Abstract
During mitosis, chromosomes are highly condensed and transcription is silenced globally. One explanation for transcriptional repression is the reduced accessibility of transcription factors. To directly test this hypothesis and to investigate the dynamics of mitotic chromatin, we evaluate the exchange kinetics of several RNA polymerase I transcription factors and nucleosome components on mitotic chromatin in living cells. We demonstrate that these factors rapidly exchange on and off ribosomal DNA clusters and that the kinetics of exchange varies at different phases of mitosis. In addition, the nucleosome component H1c-GFP also shows phase-specific exchange rates with mitotic chromatin. Furthermore, core histone components exchange at detectable levels that are elevated during anaphase and telophase, temporally correlating with H3-K9 acetylation and recruitment of RNA polymerase II before the onset of bulk RNA synthesis at mitotic exit. Our findings indicate that mitotic chromosomes in general and ribosomal genes in particular, although highly condensed, are accessible to transcription factors and chromatin proteins. The phase-specific exchanges of nucleosome components during late mitotic phases are consistent with an emerging model of replication independent core histone replacement.
Introduction
When cells enter mitosis, chromatin undergoes dramatic structural alterations, resulting in highly condensed chromosomes with a compaction ratio up to 1:10,000 (Li et al., 1998). Along with the condensation of chromatin, the majority of transcription is inhibited. The condensed chromosomes are generally viewed as inactive passengers directed by spindles to align at the metaphase plate and subsequently separate to opposite poles of the spindle, resulting in the formation of two daughter cells each with a copy of the genome.
Although a substantial amount of attention has been focused on the mechanism by which the chromosomes condense and segregate, much less is known regarding the mechanism leading to the transcription silencing during mitosis. Several possibilities could contribute to this phenotype. First, transcription factors can be inactivated by Cdks. For example, Cdk1–cyclin-B kinase is necessary to establish and maintain the suppression of ribosomal DNA (rDNA) transcription during mitosis (Heix et al., 1998; Sirri et al., 1999, 2000). Second, changes in chromatin remodeling complexes during mitosis could contribute to the silencing. Sif et al. (1998) found that components of the SWI/SNF complex are phosphorylated during mitosis and the phosphorylation can inactivate the complex in cell free assays, suggesting that phosphorylation of SWI/SNF may contribute to transcription silencing during mitosis. Third, it is also possible that the physical compaction of mitotic chromatin reduces accessibility of transcription factors to their target sites and in that way also contributes to mitotic silencing of transcription in vivo.
To examine the accessibility of DNA to transcription factors and the dynamics of chromatin structural proteins during mitotic chromatin condensation in mammalian cells, we analyzed their chromosome association kinetics using photobleaching fluorescence microscopy. We find that RNA polymerase I (RNA pol I) transcription factors rapidly exchange on and off mitotic nucleolar organizing regions (NORs). In addition, linker histone H1 shows differential exchange dynamics at various stages of mitosis with its exchange rate peaking at metaphase. The core histones appear to be stably associated with mitotic chromosomes at prophase and metaphase, but show limited and consistent exchange from chromosome during anaphase and telophase. The exchange of core histones coincides with the increase in histone H3-K9 acetylation, chromatin association of RNA pol II, and precedes the bulk reactivation of transcription.
Results
Mitotic NORs are accessible to RNA pol I transcription factors that rapidly exchange from chromatin throughout mitosis
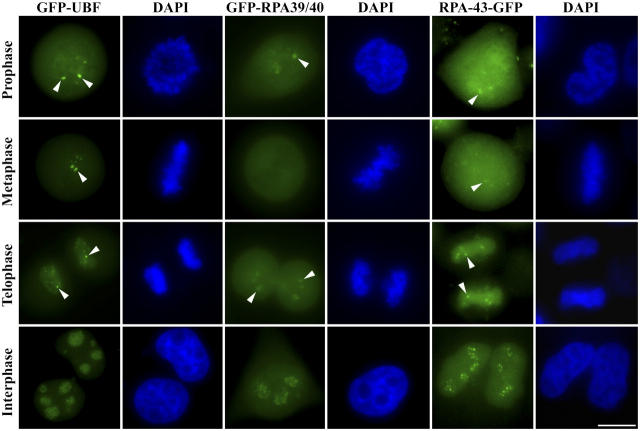
To address whether transcription factors can access their target sequences in condensed chromosomes, we first evaluated the dynamics of factors involved in RNA pol I transcription during mitosis. The RNA pol I transcription machinery is unique in comparison to RNA pol II and RNA pol III in that many of the RNA pol I transcription factors localize to the transcriptionally silent rDNA clusters and NORs during mitosis (Scheer and Rose, 1984; Roussel et al., 1993, 1996; Gilbert et al., 1995). It has not been clear whether this accumulation is due to trapping of RNA pol I components upon cessation of transcription or whether factors continuously exchange even in condensed NORs. To distinguish between these possibilities, we examined the association of a subset of RNA pol I transcription factors with mitotic NORs in living cells using FRAP microscopy. Previously characterized functional fusion proteins between the GFP and RNA pol I factors upstream-binding factor (UBF) 1, 39/40-kD subunit of RNA pol I (RPA39/40), and 43-kD subunit of RNA pol I (RPA43) were expressed in HeLa cells (Chen and Huang, 2001; Dundr et al., 2002; Leung et al., 2004). Fig. 1 summarizes their localization in interphase and mitotic cells. Identical to endogenous proteins, GFP-RNA pol I subunits and GFP-UBF1 are localized to the nucleolus in a concentrated dot pattern in interphase cells (Fig. 1; Chen and Huang, 2001; Dundr et al., 2002; Raska, 2003). In mitotic cells, GFP-UBF1 shows a characteristic association with the NORs, demonstrated by an accumulation in paired dots on chromosomes throughout all phases of mitosis (Fig. 1). In contrast, GFP-fusion proteins of RNA pol I subunits show variations in their association with NORs during mitosis. RPA43-GFP localizes to NORs throughout mitosis, whereas GFP-RPA39/40 associates with NORs during prophase, late anaphase, and telophase, but is not associated with chromosomes during metaphase and early anaphase when NORs are transcriptionally silent (Fig. 1). The GFP-RNA pol I subunits are primarily detected on NORs and are not enriched in other parts of mitotic chromosomes when colocalized with UBF. In contrast to the RNA pol I subunits, a GFP-fusion of the largest subunit of RNA pol II (Sugaya et al., 2000) is not detected at NORs on mitotic chromosomes at any phase during mitosis (see Fig. 9).
Figure 1.
Subcellular localization of GFP-fusion proteins of RNA pol I transcription factors in cells at various phases of cell cycle as labeled. Bar, 10 μM. Arrowheads indicate NOR-associated corresponding GFP-fusion proteins.
Figure 9.
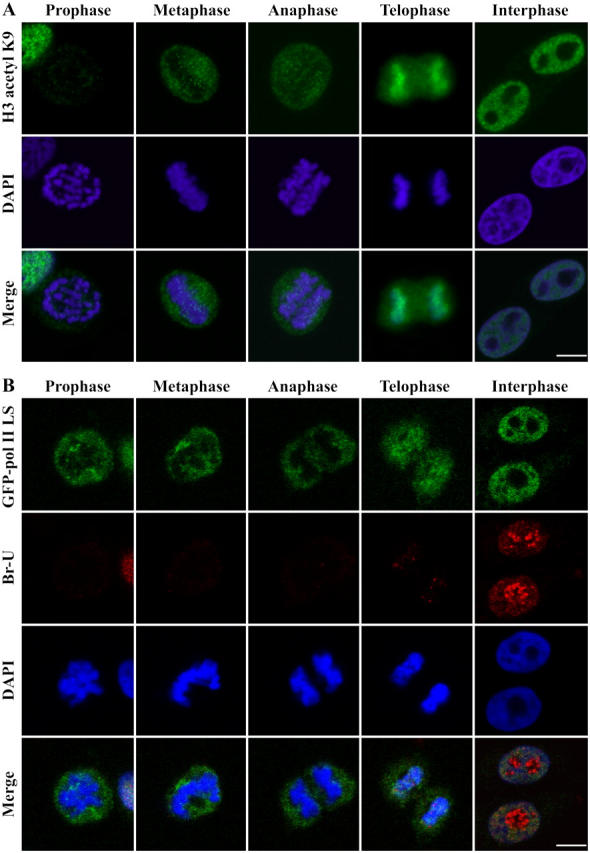
H3 acetylation and polymerase II association to chromosome take place at anaphase–telophase transition prior to the onset of bulk RNA synthesis. (A) Detection of histone H3 lysine 9 acetylation using a specific antibody at various mitotic phases and in interphase cells. (B) Simultaneous detection of GFP-pol II LS and Br-U incorporation through various phases mitosis. Bar, 10 μm.
In FRAP experiments, cells that expressed the lowest levels of GFP-fusion proteins were chosen for the analyses. The fluorescence signal of GFP-UBF1 at NORs recovers more slowly during mitosis with the time of 50% fluorescence recovery (t50) ranging from 17 to 45 s as compared with 5 s in interphase nucleoli (Fig. 2 A, top, B, and C). Remarkably, GFP-UBF1 shows different kinetics at various phases of mitosis (Fig. 2, B and C). NOR-associated GFP-UBF1 fluorescence recovers the fastest at metaphase with a t50 of 15 s and slower at prophase and telophase with t50 of 45 s and 31 s, respectively (Fig. 2, B and C). The fluorescence recoveries reflect the exchange dynamics of the fusion protein on and off chromosomes (Phair et al., 2004). The decrease in fluorescence recovery during mitosis could either be due to lower accessibility and thus a lower on rate of the protein with chromatin, or alternatively, it could be due to a slower off rate of the protein from chromosomes. To distinguish between these possibilities, we used fluorescence loss in photobleaching (FLIP) analysis to compare the rate of fluorescence loss of GFP-UBF1 in interphase cells with those on mitotic chromatin during continuous bleach pulses in the nucleoplasm or mitotic cytoplasm, respectively (Fig. 2 A, bottom and D). If accessibility is the cause of the slower FRAP rate, we expect similar FLIP rates in interphase and mitotic cells because FLIP primarily measures the off rate of the protein from DNA. The results demonstrate that the fluorescence loss of GFP-UBF1 is much slower on mitotic chromosomes as compared with that in interphase nucleoli (Fig. 2 A, bottom and D), suggesting that UBF1 has a slower off rate from mitotic than from interphase chromosomes.
Figure 2.
FRAP and FLIP of GFP-UBF1 during mitosis in HeLa cells. BL represents the first image acquired after photobleaching. The number at the bottom of each panel indicates the time after photobleaching. (A) Fluorescent images of the FRAP (top) and FLIP (bottom) processes. Box indicates the area of bleaching and fluorescence recovery. Bar, 10 μm. (B) Quantification of the fluorescence recoveries through changes in RFI over time for GFP-UBF1 in interphase and all phases of mitotic cells. (C) Graphic presentation of the time of 50% fluorescence recovery (t50) of GFP-UBF in cells at various mitotic phases and interphase. (D) FLIP of GFP-UBF1 at interphase and metaphase cells. Error bars represent averages from 12 to 16 cells ± SD.
In addition to UBF1, RPA43-GFP, which has been extensively characterized (Dundr et al., 2002), also associates with the NORs throughout mitosis. RPA43-GFP shows similar fluorescence recovery kinetics in both prophase, when NORs are transcriptionally inactive, and in interphase (Fig. 3 D), when NORs are transcriptionally active. The recovery kinetics increases to t50 around 2 s during metaphase (unpublished data) and anaphase from ∼7 s in interphase (Fig. 3). However, upon transcriptional reactivation of NORs in late anaphase/early telophase, FRAP kinetics of RPA43-GFP returns to levels similar to those observed in interphase nucleoli (Fig. 3 A, bottom and D; Dundr et al., 2002). This observation suggests that GFP-RPA43 may alter its association–dissociation kinetics at NORs and is loosely associated with NORs when rDNA transcription is shut down.
Figure 3.

FRAP of GFP-fusion proteins of RNA pol I subunits, RPA39/40 and RPA43 during anaphase–telophase transition. Bar, 10 μm. (A) Fluorescent images of the FRAP processes. Boxes indicate the areas of bleaching and fluorescence recovery. (B) Quantitation of the fluorescence recoveries through changes in RFI for GFP-RPA39/40 and (C) graphic presentation of t50 of GFP-RPA39/40 in interphase and mitotic cells. (D) FRAP of RPA43-GFP and (E) graphic presentation of t50 of RPA43-GFP. Error bars represent averages from 10 to 14 cells ± SD.
In contrast to UBF1 and RPA43, GFP-RPA39/40 shows a different association behavior with mitotic NORs. GFP-RPA39/40 associates with NORs at prophase, but is no longer detected in NORs on metaphase chromosomes, and it becomes again associated with NORs at late anaphase and early telophase (Fig. 1; Leung et al., 2004). GFP-RPA39/40 recovers at NORs slightly faster on prophase chromosomes as compared with those within interphase nucleoli (Fig. 3, B and C). Although GFP-RPA39/40 does not appear to be consistently associated with NORs throughout mitosis, it exchanges from NORs in a way similar to RPA43 at prophase and telophase.
The differences in the dynamics and associations of different RNA pol I subunits during mitosis is consistent with the different functional roles of these subunits. RPA43, a regulatory subunit, is thought to play a role in bridging the initiation complex and other RNA pol I subunits (Cavanaugh et al., 2002; Yuan et al., 2002). In contrast, RPA39/40 is thought to be assembled into the polymerase complex at a late stage (Dundr et al., 2002). Altogether, these findings demonstrate that several RNA pol I transcription factors rapidly exchange from chromosomes during mitosis in the absence of transcription and have access to NORs even in condensed mitotic chromosomes.
GFP-UBF1 accesses NORs during mitosis through DNA binding
Although RNA pol I transcription factors appear to retain their dynamic association/disassociation behavior on mitotic NORs, it is not clear if these factors bind directly to target DNA or interact with components that are associated with NORs. To distinguish between these possibilities, we analyzed GFP-UBF1 in detail because its DNA-binding properties have been extensively characterized (Bell et al., 1988; Jantzen et al., 1990). Deletion mutations from the COOH terminus of the protein showed that the NH2 terminus plus high mobility group (HMG) box 1 are sufficient for DNA binding as tested by binding to a DNA column or footprinting of the rDNA promoter (Jantzen et al., 1992). A mutant without the HMG box 1 can no longer be footprinted at the rDNA promoter (Jantzen et al., 1992). We generated a series of GFP-tagged deletion mutant from the COOH terminus similar to those made by Jantzen et al. (1992)(Fig. 4 A). The mutants were transfected into HeLa cells and the localization of the mutants on mitotic NORs was evaluated. As demonstrated in Fig. 4 B, all deletion mutants containing the NH2 terminus plus HMG box 1 were able to associate with metaphase NORs as pairs of dots (arrowheads). However, neither the GFP-fusion protein containing the NH2 terminus nor the COOH terminus alone was detected on the mitotic NORs (Fig. 4 B, bottom). These results demonstrate that the DNA-binding capacity of GFP-UBF1 is required for the association of the protein with mitotic NORs, thus suggesting that GFP-UBF1 is able to access NORs at the DNA level. In addition, treatment of cells with actinomycin D, a minor groove DNA intercalator (Perry and Kelley, 1968) that likely disrupts the DNA binding of GFP-UBF1, significantly reduced the exchange of GFP-UBF1 from mitotic NORs (unpublished data). These findings further support the notion that mitotic DNA, although highly compact and condensed, remains accessible to RNA pol I transcription factors.
Figure 4.
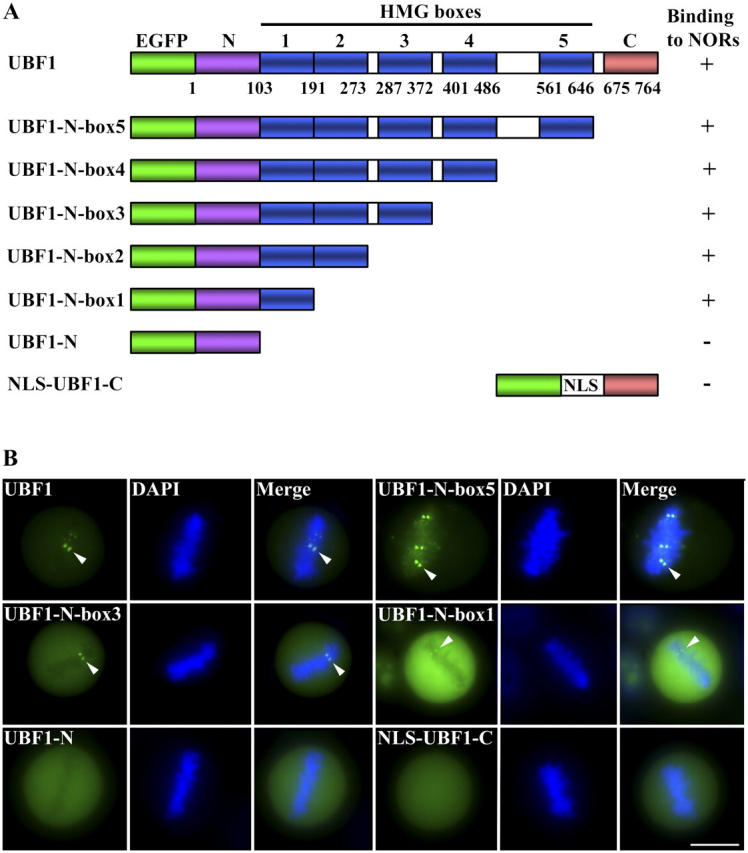
Deletion mutagenesis analyses demonstrate that DNA-binding capacity is required for the association of GFP-UBF1 with the mitotic NORs. (A) Schematic diagram of the mutants generated. (B) The localization of the UBF mutant GFP-fusion proteins on mitotic NORs (arrowheads). Bar, 10 μm.
Linker histone H1 exchanges with mitotic chromosomes with phase-specific dynamics
Because transcription factors are able to access mitotic chromatin at the DNA level, we were interested in the dynamics of structural chromatin proteins during mitosis, particularly the nucleosome components including the linker and core histones. The mammalian linker histone H1 binds to the linker region between two adjacent nucleosomes and is involved in chromatin structure, remodeling, folding, and the regulation of transcription in a gene-specific manner (Brown et al., 1997; Takami and Nakayama, 1997; Wolffe, 1998; Gunjan and Brown, 1999). To evaluate whether H1 exchanges from highly condensed and transcriptionally inactive mitotic nucleosomes, we performed FRAP and FLIP analyses of H1c-GFP during different phases of mitosis (Fig. 5). The H1c-GFP–fusion protein expressed under an inducible promoter in an NIH 3T3 cell line has been demonstrated to behave like endogenous H1c by several criteria (Lever et al., 2000; Misteli et al., 2000). The same expression construct was stably transfected into HeLa cells with an expression level of less than 5% of total H1c expression. FRAP recovery kinetics of H1c-GFP in both human and mouse cell lines were evaluated in interphase nuclei and on metaphase chromosomes. Identical recovery curves were found in mouse and human cells and the recovery curve in interphase nuclei is comparable to published studies (Lever et al., 2000; Misteli et al., 2000), suggesting that mouse H1c-GFP behaves similarly in human and mouse cells (unpublished data). Identical results were also obtained when recombinant H1 was microinjected into NIH 3T3 or HeLa cells, demonstrating that the COOH-terminal GFP tag does not interfere with protein function (unpublished data). These findings demonstrate that H1c-GFP can be used as a good marker protein for H1 in living cells.
Figure 5.

H1c-GFP rapidly exchanges with mitotic chromosome and the dynamics of the process varies throughout different mitotic phases. FRAP analyses of GFP-tagged histone H1c at various phases of mitosis (A). Fluorescent images of FRAP of H1c-GFP. (B and C) Quantification of the fluorescence recoveries through changes in RFI over time. (D) Fluorescent images of the FLIP of H1c-GFP. (E) Quantitation of the FLIP reflected by FRI over time. Bars, 10 μm. Error bars represent averages from 10 to 12 cells ± SD.
During mitosis, chromosome-associated H1c-GFP shows FRAP from prophase to telophase (Fig. 5 A). The concomitant decline in fluorescence intensity in the unbleached region with the increase in the bleached region is indicative of rapid recapture of H1c-GFP molecules on to chromosome upon dissociation, thus minimizing their residence time in a free form in the mitotic cytoplasm. Consistent with this notion, the fluorescence signal on mitotic chromosomes declines during continuous bleaching of the mitotic cytoplasm (Fig. 5, D and E).
The rate of FRAP for H1c-GFP is reduced during mitosis as compared with recovery times in interphase cells (Fig. 5, A–C). This is consistent with a previous report that H1.1-GFP showed an elevated t50 at metaphase (Lever et al., 2000). Detailed evaluation of the dynamics of H1c-GFP at various phases of mitosis demonstrates that the fluorescence recovery of H1c-GFP at metaphase is enhanced (t50 = 4.5 min) as compared with recovery during either prophase, anaphase, or telophase (t50 ranging from 10 to 15 min; Fig. 5, A–C). Our observations demonstrate that there is an active exchange of H1 to and from condensed mitotic chromosomes and that the rate of exchange is different at the various phases of mitosis, possibly reflecting changes in the association of H1 with DNA at specific phases.
Subpopulations of core histones exchange on and off anaphase and telophase chromosomes
Because linker histone continues to exchange on and off condensed chromosome during mitosis, we were interested in analyzing the association of the core histones with mitotic chromosomes. Recent evidence suggests that core histones are disrupted and replaced during transcriptional activation (Belotserkovskaya and Reinberg, 2004). The dynamics of nucleosomes during mitosis have not been explored. GFP-H2A, H2B-GFP, H3-GFP, and H4-GFP have previously been functionally characterized and demonstrated to be properly incorporated into nucleosomes in vivo (Perche et al., 2000; Kimura and Cook, 2001). Overall, the fluorescence recoveries of GFP-core histones through FRAP analyses using stably transfected human cells were very slow in non-S interphase nuclei (Kimura and Cook, 2001), with a small subpopulation of H2B-GFP that was more rapidly exchanged (Kimura and Cook, 2001). With the same stably transfected cell lines, we examined the association of these fusion proteins to condensed chromosomes throughout all phases of mitosis using FRAP analyses (Figs. 6 and 7). Although all of the GFP-core histone fusion proteins show 5% or less recovery during prophase and metaphase during 20-min observations, they show significant enhanced recovery at anaphase and telophase, reaching 13–17% recovery after 20 min (Figs. 6 and 7). H2B-GFP seemed to recover slightly more than GFP-H2A within the same period (Fig. 7, A–D), whereas H3- and H4-GFP show similar recovery curves in those phases of mitosis (Fig. 7, E–H). These findings suggest that a subpopulation of core histones is exchanged on and off anaphase and telophase chromosomes.
Figure 6.
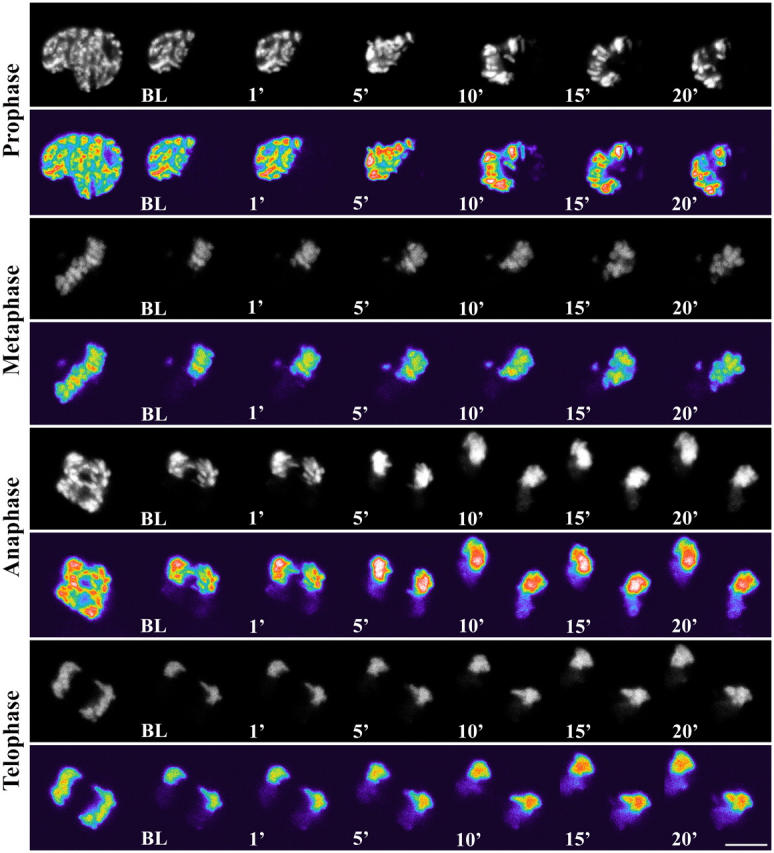
FRAP analyses of H2B-GFP at all phases of mitosis. Cells were imaged before and during recovery after bleaching of chromosome area. For each M phase cell, top panel represents original images, bottom panel represents pseudocolored images. Bar, 10 μM.
Figure 7.
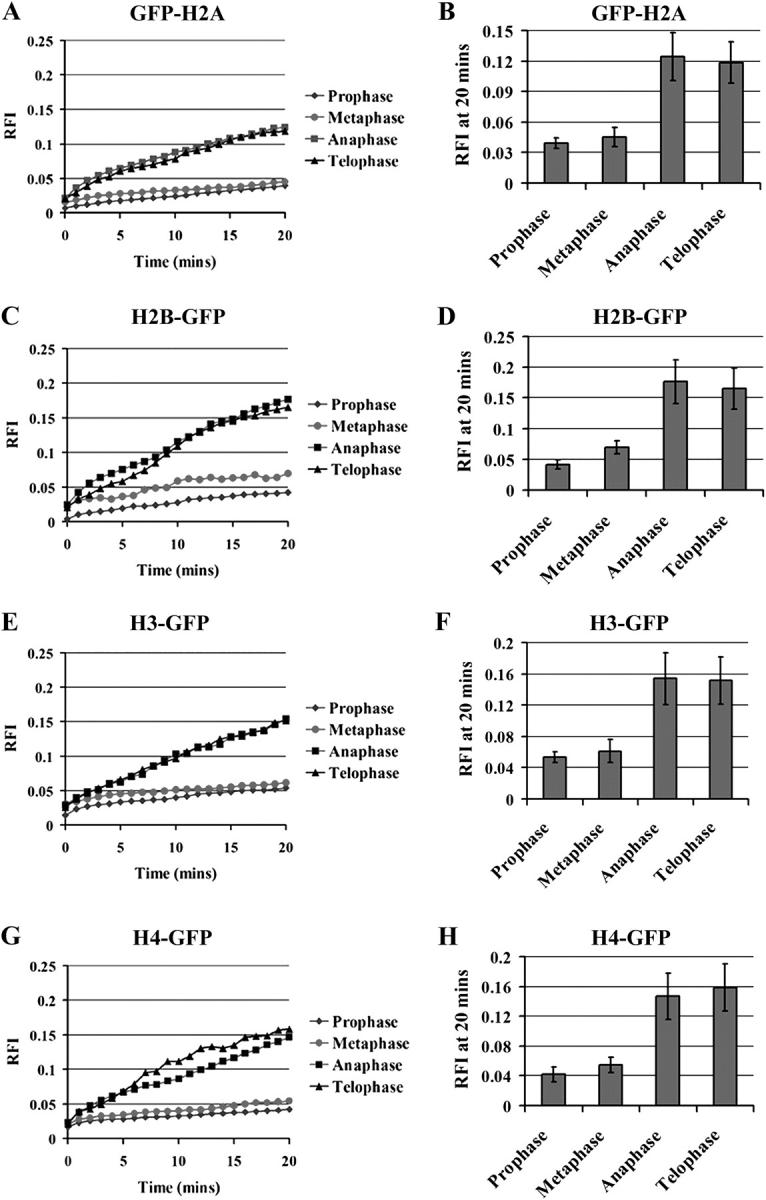
FRAP analyses of core histones at all phases of mitosis. Quantification of the fluorescence recoveries through changes in RFI over time for GFP-H2A, H2B-GFP, H3-GFP, and H4-GFP, correspondingly. Error bars represent averages from 10 cells ± SD.
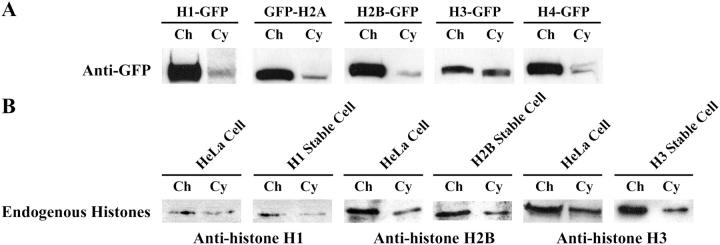
The dynamic exchange of chromatin proteins makes the prediction that a soluble unbound pool of the exchanging protein must exist. To confirm the predicted existence of a soluble pool of core histones in the mitotic cytoplasm, we fractionated metaphase cells into chromosome and cytoplasmic fractions and analyzed the distribution of endogenous and tagged histones by Western blot (Valdivia, 1998). Endogenous histones H1, H2B, and H3 can be detected both in the chromosome and cytoplasmic fraction prepared from either transfected or nontransfected cells (Fig. 8 B). Importantly, the GFP-fusion proteins of all five histones can also be detected in the cytoplasmic fraction in stably expressing cells (Fig. 8 A). These findings demonstrate the existence of a soluble cytoplasmic pool of core and linker histones in mitotic cells that is the prerequisite for any possible exchange.
Figure 8.
Western blot analyses of the presence of histones in mitotic cytoplasm. (A) Endogenous histones were detected in chromosome (Ch) and cytoplasmic fractions (Cy) prepared from both stably transfected and non transfected cells. (B) The GFP-fusion histones were detected in both the chromosome (Ch) fractions and the cytoplasmic fractions (Cy) by an anti-GFP antibody.
Core histone exchanges on anaphase and telophase chromosome temporally correlate with H3-K9 acetylation and RNA pol II polymerase association
One possible explanation for partial core histone exchange at the end of mitosis is that the replacement is linked to reactivation of transcription. To test this possibility, we evaluated the state of some of the histone tail modifications and the association of RNA pol II with chromosome at the anaphase–telophase transition, and correlated these changes with the level of transcription as determined by in situ bromo-uridine (Br-U) incorporation at the same period (Huang et al., 1998). Immunolabeling using a specific antibody against acetylated H3-K9 demonstrated that acetylated H3-K9 can be detected on segregating chromosomes as early as anaphase, whereas it is completely absent from prophase and metaphase chromosomes (Fig. 9 A). Demethylation and acetylation of H3-K9 have been correlated with chromatin remodeling associated with transcriptional activation (Fischle et al., 2003). The increase in acetylated H3-K9 suggests that chromatin remodeling begins despite the visibly persistent condensation of chromosome at this stage of mitosis. In addition, as H3-S10 phosphorylation statues has also been shown to be associated with the acetylation of H3 (Lo et al., 2000, 2001; Edmondson et al., 2002), we were interested to evaluate whether the increases in acetylation of H3-K9 correspond to the decreases in H3-S10 phosphorylation. Double labeling of acetylated H3-K9 and phosphorylated H3-S10 showed that as the labeling of acetylated H3-K9 increases on anaphase and telophase chromosomes, the signal for phosphorylated H3-S10 decreases (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200407182/DC1). These findings further support that chromatin remodeling initiates before the large-scale chromosome decondensation at mitotic exit.
To evaluate whether chromatin remodeling occurs concurrently with transcriptional reactivation, we examined the localization of RNA pol II on the anaphase–telophase chromosomes. The largest subunit of RNA pol II fused to GFP (GFP-pol II LS) has previously been shown to be fully functional in transcription (Sugaya et al., 2000; Kimura et al., 2002). The same stably transfected cells were examined for the localization of GFP-pol II LS in mitotic cells (Fig. 9 B). Although GFP-pol II LS is not detected on prophase and metaphase chromosomes, it becomes associated with chromosomes at the anaphase–telophase transition (Fig. 9 B), consistent with a report by Prasanth et al. (2003). The association is concomitant with the increased acetylation of H3-K9 (Fig. 9 A) and takes place before the onset of bulk RNA synthesis when colabeled with Br-U incorporation (Fig. 9 B). Cells expressing GPF-pol II LS were pulse labeled for 5 min with Br-U and the incorporation of nucleotide was detected using a specific antibody against Br-U (Huang et al., 1998). The results demonstrate that the most active cellular transcriptional sites at anaphase–telophase transition are in the NORs (Fig. 9 B) as shown previously (Thiry and Goessens, 1996). Outside of NORs, little Br-U labeling was detected (Fig. 9 B), suggesting that the bulk of RNA pol II transcription has yet to be initiated. These findings demonstrate that histone acetylation and RNA pol II association coincides with core-histone exchanges. These events appear to precede large-scale chromatin decondensation and general transcriptional reactivation.
Discussion
The RNA pol I transcription machinery exchanges rapidly on and off mitotic condensed NORs
Several RNA pol I transcription factors are associated with the condensed and transcriptionally silent NORs during mitosis (Scheer and Rose, 1984; Roussel et al., 1993, 1996; Scheer and Weisenberger, 1994; Gilbert et al., 1995; Moss and Stefanovsky, 1995; Jordan et al., 1996; Hernandez-Verdun et al., 2002). Although the accumulation of RNA pol I components could have been due to their stable association with NORs upon transcriptional silencing, the highly dynamic nature of these transcription factors and many chromatin proteins during interphase (Chen and Huang, 2001; Dundr et al., 2002; Phair et al., 2004) provoked the question of whether the NOR-associated proteins were dynamically exchanged rather than stably bound. In this report, we use FRAP analyses to show that the NOR-associated RNA pol I transcription machinery is highly dynamic throughout mitosis. UBF and RNA pol I subunits exchange from NORs within minutes and the association of GFP-UBF with the NORs is dependent on its DNA-binding capacity. These findings suggest that NORs are accessible at least to a subset of transcription factors at the DNA level throughout mitosis.
The presence of some RNA pol I transcription factors, including UBF and RNA pol I subunit RPA43 at transcriptionally silenced NORs throughout mitosis, suggests that chromatin condensation does not drastically change NOR accessibility. However, the exchange kinetics of the RNA pol I machinery is altered. GFP-UBF shows a slowdown in fluorescence recovery, with t50 altered from 5 s in interphase nucleoli to around 17-45 s on NORs during mitosis. Using FLIP analysis we show that these slower exchanges are most likely due to an increased residence time of UBF on NORs, suggesting that GFP-UBF1 binds to rDNA cluster more stably in mitosis than in interphase. Similarly, the fluorescence recovery of TATA-binding protein (GFP-TBP) is also significantly slower (50% fluorescence recovery cannot be reached 20 min after bleaching) when half of the entire mitotic chromosomes including NORs were bleached (unpublished data).
In comparison, polymerase subunits behave differently. GFP-RPA39/40 and RPA43-GFP both show slightly faster fluorescence recovery in mitosis than in interphase nucleoli. Although RPA43 associates with NORs throughout mitosis, RNA pol I subunits RPA39/40, RPA194, and RPA16 (Leung et al., 2004) are absent from the transcriptionally inactive NORs during metaphase and early anaphase. Therefore, it is likely that RNA pol I subunits are not efficiently recruited to the ribosomal promoters at NORs at this period. The finding that RPA43, which is responsible for recruitment of RNA pol I to the promoter through specific interactions with the preinitiation complex (Cavanaugh et al., 2002; Yuan et al., 2002), is associated with transcriptionally silenced NORs, but with significantly faster exchange kinetics, further supports the view that the recruitment of RNA pol I subunits is weak when transcription is silenced during mitosis. Together, our findings indicate that RNA pol I transcription factors have independent NOR association and dissociation kinetics. These findings also support the view that the RNA pol I transcription apparatus assembles onto promoters sequentially, rather than being recruited as a holoenzyme (Dundr et al., 2002).
It is interesting to note that factors that are part of the initiation complex, including GFP-UBF and GFP-TBP, show reduced exchange dynamics during mitosis, whereas RNA pol I subunit dynamics appear to be slightly faster during the transcription silencing of metaphase and early anaphase NORs. It is not clear what dictates the changes in the dynamics of GFP-UBF or TBP on and off mitotic chromatin when cells enter mitosis. Previous studies have shown that cell cycle–specific phosphorylation by cyclin-B kinase is essential for the silencing of RNA pol I transcription during mitosis, possibly at the initiation level (Heix et al., 1998; Kuhn et al., 1998; Klein and Grummt, 1999; Sirri et al., 1999). Perhaps either the phosphorylation of initiation complex components or the general phosphorylation state of mitotic cells influences the rate of exchange of initiation factors from NORs. In addition, it appears that all the NORs that are associated with the transcription apparatus during mitosis incorporate Br-U at the mitotic exit, supporting the idea that the NORs that are not active for transcription for the next cell cycle might not associate with the transcription apparatus during mitosis.
In addition to the dynamic association of RNA pol I transcription factors with mitotic NORs, components of initiation complex for RNA pol II, transcription factor IID, and RNA pol III transcription factor IIIB, are also associated with mitotic chromosomes (Chen et al., 2002; Christova and Oelgeschlager, 2002; Fairley et al., 2003) although the exchange kinetics is much slower (Chen et al., 2002). Most recently, other DNA-binding proteins, including GFP-fusion of topoisomerase II, HMGA1, HMGB1, B2, and RCC1 proteins, also show exchanges with mitotic chromosomes through FRAP analyses (Scaffidi et al., 2002; Tavormina et al., 2002; Harrer et al., 2004; Hutchins et al., 2004). Our observations together with studies from others demonstrate that mitotic DNA in general although highly condensed is both accessible to, and maintains dynamic interactions with, its binding factors during mitosis. Therefore, transcription silencing during mitosis is unlikely due to the inaccessibility of DNA.
Exchanges of linker histone from mitotic chromosomes
Our observations that linker and core histones can exchange from mitotic chromosomes indicate that highly compact mitotic chromosomes are accessible at the nucleosome level. Histone H1 is a linker histone that binds to the DNA sequences between two adjacent nucleosomes. In vitro and in vivo experiments have suggested its role in stabilization of condensed chromosomes, transcription regulation of a subset of genes (Carruthers and Hansen, 2000), and telomeric silencing (Jedrusik and Schulze, 2003). Although deletion of H1 in unicellular organisms has little to no effect on chromatin structure or cell viability (Shen et al., 1995; Ushinsky et al., 1997; Patterton et al., 1998; Barra et al., 2000; Ramon et al., 2000), recent studies demonstrated that H1 is essential for mouse embryonic development (Fan et al., 2003). However, the mechanism by which H1 regulates chromatin structure and gene expression remains to be clarified.
We have observed that histone H1c-GFP actively exchanges on and off mitotic chromatin, though at a slower rate than in interphase cells. The t50 of H1c-GFP varies from around 4.5 min at metaphase to 10–15 min at prophase, anaphase, and telophase. It is intriguing that the t50 of fluorescence recovery times vary at different phases of mitosis. Upon entering prophase, FRAP slows down, followed by acceleration at metaphase and a slowdown again at anaphase and telophase. The recoveries are very similar at prophase, anaphase, and telophase. The molecular mechanism behind these changes is not clear, but might involve phosphorylation because treatment of metaphase cells with staurosporine reduced the t50 for H1c-GFP from around 5 min to more than 20 min (unpublished data), consistent with the report where changes in the phosphorylation of H1.1 affected the association of H1.1 with chromatin (Hendzel et al., 2004).
Core histones exchange coincides with reactivation of transcription in telophase
Although GFP-fusion proteins of H2A, H2B, H3, and H4 show little or no FRAP of prophase and metaphase chromosomes, they showed limited but consistent dynamic exchange at later stages of mitosis. Because the FRAP analyses were performed directly on the mitotic chromosomes and there was a soluble pool of all core histones in the mitotic cytoplasm, we interpret these observations as a replacement of core histones in mitotic nucleosomes. The core histone octamer has been thought to be deposited primarily at S phase (Wolffe, 1998, 2001); however, emerging evidence indicates that core histones can be replaced, either partially or even completely, in a replication-independent manner (Ahmad and Henikoff, 2002a; Svejstrup, 2003; Vermaak et al., 2003; Belotserkovskaya et al., 2004). The identification and characterization of the facilitates chromatin transcription (FACT) complex, both in yeast and in higher eukaryotic cells, demonstrated that components of the complex interact with core histones in reconstituted cell extract assays (Belotserkovskaya et al., 2003) and are recruited to activated loci on Drosophila polytene chromosomes (Saunders et al., 2003). Loss of function of FACT homologues (spt6) in yeast induces the loss of active chromatin, suggesting that FACT is necessary for transcription-coupled nucleosome assembly (Kaplan et al., 2003). Additional studies using in vitro transcription analyses demonstrated that the passing of RNA pol II resulted in quantifiable loss of H2A: H2B (Kireeva et al., 2002). There are also several histone variants that are incorporated in a replication-independent manner. Histone variant H3.3 has been shown to incorporate only into transcriptionally active chromatin outside of S phase (Ahmad and Henikoff, 2002b). Another histone variant, H2A.Z, has also been shown to form a dimer with H2B that exchanges with the H2A: H2B dimer on an existing nucleosome array in the absence of replication (Mizuguchi et al., 2004). All these studies indicate the possibility that disruption of nucleosomes can take place independently of replication and can be coupled with transcription.
The exchange of core histones on mitotic chromatin at anaphase and telophase observed by FRAP may reflect the replacement of a subset of nucleosomes in genome regions that are transcriptionally reactivated in the earliest parts of the new cell cycle. This interpretation is consistent with evidence of chromatin remodeling and chromatin association with RNA pol II at the anaphase–telophase transition (Fig. 9; Prasanth et al., 2003). In situ incorporation of Br-U for 5 min at the same stage showed little labeling outside of NORs (Fig. 9), suggesting that the majority of transcription is yet to commence at this point. The replacement of core histones conceivably precedes transcription to allow the clearance of promoter regions for factors to engage. This interpretation is consistent with the emerging model of nucleosome disruption during transcription activation (Belotserkovskaya et al., 2004). The exchange dynamics of H3 and H4 appear to be similar, whereas those of H2B are slightly faster than H2A. These observations are consistent with the idea that H2A:H2B dimers are disrupted to a larger extent than H3 and H4 during transcription (Nacheva et al., 1989). The lesser extent of exchange of H2A in comparison with H2B could be partially due to the replacement of H2A by its variants, such as H2A.Z (Redon et al., 2002).
In summary, we have found that mitotic chromosomes in general and ribosome DNA loci in particular are accessible to transcription factors and structural proteins in spite of its high compaction ratio. Nucleosome components, including linker and core histones, also show exchanges from condensed chromosome during mitosis. These findings suggest that transcription silencing during mitosis is likely due to other reasons, such as the loss of activity and/or modifications of transcription factors, rather than to the inaccessibility of DNA. Furthermore, a small fraction of core histones undergo replacement at the anaphase–telophase transition that is temporally correlated with histone tail modification and RNA pol II association, suggesting that alterations in chromatin structure may take place before resumption of transcription as cells exit mitosis. Although the functional significance of the exchange of transcription factors and structural chromatin proteins from chromosomes during mitosis is unknown, it is tempting to speculate that the dynamic association of these proteins may help prepare the genome for its reactivation by maintaining a relatively open chromosome configuration. This may serve to minimize the requirement for decondensation when large numbers of genes must be rapidly reactivated as cells exit mitosis.
Materials and methods
Cell culture and transfection
Histone H1, H2A, H2B, H3, and H4 stable cell lines were provided by H. Kimura (Kyoto University, Japan; Kimura and Cook, 2001). Histone stable cell lines and HeLa cells were maintained in DME supplemented with 10% FBS at 37°C and 5% CO2. A clone stably expressing the largest subunit of RNA pol II tagged with EGFP was cultured in Ham's F12 medium plus 10% FBS at 39°C (Sugaya et al., 2000). Cells were diluted twice a week. Expression constructs were transiently transfected into HeLa cells by electroporation. Subconfluent cells in a 100-mm culture dish were collected by trypsinization and mixed with 20 μg of DNA, including 4 μg target DNA and 16 μg sheared salmon sperm DNA. A 280-μl mixture of cells in DME containing 10% FBS and DNA was electroporated in an electroporator (Bio-Rad Laboratories) at 250 V and 950 μF. Transfected cells and stable cell lines were subsequently seeded onto glass coverslips that were mounted on the bottom of 35-mm Petri dishes with an opening in the center (MatTek) and grown for 24 h. To inhibit phosphorylation, cells were treated with 1 μM staurosporine for 10–30 min. Staurosporine is an inhibitor of phospholipid/calcium-dependent protein kinase.
Construction of GFP-fusion proteins
pEGFP-UBF1 expression vector was described previously (Chen and Huang, 2001). RPA43-GFP expression vector was previously characterized in Dundr et al. (2002). GFP-UBF1mutants were constructed by inserting cDNA containing UBF1 mutants into the pEGFP-C1 vector (CLONTECH Laboratories, Inc.). GFP-RAP39/40 was constructed by inserting cDNA containing RPA39/40 [gi:2266928] into the BglII–KpnI restriction site of pEGFP-C1 vector (CLONTECH Laboratories, Inc.) and the HeLa stable cell lines were constructed as described in Leung et al. (2004). Expression constructs were sequenced to confirm that they contained the correct cDNA sequence.
Photobleaching and live cell imaging
Cells transfected with the GFP-fusion protein expression vector and stable cell lines were maintained in DME supplemented with 30 mM Hepes, pH 7.1, to stabilize the pH of the medium during imaging. The 35-mm dishes with coverslip bottoms were directly mounted onto a 510 confocal laser scanning microscope (Carl Zeiss MicroImaging, Inc.) equipped with an argon-krypton laser. The medium was kept at 37°C using an ASI 400 air stream incubator (Nevtek). The 488-nm laser and a 63× plan Apo lens with a 1.4 NA were used in bleaching and imaging experiments. A laser power of 0.05–1.1% of 3.75 mW was used in image acquisitions, and 100% of 3.75 mW was used in photobleaching. The time for each image acquisition ranges from 1.8 s to 1 min, which did not significantly influence the fluorescent intensity through multiple acquisitions. An area of 2 μm2 or a half area of chromosomes was bleached with an iteration of 50–150. In FRAP analyses, images were collected before, immediately after, and at 9- or 1.8-s intervals after bleaching for the nucleolar or NOR FRAP and 1-min intervals for the chromosome FRAP. At least 10 cells were analyzed for each result.
Quantitation of relative fluorescence intensity (RFI)
Fluorescence intensity was measured using Metamorph (Universal Imaging Corp.) imaging software. The average intensities of the areas of interest in images, including before and immediately after, and a series of time points after bleaching were measured under the same conditions for each dataset. The fluorescence intensity of a nonphotobleached region in the same cell was also measured. The RFI at each time point was calculated as described by Phair and Misteli (2000). RFI = (It/TNt)/(I0/TN0), where It = the average fluorescence intensity of the photobleached region at various time points after photobleaching, TNt = the average fluorescence intensity of the nonphotobleached region at the corresponding time point, I0 = the average fluorescence intensity of the photobleached region before photobleaching, and TN0 = the average fluorescence intensity of the nonphotobleached region before photobleaching. When It/TNt = I0/TN0, namely, when RFI = 1, the fluorescence recovery of the photobleached region reaches 100%.
Detection of histone proteins of chromosome and cytoplasm fractions in mitotic cells
The isolation and purification of mitotic chromosomes were performed as described previously (Valdivia, 1998). HeLa cells were treated with nocodazole (500 ng/ml; Sigma-Aldrich) for ∼16 h. Mitotic cells were then incubated in 75 mM KCl on ice for 20 min and subsequently in disruption buffer (10 mM Tris-HCl, pH 7.4, 120 mM KCl, 20 mM NaCl, 0.1% Triton X-100, 2 mM CaCl2, 5 μg/ml aprotinin, 5 μg/ml leupeptin, 0.5 μg/ml pepstatin A and 0.1 mM PMSF) for 10 min. The cells were homogenized and the resultant extract centrifuged at 3,000 g for 15 min at 4°C. The supernatant was used as the cytoplasmic fraction. The pellet, which is enriched with crude chromosomes, was further purified by centrifugation in a 36-ml linear gradient consisting of 20–60% wt/vol sucrose in disruption buffer. Fractions containing chromosomes were pooled and centrifuged at 2,500 g for 10 min at 4°C. The chromosome pellet was washed, sonicated, and resuspended in aqueous disruption buffer. To extract the histone proteins, sulfuric acid was added into purified chromosome and cytoplasmic fraction to a final concentration of 0.4 N and left on ice 30 min. After centrifugation at 1,200 g for 10 min, to the supernatants TCA was added to 20%, these were left on ice 30 min, and then centrifuged again for 10 min. The pellets were washed with acetone, 0.1% HCl, and then rewashed with acetone. After being air dried, the pellets were resuspended in Laemmli sample buffer, sonicated, and boiled. Proteins present in the chromosome and cytoplasmic fractions were separated by SDS-PAGE for Western blot analyses using antibodies directed against histones H1, H2A (Upstate Biotechnology), H2B (Cell Signaling Technology), H3 (Upstate Biotechnology), H4 (a gift from D. Allis, The Rockefeller University, New York, NY), and GFP (BD Biosciences Clontech). Immunoblotting signals were visualized by conversion of SuperSignal West Pico Chemiluminescent Substrate (Pierce Chemical Co.).
In vivo incorporation of Br-U
HeLa cells were seeded onto glass coverslips in 35-mm Petri dishes and were grown overnight. Cells were rinsed once with PBS and once with a glycerol buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 25% glycerol, 0.5 mM PMSF, and 0.5 mM EGTA). Cells were then permeabilized in the glycerol buffer containing 5 μg/ml digitonin at RT for 3 min. Subsequently, cells were incubated in transcription cocktail (100 mM KCl, 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol, 1 mM PMSF, 2 mM ATP, 0.5 Mm CTP, 0.5 mM GTP, 0.2 mM Br-UTP and 25 U/ml RNAsin) for 5 min at 37°C.
Immunolabeling
For Br-U staining, cells were gently rinsed twice with PBS and fixed in 2% PFA after the transcription reaction. For acetyl histone H3 (K9) staining, subconfluent HeLa cells grown on glass coverslips were extracted with CSK buffer containing 0.5% Triton X-100 on ice for 5 min and immediately fixed in 2% formaldehyde for 10 min. Antibodies specifically recognizing Br-U (Sigma-Aldrich), UBF (a gift from E.K.L. Chang, University of Florida, Gainesville, FL) and acetyl histone H3-K9 (Upstate Biotechnology) were incubated with cells for 1 h at RT. The immunolabeling signals were subsequently detected by incubating cells with Texas red– or FITC-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Fluorescent images were collected on a confocal laser scanning microscope (model LSM 510; Carl Zeiss MicroImaging, Inc.) equipped with META and a 405-nm laser (Carl Zeiss MicroImaging, Inc.).
Online supplemental material
Fig. S1 shows the increases of H3-K9 acetylation on telophase chromosomes temporally correlate with the decreases of H3-S10 phosphorylation. Bar, 10 μM. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200407182/DC1.
Acknowledgments
We would like to thank Drs. D. Brown (University of Mississippi, University, MS), E.K.L. Chan (University of Florida), H. Kimura, and P. Cook (University of Oxford, Oxford, UK) for their generosity in supplying cell lines and reagents. We are also grateful to Drs. S. Adam and Y. Gruenbaum for constructive discussion, and Drs. D. Leary and M. McDonough for critical reading of the manuscript.
This work is supported by a National Cancer Institute grant (R01 CA77560) to S. Huang, Keith R. Porter Endowment for Cell Biology to T. Misteli, Human Frontier Science Program and Wellcome Trust to A. Lamond, and Croucher Foundation Scholarship to A.K.L. Leung.
Abbreviations used in this paper: Br-U, bromo-uridine; FACT, facilitates chromatin transcription; FLIP, fluorescence loss in photobleaching; HMG, high mobility group; NOR, nucleolar organizing region; RNA pol I, RNA polymerase I; rDNA, ribosomal DNA; RFI, relative fluorescence intensity; RPA39/40, 39/40-kD subunit of RNA pol I; RPA43, 43-kD subunit of RNA pol I; t50, time of 50% fluorescence recovery; TBP, TATA-binding protein; UBF, upstream-binding factor.
References
- Ahmad, K., and S. Henikoff. 2002. a. Epigenetic consequences of nucleosome dynamics. Cell. 111:281–284. [DOI] [PubMed] [Google Scholar]
- Ahmad, K., and S. Henikoff. 2002. b. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 9:1191–1200. [DOI] [PubMed] [Google Scholar]
- Barra, J.L., L. Rhounim, J.L. Rossignol, and G. Faugeron. 2000. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol. Cell. Biol. 20:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.P., R.M. Learned, H.M. Jantzen, and R. Tjian. 1988. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 241:1192–1197. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya, R., and D. Reinberg. 2004. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 14:139–146. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya, R., S. Oh, V.A. Bondarenko, G. Orphanides, V.M. Studitsky, and D. Reinberg. 2003. FACT facilitates transcription-dependent nucleosome alteration. Science. 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya, R., A. Saunders, J.T. Lis, and D. Reinberg. 2004. Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta. 1677:87–99. [DOI] [PubMed] [Google Scholar]
- Brown, D.T., A. Gunjan, B.T. Alexander, and D.B. Sittman. 1997. Differential effect of H1 variant overproduction on gene expression is due to differences in the central globular domain. Nucleic Acids Res. 25:5003–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers, L.M., and J.C. Hansen. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285–37290. [DOI] [PubMed] [Google Scholar]
- Cavanaugh, A.H., I. Hirschler-Laszkiewicz, Q. Hu, M. Dundr, T. Smink, T. Misteli, and L.I. Rothblum. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277:27423–27432. [DOI] [PubMed] [Google Scholar]
- Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., C.S. Hinkley, R.W. Henry, and S. Huang. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol. Biol. Cell. 13:276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79–82. [DOI] [PubMed] [Google Scholar]
- Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L.I. Rothblum, R.D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 298:1623–1626. [DOI] [PubMed] [Google Scholar]
- Edmondson, D.G., J.K. Davie, J. Zhou, B. Mirnikjoo, K. Tatchell, and S.Y. Dent. 2002. Site-specific loss of acetylation upon phosphorylation of histone H3. J. Biol. Chem. 277:29496–29502. [DOI] [PubMed] [Google Scholar]
- Fairley, J.A., P.H. Scott, and R.J. White. 2003. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 22:5841–5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Y., T. Nikitina, E.M. Morin-Kensicki, J. Zhao, T.R. Magnuson, C.L. Woodcock, and A.I. Skoultchi. 2003. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 23:4559–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle, W., Y. Wang, and C.D. Allis. 2003. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15:172–183. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., L. Lucas, C. Klein, M. Menager, N. Bonnet, and D. Ploton. 1995. Three-dimensional co-location of RNA polymerase I and DNA during interphase and mitosis by confocal microscopy. J. Cell Sci. 108:115–125. [DOI] [PubMed] [Google Scholar]
- Gunjan, A., and D.T. Brown. 1999. Overproduction of histone H1 variants in vivo increases basal and induced activity of the mouse mammary tumor virus promoter. Nucleic Acids Res. 27:3355–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer, M., H. Luhrs, M. Bustin, U. Scheer, and R. Hock. 2004. Dynamic interaction of HMGA1a proteins with chromatin. J. Cell Sci. 117:3459–3471. [DOI] [PubMed] [Google Scholar]
- Heix, J., A. Vente, R. Voit, A. Budde, T.M. Michaelidis, and I. Grummt. 1998. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 17:7373–7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendzel, M.J., M.A. Lever, E. Crawford, and J.P. Th'ng. 2004. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J. Biol. Chem. 279:20028–20034. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun, D., P. Roussel, and J. Gebrane-Younes. 2002. Emerging concepts of nucleolar assembly. J. Cell Sci. 115:2265–2270. [DOI] [PubMed] [Google Scholar]
- Huang, S., T.J. Deerinck, M.H. Ellisman, and D.L. Spector. 1998. The perinucleolar compartment and transcription. J. Cell Biol. 143:35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, J.R., W.J. Moore, F.E. Hood, J.S. Wilson, P.D. Andrews, J.R. Swedlow, and P.R. Clarke. 2004. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol. 14:1099–1104. [DOI] [PubMed] [Google Scholar]
- Jantzen, H.M., A. Admon, S.P. Bell, and R. Tjian. 1990. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 344:830–836. [DOI] [PubMed] [Google Scholar]
- Jantzen, H.M., A.M. Chow, D.S. King, and R. Tjian. 1992. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 6:1950–1963. [DOI] [PubMed] [Google Scholar]
- Jedrusik, M.A., and E. Schulze. 2003. Telomeric position effect variegation in Saccharomyces cerevisiae by Caenorhabditis elegans linker histones suggests a mechanistic connection between germ line and telomeric silencing. Mol. Cell. Biol. 23:3681–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, P., M. Mannervik, L. Tora, and M. Carmo-Fonseca. 1996. In vivo evidence that TATA-binding protein/SL1 colocalizes with UBF and RNA polymerase I when rRNA synthesis is either active or inactive. J. Cell Biol. 133:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, C.D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 301:1096–1099. [DOI] [PubMed] [Google Scholar]
- Kimura, H., and P.R. Cook. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H., K. Sugaya, and P.R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva, M.L., W. Walter, V. Tchernajenko, V. Bondarenko, M. Kashlev, and V.M. Studitsky. 2002. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 9:541–552. [DOI] [PubMed] [Google Scholar]
- Klein, J., and I. Grummt. 1999. Cell cycle-dependent regulation of RNA polymerase I transcription: the nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA. 96:6096–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A., A. Vente, M. Doree, and I. Grummt. 1998. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 284:1–5. [DOI] [PubMed] [Google Scholar]
- Leung, A.K., D. Gerlich, G. Miller, C. Lyon, Y.W. Lam, D. Lleres, N. Daigle, J. Zomerdijk, J. Ellenberg, and A.I. Lamond. 2004. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166:787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever, M.A., J.P. Th'ng, X. Sun, and M.J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 408:873–876. [DOI] [PubMed] [Google Scholar]
- Li, G., G. Sudlow, and A.S. Belmont. 1998. Interphase cell cycle dynamics of a late-replicating, heterochromatic homogeneously staining region: precise choreography of condensation/decondensation and nuclear positioning. J. Cell Biol. 140:975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, W.S., R.C. Trievel, J.R. Rojas, L. Duggan, J.Y. Hsu, C.D. Allis, R. Marmorstein, and S.L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 5:917–926. [DOI] [PubMed] [Google Scholar]
- Lo, W.S., L. Duggan, N.C. Emre, R. Belotserkovskya, W.S. Lane, R. Shiekhattar, and S.L. Berger. 2001. Snf1–a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 293:1142–1146. [DOI] [PubMed] [Google Scholar]
- Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D.T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature. 408:877–881. [DOI] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W.H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 303:343–348. [DOI] [PubMed] [Google Scholar]
- Moss, T., and V.Y. Stefanovsky. 1995. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 50:25–66. [DOI] [PubMed] [Google Scholar]
- Nacheva, G.A., D.Y. Guschin, O.V. Preobrazhenskaya, V.L. Karpov, K.K. Ebralidse, and A.D. Mirzabekov. 1989. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 58:27–36. [DOI] [PubMed] [Google Scholar]
- Patterton, H.G., C.C. Landel, D. Landsman, C.L. Peterson, and R.T. Simpson. 1998. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 273:7268–7276. [DOI] [PubMed] [Google Scholar]
- Perche, P.Y., C. Vourc'h, L. Konecny, C. Souchier, M. Robert-Nicoud, S. Dimitrov, and S. Khochbin. 2000. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr. Biol. 10:1531–1534. [DOI] [PubMed] [Google Scholar]
- Perry, R.P., and D.E. Kelley. 1968. Persistent synthesis of 5S RNA when production of 28S and 18S ribosomal RNA is inhibited by low doses of actinomycin D. J. Cell Physiol. 72:235–246. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–649. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D.T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.V., P.A. Sacco-Bubulya, S.G. Prasanth, and D.L. Spector. 2003. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 14:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon, A., M.I. Muro-Pastor, C. Scazzocchio, and R. Gonzale. 2000. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in aspergillus nidulans. Mol. Microbiol. 35:223–233. [DOI] [PubMed] [Google Scholar]
- Raska, I. 2003. Oldies but goldies: searching for Christmas trees within the nucleolar architecture. Trends Cell Biol. 13:517–525. [DOI] [PubMed] [Google Scholar]
- Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162–169. [DOI] [PubMed] [Google Scholar]
- Roussel, P., C. Andre, C. Masson, G. Geraud, and D. Hernandez-Verdun. 1993. Localization of the RNA polymerase I transcription factor hUBF during the cell cycle. J. Cell Sci. 104:327–337. [DOI] [PubMed] [Google Scholar]
- Roussel, P., C. Andre, L. Comai, and D. Hernandez-Verdun. 1996. The rDNA transcription machinery is assembled during mitosis in active NORs. J. Cell Biol. 133:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, A., J. Werner, E.D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J.T. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 301:1094–1096. [DOI] [PubMed] [Google Scholar]
- Scaffidi, P., T. Misteli, and M.E. Bianchi. 2002. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 418:191–195. [DOI] [PubMed] [Google Scholar]
- Scheer, U., and K.M. Rose. 1984. Localization of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immuncytochemistry. Proc. Natl. Acad. Sci. USA. 81:1431–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, U., and D. Weisenberger. 1994. The nucleolus. Curr. Opin. Cell Biol. 6:354–359. [DOI] [PubMed] [Google Scholar]
- Shen, X., L. Yu, J.W. Weir, and M.A. Gorovsky. 1995. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 82:47–56. [DOI] [PubMed] [Google Scholar]
- Sif, S., P.T. Stukenberg, M.W. Kirschner, and R.E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri, V., P. Roussel, and D. Hernandez-Verdun. 1999. The mitotically phosphorylated form of the transcription termination factor TTF-1 is associated with the repressed rDNA transcription machinery. J. Cell Sci. 112:3259–3268. [DOI] [PubMed] [Google Scholar]
- Sirri, V., P. Roussel, and D. Hernandez-Verdun. 2000. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 148:259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugaya, K., M. Vigneron, and P.R. Cook. 2000. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 113:2679–2683. [DOI] [PubMed] [Google Scholar]
- Svejstrup, J.Q. 2003. Transcription. Histones face the FACT. Science. 301:1053–1055. [DOI] [PubMed] [Google Scholar]
- Takami, Y., and T. Nakayama. 1997. A single copy of linker H1 genes is enough for proliferation of the DT40 chicken B cell line, and linker H1 variants participate in regulation of gene expression. Genes Cells. 2:711–723. [DOI] [PubMed] [Google Scholar]
- Tavormina, P.A., M.G. Come, J.R. Hudson, Y.Y. Mo, W.T. Beck, and G.J. Gorbsky. 2002. Rapid exchange of mammalian topoisomerase II alpha at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiry, M., and G. Goessens. 1996. The Nucleolus During the Cell Cycle. Springer-Verlag, Heidelberg. 146 pp.
- Ushinsky, S.C., H. Bussey, A.A. Ahmed, Y. Wang, J. Friesen, B.A. Williams, and R.K. Storms. 1997. Histone H1 in Saccharomyces cerevisiae. Yeast. 13:151–161. [DOI] [PubMed] [Google Scholar]
- Valdivia, M.M. 1998. Chromosome isolation for biochemical and morphological analysis. Cells: A Laboratory Manual. D.L. Spector, R.D. Goldman, and L.A. Leinwand, editors. Cold Spring Harbor Laboratory Press, New York. 49.1–49.12.
- Vermaak, D., K. Ahmad, and S. Henikoff. 2003. Maintenance of chromatin states: an open-and-shut case. Curr. Opin. Cell Biol. 15:266–274. [DOI] [PubMed] [Google Scholar]
- Wolffe, A. 1998. Chromatin. London Academic Press, London. 447 pp.
- Wolffe, A. 2001. Transcriptional regulations in the context of chromatin structure. Essays Biochem. 37:45–57. [DOI] [PubMed] [Google Scholar]
- Yuan, X., J. Zhao, H. Zentgraf, U. Hoffmann-Rohrer, and I. Grummt. 2002. Multiple interactions between RNA polymerase I, TIF-IA and TAF(I) subunits regulate preinitiation complex assembly at the ribosomal gene promoter. EMBO Rep. 3:1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]