Abstract
Mitotic cell division involves the equal segregation of all chromosomes during anaphase. The presence of ribosomal DNA (rDNA) repeats on the right arm of chromosome XII makes it the longest in the budding yeast genome. Previously, we identified a stage during yeast anaphase when rDNA is stretched across the mother and daughter cells. Here, we show that resolution of sister rDNAs is achieved by unzipping of the locus from its centromere-proximal to centromere-distal regions. We then demonstrate that during this stretched stage sister rDNA arrays are neither compacted nor segregated despite being largely resolved from each other. Surprisingly, we find that rDNA segregation after this period no longer requires spindles but instead involves Cdc14-dependent rDNA axial compaction. These results demonstrate that chromosome resolution is not simply a consequence of compacting chromosome arms and that overall rDNA compaction is necessary to mediate the segregation of the long arm of chromosome XII.
Introduction
The processes by which cells ensure that all chromosomes are accurately split and distributed during cell division are of interest because they underlie normal development and inheritance of genetic traits. During S phase, replicated sister chromatids are first linked together or cohesed (Nasmyth, 2001). Cohesion is required to promote the bipolar attachment of chromosomes to the mitotic spindle, necessary to ensure equal partitioning at anaphase (Tanaka et al., 2000). However, before sister chromatid separation occurs, cohesed chromosomes undergo a dramatic structural reorganization, whereby the chromosome becomes organized into a seemingly packaged state (Swedlow and Hirano, 2003). This reorganization occurs during mitosis and is referred to as chromosome condensation. The functional significance of condensation is thought to be twofold: (1) to mediate the resolution of sister chromatids from each other, and (2) to ensure that the linear length of chromosome arms is sufficiently reduced (axial compaction) as to avoid severing of chromosomes by cytokinesis (Holm, 1994; Koshland and Strunnikov, 1996; Swedlow and Hirano, 2003; Hirano, 2000). In S. cerevisiae, mitotic chromosome condensation is largely dependent on a multi-subunit complex named condensin (Strunnikov et al., 1995; Freeman et al., 2000; Ouspenski et al., 2000; Lavoie et al., 2000, 2004; Bhalla et al., 2002). Yeast condensin becomes enriched on the repetitive ribosomal DNA (rDNA) array during mitosis (Freeman et al., 2000; Bhalla et al., 2002), suggesting that segregation of this locus requires condensin function (Freeman et al., 2000).
Chromosome XII is the largest chromosome, with 1 Mb plus the rDNA array (RDN1), which can vary in size, from 100 to 200 U of a 9.1-kb repeat (Petes, 1979), thus, reaching a total chromosome size of 2–3 Mb (Mortimer and Johnston, 1986). RDN1 is located on the right arm ∼300 kb away from the centromere and 600 kb from the right telomere. The rDNA array not only organizes the nucleolus, the center for ribosomal RNA synthesis (Shaw and Jordan, 1995), but it also holds a key regulator of mitotic exit named the Cdc14 protein phosphatase (Garcia and Pillus, 1999; Shou et al., 1999; Visintin et al., 1999). Cdc14p is kept inactive in the nucleolus until mid-anaphase when it is released throughout the cell to reach its targets (Pereira et al., 2002; Stegmeier et al., 2002). Cdc14p participates in the segregation of rDNA (Granot and Snyder, 1991; Buonomo et al., 2003; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004) by targeting the condensin complex to rDNA and hence promoting both resolution and compaction of rDNA (Guacci et al., 1994; D'Amours et al., 2004; Sullivan et al., 2004). Besides its role in rDNA segregation, Cdc14 is also required to promote spindle stability during anaphase (Pereira and Schiebel, 2003).
An open question in the rDNA segregation process is how sister rDNA chromatids are partitioned during anaphase and whether rDNA compaction precedes resolution or whether, on the contrary, sister rDNAs resolve before overall compaction takes place. In the present report, we demonstrate that rDNA inheritance requires two distinct steps separated in time. First, at the anaphase onset, unzipping of sister rDNAs generates an intermediate stage, characterized by largely resolved sister rDNA chromatids across the mother and daughter cells. Second, the linear length of rDNA is reduced, by Cdc14-dependent rDNA axial compaction, to mediate segregation of sister rDNAs to opposite poles in late anaphase. Surprisingly, the second step does not require spindles, suggesting that the main biological function of rDNA compaction is to mediate segregation of this long chromosome arm. This two-stepped mode of inheritance allows us to propose that rDNA resolution and compaction are two functionally distinct processes separated in time, with the resolution of sister rDNA arrays preceding their overall compaction.
Results
Defining the “rDNA bridge” as a stage during anaphase after the rDNA metaphase loop
The morphology of the rDNA array of budding yeast throughout the cell cycle is highly dynamic. In a recent study, it was shown that, in cells transiting from late mitosis to early mitosis of the next cell cycle, the rDNA undergoes several morphological changes that culminate in a loop-like structure (Lavoie et al., 2004). Furthermore, it is also known that in cells arrested in late anaphase, the rDNA is highly compacted (Guacci et al., 1994). Therefore, the rDNA must change from an extended loop to a compacted state as it separates and transits through mitosis. To visualize specific structural stages of the rDNA array during mitotic division we used a live-cell approach using GFP/CFP fusions of the rDNA-specific proteins Net1 and Fob1 as in vivo markers for rDNA. First, we validated these markers as bona fide rDNA labels by after the localization of Net1p/Fob1p-GFP/CFP fusions during an entire cell cycle in cells where chromosomal lacO or tetO tags (Straight et al., 1996; Michaelis et al., 1997) had been also inserted in regions either flanking or within the rDNA. This analysis allowed us to test directly whether the localization of these markers was restricted to rDNA arrays during different stages of the cell cycle. We found that both Net1p and Fob1p were always restricted to rDNA regions and localized between the chromosomal tag insertions flanking the locus (unpublished data; see Fig. 2 and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200408087/DC1). Furthermore, cells simultaneously expressing Net1p-CFP and Fob1p-GFP showed colocalization of the two proteins at all stages of the cell cycle (Fig. 1 B, not depicted).
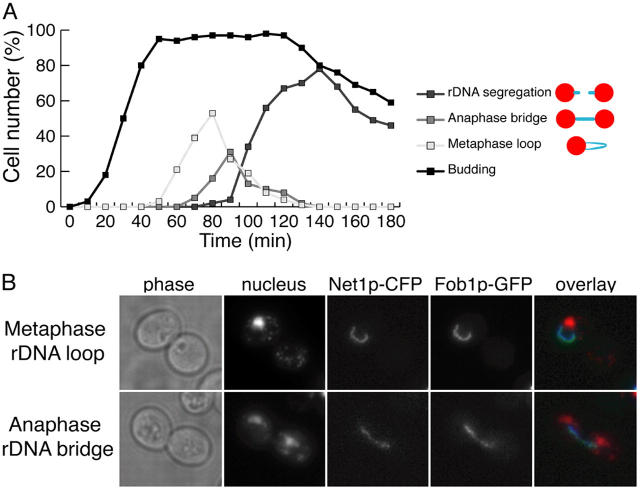
Figure 1.
A defined “anaphase rDNA bridge” stage occurs after the rDNA loop in synchronous cultures. (A) Strain CCG1580 (Mata FOB1-GFP NET1-CFP) was arrested in G1 using α-factor and released into fresh medium at 30°C. Time-points collected every 10 min were analyzed for the organization of rDNA arrays. Categories scored are shown in the figure legend and include rDNA metaphase loops, “anaphase bridges” and segregated rDNA. (B) Strain CCG1580 (Mata FOB1-GFP NET1-CFP) was arrested in metaphase using nocodazole and released into fresh medium at 30°C. Micrographs representative of metaphase rDNA loops and anaphase rDNA bridges are shown.
In previous reports the rDNA array has been shown to form a distinct loop during metaphase (Guacci et al., 1994; Lavoie et al., 2004). We were able to detect and quantify the occurrence of these looped arrays in synchronous cultures as the cells transited through metaphase (Fig. 1 A), further validating the use of our cytological approach. We have previously reported an intermediate stage during rDNA segregation where the rDNA becomes elongated across the bud-neck forming a bridge between the separated nuclear masses (Fig. 1 B; Torres-Rosell et al., 2004). We refer to this stage as the “rDNA bridge”. We quantified the occurrence of “rDNA bridges” in relation to the appearance of metaphase loops in synchronous populations and confirmed that it represents a defined period during yeast anaphase, immediately after metaphase loops (Fig. 1, A and B).
Sister-chromatid rDNAs are resolved at centromere-proximal but not centromere-distal regions in rDNA bridges
To examine how sister chromatids are organized in the rDNA-containing arm of chromosome XII, we visualized chromosomal tags inserted at various positions along this arm using the tetO/tetR-YFP system (Michaelis et al., 1997) in cells that also expressed Net1p-CFP. The tag insertions chosen were: ∼45 kb right of CEN12 (tetO:194), the centromere-proximal edge of the rDNA array (tetO:450), the centromere-distal edge of the rDNA array (tetO:487), or 20 kb away from the right telomere of chromosome XII (tetO:1061). First, we looked at cells undergoing metaphase. The analysis revealed that DNA regions flanking the rDNA locus (tetO:450 and tetO:487) were always located at the base of the loop with the rest of the nuclear mass (Fig. S1). Chromosome tags inserted close to the centromere (tetO:194) or telomere (tetO:1061) were also located within the main nuclear mass. In contrast, chromosomal tags inserted in the middle of the rDNA array (RDN1:lacO) appeared clearly separated from the nuclear mass within the rDNA loop (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200408087/DC1). RDN1:lacO tags appeared as a single GFP dot within the loop which indicates that rDNA sister chromatids are fully cohesed at this stage. These results demonstrate that rDNA metaphase loops are restricted to rDNA sequences and sisters are fully cohesed throughout the locus.
We then extended our analysis to cells undergoing the “rDNA bridge” period, when rDNA arrays are stretched across the bud-neck (Fig. 1 B and Fig. 2). Centromere tags (tetO:194) and tags in the proximal flank of rDNA (tetO:450) were fully segregated in these cells (Fig. 2 A, tetO:194 and tetO:450 columns). Interestingly, tags in the distal flank of rDNA (tetO:487) were not separated and localized to the middle of the elongated rDNA (Fig. 2 A, tetO:487 column). Telomere tags (tetO:1061) were last to separate and always trailed sister rDNA arrays to the poles (Fig. 2 A, tetO:1061 column). Therefore, the 600-kb region between the distal flank of rDNA and the right telomere follows behind the rDNA arrays during segregation. Furthermore, we asked to what extent rDNA unzips in the bridge stage. To do so, we measured the average distance between separated sister tags in the centromere-proximal edge of the rDNA (tetO:450) during this stage and found the distance to be 5.2 μm (SD 1.4 μm; Fig. 2 B). This separation indicates that a large proportion of rDNA repeats within the array is resolved when cells reach the bridge. This is also in agreement with the observation that the distal edge of the rDNA lays around the middle of the bridge (Fig. 2 A, tetO:487). However, the fact that this distal tag appears as a single focus implies that not all the rDNA repeats in the array are resolved. We have determined the percentage of sister resolution in the rDNA array at the bridge stage by making use of a strain that bears tags in the middle of the rDNA (lacO:RDN1) (Torres-Rosell et al., 2004). The average distance separation between lacO:RDN1 tags in the bridge stage was 1.8 μm (SD 1.5 μm), thus, ∼77% of the array is already resolved at the bridge stage (Fig. 2 B, 2.6 μm from a total chromatid length of 3.4 μm). These results demonstrate that, during anaphase, sister rDNAs unzip from centromere-proximal to centromere-distal regions before they segregate to opposite poles. Furthermore, the directionality of the unzipping process demonstrates that the partitioning of sister telomeres is delayed compared with the rest of the chromosome arm.
Figure 2.
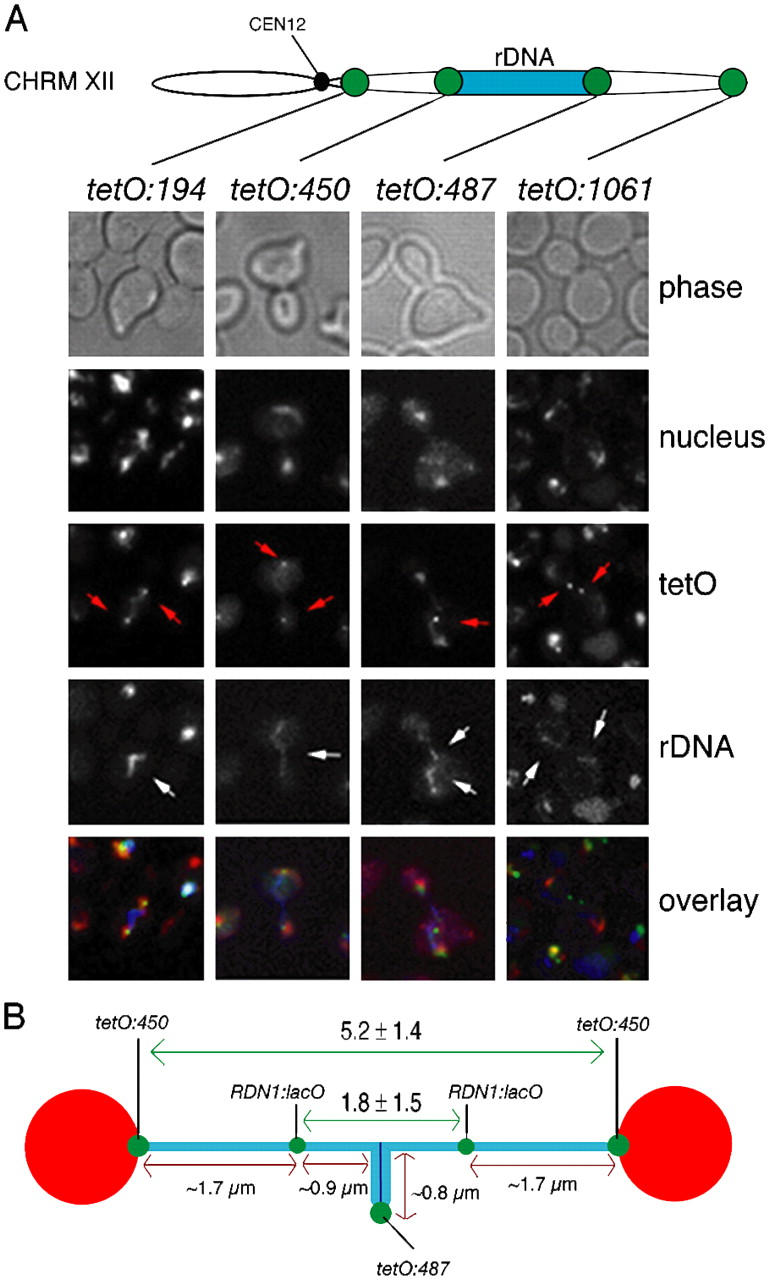
Sister rDNA arrays unzip from centromere-proximal to centromere-distal regions during bridge the stage before segregation to opposite poles. (A) Strains CCG1326 (Mata TetR-YFP tetO:194Kb-ChrXII NET1-CFP), CCG1327 (Mata TetR-YFP tetO:450Kb-ChrXII NET1-CFP), CCG1328 (Mata TetR-YFP tetO:487Kb-ChrXII NET1-CFP), and CCG1329 (Mata TetR-YFP tetO:1061Kb-ChrXII NET1-CFP), were released from a G1 block and imaged as cells underwent segregation using nuclear separation as a reference for anaphase entry. All the cells analyzed during the bridge period showed the pattern displayed in the representative cells of each category. Chromosome tags (red arrows) and rDNA arrays (white arrows) are indicated. Sister rDNA arrays are separated at their centromeric proximal flanks yet are still connected through their telomeric flanks during the anaphase bridge period. Note that the tetO:1061 panel shows a cell already transiting from the bridge to fully segregated rDNA in order to show that sister chromosome XII right telomeres trail rDNA during the segregation. (B) Distances between tags at the rDNA bridge stage were measured for strains CCG1327 (Mata TetR-YFP tetO:450Kb-ChrXII NET1-CFP) and CCG1191 (Mata LacI-GFP RDN1:lacO TUB4-CFP NET1-CFP) after a G1 block release. The position of the lacO in the middle of the rDNA (Torres-Rosell et al., 2004) allowed to estimate the percentage of rDNA array resolved at this stage. We found it to be ∼77% (1.7 μm of resolved rDNA chromatids/3.4 μm of total rDNA chromatid length).
Compaction of rDNA occurs after the “anaphase rDNA bridge”
Chromosome condensation is thought to serve two essential functions: (1) the resolution of sisters into two separable units, and (2) the shortening/compacting of the chromosome arms along their linear axis, to prevent bisection by cytokinesis. Different studies have shown that by late anaphase segregated rDNAs are hyper compacted (Guacci et al., 1994; Lavoie et al., 2004; Sullivan et al., 2004). We compared the compaction status of rDNA arrays between cells at metaphase (loops), anaphase (rDNA bridges), or with fully segregated rDNAs by looking at Net1p-CFP/Fob1p-GFP. We measured the axial length of rDNA chromatids, and found the average axial length of rDNA to be 3.72 μm (SD 1.07 μm) in metaphase loops, 5.9 μm (SD 1.45 μm) in anaphase bridges, and 1.17 μm (SD 0.3 μm) in segregated chromatids (Fig. 3 A). The value of the anaphase bridge in this experiment, 5.9 ± 1.5 μm, was comparable to that seen for tetO:450, 5.2 ± 1.4 μm (Fig. 2 B). Since the measurements of anaphase bridges represent the length of two sister arrays (as they are largely unzipped; Fig. 2), the average axial length of each resolved rDNA chromatid is ∼2.95 μm. Taking into account that ∼23% of the array is not seen as part of the bridge because they are not yet resolved (Fig. 2 B), the actual length of each rDNA chromatid at the anaphase bridge is ∼3.85 μm. Therefore, the transition from the metaphase loop to the rDNA bridge does not involve a significant shortening in the axial length of the rDNA (from ∼3.72 to ∼3.85 μm). In contrast, the transition from rDNA bridge to fully segregated rDNA shortens the axial length of each sister rDNA chromatid by a factor of 3.3 (from ∼3.85 to ∼1.17 μm).
Figure 3.
Axial compaction of rDNA arrays occurs after the anaphase bridge period and is not a consequence of nuclear envelop fission. (A) The average axial length of the rDNA arrays was measured in metaphase loops, anaphase bridges, or segregated rDNA arrays in the CCG1580 strain. Note that the average axial length of rDNA loops represents cohesed chromatids, the average length of anaphase rDNA bridges represents two chromatids that are separated at their centromeric proximal flanks but still connected through their telomeric flanks; and the average length of segregated rDNA chromatids represents that of each chromatid (i.e., the value of the anaphase rDNA bridge should be divided by a factor of two for direct comparison to the value of metaphase loops and segregated chromatids, which represent a single chromatid length). Overall, rDNA compaction takes place after the anaphase bridge period. (B) Representative micrograph of a cell in telophase from strain CCG1582 (Mata NUP49-GFP NET1-CFP) which has segregated both bulk DNA and rDNA. Note that the nuclear envelop still connects the divided nuclear masses.
Axial length shortening of rDNA is likely to represent compaction of the locus; however, it is possible that the shortening simply reflects the gathering of the rDNA by the nuclear envelop as the nuclear membrane changes from an extended shape in anaphase to a small post-anaphase sphere. To exclude this possibility, we investigated whether rDNA segregation and axial shortening precedes nuclear envelop fission. The nucleoporin Nup49p has been shown to localize exclusively to the nuclear envelope (Heun et al., 2001). Anaphase cells simultaneously expressing Nup49p-GFP and Net1p-CFP showed that axial shortening of rDNA (and segregation) precedes nuclear envelope fission (Fig. 3 B). This is in agreement with previous reports showing that nuclear envelop fission is linked to cytokinesis and not chromosome segregation (Lippincott and Li, 2000). Therefore, the axial shortening of rDNA represents linear compaction. Our data show that, during the bridge stage, rDNA arrays are not yet compacted, and that compaction of this locus occurs only when cells fully separate sister rDNAs and transit towards telophase. The implication of these findings is that resolution and compaction of rDNA are temporally uncoupled processes with the resolution of rDNA sister arrays preceding their overall compaction. This is in contrast to the view that resolution is simply the consequence of compacting chromosome arms.
Pausing at and progressing out of the rDNA bridge stage
During our experiments with synchronous cultures, we noticed that as cells enter anaphase there is a rapid transition from loop to bridge followed by a several minute-long pause at the rDNA bridge (see Fig. 6, not depicted). We reasoned that this might suggest that progression out of bridges could be under specific cell cycle regulation. Therefore, we searched for mutants that would confer arrest specifically at the bridge stage.
Recently it has been shown that inactivation of FEAR network, Cdc14p or condensin causes nucleolar segregation defects (Freeman et al., 2000; Sullivan and Uhlmann, 2003; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004). We evaluated which mutants or combinations provided us with the closest arrest to the bridge stage observed in wild-type cells, with the rDNA stretched across the mother and daughter cells (Figs. 1 and 2). We found that FEAR and MEN double mutants (cdc15-2 spo12Δ and cdc15-2 slk19Δ), despite their known impairments in full segregation of rDNA to the poles (D'Amours et al., 2004; Torres-Rosell et al., 2004), did not arrest with stretched rDNA. Instead, most of the cells with misegregated rDNAs showed separated rDNA arrays in the same cell body (Fig. 4 A). Polo kinase, Cdc5p, is the only common member of both FEAR and MEN networks (Stegmeier et al., 2002). Inactivation of polo through the cdc5-1 allele produced a severe impairment in both nuclear and nucleolar segregation (Fig. 4 A). A double mutant of condensin, smc2-8, and cdc15-2 also arrested before the anaphase bridge stage with severe impairment in the segregation of the main nuclear mass (Fig. 4 A). Cdc14p activity in anaphase triggers numerous events including segregation of rDNA (Granot and Snyder, 1991), targeting of Ipl1-Sli15 complex to spindle midzones (Pereira and Schiebel, 2003) and timely coordination of mitotic exit. We used the cdc14-1 mutant allele to test whether a cdc14-1 block resembles the bridge stage of wild-type cells since this allele has been reported to arrest with stretched nucleoli between mother and daughter cells (Granot and Snyder, 1991). We found that nucleoli did not segregate in >80% of cells, whereas nuclear masses appear fully separated in a similar percentage (Fig. 4 A). cdc14-1 blocked cells with unsegregated nucleoli could be divided into two categories, representing ∼50% of the cells each. These were: (a) cells with rDNA stretched across the mother and daughter bodies, similar to the bridge stage in wild-type cells (Fig. 4 A), and (b) cells with a single rDNA signal, in either mother or daughter cell body, connecting two separated nuclear masses in the same cell body (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200408087/DC1). In these cells, although the nucleolus is not stretched across the neck, it still seems to form a bridge between the DAPI-stained bulk DNAs.
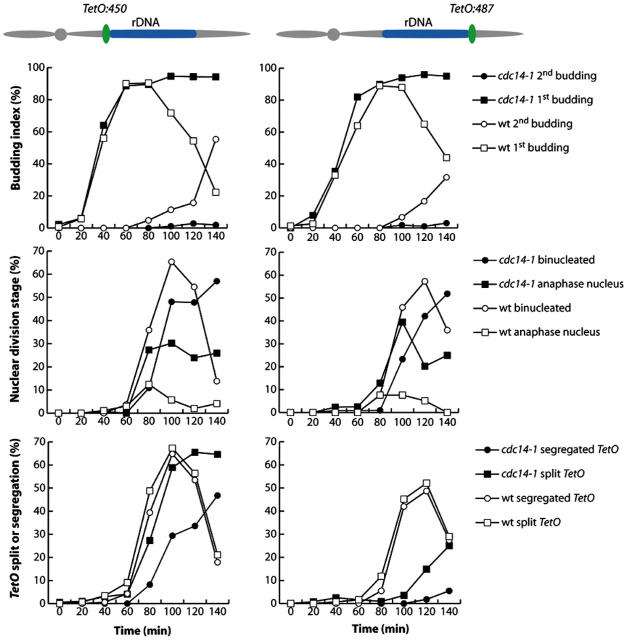
Figure 4.

cdc14-1 mutants arrest at the anaphase bridge stage. (A) Strains CCG938 (cdc15-2 NET1-GFP), CCG975 (cdc5-1 NET1-GFP), CCG1168 (cdc15-2 NET1-GFP slk19Δ), CCG1222 (cdc15-2 NET1-GFP spo12Δ), CCG1248 (cdc15-2 NET1-GFP smc2-8), and CCG1622 (cdc14-1 NET1-GFP) were arrested in G1 using α-factor and released into fresh medium at 37°C. Cells were collected after 150 min and scored for the following categories (representative micrographs and diagrams are shown): (1) undivided nuclei and rDNA (majority of the scored nucleus were stretched across the neck); (2) anaphase rDNA bridge; (3) fully segregated nuclei with unique rDNA in one cell body; (4) fully segregated nuclei with separated but not fully segregated rDNA (majority of the scored cells had one rDNA sister stretched across the neck); and (5) fully segregated nuclei and rDNA. Note that cdc14-1 cells arrested with segregated nuclear masses but unseparated rDNA arrays, which usually extended between mother and daughter cells. (B) Strains CCG1402 (Mata TetR-YFP tetO:487Kb-ChrXII NET1-CFP cdc14-1), CCG1605 (Mata TetR-YFP tetO:194Kb-ChrXII NET1-CFP cdc14-1), CCG1607 (Mata TetR-YFP tetO:450Kb-ChrXII NET1-CFP cdc14-1), and CCG1609 (Mata TetR-YFP tetO:1061Kb-ChrXII NET1-CFP cdc14-1), were released from a G1 block to fresh media at 37°C. Cells were collected after 120 min and imaged for separation of tags. Representative micrographs for each chromosome tag insertion are shown. Chromosome tags (red arrows) and rDNA arrays (white arrows) are indicated. Note that cells arrest with their centromere proximal flank of the rDNA (tetO:450) resolved and greatly separated whereas neither the distal flank of the rDNA (tetO:487) nor the chromosome XII right telomere (tetO:1061) are resolved. (C) Percentage of cdc14-1 cells with different chromosome tag insertions on the right arm of chromosome XII where two individual chromosome tags were visible (i.e., resolved sister chromosome tags). Cells were treated as B. 100 cells were scored for each tag insertion. (D) Average distance between resolved sister chromosome tags inserted on the right flank of CEN12 (tetO:194) and centromere-proximal flank of the rDNA array (tetO:450). Only cells with resolved tags were scored. The error bars represent the SD.
A recent report using TEV-induced anaphase shows that, in the absence of separase and Cdc14 activation, the rDNA sequences in sister chromatids remain joined (Sullivan et al., 2004), demonstrating that Cdc14 participates in the resolution of sister rDNAs. This raises the possibility that in cdc14-1 blocked cells with stretched rDNAs, sister chromatids are still zipped along the rDNA, which would be clearly different from the bridge stage of wild-type cells, where sister rDNAs are resolved at their proximal flank (Fig. 2). To further investigate this, we expressed FP-tagged nucleolar markers and used chromosomal tag insertions in cdc14-1. Cells were released from G1 at nonpermissive temperature and the final block was analyzed 150 min after release. Chromosome tags in the centromere-distal flank of rDNA separated only in 10% of cells, and telomere tags in <5% (Fig. 4, B and C). In contrast to telomeric regions, chromosome tags in the centromere-proximal flank of rDNA (tetO:450) were separated in ∼80% cells (Fig. 4, B and C) with an average distance between the tags of 3.9 μm (SD 1.7 μm; Fig. 4 D). Even in cdc14-1 blocked cells with a single rDNA signal on either mother or daughter cell bodies, sister tetO:450 tags were clearly separated (Fig. S3, arrow). We observed a reduced distance between sister tetO:450 tags in cdc14-1 bridges compared with wild type (Fig. 2 B and Fig. 4 D). However, we also found that many cdc14-1 arrested cells showed problems in the segregation of separated nuclear masses to the mother and daughter cells (Fig. S3, arrow). In such cells the distance between nuclear masses and separated tetO:450 tags is necessarily shorter that in bridged wild-type cells. Therefore, the reduced distance between sister tetO:450 tags in cdc14-1 bridges is partially due to nuclear segregation defects. We have also quantified the kinetics of the resolution and segregation of proximal and distal edges of rDNA in a synchronous culture of cdc14-1 relative to wild-type cells (Fig. 5). As expected, cdc14-1 progresses throughout G1/S phases as wild type, entering anaphase on schedule. However, cdc14-1 exhibited both a delay in the appearance of binucleated cells and a higher frequency of stretched nuclei (Fig. 5, second row of panels) suggesting nuclear segregation problems. These phenotypes can be attributed to the known role of Cdc14p in the segregation of telomeric regions (D'Amours et al., 2004). The resolution of centromere-proximal flanks of rDNA (tetO:450) was slightly delayed in cdc14-1 (∼5–10 min; Fig. 5, third row of panels). Segregation of the tags, measured as two foci located in different cell bodies, was clearly delayed for tetO:450. In contrast, both resolution and segregation of the centromere-distal flanks of rDNA (tetO:487) were severely delayed (Fig. 5, third row of panels). Similar separation kinetics for rDNA centromere-distal flanks have been recently reported for another mutant allele of CDC14, cdc14-3 (D'Amours et al., 2004). We conclude that the cdc14-1 arrest is similar to the anaphase bridge period in wild-type cells, with separated nuclear masses and sister rDNAs largely unzipped but not segregated, since the majority of tetO:450 tags and lacO:RDN1 tags (in the middle of rDNA), but not tetO:487 tags, are separated (Fig. 4, B and C; Fig. 5; Torres-Rosell et al., 2004). Together, these results demonstrate that cells can be genetically arrested in the bridge stage using the CDC14 mutant allele cdc14-1.
Figure 5.
cdc14-1 mutant is delayed in the resolution of rDNA centromere distal regions. Strains CCG1298 (Mata TetR-YFP tetO:450Kb-Chrm XII), CCG1300 (Mata TetR-YFP tetO:450Kb-Chrm XII), CCG1677 (Mata TetR-YFP tetO:450Kb-Chrm XII cdc14-1), and CCG1679 (Mata TetR-YFP tetO:487Kb-Chrm XII cdc14-1) were released from a G1 arrest into fresh YPD medium at 37°C. Cells were collected every 20 min for ∼2.5 h and scored for budding/rebudding, nuclear morphology, tag resolution, and segregation of the tags (resolved tags located in different cell bodies). Centromere-proximal flank tags for the rDNA (tetO:450) resolve early in anaphase whereas distal flank tags resolution is severely delayed (tetO:487).
Segregation of sister rDNA arrays after the bridge stage does not require a mitotic spindle
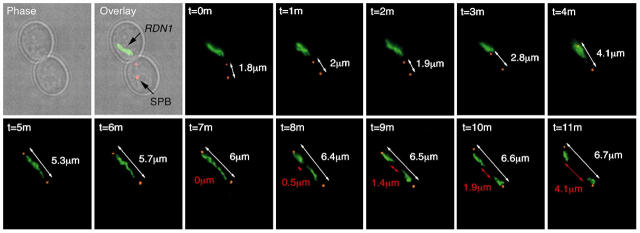
We have shown that rDNA compaction occurs after the anaphase bridge period (Fig. 3 A). To test whether rDNA compaction is sufficient for rDNA segregation after the bridge stage, or whether spindle pull also contributes to the poleward movement of sister rDNA arrays, we monitored rDNA arrays and spindle pole bodies (SPBs) in live cells undergoing anaphase. We reasoned that if rDNA segregation after the bridge stage depends on spindles pulling the chromatids, it could be predicted that as the sister rDNA arrays separate, a similar increase in spindle length would be observed (compared with the increase in separation of rDNA arrays). To this aim, we imaged rDNAs in cells undergoing anaphase and incorporated spindle length (measured as distance between SPBs) into our analysis (Fig. 6). At the onset of anaphase, before spindle elongation, the rDNA loop is often found in a different cell body from the nucleus (most frequently the mother cell; unpublished data). As spindles elongated, the rDNA arrays in the metaphase loop unzipped into the “rDNA bridge” stage. Interestingly, the transition from rDNA loop to bridge closely follows the elongation of the spindle, suggesting that the unzipping of sister rDNAs is largely driven by the early anaphase spindle force (Fig. 6, time = 4–6 min). Once cells reached the bridge stage, nucleolar markers appeared as elongated lines across the bud-neck (Fig. 6, time = 5–6 min). After the bridge formation, the segregation of rDNA arrays took place (Fig. 6, time = 8–11 min). As the separated sister rDNAs migrated to opposite poles, the distances between sister rDNAs increased from 0 (red measurement, Fig. 6, time = 8 min) to 4.1 μm (red measurement, Fig. 6, time = 11 min), in contrast to the distances between SPBs, which only increased by 0.7 μm (white measurement, Fig. 6, from 6 μm at time = 8 min to 6.7 μm at time = 11 min) in the same time period. Therefore, the increase in distance between separating sister rDNA arrays is 5.8 times greater than the increase in distance between the SPBs, suggesting mechanisms additional to spindle pulling as the main driving force behind the poleward segregation of sister rDNAs. Moreover, sister rDNAs compacted 2.7-fold from the anaphase bridge (Fig. 6, time = 7 min) to the time when they were fully segregated (Fig. 6, time = 11 min), suggesting that compaction substantially contributes to the final segregation of this chromosome arm.
Figure 6.
Transition from anaphase rDNA bridge to segregated rDNA sisters does not require further mitotic spindle elongation but axial compaction of the sister arrays. Strain CCG919 (Mata NET1-GFP TUB4-CFP) was imaged during anaphase. A representative cell from a total of nine is shown. SPBs (TUB4-CFP) have been pseudocolored in red. Measurements are shown in white (white arrows) for SPBs and red (red arrows) for rDNA separation. The anaphase bridge stage characterized by rDNA chromatin extended across the bud neck of the dividing cell is seen (from time 5 to 8 min) occurring before rDNA separation. Note that rDNA separates by 4.1 μm (from time 7 to 11 min), whereas spindle length increases only by 0.7 μm in the same time period.
Our data strongly support that active rDNA compaction after bridge formation is responsible for the observed poleward segregation of rDNA from the anaphase bridge to fully segregated arrays. Moreover, there are two other possible explanations: (1) spindle de/polymerization reactions that, while maintaining a constant spindle length could generate the pulling force behind segregation, or (2) a delay in the resolution of some sister chromatid linkages within or downstream of the rDNA arrays. The latter would generate a situation where sister rDNA arrays would recoil after being stretched due to spindle-generated tension. Intact spindles should be dispensable only if active compaction is the driving force behind rDNA array segregation.
To test whether spindles are required for the segregation of rDNA arrays after the bridge stage, we took advantage of the fact that a cdc14-1 block is similar to the anaphase bridge stage (Fig. 4, B and C). We exposed cdc14-1 cells to nonpermissive conditions and, after the block, the culture was separated in two and nocodazole was added to one half. After a further 10 min of incubation at 37°C, both cultures were returned to 23°C to reactivate Cdc14p and we took samples every 30 min for 2 h. The level of rDNA segregation (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200408087/DC1) and tetO:487 segregation (Fig. 7) followed identical kinetics in both experimental conditions. Therefore, the presence of nocodazole, i.e., absence of spindles, did not prevent the poleward segregation of rDNA arrays, demonstrating that the transition from the anaphase bridge stage to the full segregation of sister rDNAs does not require spindles. We conclude that after the anaphase bridge period Cdc14-dependent compaction of rDNA is required for the segregation of the long RDN1 locus.
Figure 7.
rDNA segregation during late anaphase does not require mitotic spindles. (A) Strain CCG1402 (Mata NET1-CFP ChrmXII:tetO:487 TetR-YFP cdc14-1) was grown and incubated at 37°C (nonpermissive conditions) for 150 min. cdc14-1 mutant cells arrested with segregated nucleus but with the rDNA across the bud-neck and tetO:487 tags unresolved (Fig. 4). The culture was divided in two and the microtubule poison nocodazole was added to one half (time = 0′) 10 min before the cells were returned to permissive temperature (23°C). Time points were collected and analyzed for segregation of tetO:487 chromosome tags. More than 100 cells were scored for every time point. (B) Representative micrographs of nocodazole treated cells at time 0 min (budded cell, top row) and 120 min (rebudded cell, bottom row) are shown. Nucleus is shown in red, rDNA arrays in blue, and tetO:487 tags in green.
Discussion
The nucleolus and telomeres in yeast segregate late during mitosis (Alexandru et al., 2001; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004). This is independent of the cohesin complex but requires FEAR-mediated activation of Cdc14 and the condensin complex (D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004; Wang et al., 2004). In this work, we have used in vivo microscopy to analyze in detail the segregation of the rDNA arrays during anaphase. Our study demonstrates that: (a) rDNA segregation involves two temporally separated steps, unzipping (or resolution) and compaction, and (b) rDNA compaction in late anaphase is responsible for the segregation of sister rDNAs in a spindle-independent manner. We also discuss the contribution of Cdc14p to these processes.
rDNA resolution and compaction are temporally separated during anaphase
The structure of the long arm of chromosome XII is different from most other chromosomes as it harbors the rDNA. We have analyzed the configuration of rDNA in metaphase and anaphase cells using a combination of chromosome tags along the arm and fluorescent rDNA-binding proteins. During metaphase, rDNA forms a loop as previously described by others (Guacci et al., 1994; Lavoie et al., 2004). The loop is restricted to rDNA repeats as chromosome tags at the proximal (tetO:450) and distal (tetO:487) edges of rDNA are located at the base of the rDNA loop in the nucleus (Fig. S1). A similar configuration of the rDNA in metaphase has been suggested in a recent report while writing of this manuscript was in progress (Fuchs and Loidl, 2004).
Previously, we identified a stage during mid-anaphase when the rDNA is stretched across the mother and daughter cells (Torres-Rosell et al., 2004). In the present report, we show that cells undergoing this “anaphase rDNA bridge” have sister rDNAs unzipped from each other and, thus, largely resolved (Fig. 2). Sister chromosome tags at the centromere-proximal edge (tetO:450) of rDNA are separated, with an average distance between the tags of ∼5.2 and 5.9 μm (Fig. 2 B and Fig. 3 A), and locate within the main nuclear masses which are already segregated. In contrast, sister chromosome tags in the centromere-distal edge (tetO:487) of rDNA are unresolved and locate to the middle of the nucleolar bridge (Fig. 2). The large distances between sister tetO:450 tags and the localization of tetO:487 tags to the middle of the bridge demonstrate that sister chromatids have resolved most of the rDNA array by the time they arrive at this anaphase rDNA bridge. Furthermore, we have also observed that a chromosome tag inserted in the middle of the RDN1 locus (RDN1:lacO) separates by ∼2 μm in bridged cells, demonstrating that around three fourths of the arrays are fully resolved at the bridge stage (Fig. 2 B). Interestingly, we have also shown that the rDNA compaction in bridged cells is not significantly different from that of the metaphase loops (Fig. 3 A) and that overall rDNA compaction occurs during late anaphase after the bridge stage (Fig. 3 A and Fig. 6). Therefore, we conclude that the rDNA resolution and compaction processes occur separately and consecutively during anaphase and that there is a narrow window in the anaphase timing (the “rDNA bridge stage”) when sister rDNAs are largely resolved yet not compacted.
Hyper-compaction of rDNA is required during segregation because anaphase spindles are not sufficiently long
The presence of the rDNA arrays in the right arm of the chromosome XII makes this arm the longest of the genome (2–3 Mbp, which is at least 1 Mbp longer than the right arm of chromosome IV, the next in length). During mitosis, repetitive (rDNA) and unique regions in yeast chromosomes are 115–140 fold more compacted than B-form DNA, yielding an average compaction of 2.54 μm/Mb (Guacci et al., 1994). Based on this packaging, every chromosome arm in the genome except for the long arm of chromosome XII can be fully segregated with an 8 μm anaphase spindle (Fig. 8 A). However, the segregation of the long arm of chromosome XII would require a spindle length of between 9.8 and 14.9 μm depending on the size of the array (Fig. 8 A). Therefore, yeast cells would be faced with a problem if they were to execute cytokinesis with the chromosome folding levels of metaphase. Instead, they must require additional compaction during anaphase to ensure that the linear length of the right arm of chromosome XII is sufficiently reduced as to be segregated and avoid severing of this arm by cytokinesis. Interestingly, the coccidian parasite Aggregata eberthi, which also uses an intranuclear spindle during mitosis (like that of S. cerevisiae), has an abnormally reduced spindle size relative to the nucleus (Darlington, 1937). During metaphase A. eberthi chromosomes retain the length and appearance normally seen in prophase contracting further only in late anaphase (Darlington, 1937). Sometimes the anaphase compaction is delayed and the ends of longer chromosomes appear unseparated (Darlington, 1937). Therefore, hyper-compaction–mediated segregation might be a strategy to cope with long chromosome arms and a relative reduced spindle size. Here, we have presented strong evidence supporting the idea that rDNA compaction is the driving force behind the poleward segregation of the long arm of chromosome XII in late anaphase. First, live cell analysis revealed that rDNA segregation is accompanied by substantial axial compaction of sister rDNA but little increment in the spindle length (Fig. 3 A and Fig. 6). Second, cells arrested in the anaphase bridge can segregate rDNA in the absence of spindles when allowed to progress out of the arrest (Fig. 7).
Figure 8.
Model for the rDNA segregation in anaphase. (A) Spindle length required for the segregation of each chromosome arm on the yeast genome (L, left; R, right) based on metaphase compaction ratio of 2.54 μm/Mb. This compaction ratio was calculated as the mean of previously reported ratios observed for cells arrested with either nocodazole or Cdc20 depletion (Guacci et al., 1994). The required spindle length was calculated according to the formula: spindle length (μm) = 2 × 2.54× arm length (Mb). Note that the right arm of chromosome XII would require greater spindle lengths than those observed for typical late anaphase cells (dotted line, 8 μm). Two spindle length values for the right arm of chromosome XII are given, flanking the typical size of the RDN1 locus. (B) Schematic representation of the organization of rDNA arrays in metaphase and anaphase cells obtained from data shown in previous figures. The position of different chromosome tags and the rDNA array along the long arm of chromosome XII are shown and labeled in each stage. During metaphase, rDNA arrays are organized as a loop. As cells enter mid-anaphase, rDNA loops become anaphase bridges and sister rDNAs unzip from centromere-proximal to centromere-distal regions, thus, resolving from each other. The bridge period is followed by the movement of sister rDNA arrays to the poles and this transition involves axial compaction of rDNA arrays. Therefore, rDNA resolution takes places during mid-anaphase (during the loop-to-bridge transition), before rDNA compaction is induced in late anaphase (during the bridge-to-pole transition), demonstrating that these two processes are temporally uncoupled. The rDNA compaction leads to the final segregation of the right arm of chromosome XII.
Cdc14 role during rDNA segregation
Cdc14 plays an important role throughout the rDNA segregation process (Granot and Snyder, 1991; Buonomo et al., 2003; D'Amours et al., 2004; Sullivan et al., 2004; Torres-Rosell et al., 2004). Cdc14 activity is required to target the condensin complex to rDNA (D'Amours et al., 2004; Wang et al., 2004), hence mediating the resolution and compaction of this locus in mitosis. Here, we have also shown that cdc14-1 mutants arrest in a stage equivalent to the rDNA bridge of wild-type cells, with partially unzipped sister rDNAs (Figs. 4 and 5). The cdc14-1 block at nonpermissive temperatures is complex and probably represents the sum of several defects associated with this key phosphatase. Aside from the most evident defect in mitosis exit, we found that cdc14-1 arrested cells exhibited problems in the migration of the separated nuclear masses to the poles (Fig. S3, arrow), shorter spindles and nuclear masses still connected, as seen by DAPI. All these phenotypes might be related, or alternatively might be due to independent processes regulated by Cdc14, as it has been suggested (Pereira and Schiebel, 2003; D'Amours et al., 2004; Ross and Cohen-Fix, 2004). The final cdc14-1 arrest is, thus, conceptually equivalent to the anaphase rDNA bridge of wild-type cells, however there are some noteworthy differences: (a) the overall distance between the tetO:450 tags is about one fourth smaller in cdc14-1 bridges than that of the bridge in the wild type (Fig. 2 B and Fig. 4 C), and (b) unlike wild type, the cdc14-1 bridge is not entirely symmetrical. In cdc14-1 bridges, the core of Net1p or Fob1p signals and tetO:487 tags sometimes appear closer to one of the nuclear masses (Fig. 4 B and Fig. 7 B). The small differences between wild-type and cdc14-1 bridges are most likely explained by the nuclear segregation impairments observed in cdc14-1 (Fig. 5, second row panel; Fig. S3; D'Amours et al., 2004).
Recently, it has been reported that in a TEV-induced anaphase, the absence of Cdc14p activity generates a situation where tags in the proximal and distal edges of rDNA do not resolve, but tags in the centromeric and telomeric regions are not only resolved but also fully segregated to the poles (Sullivan et al., 2004). These findings raise the possibility that in wild-type cells the telomere of the long arm of chromosome XII might also segregate before and independent of the rest of arm itself. Nevertheless, in our investigation, we have never observed, neither in wild-type cells nor in the cdc14-1 mutant analyzed, that distal regions to rDNA (i.e., in particular, telomeric regions) segregate before the entire rDNA has been fully resolved (Figs. 2 and 4). The difference, with respect to tetO:450 separation, between a cdc14-1 block and the TEV-induced anaphase (i.e., resolution in cdc14-1 blocked cells but not in TEV-induced anaphases) could be due to problems in spindle stabilization in the TEV-anaphase due to the absence of separase activity (Sullivan et al., 2001). In contrast, anaphase spindles are fully stable in cdc14 mutants because of residual Slk19p function (Pereira and Schiebel, 2003). In cdc14-1 cells there was a minor delay in the separation of tetO:450 tags (Fig. 5), suggesting that Cdc14p activity is involved at the onset of rDNA resolution. However, Cdc14p is not essential for rDNA resolution since cells arrested by cdc14-1 can reach the bridge stage of wild-type cells with tetO:450 separated an average distance of ∼4 μm (Fig. 4 D).
We have shown that Cdc14-mediated compaction of rDNA is necessary for the resolution and segregation of tetO:487 tags independent of spindles (Fig. 7). Interestingly, the resolution of tetO:487 tags can also be induced in the absence of spindles by overexpressing Cdc14 in metaphase arrested cells (D'Amours et al., 2004). This demonstrates that rDNA resolution requires additional Cdc14-mediated mechanisms independent of spindles (D'Amours et al., 2004). Additionally, overexpression of Cdc14p in metaphase cells also has been shown to induce rDNA compaction (Sullivan et al., 2004). Our data and these studies support the idea that the separation and segregation of distal regions of chromosome XII require rDNA compaction. Together, the data demonstrate that progression out of the bridge stage is under specific cell cycle regulation, requiring Cdc14p activity. Furthermore, it strongly suggests that stable anaphase spindles are important during the unzipping of the rDNA array observed during the bridge period.
Conclusions
Here, we have shown that late anaphase compaction of rDNA is necessary for its segregation independent of spindles. The data demonstrate that rDNA unzipping/resolution and compaction occur at different times in wild-type cells, with the resolution of sister rDNAs preceding their compaction. Also we found that these two steps needed for rDNA segregation require distinct mechanisms. The first step occurs from metaphase to mid-anaphase and involves unzipping of sister rDNAs from centromere-proximal to centromere-distal regions. The second step takes place from mid- to late anaphase and involves the compaction of sister rDNAs (Fig. 8 B). These findings show that the biological function of rDNA compaction is to ensure full segregation and demonstrate that sister chromatid resolution does not simply result from the compaction of individual chromosome arms, at least for yeast rDNA.
Materials and methods
Yeast strains, plasmids, and experimental conditions
A list of the strains used in this study is provided in Table I. Net1p was tagged at its COOH terminus with GFP using the plasmid pDM266a (provided by D. Moazed, Massachusetts Institute of Technology, Cambridge, MA) digested with BglII. Tub4p was tagged at its COOH terminus with CFP using pTUB4-CFP plasmid (provided by G. Goshima and M. Yanagida, Kyoto University, Kyoto, Japan) digested with BamHI. Nup49-GFP has been described before (provided by S. Gasser, Friedrich Miescher Institute, Basel, Switzerland; Heun et al., 2001). Epitope tagging for Net1p-CFP and Fob1p-GFP at their COOH terminus were performed using PCR-based methods (provided by T. Davis, University of Washington, Seattle, WA, and E. Schiebel, University of Manchester, Manchester, UK; Knop et al., 1999; Hailey et al., 2002). cdc14-1 (provided by F. Cross, The Rockefeller University, New York, NY; V. Cid, University of Madrid; and C. Nombela, University of Madrid, Madrid, Spain), and smc2-8 (provided by A. Strunnikov, National Institutes of Health [NIH], Bethesda, MD) allele replacements were also performed using PCR-based methods. cdc5-1 allele was provided by K. Lee (NIH). Constructs to target 5.6 kb of tetO arrays to different sites on the long arm of chromosome XII were generated by cloning 1-kb fragments of the desired target site into p3939 (provided by T. Tanaka, University of Dundee, Dundee, UK). Tag integrations were confirmed by PCR.
Table I. Yeast strains used in this study.
| Strain | Relevant genotype | Reference |
|---|---|---|
| AS499 | MATa bar1Δ leu2-3,112 ura3-52 his3-Δ200 trp1-Δ63 ade2-1 lys2-801 pep4 | A. Strunnikov |
| DOM0114 | MATa bar1:hisG ura3-1 trp1-1 leu2-3,112 his3-11 ade2-1 can1-100 GAL+ cdc15-2 | A. Strunnikov |
| KLY1546 | MATa cdc5-1 leu2 ura3 his3 trp1 | K. Lee (NIH) |
| CCG771 | AS499 NET1-GFP LEU2 | L. Aragon |
| CCG919 | AS499 NET1-GFP LEU2 TUB4-CFP TRP1 | L. Aragon |
| CCG938 | DOM0114 NET1-GFP LEU2 | L. Aragon |
| CCG975 | KLY1546 NET1-GFP LEU2 | L. Aragon |
| CCG1145 | AS499 RDN1:lacO:klLEU2 lacI-GFP URA3 TUB4-CFP TRP1 | L. Aragon |
| CCG1168 | CCG938 slk19::KanMX4 | L. Aragon |
| CCG1191 | CCG1145 NET1-CFP HygB | This study |
| CCG1222 | CCG938 spo12::KanMX4 | L. Aragon |
| CCG1248 | CCG938 smc2-8:URA3 | This study |
| CCG1298 | AS499 TetR-YFP ADE2 TetO(5.6Kb):450Kb-ChrXII URA3 | This study |
| CCG1300 | AS499 TetR-YFP ADE2 TetO(5.6Kb):487Kb-ChrXII HIS3 | This study |
| CCG1326 | AS499 TetR-YFP ADE2 TetO(5.6Kb):194Kb-ChrXII HIS3 NET1-CFP KanMX5 | This study |
| CCG1327 | AS499 TetR-YFP ADE2 TetO(5.6Kb):450Kb-ChrXII URA3 NET1-CFP KanMX5 | This study |
| CCG1328 | AS499 TetR-YFP ADE2 TetO(5.6Kb):487Kb-ChrXII HIS3 NET1-CFP KanMX5 | This study |
| CCG1329 | AS499 TetR-YFP ADE2 TetO(5.6Kb):1061Kb-ChrXII HIS3 NET1-CFP KanMX5 | This study |
| CCG1402 | CCG1328 cdc14-1 TRP1 | This study |
| CCG1549 | AS499 FOB1-GFP KanMX5 | This study |
| CCG1580 | AS499 FOB1-GFP KanMX5 NET1-CFP HygB | This study |
| CCG1582 | AS499 NUP49-GFP URA3 NET1-CFP HygB | This study |
| CCG1605 | CCG1326 cdc14-1 TRP1 | This study |
| CCG1607 | CCG1327 cdc14-1 TRP1 | This study |
| CCG1609 | CCG1329 cdc14-1 TRP1 | This study |
| CCG1622 | CCG771 cdc14-1 TRP1 | This study |
| CCG1677 | CCG1298 cdc14-1 TRP1 | This study |
| CCG1679 | CCG1300 cdc14-1 TRP1 | This study |
The 1.5kb lacO array (provided by K. Bloom, University of North Carolina, Chapel Hill, NC) inserted within the RDN1 has been characterized before (Torres-Rosell et al., 2004). We have noticed that the chromosome tag inserted in the middle of the array (lacO:RDN1) is not fully stable through many generation, possibly due to recombination events. Therefore, in our experiments we worked from single colonies and we checked that the number and location of the tag was correct. In the experiment shown in Fig. 2 B <10% of the cells in the population showed changes in tag number and location. Growth conditions are described in figure legends. When G1 release into 37°C experiments were performed, pretreatment at 37°C for 30′ was performed before removing the α-factor pheromone.
Microscopy
Yeast cells with CFP- or GFP-tagged proteins were analyzed by fluorescence microscopy after DAPI staining. Series of z-focal plane images were collected on a microscope (model IRB; Leica) using a C4742-95 digital camera (Hamamatsu) and OpenLab software (Improvision). A tunable light source (Polychrome IV) with a Xenon lamp was used. Images in different z-axis planes were flattened into a two-dimensional projection and processed in Openlab. Live cell imaging (Fig. 6) was done using 63×/1.4 or 100×/1.35 lenses and a Polychrome IV device, tunable light source optimized for live imaging that minimizes cell exposure. The viability of cells after imaging was comparable to nonimaged cells under the same conditions. DNA was stained using DAPI (Molecular Probes) at 1 μg/ml final concentration after short treatment of the cells with 1% Triton X-100. Imaging was done in antifade/DAPI medium (Molecular Probes) at RT.
Online supplemental material
Fig. S1 shows that the right arm of chromosome XII forms a loop restricted to rDNA during metaphase. Fig. S2 shows that rDNA sister chromatid cohesion is maintained in metaphase loops. Fig. S3 shows that cdc14-1 mutants arrest with separated rDNA proximal regions. Fig. S4 shows that nucleolar segregation during late anaphase does not require mitotic spindles. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200408087/DC1.
Acknowledgments
We are grateful to S. Gasser, C. Nombela, V. Cid, T. Davis, F. Cross, D. Moazed, K. Bloom, G. Goshima, M. Yanagida, T. Tanaka, A. Strunnikov, and E. Schiebel for reagents, plasmids, and strains. We thank S. Farmer for helpful comments on the manuscript. J. Torres-Rosell was supported by the EC (Marie Curie Training Fellowship).
This work was supported by the Medical Research Council (MRC) UK.
Abbreviations used in this paper: rDNA, ribosomal DNA; SPB, spindle pole body.
References
- Alexandru, G., F. Uhlmann, K. Mechtler, M.A. Poupart, and K. Nasmyth. 2001. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 105:459–472. [DOI] [PubMed] [Google Scholar]
- Bhalla, N., S. Biggins, and A.W. Murray. 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 13:632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo, S.B., K.P. Rabitsch, J. Fuchs, S. Gruber, M. Sullivan, F. Uhlmann, M. Petronczki, A. Toth, and K. Nasmyth. 2003. Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Dev. Cell. 4:727–739. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., F. Stegmeier, and A. Amon. 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 117:455–469. [DOI] [PubMed] [Google Scholar]
- Darlington, C.D. 1937. Recent Advances in Cytology. J&A Churchill Ltd., London. 356 pp.
- Freeman, L., L. Aragon-Alcaide, and A. Strunnikov. 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149:811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, J., and J. Loidl. 2004. Behaviour of nucleolus organizing regions (NORs) and nucleoli during mitotic and meiotic divisions in budding yeast. Chromosome Res. 12:427–438. [DOI] [PubMed] [Google Scholar]
- Garcia, S.N., and L. Pillus. 1999. Net results of nucleolar dynamics. Cell. 97:825–828. [DOI] [PubMed] [Google Scholar]
- Granot, D., and M. Snyder. 1991. Segregation of the nucleolus during mitosis in budding and fission yeast. Cell Motil. Cytoskeleton. 20:47–54. [DOI] [PubMed] [Google Scholar]
- Guacci, V., E. Hogan, and D. Koshland. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey, D.W., T.N. Davis, and E.G. Muller. 2002. Fluorescence resonance energy transfer using color variants of green fluorescent protein. Methods Enzymol. 351:34–49. [DOI] [PubMed] [Google Scholar]
- Heun, P., T. Laroche, K. Shimada, P. Furrer, and S.M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science. 294:2181–2186. [DOI] [PubMed] [Google Scholar]
- Hirano, T. 2000. Chromosome cohesion, condensation, and separation. Annu. Rev. Biochem. 69:115–144. [DOI] [PubMed] [Google Scholar]
- Holm, C. 1994. Coming undone: how to untangle a chromosome. Cell. 77:955–957. [DOI] [PubMed] [Google Scholar]
- Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 15:963–972. [DOI] [PubMed] [Google Scholar]
- Koshland, D., and A. Strunnikov. 1996. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 12:305–333. [DOI] [PubMed] [Google Scholar]
- Lavoie, B.D., E. Hogan, and D. Koshland. 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 18:76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, B.D., K.M. Tuffo, S. Oh, D. Koshland, and C. Holm. 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell. 11:1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 2000. Nuclear envelope fission is linked to cytokinesis in budding yeast. Exp. Cell Res. 260:277–283. [DOI] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 91:35–45. [DOI] [PubMed] [Google Scholar]
- Mortimer, R.K., and J.R. Johnston. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics. 113:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673–745. [DOI] [PubMed] [Google Scholar]
- Ouspenski, I.I., O.A. Cabello, and B.R. Brinkley. 2000. Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol. Biol. Cell. 11:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., C. Manson, J. Grindlay, and E. Schiebel. 2002. Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J. Cell Biol. 157:367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124. [DOI] [PubMed] [Google Scholar]
- Petes, T.D. 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA. 76:410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, K.E., and O. Cohen-Fix. 2004. A role for the FEAR pathway in nuclear positioning during anaphase. Dev. Cell. 6:729–735. [DOI] [PubMed] [Google Scholar]
- Shaw, P.J., and E.G. Jordan. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93–121. [DOI] [PubMed] [Google Scholar]
- Shou, W., J.H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z.W. Chen, J. Jang, H. Charbonneau, and R.J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 97:233–244. [DOI] [PubMed] [Google Scholar]
- Stegmeier, F., R. Visintin, and A. Amon. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 108:207–220. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., A.S. Belmont, C.C. Robinett, and A.W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A.V., E. Hogan, and D. Koshland. 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9:587–599. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., T. Higuchi, V.L. Katis, and F. Uhlmann. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell. 117:471–482. [DOI] [PubMed] [Google Scholar]
- Sullivan, M., C. Lehane, and F. Uhlmann. 2001. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nat. Cell Biol. 3:771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., and F. Uhlmann. 2003. A non-proteolytic function of separase links the onset of anaphase to mitotic exit. Nat. Cell Biol. 5:249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swedlow, J.R., and T. Hirano. 2003. The making of the mitotic chromosome: modern insights into classical questions. Mol. Cell. 11:557–569. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492–499. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell, J., F. Machin, A. Jarmuz, and L. Aragon. 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle. 3:496–502. [PubMed] [Google Scholar]
- Visintin, R., E.S. Hwang, and A. Amon. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 398:818–823. [DOI] [PubMed] [Google Scholar]
- Wang, B.D., V. Yong-Gonzalez, and A.V. Strunnikov. 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 3:960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]