Abstract
CLIP-associating protein (CLASP) 1 and CLASP2 are mammalian microtubule (MT) plus-end binding proteins, which associate with CLIP-170 and CLIP-115. Using RNA interference in HeLa cells, we show that the two CLASPs play redundant roles in regulating the density, length distribution and stability of interphase MTs. In HeLa cells, both CLASPs concentrate on the distal MT ends in a narrow region at the cell margin. CLASPs stabilize MTs by promoting pauses and restricting MT growth and shortening episodes to this peripheral cell region. We demonstrate that the middle part of CLASPs binds directly to EB1 and to MTs. Furthermore, we show that the association of CLASP2 with the cell cortex is MT independent and relies on its COOH-terminal domain. Both EB1- and cortex-binding domains of CLASP are required to promote MT stability. We propose that CLASPs can mediate interactions between MT plus ends and the cell cortex and act as local rescue factors, possibly through forming a complex with EB1 at MT tips.
Introduction
Microtubules (MTs) are polar filaments, which can undergo alternating phases of growth and shortening, a behavior termed dynamic instability (Desai and Mitchison, 1997). In living cells, a large number of protein factors regulate dynamic instability, thus determining the shape of the MT network in different phases of the cell cycle and in different cell regions. An interesting group of MT-associated factors, involved in regulation of MT dynamics, binds specifically to the ends of growing MTs. These factors, named +TIPs (plus-end tracking proteins), include members of structurally unrelated protein families, such as end binding proteins EB1, EB2 (RP1), and EB3, cytoplasmic linker proteins CLIP-170 and CLIP-115, dynactin large subunit p150Glued, adenomatous polyposis coli (APC), and CLIP-associating proteins (CLASPs; for reviews see Schuyler and Pellman, 2001; Carvalho et al., 2003; Howard and Hyman, 2003; Mimori-Kiyosue and Tsukita, 2003).
CLASPs are a family of MT-associated proteins, conserved in animals and fungi (Inoue et al., 2000; Lemos et al., 2000). A distant member of this family, the budding yeast Stu1p, was isolated as a suppressor of a cold-sensitive tubulin mutation and an essential component of the mitotic spindle (Pasqualone and Huffaker, 1994; Yin et al., 2002). The first member of this family from higher organisms to be characterized in detail, the Drosophila Orbit/MAST, was also shown to be essential for mitosis (Inoue et al., 2000; Lemos et al., 2000), and the same holds true for one of the three C. elegans homologues, cls-2 (R106.7; Gonczy et al., 2000). Mammalian homologues, CLASP1 and 2, were initially characterized through their ability to bind to CLIP-170 and CLIP-115 (Akhmanova et al., 2001). The interaction between the CLASP and CLIP counterparts was also observed in flies (Mathe et al., 2003).
CLASP family members participate in generating polarized MT networks: in migrating fibroblasts, CLASP2 is involved in stabilizing MTs specifically at the leading edge (Akhmanova et al., 2001), whereas during fly oogenesis, Orbit/MAST organizes MTs, which interconnect the oocyte and the nurse cells (Mathe et al., 2003). During mitosis, Orbit/MAST plays a role in maintaining the bipolarity of the mitotic spindle, in the MT-kinetochore attachment and chromosome congression (Maiato et al., 2002). Similar functions were ascribed to the human CLASP1 (Maiato et al., 2003a,b). In addition, Orbit/MAST is needed for cytokinesis, because it stabilizes the interior MTs of the central spindle, necessary for the progression of the cleavage furrow (Inoue et al., 2004). Although the MT-stabilizing role of CLASP homologues has been established, the effect of CLASPs on the parameters of MT dynamic instability has not been analyzed, and the functional redundancy of the two mammalian CLASPs has not been addressed.
In this study, we show that CLASP1 and 2 have similar and redundant roles in organizing the interphase MTs in HeLa cells. CLASPs stabilize MTs, by retaining their plus ends in the peripheral cortical region, where they are pausing or undergo short polymerization–depolymerization excursions. To get insight into the mechanism of CLASP action, we have analyzed the targeting domains of the CLASP proteins. We show that a short repetitive region in the middle part of CLASP1 and 2 can bind to EB1 and EB3 and recognize growing MT tips, whereas the COOH-terminal domain of CLASP2 associates with the Golgi apparatus and cell cortex. We propose that the MT-stabilizing activity of CLASPs depends on the interaction with the EB proteins.
Results
Characterization of siRNAs, specific for CLASP1 and CLASP2
To detect CLASP1 protein, we have raised two new antibodies against CLASP1, #402 and #2292, which strongly react with GFP-CLASP1α, and display some cross-reactivity with GFP-CLASP2α (Fig. 1 A). To detect CLASP2 we have used the previously described antibody #2358 (Akhmanova et al., 2001), which strongly reacts with GFP-CLASP2α and cross-reacts to some extent with GFP-CLASP1α (Fig. 1 A). All three antibodies recognized endogenous HeLa cell proteins of ∼160 kD, in agreement with the predicted molecular weight of CLASP1/2α isoforms.
Figure 1.
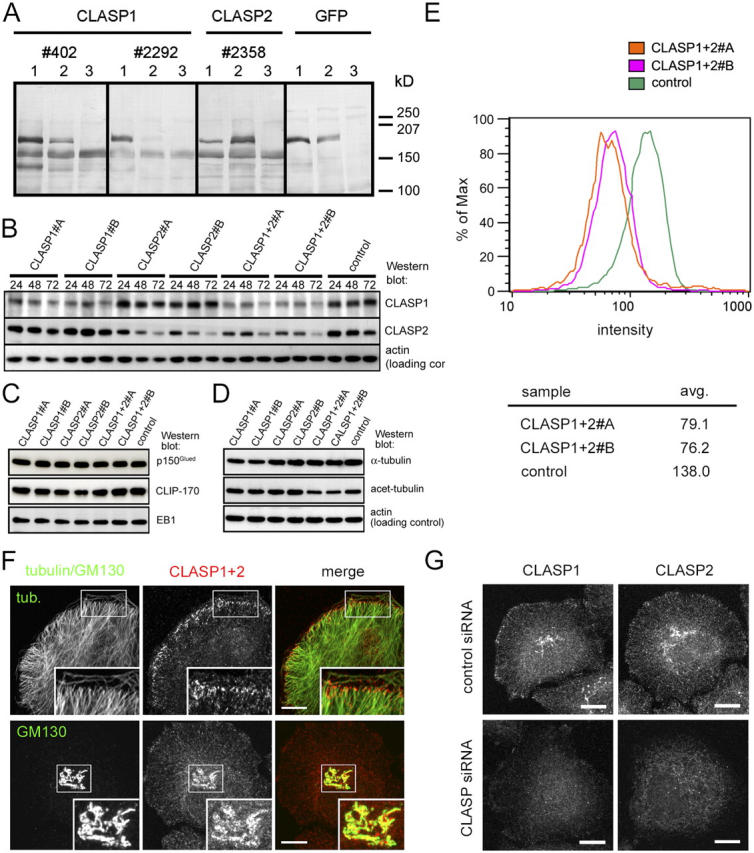
Characterization of CLASP-specific antibodies and RNAi tools. (A) Lysates of HeLa cells, transfected with GFP-CLASP1α (lane 1), GFP-CLASP2α (lane 2), or mock transfected (lane 3), were analyzed by Western blotting with the indicated antibodies. (B) Lysates of HeLa cells, transfected with the indicated siRNAs, were prepared 24, 48, or 72 h after transfection and analyzed by Western blotting with the indicated antibodies. (C and D) Lysates of HeLa cells, transfected with the indicated siRNAs, were prepared 72 h after transfection and analyzed by Western blotting with the indicated antibodies. (E) HeLa cells, transfected either with the control siRNA, CLASP1+2#A or #B siRNAs, were stained with the mixture of antibodies #402 or #2358 72 h after transfection and analyzed by FACS. (F) HeLa cells were stained with a mixture of antibodies #402 (CLASP1) and #2358 (CLASP2) and either α-tubulin or the Golgi marker GM130. Bars, 10 μm. (G) HeLa cells, transfected either with the control siRNA or the CLASP1+2#B siRNAs, were stained with antibodies #402 or #2358 72 h after transfection. Bars, 10 μm.
To knock down CLASP1 and 2, we have selected two different small interfering RNAs (siRNAs), specific for CLASP1 (these will be referred to as CLASP1#A and CLASP1#B) and two siRNAs, specific for CLASP2 (CLASP2#A and CLASP2#B). The siRNAs were transfected into HeLa cells separately, or as combinations of the two “A” siRNAs (CLASP1+2#A) or the two “B” siRNAs (CLASP1+2#B). Three days after transfection, the levels of both CLASP1 and CLASP2, estimated by Western blotting, were reduced by ∼70%, which can be regarded as a hypomorphic state (Fig. 1 B). Control siRNAs had no effect on CLASP levels (Fig. 1 B and not depicted).
The partial down-regulation of CLASPs, separately or together, had no effect on the levels of tubulin or other +TIPs (Fig. 1, C and D). However, when CLASP1 and CLASP2 were knocked down simultaneously, the amount of acetylated tubulin (a marker of stable MTs; Bulinski and Gundersen, 1991) was diminished by ∼40%, suggesting that MT stability was reduced (Fig. 1 D), confirming our previous findings in 3T3 fibroblasts (Akhmanova et al., 2001).
CLASP1- and CLASP2-directed antibodies #402 and #2358 produced similar staining patterns in interphase HeLa cells: in line with previous observations (Akhmanova et al., 2001; Maiato et al., 2003a), they decorated the Golgi apparatus, the centrosome, and MT plus ends (Fig. 1 F and not depicted). The MT tip staining, which nicely colocalized with other +TIPs (EB1, CLIP-170, and p150Glued), was especially prominent in the ∼1-μm region at the cell edges, which were not in contact with other cells (Fig. 1 F; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1). Specific accumulation of CLASPs at the cell edge was confirmed by expressing GFP-tagged CLASPs in HeLa cells (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1).
After CLASP1+2 siRNA treatment, all CLASP-specific signals were strongly reduced (Fig. 1, E and G). Analysis of the CLASP1+2 knockdown cells by FACS after staining with CLASP1 and 2 antibodies showed that they represent a reasonably homogeneous population (Fig. 1 E). These cells displayed a dim diffuse pattern, which was likely in part due to background staining with anti-CLASP antibodies, whereas the specific signals at the MT tips, Golgi, and the centrosome were greatly diminished (Fig. 1 G).
Organization of the MT network after CLASP knockdown
Simultaneous knockdown of the two CLASPs caused mitotic defects, which were similar to those observed in the Orbit/MAST mutants (Maiato et al., 2002; Inoue et al., 2004) and will be described elsewhere (unpublished data). In interphase cells, a significant decrease in density of the MT network was observed (Fig. 2, A and B). This effect was particularly obvious in cells, which had no contacts with their neighbors and, therefore, we confined our analysis to such isolated cells.
Figure 2.
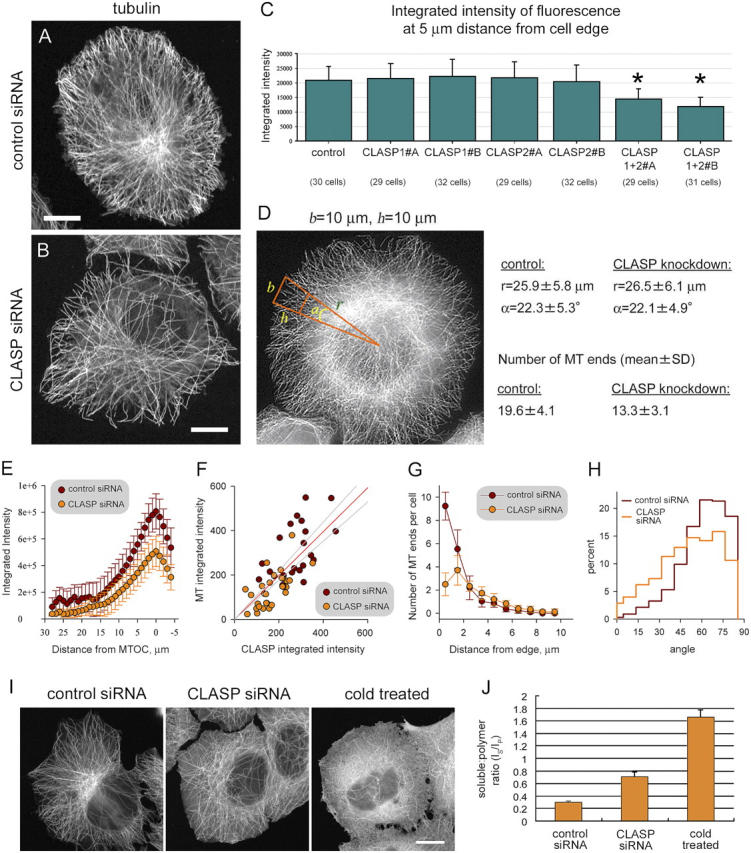
Effect of CLASP knockdown on the interphase MT network. (A and B) HeLa cells 72 h after transfection with the control siRNA (A) or CLASP1+2#B siRNAs (B), fixed with methanol, and stained for α-tubulin. Bars, 10 μm. (C) Plots of integrated intensity of fluorescence of α-tubulin staining, which was performed as in A and B, in HeLa cells 72 h after transfection with the indicated siRNAs. The integrated intensity was measured within a 5-μm2 box at 5 μm distance from the cell edge, with subtracting the background. Measurements were performed in ∼30 cells per siRNA in three different cell regions. Values significantly different from the control (P < 0.001) are indicated by asterisks. (D) Number of MT ends in a trapezoid part of the cell sector (with a base b and side h) in HeLa cells 72 h after transfection with the control siRNA (30 cells, n = 587) or CLASP1+2#B siRNAs (30 cells, n = 401). (E) Integrated intensity within a 5-μm2 box was measured along the cell radius in the same cells as described in D. MTOC, the MT organizing center, was determined as the site of radial gathering of MTs. (F) HeLa cells, transfected either with the control siRNA or the CLASP1+2#B siRNAs, were methanol fixed and stained for CLASPs (using with the mixture of antibodies #402 or #2358) and for tubulin 72 h after transfection. Representative cells were selected to include examples with both high and low levels of CLASP staining. Integrated intensity of CLASP staining was plotted versus integrated intensity of tubulin staining. Each dot represents an average of five independent measurements in different areas at the periphery of one cell. The regression line is shown in red and the 95% confidence interval is indicated by gray lines. (G) Distribution of distances from MT ends to the cell edge within the trapezoid part of the cell sector shown in D. Analyzed images were the same as in D. (H) Distribution of angles of distal segments of MTs to the cell radius. Analyzed images were the same as in D. (I) Confocal microscope images (1-μm optical sections) of HeLa cells, fixed with formaldehyde 72 h after transfection with the control or CLASP1+2#B siRNAs and stained for α-tubulin. Control cells were also incubated for 1 h at 4°C before fixation, to depolymerize MTs (“cold treated”). Bar, 10 μm. (J) Plots of the ratio of the intensity of soluble tubulin staining IS (after subtracting background) to the intensity of polymer staining IP (after subtracting background and IS), obtained from the images as shown in H. Measurements were performed in 20 control cells (n = 158), 35 CLASP-knockdown cells (n = 225), and 18 cold-treated cells (n = 130). Error bars represent the SD.
To get a measure of MT density, we have determined integrated intensity of α-tubulin staining after methanol fixation, which preserves MT polymer, while soluble tubulin is extracted. This analysis indicated that MT density was significantly decreased, only when combinations of CLASP1 and CLASP2-specific siRNAs were used, whereas partial knockdown of either CLASP separately had no visible effect (Fig. 2 C). The fact that integrated intensity was a valid measure of MT density was confirmed by counting the number of MTs in a cell sector, which was decreased by ∼30% after both CLASPs were knocked down (Fig. 2 D). A decrease in integrated intensity of tubulin staining after partial CLASP1/2 knockdown was especially obvious in the central part of the cell, around the Golgi apparatus (Fig. 2 E). It should be noted that the morphology of the Golgi complex was not significantly affected by CLASP knockdown (unpublished data).
Because our siRNA treatment resulted only in a partial depletion of the two CLASP proteins, we analyzed the dependence of the severity of the observed phenotype on the CLASP1/2 levels. The density of MTs was proportional to the level of CLASP1/2 proteins (Fig. 2 F), indicating that CLASPs regulate MT number in a concentration-dependent manner.
Because the amount of total tubulin was not affected by CLASP1+2 siRNA treatment (Fig. 1 D), the diminished number of MTs should have resulted in increased levels of nonpolymerized tubulin. This was confirmed by the analysis of confocal microscope images of tubulin staining in formaldehyde-fixed cells, in which soluble tubulin is maintained during the staining procedure (Fig. 2, I and J).
Most strikingly, the distribution of MT plus ends near the cell margin was changed after CLASP depletion. In control cells, most MTs terminated at ∼1 μm distance from the cell edge. This population of MTs was selectively lost after CLASP knockdown, whereas a small peak of plus ends was observed at ∼2 μm distance from the edge (Fig. 2 G). This change in MT plus-end distribution was also apparent in cells, stained for EB1, CLIP-170, and p150Glued (Fig. S1). The distribution of MT ends in the internal cytoplasm was almost identical in control and CLASP1+2#B siRNA-treated cells (Fig. 2 G).
We have also noticed that after CLASP knockdown, MTs were often oriented along the cell margin, whereas in control cells most MTs were perpendicular to the cell edge (Fig. 2 H). Analysis of the angles between the MTs and the cell margin indicated that the number of MTs, deviating by >45° from the cell radius, was increased by a factor of 2.6, when both CLASPs were partially depleted.
HeLa cells do not migrate, but their membranes exhibit protrusion activity and retrograde flow. The rate of the retrograde flow was reduced almost by half in CLASP1+2#B siRNA-treated cells, compared with control (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1). It is possible that CLASPs directly affect the membrane flow, by influencing the process of secretion or the dynamics of the actin cytoskeleton; the observed effect, however, may also be a consequence of the altered MT organization.
MT dynamics after CLASP knockdown
To confirm that the observed effects on the MT network result from the knockdown of CLASP proteins, rather than unspecific effects of the used RNA duplexes, we transfected cells, treated with CLASP1+2 siRNA combinations, with rescue constructs, encoding GFP or monomeric RFP (mRFP) fusions of CLASP1α or CLASP2α, resistant to the siRNAs due to silent substitutions in the siRNA target regions. Both CLASP1α and 2α fusions could restore MT density and the alignment of MT tips along the cell cortex (Fig. 3 A; Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1). After the rescue with mRFP-CLASP2α the number of MT ends in a sector increased from 14.0 ± 4.3 in surrounding CLASP knockdown cells to 27.9 ± 2.6 in cells, expressing the rescue construct. This increase was above the levels in control cells (19.6 ± 4.1), probably due to overexpression of the rescue construct by a factor of 3–4 compared with endogenous CLASPs (unpublished data). Remarkably, the time the MT plus ends stayed in the vicinity (1 μm distance) from the cell edge was significantly reduced after both CLASPs were knocked down and restored after the rescue with mRFP-CLASP2α (Fig. 3 B).
Figure 3.
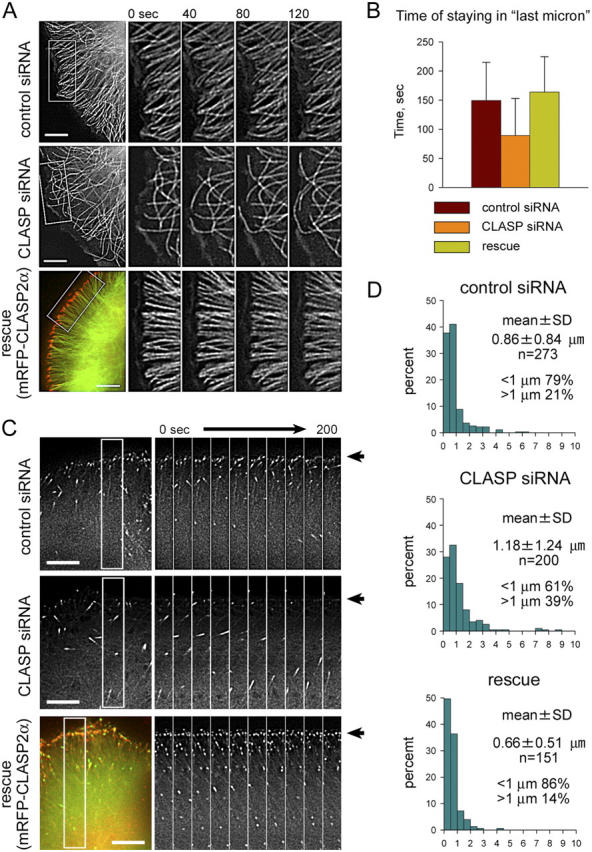
Effect of CLASP knockdown on MT dynamics. (A) Time-lapse images of HeLa cells, stably expressing GFP-α-tubulin, 72 h after transfection with the control or CLASP1+2#A siRNAs. For rescue, CLASP1+2#A siRNA-treated cells were transfected with mRFP-CLASP2α 48 h after the siRNA transfection and observed 24 h later. In the “rescue” panel mRFP-CLASP2α is shown in red and GFP-α-tubulin is shown in green. Bars, 5 μm. (B) Time of staying of MT ends within 1 μm distance from the cell edge, determined from the time-lapse image series as shown in A (cells were imaged with 2-s interval). The difference between the control and CLASP knockdown is statistically significant (P < 0.001), the difference between control and rescue is not significant. (C) Time-lapse images of HeLa cells, stably expressing EB1-GFP, 72 h after transfection with the control or CLASP1+2#A siRNAs. Rescue was performed as described in A. In the “rescue” panel mRFP-CLASP2α is shown in red and EB1-GFP is shown in green. Cell margin is indicated by an arrow. Bars, 5 μm. (D) Length of the shortening excursions from the cell edge, measured from the same dataset as in A and B. Error bars represent the SD.
We have also analyzed HeLa cells stably expressing a GFP fusion of EB1, which binds predominantly to the ends of growing MTs (Mimori-Kiyosue et al., 2000). In control cells a large number of MT ends at the cell periphery was strongly positive for EB1, suggesting that these MTs were growing (Fig. 3 C). This accumulation of EB1-positive ends was clearly reduced after the partial CLASP depletion and restored after the knockdown cells were rescued with mRFP-CLASP2α (Fig. 3 C; Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1). In many cases, EB1 signals at the periphery of control cells appeared rather static, while their intensity varied greatly. This could be explained if pausing MTs or MTs, which started to depolymerize, retained small amounts of EB1, whereas the enhancement of EB1 signal would correlate with MT growth.
We conclude that when CLASP levels are normal, MT ends are maintained in the proximity of cell margin, but remain dynamic. This can be most easily accounted for by very short alternating bursts of MT growth and shrinkage. Indeed, the number of MTs, displaying longer (>1 μm) depolymerization episodes, was increased after the partial CLASP knockdown (Fig. 3 D). This effect was reversed by introducing the mRFP-CLASP2α rescue construct.
The analysis of parameters of MT dynamic instability (Table I) revealed that in the cortical 1-μm-wide region MTs spent more time pausing and exhibited less frequent transitions to growth or shortening in control cells, than in cells with partially depleted CLASPs. In addition, shortening and growing MTs underwent more frequent reverse transitions (rescues or catastrophes, respectively); as a result, dynamic MT ends were maintained in the vicinity of the cell margin, accounting for the observed longer time of staying of MT ends at the cell periphery in control cells, compared with CLASP knockdown cells. Treatment with CLASP1+2 siRNAs had no significant effect on transition frequencies in the internal cytoplasm, proving that CLASPs are truly local regulators of MT dynamics. However, MT growth rate was higher, and the MT shrinkage rate lower in CLASP knockdown cells, compared with control cells (Table I). This can be explained by the increased level of soluble tubulin after the partial CLASP depletion (Fig. 2, I and J). It should be noted that the large contribution of the pausing to growth and shortening episodes at the cell margin makes the differences in growth and shrinkage rates at the cell periphery much less apparent, than those in the internal cytoplasm.
Table I. Parameters of dynamic instability.
| Control | CLASP knockdown | |||
|---|---|---|---|---|
| 1-μm region at the cell edge | Cell interior | 1-μm region at the cell edge | Cell interior | |
| Rate of growth, μm/min | ||||
| Direct MT observation | 9.9 ± 3.5 | 11.7 ± 4.8 | 11.1 ± 1.4 | 12.6 ± 5.1 |
| Subtraction analysis | 14.4 ± 3.9 | 18.2 ± 5.8 | ||
| EB1 tracks | 15.9 ± 6.0 | 22.8 ± 7.3 | ||
| Rate of shortening, μm/min | ||||
| Direct MT observation | −12.0 ± 6.2 | −20.7 ± 11.6 | −12.6 ± 6.5 | −17.8 ± 11.3 |
| Subtraction analysis | −35.6 ± 11.2 | −28.2 ± 9.8 | ||
| Transition frequencies | ||||
| Growth–Shortening, s−1 | 0.078 | 0.035 | 0.030 | 0.045 |
| Growth–Pause, s−1 | 0.402 | 0.238 | 0.360 | 0.250 |
| Shortening–Growth, s−1 | 0.066 | 0.085 | 0.019 | 0.072 |
| Shortening–Pause, s−1 | 0.275 | 0.213 | 0.217 | 0.210 |
| Pause–Growth, s−1 | 0.016 | 0.053 | 0.032 | 0.046 |
| Pause–Shortening, s−1 | 0.026 | 0.038 | 0.038 | 0.040 |
| Time in growth, % | 6.0 | 16.7 | 13.3 | 14.5 |
| Time in pauses, % | 88.9 | 69.3 | 80.1 | 70.7 |
| Time in shortening, % | 5.1 | 14.0 | 6.6 | 14.8 |
HeLa cells, stably expressing GFP-α-tubulin or EB1-GFP, were imaged with a 2-s interval 72 h after transfection with the control or CLASP1+2#B siRNAs. Life history plots of 65 MTs in five control cells and 75 MTs in five CLASP knockdown cells were analyzed as described by Shelden and Wadsworth (1993). In addition, to obtain more accurate values of growth and shortening rates in the internal cytoplasm, subtraction analysis (Vorobjev et al., 1999) and measurements of displacements of EB1-GFP comets were used. The differences in rates, obtained by different methods, can be explained by a higher contribution of the pausing state to the growth/shortening episodes at the cell periphery, which can be detected by direct observation, as compared with those inside the cells, which can be identified by subtraction analysis. EB1 associates predominantly with growing MTs (Mimori-Kiyosue et al., 2000) and, therefore, MT pauses hardly contribute to the growth episodes, which are detected by this method. The data are presented in format: mean ± SD. Rates are instantaneous (measured within one interval between successive frames).
CLASP1 and 2 bind to EB1 and EB3
To get a better insight into the mechanism of CLASP action at the MT tip, we have determined domains of CLASP1 and 2, responsible for their plus-end accumulation. Our previous study has shown that the COOH-terminal domains of CLASP1 and 2 bind to CLIP-170 and CLIP-115, but are not essential for MT tip targeting by CLASPs (Akhmanova et al., 2001). Deletion mapping of CLASP2γ (the shortest splice form of CLASP2, which associates efficiently with the MT tips; Akhmanova et al., 2001) identified a region of ∼140 aa in the middle of CLASP2, CLASP2-M, which was sufficient to target GFP to the ends of growing MTs (Fig. 4, A–E). The corresponding region of CLASP1 (CLASP1-M) could also direct GFP to the MT tips (Fig. 4, F and G). This region is extremely rich in serines, arginines, and lysines, and contains a number of short direct repeats of several types, which are conserved between CLASP1, CLASP2, and their Drosophila homologue, Orbit/MAST (Fig. 4 H).
Figure 4.
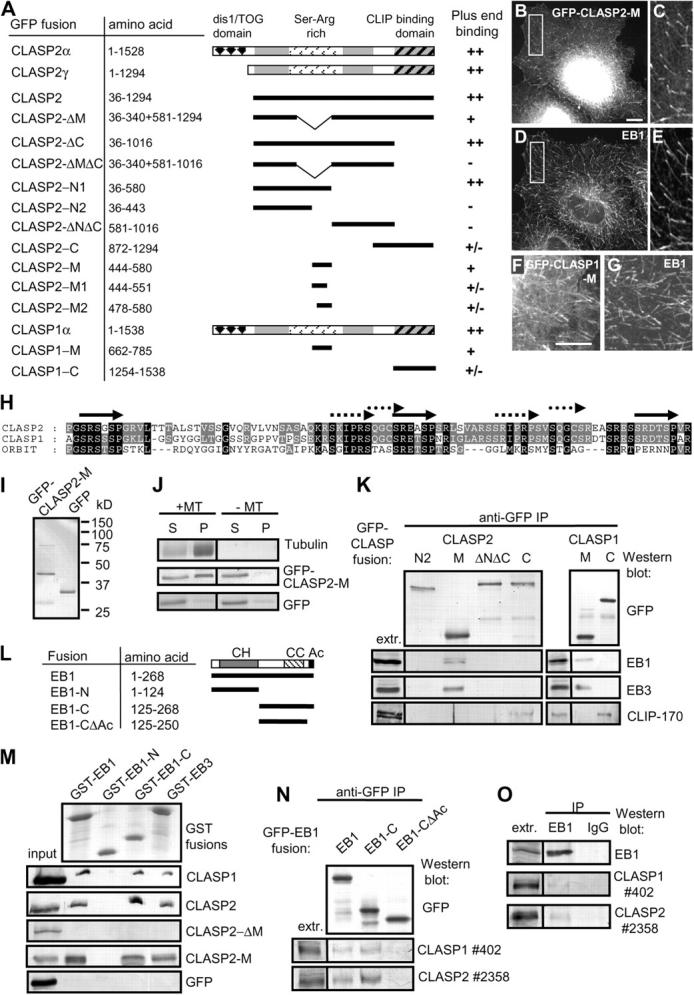
Identification of the MT plus-end binding and EB1-binding domains in CLASP1 and 2. (A–G) GFP fusions of CLASP2α, CLASP2γ, different CLASP2γ deletion mutants, CLASP1α and its deletion mutants were transfected in COS-1 or COS-7 cells, fixed and counterstained for EB1, and the MT plus-end accumulation was assessed as: ++, strong; +, clearly visible; +/−, weakly detectable, −, undetectable. B, C, and F, GFP signal; D, E, and G, EB1 staining. Bars, 10 μm. (H) Alignment of the repetitive part of the CLASP2-M fragment with the corresponding regions of CLASP1 and D. melanogaster Orbit/MAST. Repeats of different types are indicated by different arrows. (I) Coomassie-stained gel, showing purified HIS-tagged GFP-CLASP2-M and GFP. (J) MT pelleting assay with purified GFP-CLASP2-M and GFP. Coomassie-stained gel is shown for tubulin and Western blots with anti-GFP antibodies for the GFP fusions. S, supernatant; P, pellet. (K) Immunoprecipitation with anti-GFP antibodies from COS-1 cells, transfected with the indicated GFP-CLASP1 and 2 deletion mutants. (L) Schematic representation of EB1 structure and the deletion mutants used in this study. CH, calponin homology domain; CC, coiled coil; Ac, acidic tail domain. (M) GST pull-down assays with the indicated GST fusions. Purified GFP-CLASP2-M and GFP or extracts of COS-1 cells, overexpressing GFP-CLASP1α, GFP-CLASP2, and GFP-CLASP2-ΔM were used. Coomassie-stained gel is shown for the GST fusions and Western blots with anti-GFP antibodies for the GFP fusions. 10% of the input and 25% of the material, bound to the beads, were loaded on gel. (N) Immunoprecipitation with anti-GFP antibodies from COS-1 cells, transfected with the indicated GFP-EB1 fusions. (O) Immunoprecipitation with a rabbit polyclonal antibody against EB1 or a control rabbit IgG from untransfected COS-1 cells.
To understand how the CLASP2-M fragment targets MT plus ends, we have purified 6x histidine (HIS)–tagged GFP-CLASP2-M and GFP from bacteria (Fig. 4 I). In vitro pelleting assays demonstrated that GFP-CLASP2-M, but not GFP alone, could bind to taxol-stabilized MTs in vitro (Fig. 4 J). It is possible that the interaction between the MTs and CLASP2-M is at least partly due to the positively charged character of this fragment and the presence of highly acidic COOH termini in α- and β-tubulin. Interestingly, +TIPs, which belong to the EB family, such as EB1 and EB3, also possess highly acidic COOH termini, similar to those of tubulins. Therefore, we tested if CLASP2-M could associate with EB1 and EB3, and found that it was indeed the case. Among the CLASP2 fragments, only CLASP2-M could coprecipitate EB1 and EB3 (Fig. 4 K). The corresponding region of CLASP1 could also coprecipitate EB1 and EB3 (Fig. 4 K), suggesting that this property is conserved between the two CLASPs. In vitro, purified GFP-CLASP2-M bound to purified EB1 and EB3, fused to GST. It associated with the COOH-, but not the NH2-terminal half of the EB1 protein (Fig. 4, L and M). This binding was highly specific, because a number of other basic proteins tested, such as histone H1, did not bind to EB1 COOH terminus (unpublished data).
Also the full-length CLASP1α and CLASP2γ, expressed in COS-1 cells, associated with the GST fusions of EB1, EB3 and EB1 COOH terminus (Fig. 4 M). In contrast, the CLASP2-ΔM mutant, which contains an in-frame deletion removing the EB1-binding fragment, was no longer pulled down by GST-EB1 or EB3 fusions (Fig. 4 M). Endogenous CLASP1 and CLASP2 coprecipitated with EB1-GFP fusion, as well as with the GFP-EB1 COOH terminus, but only when the acidic tail of this protein (the last 18 aa) was intact (Fig. 4 N). The validity of interaction CLASP1/2–EB1 was further confirmed by coprecipitation, albeit a weak one, of endogenous CLASP1 and 2 proteins with endogenous EB1 (Fig. 4 O). Together, these data establish that CLASP1 and 2 bind to EB1 and EB3. This interaction is likely to be transient and regulated, and may occur preferentially in the context of MT tip binding, which would account for the weak coprecipitation of endogenous proteins.
Although GFP-CLASP2-M associated with MT ends (Fig. 4, B–E), unexpectedly, the in-frame deletion of this fragment did not preclude the binding of the rest of the CLASP2γ protein (CLASP2-ΔM mutant) to the MT tips (Fig. 5, A and B). In fact, the COOH-terminal region of CLASP2, containing the CLIP-binding domain, could also weakly target MT tips, suggesting that CLIPs can recruit CLASP to the growing MT ends. We have used a dominant negative CLIP-170 mutant, overexpression of which removes endogenous CLIPs, but not EB1 and EB3 from the MT tips (Fig. 5 C; Komarova et al., 2002), to determine if the binding of different CLASP mutants to MT ends is CLIP dependent. We found that GFP-CLASP2-ΔM mutant depended on the presence of CLIPs for the association with MT ends, whereas GFP-CLASP2-M did not (Fig. 5, D–I). It is possible that the interaction with EB1/EB3, which occurs in addition to binding directly to MTs, increases the affinity of CLASPs for the growing MT ends and contributes to the specific CLASP accumulation at these sites.
Figure 5.
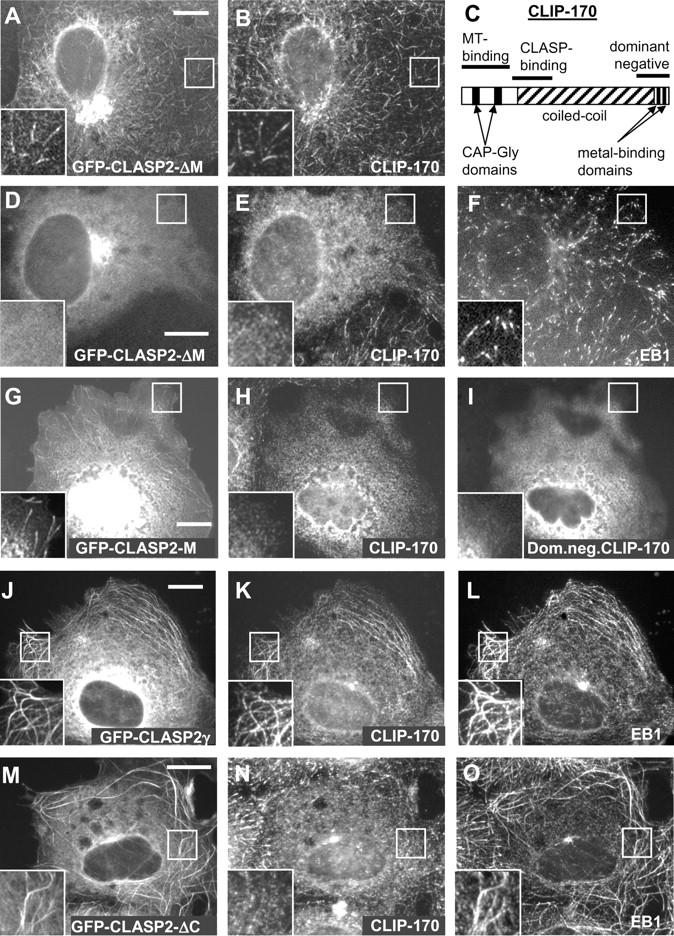
Dependence of CLASP2 deletion mutants on CLIP-170 for their localization to the MT tips and EB1 recruitment to the MTs. (A and B) COS-7 cells were transfected with GFP-CLASP2-ΔM and stained for CLIP-170. (C) Schematic representation the structure of CLIP-170 and the dominant negative construct, used in this study. (D–F) COS-7 cells were cotransfected with GFP-CLASP2-ΔM and the dominant negative CLIP-170, and stained for EB1 and the endogenous CLIP-170, using the antibody against its NH2 terminus. The cells, expressing the dominant negative CLIP-170 construct, can be recognized by the diffuse pattern of endogenous CLIP-170 staining. (G–I) COS-7 cells were cotransfected with GFP-CLASP2-M and the dominant negative CLIP-170, and stained for EB1, the endogenous CLIP-170 (using the antibody against its NH2 terminus) and the dominant negative CLIP-170 (using the antibody against its COOH terminus). (J–L) COS-7 cells were transfected with GFP-CLASP2γ (J) and stained for CLIP-170 (K) and EB1 (L). (M–O) COS-7 cells were transfected with GFP-CLASP2-ΔC (M) and stained for CLIP-170 (N) and EB1 (O). Enlarged portions of the boxed areas are shown in the insets. Bars, 10 μm.
Next, we examined if CLASPs can recruit EB1 to MTs and found, that indeed, overexpressed CLASP1α and CLASP2γ induced strong accumulation of EB1 along the MTs (Fig. 5, J and L; not depicted). In these conditions, CLIP-170 was also accumulated on the MT bundles (Akhmanova et al., 2001; Fig. 5 K and not depicted). However, EB1 recruitment was CLIP independent, because it was also observed with the CLASP2-ΔC mutant, which cannot bind to CLIPs and, when present at high levels, actually displaced CLIP-170 from the MTs (Fig. 5, M–O). CLASP2-ΔM mutant did not accumulate efficiently along the MTs and did not influence EB1 localization (unpublished data). Although we cannot exclude that CLASPs cause accumulation of EB1 at MT bundles by modifying the structure of the MT lattice, the EB1 recruitment is most likely due to direct CLASP–EB interaction.
CLASP2 COOH-terminal domain is responsible for targeting to the cell cortex and the Golgi complex
The observation that MT ends of control HeLa cells localized in the close proximity of the cell margin raised a possibility that they come in contact with the cell cortex. To support this idea, we used total internal reflection fluorescence (TIRF) microscopy, which visualizes only a very narrow region (∼200 nm) at the bottom of the cell. Indeed, in live cells, transfected with either GFP-CLASP1α or GFP-CLASP2γ, the accumulation of these proteins at the distal MT ends at the cell periphery was readily observed by TIRF, indicating that they are very close to the substrate (Fig. 6, A and B). In control cells, expressing GFP-α-tubulin or EB3-GFP (a plus-end marker, similar to EB1-GFP; Stepanova et al., 2003), a large proportion of the MT ends at the cell margin was visualized by TIRF (Fig. 6, C and E; Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1), again suggesting their proximity to the cortical region under the cell. In striking contrast, CLASP1+2 siRNA-treated cells displayed only a few peripheral MT ends, visible by TIRF microscopy (Fig. 6, D and F; Video 4), indicating that most of the MT tips at the cell margin are not touching the underlying cortex. Therefore, we conclude that CLASPs bring peripheral MT segments in close contact with the cortex underneath the cell.
Figure 6.
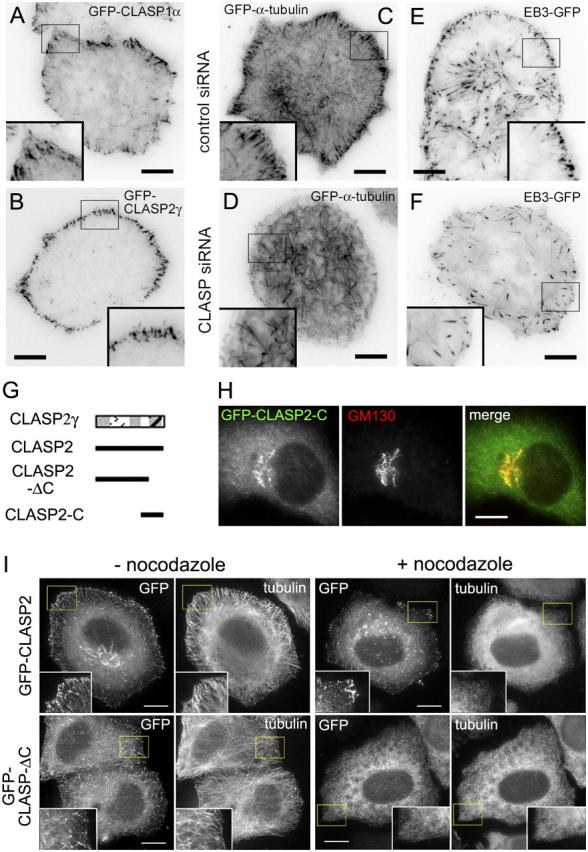
The COOH-terminal domain of CLASP2 is responsible for association with the cell cortex and Golgi complex. (A–F) TIRF microscopy images of live HeLa cells, expressing GFP-CLASP1α (A), GFP-CLASP2γ (B), GFP-α-tubulin (C and D), or EB3-GFP (E and F). Cells were either not treated with siRNAs (A and B), or treated for 72 h with control (C and E) or CLASP1+2#B siRNAs (D and F). The contrast is inverted. Bars, 10 μm. (G) Schematic representation of CLASP2γ and the relevant deletion mutants. (H) HeLa cells were transfected with GFP-CLASP2-C and stained for the Golgi marker GM130. Bar, 10 μm. (I) HeLa cells were transfected with GFP-CLASP2 or GFP-CLASP2-ΔC and were either fixed directly or treated with 10 μM nocodazole for 1 h before fixation and stained for α-tubulin. Bars, 10 μm.
These observations prompted us to investigate which CLASP2 domains are responsible for cortical and/or membrane targeting. Deletion analysis demonstrated that the COOH-terminal portion of CLASP2 alone was sufficient to target GFP to the Golgi, whereas the GFP-CLASP2-ΔC mutant, lacking the COOH-terminal 278 aa, was unable to accumulate at the Golgi (Fig. 6, G and H; not depicted). To investigate the cortical localization of CLASP2 deletion mutants, we made use of the fact that the peripheral accumulation of the full-length CLASP1 and 2 was partially maintained after the MTs were depolymerized with nocodazole (Fig. 6 I and not depicted). This MT-independent localization was not observed with GFP-CLASP2-ΔC mutant, indicating, that, similar to the Golgi localization, it depends on the COOH-terminal domain of CLASP2. It should be noted that our previous studies identified the COOH-terminal ∼280 aa of CLASP2 as the CLIP-binding region (Akhmanova et al., 2001). However, CLIPs are not present at the Golgi and do not accumulate at the cell cortex in MT-independent manner (unpublished data), suggesting that association of CLASP2 with these structures is not dependent on CLIPs.
Both the EB1-binding and the COOH-terminal domains of CLASP2 are required to rescue the organization of the MT network after CLASP knockdown
We have established that the middle (EB1-binding) and COOH-terminal domains of CLASP2 target the protein to the MT plus ends and the cell cortex, respectively. To clarify, which CLASP domains are critical to stabilize MTs at the cell edge, we have performed rescue experiments using GFP fusions of the full-length CLASP1 and 2 or CLASP2 deletion mutants (Fig. 7, A–D, G, and H). We have examined the ratio between the MT and GFP signal, to determine, which proteins can restore MT density at low expression levels (Fig. 7 H). From these experiments, it became clear that both full-length CLASPs could restore MT density more efficiently than the CLASP2 deletion mutants CLASP2-ΔC or -ΔM, which lack either the COOH-terminal or the EB1-binding motif. A shorter deletion mutant, CLASP2-N1, displayed no rescue activity in this assay (Fig. 7 G), and was used as a negative control in this experiment. It should be noted that these deletion mutants associated with MT tips in CLASP1+2 siRNA-treated cells as efficiently as in control cells, suggesting that they do not depend on heterodimerization with endogenous CLASPs for their plus-end localization (although due to the partial character of the knockdown, we cannot fully rule out this possibility). In addition, full-length CLASPs, but not the CLASP2-ΔC or -ΔM deletion mutants were able to rescue the characteristic enrichment of the EB1-positive tips at the cell margin (Fig. 7, E and F; not depicted). These results indicate that both the EB1-binding and the cortex-binding COOH-terminal domains of CLASP2 are needed to stabilize MT ends at the cell periphery.
Figure 7.
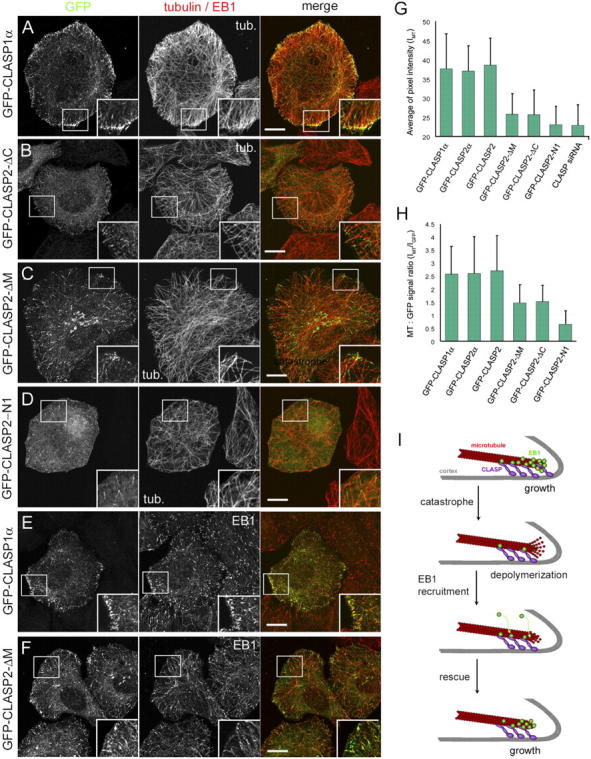
Rescue of CLASP knockdown with CLASP2 deletion mutants and a model for CLASP action at the cell cortex. (A–F) HeLa cells, depleted for CLASPs as described in Fig. 3 A, were transfected with different rescue constructs (indicated on the left), fixed with methanol and stained for α-tubulin (A–D) or EB1 (E and F). Cell images were collected with the confocal microscope (1-μm-thick optical sections). GFP (green) and tubulin/EB1 (red) signals are superimposed on the right. Enlarged portions of the boxed areas are shown in the insets. Bars, 10 μm. (G) Average of pixel intensity of α-tubulin staining (∼10 cells per construct). (H) Ratio of the average pixel intensity of MT fluorescence (IMT) to the integrated intensity of GFP fluorescence (IGFP; ∼10 cells per construct). (I) Proposed model of CLASP rescue activity at the cell cortex. CLASPs can interact with the plus end of a growing MT directly and/or through the association with EB1 and the same time make contact with the cell cortex. After the MT undergoes a catastrophe, most of the EB1 proteins are lost from the tip, but CLASPs remain associated with the cortex and the peripheral stretches of the depolymerizing MT. By enhancing the affinity of EB1–MT interaction at these sites, CLASPs help to retain and/or recruit EB1 to such MTs, leading to their rescue. The oscillations of EB1 signals at the cortical MT tips in control cells support this idea (with strong EB1 accumulation corresponding to growth episodes, and weak EB1 accumulation observed during pausing/depolymerization events). Error bars represent the SD.
Discussion
In this study, we have used an RNAi approach to reduce the levels of CLASP1 and CLASP2 in HeLa cells. The specificity of the used siRNAs is supported by the fact that two different oligonucleotides for each CLASP produced similar effects on the MT network, which were not observed with control siRNAs. Moreover, significant defects in MT density and stability were only observed when combinations of siRNAs, specific for the two CLASPs, were transfected into cells simultaneously. The phenotype, caused by CLASP1/2-specific siRNAs could be rescued by introducing into cells CLASP1 or 2 expression constructs, insensitive to the used siRNA duplexes. These data indicate that the observed defects were due to reduction of the CLASP1/2 expression, rather than the off target activities of the used siRNAs.
HeLa cells express CLASP1α and CLASP2α isoforms, which are ∼77% similar in their protein sequence. Our data, based on the antibody staining, expression of GFP fusion proteins, RNAi-mediated phenotypes and their rescues, indicate that these two CLASPs display very similar localizations and functions in interphase cells. These observations do not rule out, however, that in the context of the whole organism these proteins may have some unique and specific functions. The depletion of CLASPs in our system was only partial, and it is likely that additional and more severe phenotypes would be observed after a complete loss of both CLASPs. Also, we cannot exclude that a complete inactivation of one of the two CLASPs would have different effects compared with the ∼70% inactivation of both, although our results have provided no indication for such a possibility.
Our previous study, based on the analysis of tubulin modifications, has shown that CLASPs are involved in locally stabilizing MTs at the leading edge of motile fibroblasts (Akhmanova et al., 2001). Our new findings fully support the role of CLASPs in stabilizing MTs, by showing that the MT density and the amount of acetylated tubulin are reduced after CLASP knockdown. Detailed analysis of MT behavior shows that CLASPs strongly affect MT plus-end dynamics locally, in the 1-μm-broad region at the cell margin. This is in line with the accumulation of CLASP proteins at the distal segments of MTs specifically in this region of the cell cortex. In the presence of CLASPs, MTs at the cell edge remain dynamic, but keep undergoing alternating frequent rescues and catastrophes. MT pausing, apparently stimulated by CLASPs, might in fact be not a truly nondynamic state, but rather very short alternating growth and shrinking episodes, with amplitudes below the resolution of the fluorescent microscope. We propose that the main activity of CLASPs is MT rescue at the cell edge, which reduces the number of long depolymerization episodes and increases MT longevity. It is possible, that interaction with EB1 is mechanistically important for the CLASP rescue activity by stimulating a switch to the growth phase, because EB proteins promote MT growth (Tirnauer et al., 2002; Busch and Brunner, 2004). The capacity of CLASPs to enhance EB1 accumulation at the MTs is fully in line with this idea, which is also supported by the recent findings showing the essential role of EB1 for generation of stable interphase MTs (Wen et al., 2004). The importance of CLASP–EB interaction is underscored by a recently published report, showing that the Drosophila CLASP homologue, Orbit/MAST is present among the proteins, pulled down by GST-EB1 (Rogers et al., 2004). We propose that CLASPs, associated with the distal segments of MTs at the cell cortex, are partially retained on MTs when they start shrinking and can rescue MTs by enhancing their affinity for EB1 (Fig. 7 I).
CLASP-dependent rescues are quickly followed by catastrophes, which we believe to be caused by the cell edge acting as an impermeable boundary (Janson et al., 2003). If our assumption is correct, how can we explain that the MTs, rescued by CLASP, do not make a turn and start to grow along the cell edge, as observed after the CLASP1/2 knockdown? We propose that CLASPs, bound to the growing MT tip and at the same time associated with the cell cortex, bring the MT end in very close proximity to the cell edge. An abrupt MT turning at this point would require a considerable distortion of the MT lattice, which would be energetically unfavorable; therefore, a catastrophe occurs instead. Catastrophes at the cell edge may also be caused by MT-destabilizing factors, such as depolymerizing kinesins or stathmin, and we cannot rule out that there is a functional interplay between CLASPs and such MT-destabilizing proteins.
The effect of CLASPs on MT rescue is local, whereas the transition frequencies in the internal cytoplasm are not significantly affected. Therefore, CLASP function is different from that of CLIPs, which affect MT rescue frequency throughout the cell (Komarova et al., 2002). It is possible that CLIP–CLASP interaction can contribute for the CLASP rescue activity. However, we cannot exclude that the interaction with CLIPs regulates CLASP accumulation at the MT tips, as opposed to other cellular structures, such as the cortex or the Golgi apparatus, whereas the effect of CLASPs on MT dynamics is CLIP independent.
The rescue activity of CLASPs is likely to be relevant to the mitotic function of these proteins. CLASPs/Orbit/MAST are present at the kinetochores, and their inactivation results in monopolar spindles with chromosomes buried in the interior of the aster (Maiato et al., 2002, 2003a). If CLASPs exhibit a local MT rescue activity at the kinetochore, similar to that shown by us at the cell cortex, this would explain why in the absence of CLASPs/Orbit/MAST the kinetochore fibers are unable to regrow, once they start shortening, causing the chromosomes to be “reeled in” to the pole (Maiato et al., 2002, 2003a).
We have observed very few depolymerizing minus ends either in control or in CLASP knockdown cells (unpublished data), indicating, that in HeLa cells MT renewal is accounted for by plus-end dynamics and that CLASPs do not play a significant role in stabilizing MT minus ends. However, the striking effect of CLASP depletion on MT density indicates, that in addition to regulating plus-end dynamics, CLASPs might play a role in MT nucleation.
Recent studies have identified two other large +TIPs, spectraplakin ACF7 and APC, as MT-stabilizing proteins, which, similar to CLASPs, act the leading edge of motile cells (Kodama et al., 2003; Wen et al., 2004). Similar to CLASPs, APC also binds to EB1 and is negatively regulated by GSK3β (Su et al., 1995; Zumbrunn et al., 2001). ACF7 might be an EB1-binding protein, too, because its Drosophila homologue, Short stop, interacts with the fly homologue of EB1 (Subramanian et al., 2003). CLASPs and APC can bind the cell cortex (this study; Mimori-Kiyosue and Tsukita, 2001), and ACF7 can directly associate with actin filaments (Kodama et al., 2003), suggesting that these proteins can link MTs to these structures. Studies of MT organization at the muscle–tendon junction in fly larvae indicate that Short stop and APC are acting together (Subramanian et al., 2003). No similar data are available for their mammalian counterparts, so the analysis of a potential functional synergism or redundancy between CLASPs, APC, and the spectraplakins presents an important challenge for future studies.
Materials and methods
Cell lines and transfection of plasmids and siRNAs
HeLa, COS-1, and COS-7 cells were grown as described previously (Akhmanova et al., 2001). Effectene or Superfect transfection reagents (QIAGEN) were used for plasmid transfection. Mass transfection of COS-1 cells was performed by DEAE-dextran method (Akhmanova et al., 2001). Stable clones were selected in the presence of 0.3–0.4 mg/ml G418 (Calbiochem). Synthetic siRNAs (Proligo) were transfected, using Oligofectamine (Invitrogen). SiRNAs were directed against the following target sites: CLASP1#A, GCCATTATGCCAACTATCT; CLASP1#B, GGATGATTTACAAGACTGG; CLASP2#A, GTTCAGAAAGCCCTTGATG; CLASP2#B, GACATACATGGGTCTTAGA; control (scrambled CLASP1#A), GCACTCATTATGACTCCAT. 7–10% confluent HeLa cells were transfected, using Oligofectamine (Invitrogen) with siRNAs at the minimal effective concentration (10 nM for CLASP1#A and CLASP1#B, 200 nM for CLASP2#A and CLASP2#B), whereas the control siRNA was used at 200 nM concentration.
Expression constructs
We used the previously described expression vectors for GFP-CLASP1α, GFP-CLASP2γ, GFP-CLASP2 (Akhmanova et al., 2001), EB1-GFP, EB3-GFP (Stepanova et al., 2003), and GFP-α-tubulin (CLONTECH Laboratories, Inc.). GFP-CLASP2α was constructed in pEGFP-C1 by linking the 5′ portion of the human truncated EST clone 7k43h10.x1 (IMAGE:3478506) to the nucleotides 194–5614 of the KIAA0627 cDNA. All CLASP2 deletion mutants were derived from the GFP-CLASP2 construct, using restriction sites (Fig. 4 A), with the exception of CLASP2-C, M1 and M2, which were generated by PCR, as well as CLASP1 deletion mutants. Dominant negative CLIP-170 construct, used in this study, contained nucleotides 2871–4597 of the rat brain CLIP-170 cDNA (GenBank/EMBL/DDBL accession no. AJ237670). CLASP1/2 rescue constructs were prepared by a PCR-based strategy, by introducing five silent substitutions in the target site of CLASP1#A siRNA (resulting in a sequence GCTATCATGCCTACCATT) and six silent substitutions in the target site of CLASP2#A siRNA (resulting in a sequence GTCCAAAAGGCTCTCGAC). GFP was substituted for mRFP (gift of R. Tsien, University of California, San Diego, La Jolla, CA; Campbell et al., 2002) to generate red fluorescent fusions.
Protein purification, in vitro binding assays, immunoprecipitation, and Western blotting
To produce HIS-tagged proteins, GFP (derived from pEGFP-C1; CLONTECH Laboratories, Inc.) or GFP-CLASP2-M fragment of CLASP2 were subcloned into pET-28a, expressed in Rosetta (DE3) pLysS E. coli and purified using Ni-NTA agarose (QIAGEN). GST-tagged fusions of mouse EB1 (GenBank/EMBL/DDBL accession no. NM_007896) and human EB3 (Stepanova et al., 2003) were produced in BL21 E. coli and purified using glutathione-Sepharose 4B (Amersham Biosciences). GST pull-down assays, immunoprecipitations and Western blotting were performed as described previously (Akhmanova et al., 2001). To estimate the degree of protein knockdown after RNAi, the signals in experimental lanes were compared with serial dilutions of the control extract, present on the same Western blot. MT pelleting assays were performed using the MT-associated protein spin-down assay kit (Cytoskeleton, Inc.).
Antibodies and immunofluorescent staining
Rabbit antibodies against CLASP1 (#402 and #2292) and EB1 were raised as described previously (Akhmanova et al., 2001), using GST fusions of the mouse CLASP1 COOH terminus (GenBank/EMBL/DDBL accession no. AJ288061) and the mouse full-length EB1. We used mouse mAbs against EB1 and p150Glued (Transduction Laboratories), α- and β-tubulin and acetylated tubulin (Sigma-Aldrich), actin (CHEMICON International, Inc.); rat mAb against α-tubulin (YL1/2; Abcam), rabbit pAbs against GFP (MBL; CHEMICON International Inc., and Abcam), CLASP2 (Akhmanova et al., 2001) and CLIP-170 (Coquelle et al., 2002), EB3 (Stepanova et al., 2003) and chicken pAb against GFP (CHEMICON International, Inc.). For secondary antibodies, Cy2-conjugated anti–mouse IgG and anti–rabbit IgG pAbs, Texas red–conjugated anti–mouse IgG, anti–rat IgG, and anti–rabbit IgG pAb, Cy5-conjugated anti–mouse IgG, anti–rat IgG, and anti–rabbit IgG pAbs were purchased from Jackson ImmunoResearch Laboratories. Fresh medium was added to cells ∼2 h before fixation. Cell fixation and staining were performed as described previously (Mimori-Kiyosue et al., 2000).
Fluorescence microscopy and image analysis
Images of cells were collected with a DeltaVision optical sectioning system using PlanApo 100×/1.40 NA oil, PlanApo 60×/1.40 NA oil ph3 or UPlanApo 20×/0.70 NA dry objectives (Olympus), using a cooled CCD camera (Series300 CH350; Photometrics). Fluorescence signals were visualized using a quad filter set (86000; Chroma Technology Corp.) for multiple color imaging, or Endow GFP bandpass emission filter set (41017; Chroma Technology Corp.) for GFP imaging. The out-of-focus signals were removed using the deconvolution technique with the DeltaVision system. Confocal imaging was performed using LSM510 confocal laser scanning microscope (v. 2.3; Carl Zeiss MicroImaging, Inc.). TIRF microscopy was performed on an Olympus IX70 (PlanApo 100×/1.45 NA, oil TIRFM objective), equipped with a 488-nm argon laser line (MELLES GRIOT), an objective-type TIRF illuminator (Olympus), and an OrcaER cooled CCD camera (Hamamatsu Photonics). The system was controlled by Aquacosmos software (Hamamatsu Photonics). The quantification and analysis of fluorescent signals was performed using MetaMorph software (Universal Imaging Corp.). Images were prepared for publication using Photoshop (Adobe). Statistical analysis was performed using with a SigmaPlot (SPSS Inc.) and Statistica for Windows (StatSoft Inc.). Unless stated differently, the statistical significance of the observed differences was evaluated using Kolmogorov-Smirnov two-sample test and in all plots SD is indicated. For FACS analysis, suspended cells were fixed in cold ethanol, labeled with anti-CLASP1/2 antibodies and Cy5-conjugated secondary antibody, and analyzed using FACSCalibur (Becton Dickinson).
Online supplemental material
Fig. S1 demonstrates the changes in distribution of +TIPs after CLASP knockdown. Fig. S2 illustrates the effect of CLASP knockdown on the retrograde flow. The videos show (Video 1) the behavior of GFP-CLASP1 in interphase cells; (Videos 2 and 3) time-lapse series of control, CLASP1+2 siRNA-treated and mRFP-CLASP2-rescued cells, depicting the dynamics of MTs and EB1-GFP, and (Video 4) the behavior of EB3-GFP in the control or CLASP1+2 siRNA cells observed with TIRF microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200405094/DC1.
Acknowledgments
We are grateful to Dr. H. Yamauchi for continuous encouragement, to M. Nihsimura for excellent assistance with FACS analysis, to S. Dekker for preparing purified GFP protein, to Y. Komarova for fruitful discussions, and to Dr. R. Tsien for providing mRFP.
This study was supported by Russian Foundation for Basic Research, grant 02-04-48839 to I. Vorobiev, and by the Netherlands Organization for Scientific Research and Dutch Cancer Society grants to A. Akhmanova and N. Galjart.
Abbreviations used in this paper: APC, adenomatous polyposis coli; CLASP, CLIP-associating protein; HIS, 6x histidine; mRFP, monomeric RFP; MT, microtubule; siRNA, small interfering RNA; +TIPs, plus-end tracking proteins; TIRF, total internal reflection fluorescence.
References
- Akhmanova, A., C.C. Hoogenraad, K. Drabek, T. Stepanova, B. Dortland, T. Verkerk, W. Vermeulen, B.M. Burgering, C.I. De Zeeuw, F. Grosveld, and N. Galjart. 2001. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 104:923–935. [DOI] [PubMed] [Google Scholar]
- Bulinski, J.C., and G.G. Gundersen. 1991. Stabilization of post-translational modification of microtubules during cellular morphogenesis. Bioessays. 13:285–293. [DOI] [PubMed] [Google Scholar]
- Busch, K.E., and D. Brunner. 2004. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14:548–559. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, P., J.S. Tirnauer, and D. Pellman. 2003. Surfing on microtubule ends. Trends Cell Biol. 13:229–237. [DOI] [PubMed] [Google Scholar]
- Coquelle, F.M., M. Caspi, F.P. Cordelieres, J.P. Dompierre, D.L. Dujardin, C. Koifman, P. Martin, C.C. Hoogenraad, A. Akhmanova, N. Galjart, et al. 2002. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, A., and T.J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., C. Echeverri, K. Oegema, A. Coulson, S.J. Jones, R.R. Copley, J. Duperon, J. Oegema, M. Brehm, E. Cassin, et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 408:331–336. [DOI] [PubMed] [Google Scholar]
- Howard, J., and A.A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature. 422:753–758. [DOI] [PubMed] [Google Scholar]
- Inoue, Y.H., M. do Carmo Avides, M. Shiraki, P. Deak, M. Yamaguchi, Y. Nishimoto, A. Matsukage, and D.M. Glover. 2000. Orbit, a novel microtubule-associated protein essential for mitosis in Drosophila melanogaster. J. Cell Biol. 149:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, Y.H., M.S. Savoian, T. Suzuki, E. Mathe, M.T. Yamamoto, and D.M. Glover. 2004. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 166:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M.E., M.E. de Dood, and M. Dogterom. 2003. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 161:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, A., I. Karakesisoglou, E. Wong, A. Vaezi, and E. Fuchs. 2003. ACF7: an essential integrator of microtubule dynamics. Cell. 115:343–354. [DOI] [PubMed] [Google Scholar]
- Komarova, Y.A., A.S. Akhmanova, S. Kojima, N. Galjart, and G.G. Borisy. 2002. Cytoplasmic linker proteins promote microtubule rescue in vivo. J. Cell Biol. 159:589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, C.L., P. Sampaio, H. Maiato, M. Costa, L.V. Omel'yanchuk, V. Liberal, and C.E. Sunkel. 2000. Mast, a conserved microtubule-associated protein required for bipolar mitotic spindle organization. EMBO J. 19:3668–3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., P. Sampaio, C.L. Lemos, J. Findlay, M. Carmena, W.C. Earnshaw, and C.E. Sunkel. 2002. MAST/Orbit has a role in microtubule-kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 157:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato, H., E.A. Fairley, C.L. Rieder, J.R. Swedlow, C.E. Sunkel, and W.C. Earnshaw. 2003. a. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 113:891–904. [DOI] [PubMed] [Google Scholar]
- Maiato, H., C.L. Rieder, W.C. Earnshaw, and C.E. Sunkel. 2003. b. How do kinetochores CLASP dynamic microtubules? Cell Cycle. 2:511–514. [DOI] [PubMed] [Google Scholar]
- Mathe, E., Y.H. Inoue, W. Palframan, G. Brown, and D.M. Glover. 2003. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oogenesis. Development. 130:901–915. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., and S. Tsukita. 2001. Where is APC going? J. Cell Biol. 154:1105–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., and S. Tsukita. 2003. “Search-and-capture” of microtubules through plus-end-binding proteins (+TIPs). J. Biochem. (Tokyo). 134:321–326. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 10:865–868. [DOI] [PubMed] [Google Scholar]
- Pasqualone, D., and T.C. Huffaker. 1994. STU1, a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J. Cell Biol. 127:1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, U. Hacker, C. Turck, and R.D. Vale. 2004. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14:1827–1833. [DOI] [PubMed] [Google Scholar]
- Schuyler, S.C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 105:421–424. [DOI] [PubMed] [Google Scholar]
- Shelden, E., and P. Wadsworth. 1993. Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J. Cell Biol. 120:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, T., J. Slemmer, C.C. Hoogenraad, G. Lansbergen, B. Dortland, C.I. De Zeeuw, F. Grosveld, G. van Cappellen, A. Akhmanova, and N. Galjart. 2003. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). J. Neurosci. 23:2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, L.K., M. Burrell, D.E. Hill, J. Gyuris, R. Brent, R. Wiltshire, J. Trent, B. Vogelstein, and K.W. Kinzler. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977. [PubMed] [Google Scholar]
- Subramanian, A., A. Prokop, M. Yamamoto, K. Sugimura, T. Uemura, J. Betschinger, J.A. Knoblich, and T. Volk. 2003. Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr. Biol. 13:1086–1095. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., S. Grego, E.D. Salmon, and T.J. Mitchison. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev, I.A., V.I. Rodionov, I.V. Maly, and G.G. Borisy. 1999. Contribution of plus and minus end pathways to microtubule turnover. J. Cell Sci. 112:2277–2289. [DOI] [PubMed] [Google Scholar]
- Wen, Y., C.H. Eng, J. Schmoranzer, N. Cabrera-Poch, E.J. Morris, M. Chen, B.J. Wallar, A.S. Alberts, and G.G. Gundersen. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830. [DOI] [PubMed] [Google Scholar]
- Yin, H., L. You, D. Pasqualone, K.M. Kopski, and T.C. Huffaker. 2002. Stu1p is physically associated with beta-tubulin and is required for structural integrity of the mitotic spindle. Mol. Biol. Cell. 13:1881–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbrunn, J., K. Kinoshita, A.A. Hyman, and I.S. Nathke. 2001. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr. Biol. 11:44–49. [DOI] [PubMed] [Google Scholar]


