock dep) or anti-TACC3 antibody (lanes 3–6; Δ TACC3) with adding back of TACC3 (lanes 5 and 6) or control buffer (lanes 1–4). The blots of interphase (lanes 1, 3, and 5) and mitotic extracts (lanes 2, 4, and 6) were probed by anti-TACC3 (top; TACC3) and anti-XMAP215 (bottom; XMAP215). (D) Microtubules assembled around mitotic centrosomes in immunodepleted extracts with add back of recombinant proteins. Purified recombinant TACC3 or control buffer were added in mock-depleted or TACC3-depleted extracts. Bar, 10 μm. Insets show immunolocalization of TACC3 on centrosomes. Bar, 1 μm. (E) Quantification of tubulin fluorescence in D. (F) Inhibition of MCAK in TACC3-depleted extracts. Microtubules were assembled around centrosomes in mock-depleted extracts plus control buffer (left), TACC3-depleted extracts plus control buffer (middle), or anti-MCAK antibody (right). The addition of inhibitory MCAK antibody rescued the effect of TACC3 depletion on microtubule assembly around mitotic centrosomes. Bar, 10 μm. (G) Quantification of tubulin fluorescence in F. (B, E, and G) Relative fluorescence intensity of labeled tubulin around mitotic centrosomes (within 21.079 μm radius) was measured. Values shown are the means plus SD.
The in vitro characterization of TACC3 activity suggested the possibility that in the absence of TACC3, microtubules cannot grow from mitotic centrosomes because of high MCAK activity. To test this idea, we examined the effect of MCAK inhibition on microtubule growth in TACC3-depleted extracts. We found that the addition of inhibitory MCAK antibodies to TACC3-depleted extracts stimulated the growth of microtubules from mitotic centrosomes (Fig. 3, F and G). This led to the conclusion that the likely function of TACC3 is to antagonize the microtubule destabilization by MCAK, presumably by increasing XMAP215 activity on centrosomes in M phase but not in interphase.
Aurora A–phosphorylated TACC3 is enriched at mitotic centrosomes
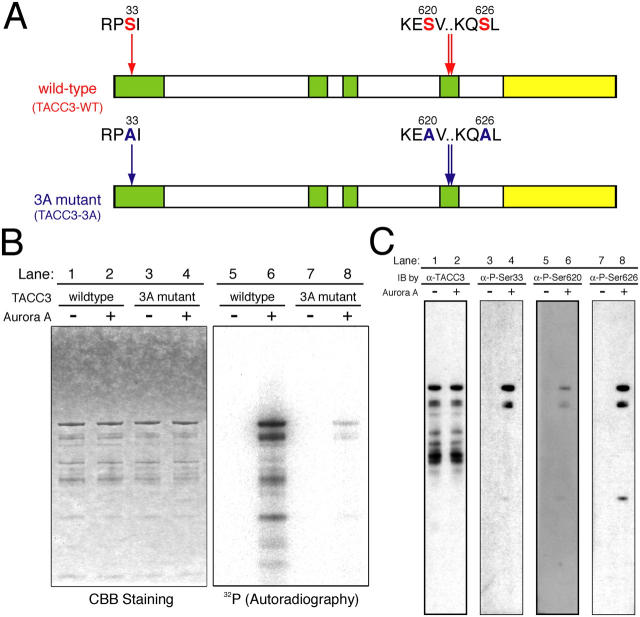
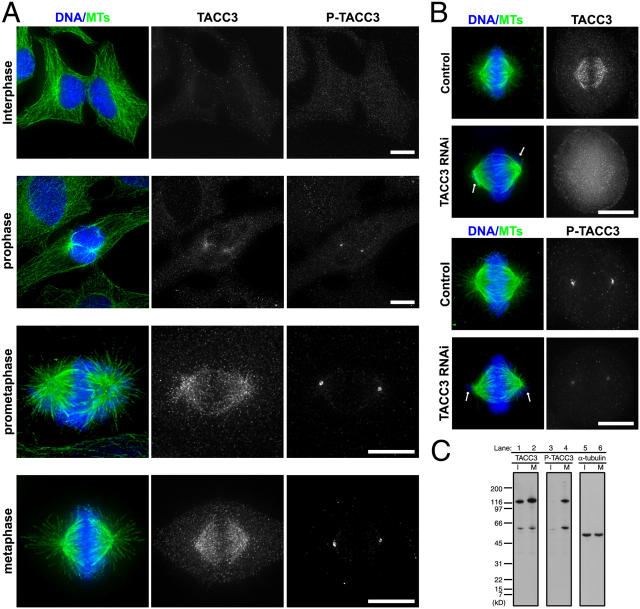
The results argue that TACC3 activity at centrosomes is specifically required for M phase microtubule growth and poses the question as to whether cell cycle–specific modulation of TACC3 activity could regulate the growth of microtubules from centrosomes. Experiments in D. melanogaster and C. elegans suggested a role of the mitotic kinase Aurora A in TACC3 localization to centrosomes (Giet et al., 2002; Bellanger and Gönczy, 2003; Le Bot et al., 2003) but could not distinguish between a specific role of the kinase in modulating TACC3 activity at centrosomes and a more general role in regulating centrosome assembly and maturation. Therefore, we decided to investigate the specific role of Aurora A in the regulation of TACC3 localization and activity. TACC3 has three consensus Aurora A sites (Cheeseman et al., 2002) that are conserved between Xenopus (Ser33, Ser620, and Ser626) and humans (Ser34, Ser552, and Ser558; Fig. 4 A). Recent experiments have shown that TACC3 is an in vitro substrate of Aurora A in Xenopus (at Ser626; Pascreau et al., 2005) and humans (Tien et al., 2004). We confirmed by mass spectrometry that Xenopus TACC3 is indeed phosphorylated by Aurora A at these three consensus sites (Fig. S2, available at http://jcb.org/cgi/content/full/jcb.200503023/DC1). Furthermore, we mutated these three conserved sites to alanine and showed that the incorporation of labeled 32P into the alanine-mutated protein (3A mutant; TACC3-3A) is reduced to 3% compared with the wild-type (WT) protein (TACC3-WT; Fig. 4 B). We raised phosphospecific antibodies to the three conserved Aurora A sites (Fig. 4 C). Because of the limitations of cytology in Xenopus systems and in order to use RNA interference as a control for antibody specificity, we examined the localization of phosphorylated TACC3 in human tissue culture cells. Interestingly, phospho-TACC3, which was stained by antibodies against P-Ser626 (in Xenopus; Ser558 in humans), localized specifically to mitotic centrosomes, whereas the general population of TACC3, which was stained by polyclonal antibodies against the fragment of TACC3 protein (73–265 amino acids), localized throughout the mitotic spindle as previously reported (Fig. 5 A; Gergely et al., 2000, 2003). siRNA of TACC3 greatly reduced the staining of phospho-TACC3 as well as that of nonspecific TACC3 (Fig. 5 B). Phospho-TACC3 antibodies recognized a band of ∼130 kD molecular mass specifically in mitosis-arrested tissue culture cells (Fig. 5 C) as well as a band of the same molecular mass that disappeared after the immunodepletion of either TACC3 (see Fig. 6 D) or Aurora A (Fig. S3, available at http://jcb.org/cgi/content/full/jcb.200503023/DC1) from Xenopus egg extracts in mitosis. Therefore, we conclude that TACC3 protein on centrosomes is specifically phosphorylated during M phase by Aurora A.
Figure 4.
Determination of Aurora A phosphorylation sites of TACC3 in vitro . (A) Consensus sequences for Aurora A phosphorylation in Xenopus TACC3/maskin. Yellow boxes indicate conserved domains among the TACC family, whereas green boxes are the domains that are highly conserved with TACC3 homologues only. The 3A mutant protein (TACC3-3A) has mutations of alanine substitution on three serine residues (Ser33, Ser620, and Ser626) in the consensus sequences of Aurora A phosphorylation. Numbers represent the positions for amino acid residues of Aurora A phosphorylation target sites in the amino acid sequence of TACC3. (B) Aurora A kinase assay with recombinant WT versus alanine mutant TACC3 proteins. WT or 3A mutant TACC3 was incubated with or without recombinant Aurora A (−/+ Aurora A) in the presence of γ−[32P] ATP (see Materials and methods). The reaction mixture was loaded onto SDS-PAGE, and the gel was stained with Coomassie brilliant blue (CBB; lanes 1–4). The incorporation of 32P into TACC3 in the gel was measured by autoradiography (32P; lanes 5–8). (C) Characterization of phosphospecific antibodies. TACC3-WT was incubated with or without Aurora A (−/+ Aurora A), and the reaction mixture was loaded onto SDS-PAGE. The blots were probed with anti-TACC3 antibody (lanes 1 and 2), antiphospho-Ser33 (lanes 3 and 4), antiphospho-Ser620 (lanes 5 and 6), and antiphospho-Ser626 (lanes 7 and 8).
Figure 5.
Aurora A phosphorylated TACC3 is enriched at mitotic centrosomes. (A) Immunolocalization of TACC3 and phospho-TACC3. Deconvolved images of human tissue culture cells stained for DNA (blue)/microtubules (MTs; green), TACC3, and phospho-TACC3 (P-TACC3; stained by antiphospho-Ser626 antibodies) in different cell cycle stages. Bars, 10 μm. (B) Immunolocalization of TACC3 and phospho-TACC3 in siRNA-treated cells. Deconvolved images of either control or TACC3 RNA interference–treated cells stained for DNA (blue)/microtubules (MTs; green), TACC3 (top), and phospho-TACC3 (P-TACC3; bottom). Arrows indicate misaligned chromosomes that are indicative of the TACC3 RNA interference phenotype. Bar, 10 μm. (C) Phosphorylation of TACC3 is mitosis specific in human tissue culture cells. The TACC3 antibody detects a protein of ∼130 kD in extracts prepared from either an asynchronous culture of HeLa S3 cells (lane 1; I) or cells arrested in mitosis by nocodazole treatment (lane 2; M). The phosphospecific TACC3 antibody (antiphospho-Ser626) recognizes a band with the same molecular mass size in mitotic cells (lane 4) but not in asynchronously cultured cells (lane 3). The blot probed by α-tubulin antibody is a loading control (lane 5, asynchronous cultured cells; lane 6, mitotic arrested cells).
Figure 6.
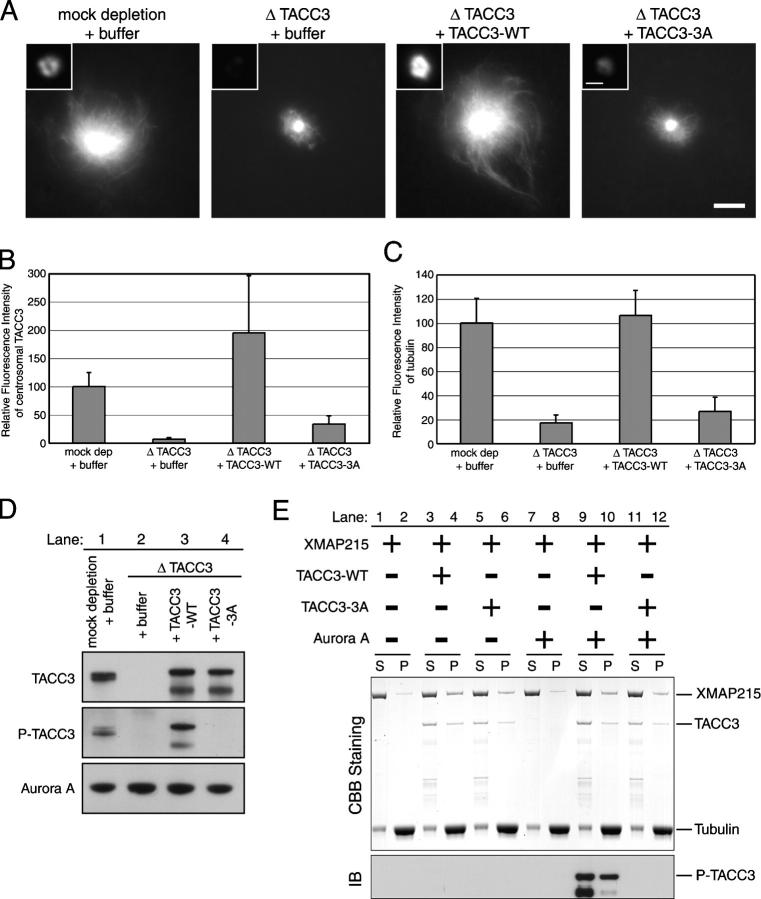
Phosphorylation of TACC3 by Aurora A is required for centrosomal localization of TACC3 and its microtubule-stimulating activity. (A) Localization of WT versus phosphorylation mutant of TACC3 on mitotic centrosomes. Microtubules around mitotic centrosomes were visualized by the addition of fluorescent tubulin in add-back experiments. Bar, 10 μm. Insets show immunolocalization of TACC3 on centrosomes in immunodepleted extracts with add back of WT versus the phosphorylation mutant of TACC3. Bar, 1 μm. (B) Quantification of immunostaining of TACC3 on centrosomes in A. Relative fluorescence intensity of TACC3 staining on centrosomes (within 1.079 μm radius) was measured. (C) Quantification of tubulin fluorescence in A. Relative fluorescence intensity of labeled tubulin around mitotic centrosomes (within 21.079 μm radius) was measured. (B and C) Values shown are the means plus SD. (D) Immunoblots of depleted extracts in add-back experiments. Mitotic extracts were immunodepleted with control IgG (lane 1; mock dep) or anti-TACC3 antibody (lanes 2–4; Δ TACC3) with adding back of control buffer (lanes 1 and 2; + buffer), TACC3-WT (lane 3; + TACC3-WT), or phosphorylation mutant TACC3 (lanes 4; + TACC3-3A). (E) Sedimentation assay to monitor XMAP215 affinity to microtubules in the presence of WT or phosphorylation mutant TACC3. 2.5 μM GMPCPP microtubules were mixed with 0.5 μM XMAP215 alone (lanes 1, 2, 7, and 8), 0.5 μM XMAP215 + 1.0 μm TACC3-WT (lanes 3, 4, 9, and 10), or 0.5 μM XMAP215 + 1.0 μm TACC3-3A (lanes 5, 6, 11, and 12) in the absence (lanes 1–6) or presence (lanes 7–12) of Aurora A. Mixtures were incubated for 38 min at 30°C and sedimented, and supernatants (S) and pellets (P) were subjected to SDS-PAGE. The gels were stained with Coomassie brilliant blue (top; CBB staining) or transferred onto nitrocellurose membrane for immunoblotting (IB) using the phosphospecific TACC3 antibody (antiphospho-Ser626; bottom).
Phosphorylation of TACC3 by Aurora A is required for its targeting to mitotic centrosomes
To test whether phosphorylation is required to target TACC3 to centrosomes, we examined the localization of the nonphosphorylatable mutant of TACC3 in Xenopus egg extracts. By adding back WT (TACC3-WT) versus alanine-mutated TACC3 (TACC3-3A) to the depleted extracts (Fig. 6 D), we found that centrosomes contained 10% of TACC3-3A compared with TACC3-WT (Fig. 6 A, insets; and B). We confirmed that the TACC3-3A mutant could still interact with XMAP215 both in vitro and in Xenopus egg extracts (Fig. S4, available at http://jcb.org/cgi/content/full/jcb.200503023/DC1). Furthermore, TACC3-3A still enhanced the affinity of XMAP215 for microtubules (Fig. 6 E). Importantly, TACC3-3A cannot target XMAP215 to mitotic centrosomes as efficiently as TACC3-WT (Fig. S5, available at http://jcb.org/cgi/content/full/jcb.200503023/DC1), and this argues that TACC3 phosphorylation by Aurora A is required for efficient centrosomal localization of the TACC3–XMAP215 complex in M phase.
To test whether targeting of TACC3 to centrosomes is required for its activity, we assayed centrosome-mediated microtubule assembly in TACC3-depleted extracts that were reconstituted with either WT or alanine-mutated TACC3. Compared with the WT protein, the alanine-mutated TACC3 could only poorly rescue the effect of TACC3 depletion (Fig. 6, A and C). We confirmed that, after addition to the extract, TACC3-WT was phosphorylated, whereas TACC3-3A was not (Fig. 6 D). Thus, the targeting of TACC3 to centrosomes by Aurora A phosphorylation is required to stimulate microtubule growth from mitotic centrosomes. We were unable to detect any effect of Aurora A phosphorylation of TACC3-WT on its ability to enhance the affinity of XMAP215 for microtubules (Fig. 6 E). This implies that the likely function of Aurora A phosphorylation of TACC3 is targeting of the TACC3–XMAP215 complex to mitotic centrosomes rather than directly regulating its activity toward microtubules.
Discussion
Activation of microtubule assembly by the TACC3–XMAP215 complex on mitotic centrosomes
Previous studies have shown that the combination of XMAP215 and XKCM1/MCAK is essential to promote the dynamic properties of mitotic microtubule assembly (Kinoshita et al., 2001; Noetzel et al., 2005). We show that in vitro TACC3 and XMAP215 form a one-to-one complex that enhances the ability of XMAP215 to oppose the inhibitory activity of MCAK by increasing the affinity of XMAP215 for the microtubule lattice. Therefore, TACC3 appears to have a specific role in modulating the dynamic behavior of microtubules by modifying the activity of XMAP215.
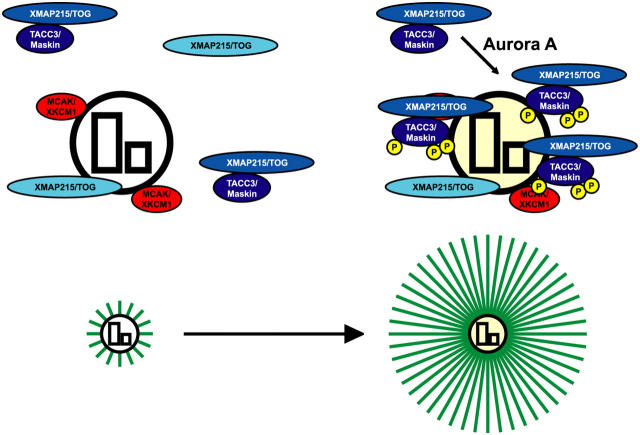
Microtubule growth from centrosomes or chromosomes requires the presence of XMAP215 (Tournebize et al., 2000). Because Xenopus meiotic spindle assembly is apparently unaffected in the absence of TACC3, it appears that TACC3 is not required for the global activity of XMAP215. Why is TACC3 required specifically for XMAP215 activity at mitotic centrosomes? Our data suggest that targeting of the TACC3–XMAP215 complex to mitotic centrosomes overcomes the high microtubule-destabilizing activity of MCAK at centrosomes. Consistent with this idea, MCAK is localized to centrosomes in many systems (Walczak et al., 1996; Oegema et al., 2001), and colonic and hepatic tumor overexpressed gene (TOG)/TOG protein, which is a human XMAP215 orthologue, is required to protect spindle microtubules from MCAK activity at centrosomes (Holmfeldt et al., 2004). We propose that in the absence of TACC3, XMAP215 is still able to counteract the activity of cytoplasmic MCAK. MCAK localization to centrosomes, however, generates an environment in which plus end growth is not favored, and XMAP215 cannot stabilize nascent nucleated plus ends. This could either be because MCAK activity at centrosomes is enhanced or because of an increased concentration of MCAK at centrosomes. Targeting of TACC3–XMAP215 would enhance the activity of XMAP215 at centrosomes, stabilizing nascent plus ends and allowing them to grow from centrosomes (Fig. 7). This provides an explanation for the seemingly paradoxical observation that a plus end stabilizer localizes to centrosomes where microtubules are attached via their minus ends (Gard et al., 2004; Gräf et al., 2004).
Figure 7.
A model of centrosomal microtubule assembly in M phase. XKCM1/MCAK (red) localized on centrosomes generates an environment in which plus end growth of microtubules (green) is not favored, and XMAP215/TOG (light blue) cannot stabilize nascent nucleated plus ends (left). Aurora A phosphorylation of TACC3/maskin targets the TACC3–XMAP215 complex (dark blue) to centrosomes. The targeting enhances the activity of XMAP215 at centrosomes, stabilizing nascent plus ends and allowing microtubules to grow from centrosomes (right). The encircled P (yellow) represents the serine residues that were phosphorylated by Aurora A in TACC3.
The centrosome-specific role of TACC3 could explain why the phenotype of RNA interference of TACC family members in tissue culture cells is not very severe. The chromatin-mediated pathway of spindle assembly obscures centrosome-specific effects on microtubule growth. Interestingly, in C. elegans embryos in which mitotic spindle assembly is dominated by its centrosomes, the removal of TACC results in severe microtubule-based phenotypes (Bellanger and Gönczy, 2003; Le Bot et al., 2003; Srayko et al., 2003).
Aurora A regulates centrosomal microtubule assembly in mitosis
The targeting of TACC3 to centrosomes defines a potential mechanism for regulating the polymerization of microtubules from centrosomes in mitosis. Previous data in other systems has shown that Aurora A is required to target TACC to centrosomes (Giet et al., 2002; Bellanger and Gönczy, 2003; Le Bot et al., 2003). However, the specific role of TACC3 phosphorylation has been unclear, as Aurora A has general roles in both centrosome assembly and cell cycle progression (Hannak et al., 2001; Giet et al., 2002; Hirota et al., 2003). Our data show that targeting of TACC3 to centrosomes specifically requires Aurora A phosphorylation. Furthermore, targeting is essential for TACC activity at centrosomes. Because Aurora A activity increases as cells enter M phase, this would account for the increase in microtubule polymerization that was observed as centrosomes mature during the cell cycle. Consistent with this idea, the protein level of both TACC3 and Aurora A as well as the kinase activity of Aurora A are all highly cell cycle regulated, reaching a peak during mitosis in human tissue culture cells (Bischoff et al., 1998; Gergely et al., 2003). In Xenopus egg extracts, the increase of Aurora A kinase activity may be sufficient to trigger the activation of centrosomal microtubule polymerization in mitosis. Thus, the phosphorylation of TACC3 provides a mechanism for Aurora A to specifically modulate the growth of microtubules from centrosomes. However, Aurora A regulates processes other than microtubule assembly at centrosomes. Identification of the other Aurora A substrates will be essential for understanding the mechanisms of mitotic spindle assembly.
Materials and methods
Expression and purification of recombinant proteins
Full-length Aurora A cDNA was subcloned into pET21 vector (CLONTECH Laboratories, Inc.) for Xenopus Aurora A expression in bacteria (pET21 Aurora A; a gift from J. Swedlow, University of Dundee, Dundee, UK). TACC3 was expressed both in bacteria and insect cells. Full-length TACC3 cDNA was subcloned into pET30 vector (CLONTECH Laboratories, Inc.) for expression in bacteria (pET30a maskin; a gift from J. Richter, University of Massachusetts, Worcester, MA). The mutations with an alanine substitution of Aurora A target sites were introduced by PCR. Bacterially expressed His-tagged proteins were purified by using Ni–nitrilotriacetic acid (QIAGEN). For the expression of TACC3 in insect cells, we used the Bac-to-bac baculovirus expression system (Invitrogen). Full-length TACC3 cDNA was subcloned into pFastbac vector for the preparation of bacmid DNA (NH2-terminal His6 tagged). TACC3 baculovirus-infected expresSF+ cells (Protein Sciences) were pelleted after a 42-h incubation at 27°C and were resuspended in ice-cold lysis buffer (50 mM Hepes, pH 7.5, 0.1% Triton X-100, 200 mM NaCl, 10 mM CaCl2, 25 U/ml benzonase [Novagen], 10 μg/ml nocodazole [Sigma-Aldrich], and 1× protease inhibitor mix). The inhibitor mix consisted of 10 μg/ml antipain-HCl, 10 μg/ml APMSF, 6 μg/ml chymostatin, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, 0.7 μg/ml pepstatin, and 3.6 μg/ml E64 (all protease inhibitors were purchased from Sigma-Aldrich except for E64, which was obtained from BIOMOL Research Laboratories, Inc.). The resuspended pellets were dounced 20 times on ice using a Dounce homogenizer. The lysate was cleared by centrifugation, loaded onto a column (HiTrap Q FF; GE Healthcare), and equilibrated in 20 mM anion exchange buffer (Tris, BisTris-propane, and HCl), pH 7.5. The peak containing full-length TACC3 was supplemented with 3 mM imidazole (Sigma-Aldrich) and loaded onto a HiTrap chelating HP column (GE Healthcare) that was equilibrated in binding buffer (25 mM Tris-HCl, pH 8.0, 300 mM NaCl, 3 mM imidazole, and 20% glycerol). The column was washed stepwise with binding buffer supplemented with 5 mM ATP, 1.5 M NaCl, and 30 and 60 mM imidazole and was eluted with 300 mM imidazole. The peak containing full-length TACC3 was pooled and desalted with a column (NAP25; GE Healthcare) into 20 mM anion buffer, including 330 mM NaCl, 1 mM DTT, 0.1× protease inhibitor mix, and 10% glycerol. The recombinant XMAP215 and XKCM1/XlMCAK were expressed in expresSF+ insect cells (Protein Sciences) and were purified as previously described (Tournebize et al., 2000; Kinoshita et al., 2001). All of the purified proteins were aliquoted, frozen in liquid nitrogen, and stored at −80°C. Protein concentration was determined by using the Bradford assay and the molecular extinction coefficient at OD280nm.
Immunoprecipitation, sucrose density gradient centrifugation, and gel filtration
For immunoprecipitation using insect cell lysates, protein A–Sepharose beads that bound control rabbit IgG or anti-TACC3 antibodies were incubated with lysate from insect cells that were infected either with TACC3 baculovirus or with both TACC3 and XMAP215 baculovirus. Eluted immunoprecipitates were separated by SDS-PAGE and stained by Coomassie brilliant blue.
Sucrose gradients (3–15%) were poured as step gradients (five steps of equal volume) in 1× PBS, 0.1% Tween 20, 1 mM DTT, and 0.2× protease inhibitor mix. Gradients were incubated at 4°C for 12 h to allow diffusion into a continuous gradient. Equal concentrations of TACC3 and XMAP215 were mixed in vitro, incubated for 10 min on ice, diluted into gradient buffer to a final concentration of 1% glycerol, loaded onto the gradient, and spun for 6.5 h at 4°C and 120,000 g. Gradients were fractionated from the top by hand with a cut pipette tip. Fractions were analyzed by 4–12% SDS-PAGE (NuPage; Invitrogen) and stained by Coomassie brilliant blue. Standard proteins with known sedimentation values were run in parallel and scanned, band intensities were quantified, and peak fractions were assigned. Standard curves of peak fractions versus sedimentation coefficient were used to estimate the S value of proteins. Standard proteins that were used in sucrose gradients are listed as follows (sedimentation values are indicated in parentheses): chymotrypsinogen (2.6 S); ovalbumin (3.5 S); BSA (4.6 S); aldolase (7.3 S); and catalase (11.3 S).
Gel filtration chromatography was performed using a column (model G5000PWXL; Tosoh) in 1× PBS, 0.1% Tween 20, 1 mM DTT, 5% glycerol, and 0.2× protease inhibitor mix. The column was calibrated with standards of known Stokes radii. Standard curves of peak fractions versus the logarithm of the Stokes radii were used to determine the Stokes radii of proteins. Fractions were separated by 4–12% SDS-PAGE (NuPage; Invitrogen) and stained by Coomassie brilliant blue. Standard proteins that were used in gel filtration chromatography are listed as follows (Stokes radii are indicated in parentheses): ribonuclease A (1.64 nm); chymotrypsinogen (2.09 nm); ovalbumin (3.05 nm); BSA (3.55 nm); aldolase (4.81 nm); catalase (5.22 nm); ferritin (6.1 nm); and thyroglobin (8.5 nm)
Depolymerization assay and sedimentation analysis of stabilized microtubules
Sedimentation analysis of the depolymerization of GMPCPP-stabilized microtubules was performed as described previously (Desai et al., 1999a). 2.5 μM of prepolymerized GMPCPP microtubules were added to the reaction mix that was supplemented with 125 nM MCAK or control buffer in 1× BRB80 (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) containing 125 mM KCl and 1.5 mM Mg-ATP. 500 nM XMAP215 and increasing amounts of TACC3 or control buffer were added to reactions. Reactions were sedimentated after 38 min at 30°C through a glycerol cushion. Equivalent amounts of the supernatant and pellets were analyzed by 4–12% SDS-PAGE followed by Coomassie brilliant blue staining. For sedimentation assays to monitor the affinity of XMAP215 to microtubules, 2.5 μm of prepolymerized GMPCPP microtubules were incubated with 500 nM XMAP215 or control buffer in 1× BRB80 containing 125 nM KCl in the presence of TACC3 or control buffer. In each condition, we confirmed that all proteins were in the supernatant after sedimentation without GMPCPP microtubules.
Preparation of Xenopus egg extracts and immunodepletion
Xenopus egg extracts were prepared as described previously (Murray, 1991; Desai et al., 1999b). Interphase extracts were prepared by the addition of calcium to cytostatic factor extracts. Cycloheximide was added to 100 μg/ml to avoid translation of cyclin B messenger in extracts and to arrest them in interphase. To generate mitotic extracts, a purified, nondegradable cyclin B fragment (cyclin B Δ90; a gift from H. Funabiki, The Rockefeller University, New York, NY; Glotzer et al., 1991) was added to 25 μg/ml to interphase extracts (Vorlaufer and Peters, 1998; Funabiki and Murray, 2000). Immunodepletion in Xenopus egg extracts was performed as described previously (Funabiki and Murray, 2000). All of the immunodepletion experiments were performed in the presence of 100 μg/ml cycloheximde in extracts to block translation, as maskin has been reported to be a negative regulator of translational machinery during oocyte maturation (Stebbins-Boaz et al., 1999).
Antibodies and immunoblotting
Phosphospecific antibodies were raised and purified against phosphopeptides, including phosphoserine in the consensus target sites of Aurora A phosphorylation in Xenopus TACC3 (P-Ser33; QTTGRPphospho-SILRPSQ and P-Ser620; CNSFKEphopho-SVLRKQ and P-Ser626; VLRKQphopho-SLYLKFC). Anti–Xenopus TACC3 antibodies were raised and purified against the GST fusion protein that contained an NH2-terminal fragment of TACC3 (aa 7–208). Anti–human TACC3 antibodies were previously described (Gergely et al., 2000). Anti–Aurora A and -XMAP215 antibodies were raised and purified against COOH-terminal peptides (Aurora A, CKNSQLKKKDEPLPGAQ; and XMAP215, CNIDDLKKRLERIKSSRK) as described previously (Field et al., 1998). Anti–α-tubulin antibodies (DM1A) were purchased from Sigma-Aldrich. Anti–NH2-terminal fragments of XlMCAK (XKCM1-NT) antibodies were gifts from C. Walczak (Indiana University, Bloomington, IN; Walczak et al., 1996). Immunoblotting was performed by using ECL (GE Healthcare). For quantification of phospho-TACC3 signal, the signal intensities on the blot were measured by image software (Scion Corp.).
Microtubule assembly and localization assays using Xenopus egg extracts
Recycled and fluorescently labeled tubulins (rhodamine- or Cy3-labeled) were prepared from porcine instead of bovine brain as previously described (Hyman et al., 1991; Ashford et al., 1998). Centrosomes were purified from human KE37 cells as described previously (Moudjou and Bornens, 1998). Xenopus interphase egg extracts that were supplemented with centrosomes (5 × 106/ml final) and fluorescently labeled tubulin (0.5–1 μM final) to visualize microtubules were incubated with or without cyclin B Δ90 for 20–30 min at RT. A part of the reactions was saved in SDS-PAGE sample buffer for immunoblotting analysis. For microtubule assembly assay, the extracts were fixed with extract fix (60% glycerol, 1× Marc's modified Ringer's solution, 1 μg/ml Hoechst 33342, and 10% formaldehyde; Desai et al., 1999b) and squashed onto coverslips. For the immunofluorescence of TACC3 on centrosomes, extracts were treated with 20 μM nocodazole, fixed with fixation buffer (10% glycerol, 1× BRB80, 0.1% Triton X-100, and 5% formaldehyde), and spun down through glycerol cusion (30% glycerol in 1× BRB80) onto coverslips. The samples on coverslips were postfixed in −20°C methanol and processed for immunofluorescence as described previously (Desai et al., 1999b). The images were collected using a wide-field microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) that was equipped with plan-Apochromat 100× NA 1.40 objective lens and a digital camera (model C4742-95; Hamamatsu). For quantification of fluorescence intensity, images were acquired by using a plan-Neofluor 16× NA 0.50 for the fluorescence signal of tubulin and a plan-Apochromat 100× NA 1.40 for the signal of TACC3 staining. The integrated pixel intensities within the circle (21.079 μm for tubulin; 1.079 μm for centrosomal TACC3) around centrosomes were calculated by using Metamorph software (Universal Imaging Corp.). All of the measured values were corrected by the subtraction of background signal in the same field.
In vitro protein kinase assay and mass spectometric analysis
0.4 mg/ml of bacterially expressed TACC3 was incubated with or without 0.02 mg/ml Aurora A in kinase buffer (25 mM Hepes, pH 7.7, 100 mM KCl, 5 mM MgCl2, 50 mM sucrose, and 0.1% β-mercaptoethanol) in the presence of 0.1 mM ATP containing γ-[32P] ATP for 20 min at 30°C. The reactions were stopped by the addition of SDS-PAGE sample buffer, and the samples were analyzed by SDS-PAGE. The incorporation of 32P into the TACC3 in the gel was detected by autoradiography by using a phosphoimager (model BAS-1800II; Fujifilm). Mass spectometric analysis for identification of in vitro Aurora A phosphorylation sites of TACC3 was performed as described previously (Kraft et al., 2003).
Cell culture and RNA interference
HeLa cells were grown in DME containing 10% serum supplemented with nonessential amino acids (GIBCO BRL) and were grown using standard procedures. For RNA interference, HeLa cells were seeded onto glass coverslips 16 h before transfection. Cells that were grown to ∼40% confluency were transfected for 6 h using the OligofectAMINE reagent (Invitrogen) and to a final concentration of 20 nM siRNA against either TACC3 (purchased from Ambion; Gergely et al., 2003) or a control siRNA (control siRNA#1; Ambion) according to the manufacturer's recommendations. After transfection, cells were incubated in fresh media for 72 h before analysis by immunofluorescence.
HeLa S3 cells were grown in suspension in MEM that was modified for suspension cultures (Biochrom) containing 10% serum supplemented with nonessential amino acids (Biochrom) by using standard procedures. Cells were arrested in mitosis by the addition of 90 ng/ml nocodazole (Sigma-Aldrich) 20 h before harvesting and were shown to be >95% mitotic as judged by DAPI staining.
Immunofluorescence and microscopy for culture cells
Transfected HeLa cells that were grown on coverslips were fixed and permeabilized in methanol at −20°C for 8 min. Cells were washed with PBS and incubated 10 min in PBS containing 0.2% fish skin gelatin (PBS-FSG; Sigma-Aldrich) to prevent nonspecific binding of antibodies. Cells were incubated 20 min with primary antibody. Antibodies that were used were anti–α-tubulin (DM1A; Sigma-Aldrich), anti-TACC3 (Gergely et al., 2000), and antiphospho-TACC3 (anti–P-Ser626 in this study) diluted to 1 μg ml−1 in PBS-FSG. After washing in PBS, cells were incubated for 20 min at 37°C in 1 μg ml−1 (in PBS-FSG) of donkey secondary antibodies that were labeled with either Texas red, FITC, or Cy5 (Jackson ImmunoResearch Laboratories) and were washed in PBS before mounting in the presence of 1 μg ml–1 DAPI to visualize chromatin. Three-dimensional datasets were acquired on an imaging system (DeltaVision; Applied Precision) that was equipped with a microscope (model IX70; Olympus), a camera (CoolSNAP; Roper Scientific), and a 100× NA 1.4 plan-Apochromat objective. Images were computationally deconvolved by using the SoftWork software package (Applied Precision) and were shown as two-dimensional projections.
Online supplemental material
Fig. S1 shows spindles assembled in mock-depleted versus TACC3-depleted Xenopus egg extracts. Fig. S2 shows in vitro phosphorylation sites in TACC3/maskin as determined by mass spectrometry. Fig. S3 shows TACC3 phosphorylation in Aurora A–depleted extracts. Fig. S4 shows the characterization of phosphorylation mutant TACC3 in Xenopus egg extracts and in vitro. Fig. S5 shows centrosomal localization of XMAP215-GFP in Xenopus egg extracts. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200503023/DC1.
Acknowledgments
We are grateful to Jason Swedlow and Joan Ruderman (Harvard Medical School, Boston, MA) for cDNA of Aurora A/Eg2; Joel Richter for cDNA of Xenopus TACC3/maskin; Hironori Funabiki for purified cyclin B Δ90; Claire Walczak for anti-XKCM1/MCAK antibodies; Heino Andreas (MPI-CBG) for help with the frog colony; Andrej Pozniakovski (MPI-CBG) for plasmid construction of the alanine-mutated TACC3; and Arshad Desai (University of California, San Diego, San Diego, CA), Hironori Funabiki, and Martin Srayko (MPI-CBG) for stimulating discussion and comments on the manuscript.
K. Kinoshita was supported by a research fellowship from the Uehara Memorial Foundation and a grant from the Deutsche Forschungsgemeinschaft priority program Molecular Motors. L. Pelletier was supported by a postdoctoral fellowship from the Human Frontier Science Program.
Abbreviations used in this paper: D-TACC, Drosophila melanogaster TACC; FSG, fish skin gelatin; MCAK, mitotic centromere-associated kinesin; TACC, transforming acidic coiled coil; TOG, tumor overexpressed gene; WT, wild type.
References
- Ashford, A.J., S.S. Andersen, and A.A. Hyman. 1998. Preparation of tubulin from bovine brain. Cell Biology: A Laboratory Handbook. Vol. 2. 2nd ed. J.E. Celis, editor. Academic Press/UK, London. 205–212.
- Bellanger, J.M., and P. Gönczy. 2003. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13:1488–1498. [DOI] [PubMed] [Google Scholar]
- Belmont, L.D., A.A. Hyman, K.E. Sawin, and T.J. Mitchison. 1990. Real-time visualization of cell cycle-dependent changes in microtubule dynamics in cytoplasmic extracts. Cell. 62:579–589. [DOI] [PubMed] [Google Scholar]
- Bischoff, J.R., L. Anderson, Y. Zhu, K. Mossie, L. Ng, B. Souza, B. Schryver, P. Flanagan, F. Clairvoyant, C. Ginther, et al. 1998. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17:3052–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Cullen, C.F., and H. Ohkura. 2001. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3:637–642. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. a. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. [DOI] [PubMed] [Google Scholar]
- Desai, A., A. Murray, T.J. Mitchison, and C.E. Walczak. 1999. b. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. [DOI] [PubMed] [Google Scholar]
- Field, C.M., K. Oegema, Y. Zheng, T.J. Mitchison, and C.E. Walczak. 1998. Purification of cytoskeletal proteins using peptide antibodies. Methods Enzymol. 298:525–541. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., and A.W. Murray. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 102:411–424. [DOI] [PubMed] [Google Scholar]
- Gard, D.L., B.E. Becker, and S. Josh Romney. 2004. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 239:179–272. [DOI] [PubMed] [Google Scholar]
- Gergely, F., C. Karlsson, I. Still, J. Cowell, J. Kilmartin, and J.W. Raff. 2000. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc. Natl. Acad. Sci. USA. 97:14352–14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely, F., V.M. Draviam, and J.W. Raff. 2003. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet, R., D. McLean, S. Descamps, M.J. Lee, J.W. Raff, C. Prigent, and D.M. Glover. 2002. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J. Cell Biol. 156:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M., A.W. Murray, and M.W. Kirschner. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138. [DOI] [PubMed] [Google Scholar]
- Gräf, R., C. Daunderer, and I. Schulz. 2004. Molecular and functional analysis of the dictyostelium centrosome. Int. Rev. Cytol. 241:155–202. [DOI] [PubMed] [Google Scholar]
- Hannak, E., M. Kirkham, A.A. Hyman, and K. Oegema. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak, E., K. Oegema, M. Kirkham, P. Gönczy, B. Habermann, and A.A. Hyman. 2002. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is γ-tubulin dependent. J. Cell Biol. 157:591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald, R., R. Tournebize, T. Blank, R. Sandaltzopoulos, P. Becker, A. Hyman, and E. Karsenti. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425. [DOI] [PubMed] [Google Scholar]
- Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell. 114:585–598. [DOI] [PubMed] [Google Scholar]
- Holmfeldt, P., S. Stenmark, and M. Gullberg. 2004. Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J. 23:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J., and A.A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758. [DOI] [PubMed] [Google Scholar]
- Hyman, A., D. Drechsel, D. Kellogg, S. Salser, K. Sawin, P. Steffen, L. Wordeman, and T. Mitchison. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485. [DOI] [PubMed] [Google Scholar]
- Job, D., O. Valiron, and B. Oakley. 2003. Microtubule nucleation. Curr. Opin. Cell Biol. 15:111–117. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., and I. Vernos. 2001. The mitotic spindle: a self-made machine. Science. 294:543–547. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, K., I. Arnal, A. Desai, D.N. Drechsel, and A.A. Hyman. 2001. Reconstitution of physiological microtubule dynamics using purified components. Science. 294:1340–1343. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., B. Habermann, and A.A. Hyman. 2002. XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12:267–273. [DOI] [PubMed] [Google Scholar]
- Kline-Smith, S.L., and C.E. Walczak. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 15:317–327. [DOI] [PubMed] [Google Scholar]
- Kraft, C., F. Herzog, C. Gieffers, K. Mechtler, A. Hagting, J. Pines, and J.M. Peters. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 22:6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bot, N., M.C. Tsai, R.K. Andrews, and J. Ahringer. 2003. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13:1499–1505. [DOI] [PubMed] [Google Scholar]
- Lee, M.J., F. Gergely, K. Jeffers, S.Y. Peak-Chew, and J.W. Raff. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649. [DOI] [PubMed] [Google Scholar]
- Moudjou, M., and M. Bornens. 1998. Method of centrosome isolation from cultured animal cells. Cell Biology: A Laboratory Handbook. Vol. 2. 2nd ed. J.E. Celis, editor. Academic Press/UK, London. 111–119.
- Murray, A.W. 1991. Cell cycle extracts. Methods Cell Biol. 36:581–605. [PubMed] [Google Scholar]
- Noetzel, T.L., D.N. Drechsel, A.A. Hyman, and K. Kinoshita. 2005. A comparison of the ability of XMAP215 and tau to inhibit the microtubule destabilizing activity of XKCM1. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A.A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascreau, G., J.G. Delcros, J.Y. Cremet, C. Prigent, and Y. Arlot-Bonnemains. 2005. Phosphorylation of maskin by Aurora-A participates to the control of sequential protein synthesis during Xenopus laevis oocyte maturation. J. Biol. Chem. 280:13415–13423. [DOI] [PubMed] [Google Scholar]
- Piehl, M., U.S. Tulu, P. Wadsworth, and L. Cassimeris. 2004. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA. 101:1584–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, J.W. 2002. Centrosomes and cancer: lessons from a TACC. Trends Cell Biol. 12:222–225. [DOI] [PubMed] [Google Scholar]
- Sampath, S.C., R. Ohi, O. Leismann, A. Salic, A. Pozniakovski, and H. Funabiki. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202. [DOI] [PubMed] [Google Scholar]
- Sato, M., L. Vardy, M. Angel Garcia, N. Koonrugsa, and T. Toda. 2004. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell. 15:1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko, M., S. Quintin, A. Schwager, and A.A. Hyman. 2003. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr. Biol. 13:1506–1511. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz, B., Q. Cao, C.H. de Moor, R. Mendez, and J.D. Richter. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell. 4:1017–1027. [DOI] [PubMed] [Google Scholar]
- Still, I.H., A.K. Vettaikkorumakankauv, A. DiMatteo, and P. Liang. 2004. Structure-function evolution of the transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC Evol. Biol. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, A.C., M.H. Lin, L.J. Su, Y.R. Hong, T.S. Cheng, Y.C. Lee, W.J. Lin, I.H. Still, and C.Y. Huang. 2004. Identification of the substrates and interaction proteins of aurora kinases from a protein-protein interaction model. Mol. Cell. Proteomics. 3:93–104. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., A. Popov, K. Kinoshita, A.J. Ashford, S. Rybina, A. Pozniakovsky, T.U. Mayer, C.E. Walczak, E. Karsenti, and A.A. Hyman. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2:13–19. [DOI] [PubMed] [Google Scholar]
- Usui, T., H. Maekawa, G. Pereira, and E. Schiebel. 2003. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 22:4779–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlaufer, E., and J.M. Peters. 1998. Regulation of the cyclin B degradation system by an inhibitor of mitotic proteolysis. Mol. Biol. Cell. 9:1817–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., T.J. Mitchison, and A. Desai. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47. [DOI] [PubMed] [Google Scholar]
- Walker, R.A., E.T. O'Brien, N.K. Pryer, M.F. Soboeiro, W.A. Voter, H.P. Erickson, and E.D. Salmon. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]