Abstract
In mammals, dosage compensation is achieved by X chromosome inactivation in female cells. Xist is required and sufficient for X inactivation, and Xist gene deletions result in completely skewed X inactivation. In this work, we analyzed skewing of X inactivation in mice with an Xist deletion encompassing sequence 5 KB upstream of the promoter through exon 3. We found that this mutation results in primary nonrandom X inactivation in which the wild-type X chromosome is always chosen for inactivation. To understand the molecular mechanisms that affect choice, we analyzed the role of replication timing in X inactivation choice. We found that the two Xist alleles and all regions tested on the X chromosome replicate asynchronously before the start of X inactivation. However, analysis of replication timing in cell lines with skewed X inactivation showed no preference for one of the two Xist alleles to replicate early in S-phase before the onset of X inactivation, indicating that asynchronous replication timing does not play a role in skewing of X inactivation.
Introduction
In mammalian cells, dosage compensation of X-linked genes is achieved by X chromosome inactivation in female cells. X inactivation is a complex process that is regulated by integrating several mechanisms. Genetic studies have revealed that X chromosome silencing is initiated from one location on the X chromosome, the X inactivation center (Xic; Rastan, 1983). The Xist gene, which lies within the Xic, encodes a large untranslated RNA that is required and sufficient for X inactivation (Penny et al., 1996). At the onset of X inactivation, one of the X chromosomes is chosen to remain active and Xist expression is stabilized on the future inactive X (Xi) and accumulates in cis (Brockdorff et al., 1991; Brown et al., 1991). The Xist RNA associating with the X chromosome recruits different chromatin-modifying complexes, eventually rendering the chromosome transcriptionally inactive (Silva et al., 2003).
The number of X chromosomes that will be inactivated is determined relative to the ploidy of the cell (Rastan, 1983; Rastan and Robertson, 1985). X inactivation is initiated when the number of X chromosomes exceeds one in a diploid nucleus. Each cell then makes the epigenetic choice to keep one X chromosome active (Xa) and to inactivate all supernumerary X chromosomes. These observations have led to a model that suggests the presence of a blocking factor in limited quantities that can protect only one X chromosome from inactivation per diploid set of chromosomes (Lyon, 1996).
X chromosome choice is random in the embryonic lineage of the mouse. In contrast, X inactivation in metatherian mammals such as kangaroos (Cooper et al., 1971) and in the extraembryonic tissues of some eutherian mammals including mice (Takagi and Sasaki, 1975) is imprinted, and the paternal X chromosome always undergoes inactivation. Genetic studies have revealed that X chromosome choice is influenced by the X controlling element (Xce) such that X inactivation is skewed toward the chromosome possessing the weaker Xce allele (Simmler et al., 1993). At least four Xce alleles exist in mice: Xce a, Xce b, Xce c, and Xce d, with Xce a being the weakest and Xce d being the strongest allele.
Deletion of the Xist gene also leads to nonrandom X inactivation in embryonic tissues in mice (Penny et al., 1996; Marahrens et al., 1997, 1998; Csankovszki et al., 1999). However, it is controversial whether this nonrandom X activation is achieved by primary nonrandom X inactivation or by random inactivation followed by post-choice selection (also referred to as secondary nonrandom X inactivation). Deletion of the Xist promoter and exon 1 (Xist Δprom-1) resulted in post-choice selection in which cells that choose to inactivate the mutated X retain two active X chromosomes and die (Penny et al., 1996). In contrast, a deletion extending from part of exon 1 to exon 5 (Xist Δ1-5) leaves the Xist promoter intact and results in primary nonrandom X inactivation where the wild-type allele is always chosen to be inactivated (Marahrens et al., 1997, 1998; Nesterova et al., 2003).
The choice process itself is poorly understood, and it is unclear whether or not epigenetic differences between the two alleles before the onset of X inactivation are sufficient to mediate choice. Interestingly, a clear parallel has been found between genes on the X chromosome and autosomal monoallelically expressed genes. Both display epigenetic characteristics that are already present before the choice is made of which allele will be expressed. For instance, it has been shown that X-linked and imprinted genes reveal high levels of di-methylated histone H3 Lys 4 methylation, which is restricted to the promoter region (Rougeulle et al., 2003). In contrast, bi-allelically expressed genes display equal levels of di-methylated histone H3 Lys 4 methylation in both promoter and genic regions. In addition, asynchronous replication timing has been found to correlate with loci that display monoallelic expression including imprinted genes, olfactory receptor genes, and immunoglobulin gene loci (Kitsberg et al., 1993; Chess et al., 1994; Mostoslavsky et al., 2001). A characteristic of imprinted as well as random X inactivation is that the Xi replicates late in S-phase (Takagi, 1974). Recently, asynchronous replication timing of the immunoglobulin κ-light chain locus was found to correlate with choice during VJC rearrangement, with the functional recombined allele being replicated before the unrecombined allele (Mostoslavsky et al., 2001). At this locus, asynchronous replication timing was present before one allele was chosen for recombination and may therefore play a role in determining choice (Mostoslavsky et al., 2001). However, for the X chromosomes in female cells it is unknown whether asynchronous replication timing is present before X inactivation and whether it has an effect on determining choice.
In this work, we analyzed the nature of nonrandom X inactivation in mice carrying different Xist deletions by monitoring X-linked GFP expression during early embryonic development. We then examined the role of replication timing in X chromosome choice by analyzing replication timing of the X chromosome before the onset of X inactivation in embryonic stem (ES) cell lines that show skewed X inactivation.
Results
Primary nonrandom X inactivation in cells with an Xist deletion
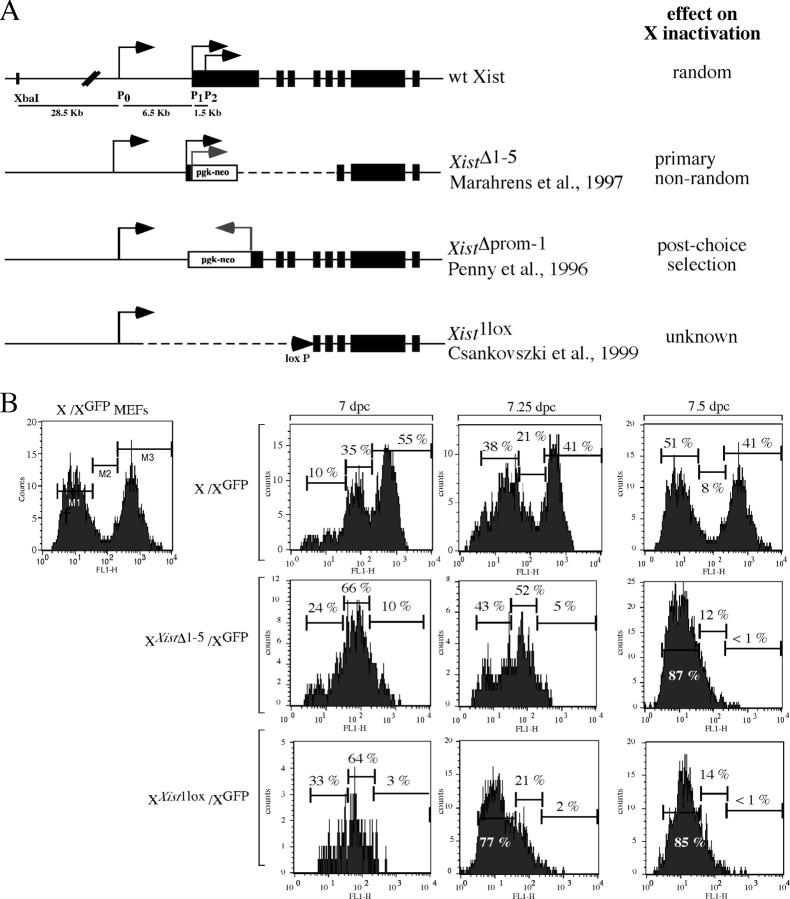
At the onset of X inactivation, one X chromosome is chosen for inactivation. Loss of Xist RNA expression leads to nonrandom X inactivation, which results in the inactivation of the wild-type X chromosome (Penny et al., 1996; Marahrens et al., 1997, 1998; Csankovszki et al., 1999). Two different Xist deletions that cover either exon 1 through exon 5, leaving the Xist promoter intact (Xist Δ1-5), or delete the Xist promoter and most of exon 1 (Xist Δprom-1), have separated primary nonrandom X inactivation from post-choice selection (Fig. 1 A). These results suggest that a region between exon 1 and 5 or the presence of residual transcription is required for choice.
Figure 1.
Primary nonrandom X inactivation in the presence of different Xist deletions. (A) Maps of the different Xist deletions and the effect of the deletion on X inactivation choice. (B) The absence of a functional Xist allele leads to primary nonrandom X chromosome choice. (left) FACS analysis of control MEFs heterozygous for the X-linked GFP used to determine cut-offs for GFP fluorescence (see text for details). (top row) FACS analysis of cells isolated from wild-type embryos heterozygous for X-linked GFP at 7, 7.25, and 7.5 dpc. (middle row) FACS analysis of cells isolated from XXist Δ1-5/XGFP embryos at 7, 7.25, and 7.5 dpc. (bottom row) FACS analysis with cells isolated from XXist 1lox/XGFP embryos at 7, 7.25, and 7.5 dpc.
To further define the sequence important for choice, we analyzed X inactivation in vivo in embryos heterozygous for the previously characterized Xist Δ1-5 allele and the Xist 1lox allele, which deletes 18 kb of the Xist locus including the promoter region and extends into intron 3 (Fig. 1 A; Csankovszki et al., 1999). The wild-type X in these mice was marked with an X-linked GFP allele that is subject to X inactivation (Hadjantonakis et al., 1998). We used FACS to monitor X inactivation as detected by a change in the number of cells expressing GFP. Based on control samples of mouse embryonic fibroblasts (MEFs) heterozygous for the X-linked GFP (Fig. 1 B, left panel) and wild-type MEFs that do not express GFP (not depicted), GFP fluorescence was arbitrarily gated into GFP-positive (Fig. 1 B, M3), GFP-negative (Fig. 1 B, M1), and intermediate GFP fluorescence (Fig. 1 B, M2). Gate M2 included cells of intermediate fluorescence that were not brightly positive or only slightly GFP positive, which represented cells that had turned off GFP expression but had not lost GFP fluorescence completely due to the half-life of the protein. All samples were compared with these exact cut-offs to evaluate relative fluorescence. In the embryo proper, random X inactivation begins at around 5.5 d past coitum (dpc) and is complete by 7.5 dpc. However, it has been shown that cells that are partially disomic for X-linked genes do not die rapidly, but that cell death continues over days until it is complete by 10 dpc (Takagi and Abe, 1990). We isolated wild-type and mutant embryos between 7.0 and 7.5 dpc, removed extraembryonic tissues for genotyping, and dissociated the embryo proper for analysis. In wild-type embryos, starting at 7.0 dpc, we observed that ∼41% of cells stayed GFP positive. The remaining cells gradually lost GFP fluorescence, which is consistent with random X inactivation that was complete by 7.5 dpc (Fig. 1 B, top). Embryos heterozygous for either Xist deletion invariably chose the wild-type chromosome for inactivation, which resulted in the loss of GFP expression in all cells over time (Fig. 1 B, middle and bottom). Heterozygous Xist Δ1-5 embryos have been shown previously to undergo primary nonrandom X inactivation (Marahrens et al., 1998). In agreement with this finding, we observed that during X inactivation all cells synchronously became GFP negative, suggesting that in every cell the wild-type chromosome was chosen to be inactivated (Fig. 1 B, middle). In XXist 1loxXGFP embryos the dynamics of X inactivation was indistinguishable from the Xist Δ1-5deletion (Fig. 1 B, bottom). These results indicate that both Xist deletions cause primary nonrandom X inactivation.
Asynchronous replication of X-linked genes before X inactivation
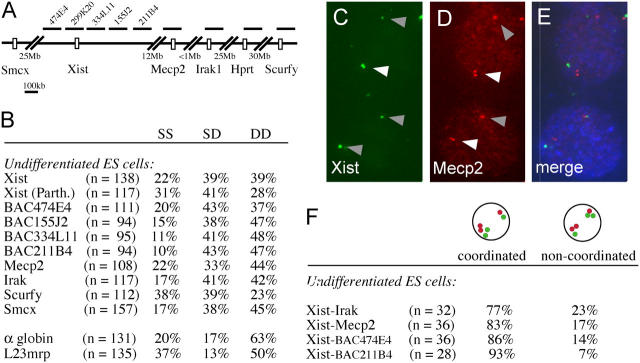
One marker associated with X inactivation is late replication of the Xi (Takagi, 1974). For many monoallelically expressed loci, asynchronous replication timing is present before gene expression and may be involved in determining which locus will be expressed. Although the X chromosomes in female cells replicate asynchronously after X inactivation, it is unknown if the X chromosomes also replicate asynchronously before X inactivation. To determine whether or not X-linked gene loci replicate asynchronously before the onset of X inactivation, we tested replication timing of several loci along the X chromosome in undifferentiated female ES cells that have two transcriptionally active X chromosomes (Fig. 2 A). Undifferentiated ES cells were BrdU pulse labeled and methanol/acetic acid fixed. In contrast to formaldehyde fixation, which retains the nuclear architecture and only resolves cohesion differences, methanol/acetic acid fixation destroys the nuclear structure that allows replication timing analysis (Azuara et al., 2003). BrdU incorporation was detected in conjunction with DNA FISH using plasmid and BAC probes. In this analysis, three different types of BrdU-positive nuclei can readily be distinguished: (1) nuclei with two single signals (single-single), which indicates that both loci have not replicated yet; (2) nuclei with two double signals (double-double), which indicates that both loci have replicated; and (3) nuclei with one single signal and one double signal (single-double [SD]), which indicates that only one locus has been replicated and the other one not. For bi-allelically expressed gene loci that replicate synchronously, the relative amount of the SD nuclei is low and varies between 10 and 20% depending on the cell type and target sequence. Asynchronously replicated gene loci display much higher relative numbers of SD nuclei in the BrdU-positive population of cells, ranging from 25 to 50% (Kitsberg et al., 1993; Chess et al., 1994; Mostoslavsky et al., 2001). We used this method to analyze replication timing of the Xic locus including the Xist gene. We found that the whole Xic region replicated asynchronously before X inactivation. Analysis of replication timing of the Xist locus with a cDNA probe and probes covering either exon 1 or 7 gave similar results (unpublished data). The high proportion of double-double nuclei (39%) relative to single-single nuclei (22%) indicates that both Xist alleles replicate relatively early in S-phase. Interestingly, we found that different X-linked loci across the X chromosome, including Mecp2, Irak1, Hprt, and Scurfy, all exhibit asynchronous replication timing in undifferentiated ES cells (Fig. 2 B). In comparison, the bi-allelically expressed autosomal loci α-globin and L23mrp showed relatively low numbers of SD nuclei (13–17%), which indicates that these loci replicated synchronously in S-phase.
Figure 2.
X-linked genes replicate asynchronously before X inactivation. (A) Map indicating the location of the X chromosome–specific BAC and plasmid probes used for the replication timing analysis. (B) Replication timing analysis of different X-linked loci in undifferentiated ES cells. SS, single-single; SD, single-double; DD, double-double. (C–E) Double label DNA FISH detecting Xist and MeCP2. Xist is shown in green (C), MeCP2 in red (D), and BrdU in blue (E). White arrowhead, replicated; gray arrowhead, not replicated. (F) Fraction of nuclei with coordinated versus noncoordinated SD nuclei for different combinations of target sequences.
Besides asynchronous replication timing, promoter-restricted high levels of H3 Lys 4 di-methylation have been found to be a specific mark for X-linked and imprinted genes (Rougeulle et al., 2003). Interestingly, Smcx, one of the very few genes that escapes X inactivation in the mouse, does not show promoter-restricted high levels of H3 Lys 4 di-methylation. To determine whether or not there is a correlation between H3 Lys 4 di-methylation levels and asynchronous replication timing we tested replication timing of the Smcx gene. We found that Smcx replicated asynchronously (Fig. 2 B), indicating that promoter-restricted high levels of H3 Lys 4 di-methylation do not correlate with asynchronous replication timing. In mice, X inactivation is imprinted in extra embryonic tissues but random in the epiblast. To answer the question of whether or not asynchronous replication timing in ES cells is due to imprinting, we analyzed replication timing of Xist in parthenogenetic ES cells, which have two maternal chromosome sets. We found that replication timing in these cells was also asynchronous (41% SD nuclei compared with 39% in wild-type ES cells), indicating that replication timing of Xist is not imprinted in undifferentiated ES cells (Fig. 2 B, Xist (Parth.)).
Asynchronous replication timing of nonimprinted autosomal monoallelically expressed genes is coordinated along the chromosome (Singh et al., 2003). To test if the same coordination is present on the X chromosome in undifferentiated ES cells, we performed double label FISH for two different loci in conjunction with BrdU staining and analyzed coordination of replication timing in nuclei with SD signals. We tested several probe combinations, including probes for Xist and Mecp2, which are separated by 12 Mb, probes for Xist and Irak1, which are 13 Mb apart, and probes for Xist and two different BACs covering the 5′ and 3′ end of the Xic (Fig. 2 A). For all probe combinations we found that in 77 to 93% of cells the SD signals were detectable on the same allele, which indicates that the replication timing along the chromosome is coordinated (Fig. 2, C–F). Analysis of probe combinations covering a greater distance was inconclusive because the signals were too far apart in the nucleus.
These results indicate that the X chromosomes in female cells replicate asynchronously before initiation of X inactivation, and replication timing seems to be coordinated along the X chromosome over relatively large distances around the Xist locus.
Replication timing and skewed X inactivation
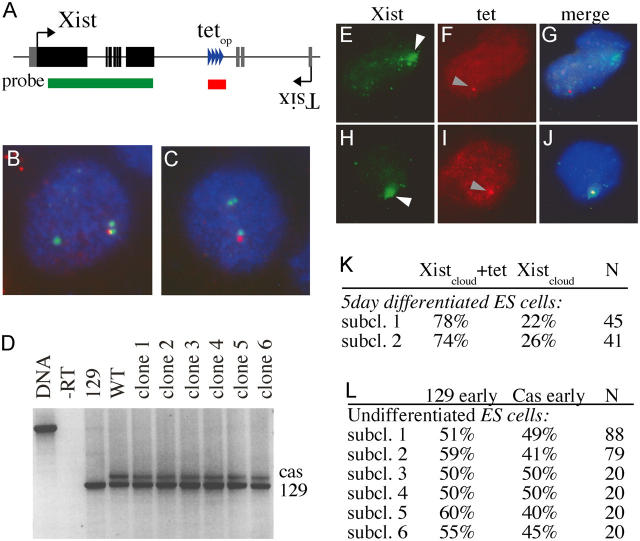
Random X inactivation is affected by the Xce locus, and X inactivation is skewed in cells carrying Xce alleles of different strength. We assessed replication timing in F1 Mus musculus (129)/Mus castaneus (cas) ES cells, which preferentially inactivate the 129 X chromosome that carries the weaker Xce a allele (relative to the Xce c allele of the cas X). To be able to distinguish the two alleles by DNA FISH analysis we introduced 56 tet operator repeats downstream of the 129 Xist locus and removed the neomycin resistance cassette (Fig. 3, A–C). We picked six independent sub-clones and induced X inactivation by differentiating the ES cells with retinoic acid. RT-PCR analysis with primers that detect a length polymorphism in the cas Xist gene showed the expected skewing toward inactivation of the 129 X chromosome after 5 d of differentiation (Fig. 3 D). Skewing of X inactivation in the subclones was comparable to skewing found in wild-type control samples, which suggests that the tet operator sequences did not affect X inactivation. In addition, we analyzed skewing of X inactivation in individual cells by RNA-DNA FISH in two independent sub-clones after 5 d of differentiation. First Xist RNA was detected with a probe that consisted of the complete cDNA sequence, followed by a brief fixation step and DNA FISH analysis with a probe specific for the tet operator DNA sequences (Fig. 3, E–K). We determined the relative number of differentiated ES cells that contained an Xist signal that colocalized with the tet operator signal relative to ES cells in which the Xist signal did not colocalize with the tet operator signal. This experiment confirmed the RT-PCR results and showed the inactivation of the 129 X chromosome in 74–78% of cells. Next, we analyzed replication timing of the cas and 129 Xist alleles before ES cell differentiation in the six ES cell sub-clones that were used for RT-PCR analysis. Using double label DNA FISH we found no preference for either the 129 or cas allele to be replicated first in S-phase (Fig. 3 L), which suggests that replication timing does not correlate with skewing of X inactivation. Because all subclones were derived from one founder clone, replication timing of the Xist locus appeared to be dynamic and switched between alleles in undifferentiated ES cells. In contrast, replication timing of X-linked genes is highly stable in cells that have undergone X inactivation (Hansen et al., 1996; Xiong et al., 1998).
Figure 3.
Replication timing of the Xist locus in ES cells with different Xce alleles. (A) Schematic representation of the integration site of the tet operator repeat and the location of the DNA FISH probes used to distinguish the two Xist alleles. (B and C) Double label DNA FISH using an Xist (FITC) and tet probe (rhodamine red) shows that the tet signal colocalizes only with one of the two Xist signals. The 129 allele of a polymorphic ES line was targeted with the tetop repeat sequence. (D) Individual subclones of the tetop-targeted ES cell line were differentiated for 5 d. Cas and 129 Xist RNA levels were determined with RT-PCR detecting a length polymorphism in exon 7. (E–J) Combined RNA-DNA FISH with cells differentiated for 5 d with two individual clones detecting the Xist RNA (FITC, white arrowheads) and tet DNA (rhodamine red, gray arrowheads; DAPI in blue) showed cells with an inactivated castaneus X chromosome (E–G) or an inactivated 129 X chromosome (H–J). (K) Quantification of RNA-DNA FISH experiment with two different subclones. (L) Replication timing analysis of individual undifferentiated ES cell subclones.
Replication timing and complete primary nonrandom X inactivation
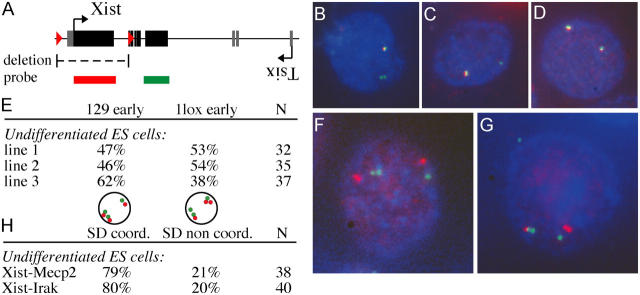
We have shown that replication timing before X inactivation does not correlate with skewing of X inactivation. We investigated whether or not replication is correlated with complete primary nonrandom X inactivation. We generated several Xist 1lox/+ heterozygous ES cell lines and analyzed replication timing of the wild-type and mutant alleles in undifferentiated ES cells before the onset of X inactivation using two different probes. One probe covered exon 1 and therefore only detected the wild-type Xist allele, and a second probe covered exon 7 and detected both the wild-type and mutant alleles (Fig. 4 A). Double label DNA FISH in conjunction with BrdU staining showed no preference for either the wild-type or mutant allele to be replicated first in S-phase, which suggests that asynchronous replication timing has no causal relation to primary nonrandom X inactivation (Fig. 4, B–E).
Figure 4.
Replication timing of the Xist locus in conditional Xist knockout ES cells. (A) Map of the Xist gene and location of the probes used for DNA FISH analysis. (B–D) BrdU detection in combination with double label DNA FISH using an exon 1 probe (rhodamine red) and an exon 7 probe (FITC) reveals nuclei with the mutant allele being replicated before (B) or after (C) the wild-type allele. (D) Control cells show two colocalizing signals. (E) Replication timing analysis with three independent Xist 1lox/+ undifferentiated ES cell lines. (F and G) BrdU detection (AMCA) and double label DNA FISH detecting the Xist (FITC) and Mecp2 (rhodamine red) loci reveals cells that have coordinated (F) and noncoordinated replication timing (G). (H) Relative amount of nuclei with coordinated versus noncoordinated replication timing.
Next, we tested if the Xist gene is required for chromosome wide coordination of replication timing. We analyzed coordination of replication timing of Xist with Irak1 and Xist with Mecp2 in Xist 1lox/+ ES cell lines and detected coordinated SD nuclei in ∼80% of the cells, similar to our findings with wild-type ES cells (Fig. 4, F–H). These results indicate that the Xist gene is not required for the coordination of replication timing along the X chromosome.
Discussion
In this work, we investigated several aspects of the X inactivation choice process. We analyzed the effect on choice of an Xist deletion that spans 5 KB upstream of the Xist promoter to intron 3 and found that this mutation results in primary nonrandom X inactivation. This result is consistent with previous reports that describe primary nonrandom X inactivation for a different deletion that encompasses part of exon 1 to exon 5. Therefore, it appears that an X chromosome can only be chosen for inactivation if Xist is intact. In addition, we studied the role of replication timing and found that asynchronous replication timing is present before X inactivation at all tested regions on the X chromosome. After completion of X inactivation, asynchronous replication timing of the two X chromosomes is stable and propagated through many cell divisions. In contrast, we found that early replication timing switches between alleles before the initiation of X inactivation. Analysis of asynchronous replication timing in ES cell lines with severe or complete skewing of X chromosome choice showed that replication timing does not correlate with choice, indicating that replication timing does not mediate skewing of the choice process.
Primary nonrandom X inactivation or post-choice selection?
In mammalian ES cell and embryonic tissues, cells randomly designate one X as the Xa and inactivate any remaining Xs. The most parsimonious model proposes the presence of an autosomally encoded blocking factor that marks the future active X and is present in limited quantities such that only one X can remain active per diploid set of autosomes (Brockdorff, 1998; Avner and Heard, 2001). Several cis-acting elements have been identified that lead to skewing of X chromosome choice, presumably by affecting the likelihood that the blocking factor binds. Evidence suggests that Xist and its negative regulator, the antisense RNA Tsix, play an important role in choice. In female cells heterozygous for an Xist deletion, only the wild-type chromosome is inactivated (Penny et al., 1996; Marahrens et al., 1997, 1998; Csankovszki et al., 1999). In addition, a chromosome with elevated sense transcription across Xist is more likely to be inactivated (Newall et al., 2001; Nesterova et al., 2003). Loss of Tsix transcription also leads to skewed choice with the mutated X being the inactive chromosome (Lee and Lu, 1999; Luikenhuis et al., 2001; Sado et al., 2001), and Tsix transcription negatively regulates Xist RNA steady-state levels in cis (Lee and Lu, 1999; Sado et al., 2001). Another genetic element implicated in choice is the X choosing element, Xce, which lies 3′ of Xist and beyond Tsix (Simmler et al., 1993). Crossing divergent mouse strains heterozygous for the Xce leads to skewing of X inactivation in the F1 female offspring such that the X chromosome bearing the stronger Xce allele is chosen more frequently as the Xa. Evidence suggests that the Xce might affect choice by modulating Tsix expression and in turn Xist RNA levels (Brockdorff et al., 1991). Recently, Xite, a cis-acting element that harbors intergenic transcription start sites and DNaseI hypersensitive sites, has been identified as a candidate locus for the Xce (Ogawa and Lee, 2003). Together, these findings suggest that choice is determined by a complex interplay of a variety of superimposed mechanisms.
Nonrandom X inactivation due to an Xist mutation can be achieved by either primary nonrandom X inactivation or random choice followed by post-choice selection. Primary nonrandom X inactivation, which leads to the inactivation of the wild-type X in a heterozygous embryo, was observed in cells carrying a deletion extending from part of exon 1 through exon 5, Xist Δ1-5 (Marahrens et al., 1997, 1998) as well as in the longer deletion (Xist 1lox) (Csankovszki et al., 1999) described here. In contrast, Penny et al. (1996) concluded that random X inactivation was followed by post-choice selection in a cell line carrying a deletion of the transcriptional start site of Xist and part of exon 1. This conclusion is similar to previous results in cells that inherit both products of the Searle's X-autosome translocation (T(X;16) 16H; McMahon and Monk, 1983). In these embryos, random choice results in either the inactivation of the wild-type X chromosome, which results in balanced cells, or in the inactivation of the translocation product that bears the Xic, which results in unbalanced cells. The cells that inactivated the translocation product are then progressively lost from the embryo with ∼75% being lost between 7 and 8 dpc (McMahon and Monk, 1983). Embryos that carry the Searle's translocation are only partially disomic for the X chromosome. We cannot exclude that embryos that are disomic for the complete X show a more severe phenotype, which may result in earlier lethality. In this case, our analysis starting at 7 dpc may have missed cells that carry two active X chromosomes. However, the Searle's translocation results in massive cell loss, and newborn pups are 20% smaller than their wild-type littermates (Lyon et al., 1964). In contrast, pups heterozygous for the Xist Δ1-5 deletion are indistinguishable from their wild type littermates (Marahrens et al., 1998). Because the two Xist mutations that do affect choice (Marahrens et al., 1997, 1998; Csankovszki et al., 1999) encompass all sequences deleted in the Penny et al. (1996) mutation (Fig. 1 A), it is difficult to define genetic elements that regulate this process. We consider the following possibilities to reconcile these differences: (a) it is possible that incomplete differentiation of the ES cells or the stability of the gene products measured by Penny et al. (1996) affected the outcome of the experiment. (b) It also cannot be excluded that the presence of the selectable marker in the antisense orientation and of exons 2 and 3 in the mutant allele somehow abrogated the role of Xist in choice.
Replication timing does not correlate with choice
Most monoallelically expressed genes display asynchronous replication timing, which, for all tested loci, is already present before expression. Interestingly, replication timing studies of the κ-light chain locus have indicated that replication timing may play a role in choosing which of the two κ-alleles will be recombined and subsequently expressed (Mostoslavsky et al., 2001). This finding prompted us to determine the role of replication timing in X inactivation choice. We have applied two-dimensional DNA FISH to analyze replication timing of sequences covering the Xic in ES cells. Although we cannot fully exclude the possibility that this method detects asynchronous chromatid separation rather than asynchronous replication timing, we and others have previously shown that two-dimensional DNA FISH analysis gives results similar to cell cycle fractionation analysis that does detect differences in replication timing (Simon et al., 1999; Azuara et al., 2003; Gribnau et al., 2003; Singh et al., 2003). We found that the Xic region replicated asynchronously before X inactivation and that replication timing along the entire Xic is coordinated. Therefore, the possibility that replication timing of a small region within the Xic is not coordinated is unlikely. In addition, we found that all other X-linked loci tested replicated asynchronously, including the Smcx gene, which escapes X inactivation. In contrast, bi-allelically expressed autosomal genes that are scattered in between coordinated asynchronously replicating monoallelically expressed genes replicate synchronously (Singh et al., 2003), which suggests that the mechanism that governs replication timing of the X may be different. Earlier studies have shown that imprinted genes and X-linked genes display promoter-restricted high levels of di-methylation of H3 Lys 4. Smcx and Xist were the only X-linked genes without promoter-restricted high levels of H3 Lys 4 di-methylation. Our studies suggest that there is no correlation between specific levels of H3 Lys di-methylation and asynchronous replication timing. The finding that the bi-allelically expressed Smcx gene replicated asynchronously could be explained by the fact that Smcx is inactivated on the Xi in the early stages of X inactivation and that bi-allelic expression is the consequence of relaxation of silencing (Lingenfelter et al., 1998). In addition, the region that escapes X inactivation in mouse is very small, roughly 30 kb. It encompasses only the Smcx gene and may lack an origin of replication (Tsuchiya et al., 2004). In contrast to promoter-restricted di-methylation of H3 Lys 4, hyperacetylation of core histones, hyper(di)methylation of H3 Lys 4, and hypo(di)methylation of Lys 9 have been found to be specific for all X-linked genes in female ES cells before X inactivation, including Smcx and Xist (O'Neill et al., 2003). The function of these specific histone modifications remains unknown; however, they correlate well with the pattern of asynchronous replication timing we have determined for X-linked genes.
Our results show that before X inactivation the proportion of asynchronously replicating (SD) X chromosomes is comparable to that of imprinted genes and nonimprinted monoallelically expressed genes (Simon et al., 1999; Gribnau et al., 2003; Singh et al., 2003). Two characteristics distinguish asynchronous replication timing before and after X inactivation. Asynchronous replication timing before X inactivation switches between alleles and is not affected by the Xce allele carried on the respective X. In contrast, in somatic cells, asynchronous replication timing of the Xa and Xi is clonal and stable through many cell divisions (Hansen et al., 1996; Xiong et al., 1998) and is influenced by the Xce. Another difference is the time window between the replication of the two alleles. Before X inactivation, we found 40% of the nuclei with an SD signal by FISH. Earlier studies that use S-phase fractionation analysis, which measures the DNA content at different stages in S-phase, have shown that this proportion of SD nuclei reflects a difference in replication timing of 1.5 to 2 h (Simon et al., 1999; Gribnau et al., 2003; Singh et al., 2003). In contrast, after X inactivation is complete, the time window is significantly larger and extends through almost the entire S-phase (Xiong et al., 1998). We conclude that replication timing before X inactivation does not correlate with skewing of the X chromosome choice. The difference between asynchronous replication timing before and after X inactivation most likely reflects the distinct chromatin states of the two X chromosomes before and after X inactivation.
Asynchronous replication timing: what does it do?
The vast majority of monoallelically expressed gene loci have been shown to replicate asynchronously in S-phase, which suggests a direct role for replication timing in the choice processes. In this work, we found that asynchronous replication timing is present throughout the X inactivation process. However, replication timing does not correlate with skewing of X inactivation choice.
Asynchronous replication timing of X-linked genes and other nonimprinted monoallelically expressed genes is random with respect to the parental origin (Mostoslavsky et al., 2001). In contrast, asynchronous replication timing of imprinted gene loci is parent specific (Simon et al., 1999). Interestingly, loss of imprinting caused by the erasure of methylation marks in the germline or after fertilization does not result in a loss of asynchronous replication timing of imprinted gene loci (Gribnau et al., 2003). In addition, at the imprinted Igf2-H19 locus, a 3-Mb inversion, which results in the establishment of a paternally imprinted Igf2-H19 locus in the female germline, does not change the replication timing characteristics of this locus (Cerrato et al., 2003). Therefore, gene expression and asynchronous replication timing appear to be separable mechanisms, raising the question about the significance of asynchronous replication. We found that asynchronous replication timing before X inactivation is random, switches between alleles, and is independent of the Xce and Xist mutations. However, we cannot exclude a role for asynchronous replication upstream of choice. It is possible that asynchronous replication timing before X inactivation reflects epigenetic differences between the two X chromosomes such as transient blocking factor binding, which may be involved in the counting process upstream of choice (Brockdorff, 1998).
Asynchronous replication timing could also be the remnant of an ancient imprinting or choice mechanism. Asynchronous replication timing could have played a role in setting up and maintaining imprints or determining choice processes for random monoallelically expressed genes. Over time, different epigenetic mechanisms, like DNA methylation or chromatin modifications, may have taken over the regulation of replication timing.
Materials and methods
Analysis of X inactivation in embryos
The appropriate genotypes were obtained by crossing Xist 1lox/+, Xist Δ1-5/+, or wild-type females to males homozygous for the X-linked GFP (Hadjantonakis et al., 1998). Pregnant females were killed at the appropriate times and embryos were collected. The embryo was separated into embryonic and extraembryonic tissues. Extraembryonic tissues were used for PCR genotyping by incubating them in 20 μl of 1× PCR buffer (GIBCO BRL) supplemented with 2 mM MgCl2 and 1 mg/ml proteinase K for 1 h at 50°C, followed by 10 min at 95°C. Standard PCR was performed by using 9 μl of the aforementioned lysate in a 20-μl reaction (30 cycles, with an annealing temperature of 52°C). For the Xist 1lox allele, the primers Xint 3R (5′-CAC TGG CAA GGT GAA TAG CA-3′), XpromL (5′-TTT CTG GTC TTT GAG GGC AC-3′), and 5′ Lox R (5′-ACC CTT GCC TTT TCC ATT TT-3′) were used, which gave a 427-bp band for the wild-type allele and a 513-bp band for the 1lox allele. For the XistKO allele, we used the primers Xist KO F (5′-AAC TGA GTG GGT GTT CAG GG-3′), Xist KO R (5′-ACC ACA AAT CAA GGC GAA TC-3′), and PGK-Pr1 (5′-GGG AAC TTC CTG ACT AGG GG-3′), which gave a 200-bp band for the wild-type allele and a 260-bp band for the knockout allele.
Embryonic tissues were washed twice in Hepes followed by trypsinization for 5 min and dissociation by pipetting. The trypsinization reaction was stopped by the addition of a small volume of DME/10% FCS. Cells were pelleted and resuspended in PBS/2% FBS supplemented with a final concentration of 1 mg/ml propidium iodine and analyzed by FACS.
Cell culture
Polymorphic Mus musculus/Mus castaneus F1-2-1 ES cells were grown on MEFs in DME (GIBCO BRL), 15% FCS (Hyclone), and 1000 U LIF/ml (Marahrens et al., 1997). ES cells were differentiated for 5 d in ES media without LIF and MEFs in the presence of 100 nM of all-trans-retinoic acid on gelatinized coverslips, and the medium was changed every day.
DNA FISH
DNA FISH was performed as in Selig et al. (1992) with minor modifications. In brief, medium of exponentially growing cells was supplemented with 10 mM BrdU and incubated for 45 min. Cells were trypsinized, washed with Hepes, and resuspended in 0.75 M KCl. After trypsinization, ES cells were incubated for 10 min on ice and all other cell types for 10 min at 37°C. Cells were fixed for 10 min in ice cold methanol/acetic acid solution (3:1 ratio), washed three times with methanol/acetic acid, and stored at 4°C or spotted onto polylysine-coated slides.
Slides were treated with 100 μg/ml RNase (2× SSC) for 30 min at 37°C, washed 3 × 5 min in 2× SSC, and dehydrated in 70, 90, and 100% ethanol. Target sequences were denatured by applying 100 μl of 70% formamide and 10 mM phosphate buffer in 2× SSC under a coverslip and incubated for 3 min on a hotplate (75°C). After removal of the coverslip, slides were washed in 2× SSC (5 min; 4°C), in 70% ethanol (5 min; −20°C), and through 90 and 100% ethanol for 3 min. Meanwhile, nick-translated BAC and cosmid probe sequences were dissolved in a hybridization mixture containing 50% formamide, 2× SSC, 50 mM phosphate buffer, pH 7.0, 10 mg/ml salmon sperm DNA, 10% dextran sulfate, and 100 ng/μl mouse Cot DNA to a final concentration of 2 ng/μl. The probe mix was denatured for 5 min, prehybridized for a minimum of 45 min, and then applied onto the slide. Slides were incubated overnight in a humidified chamber at 37°C.
BAC probes covering the Xic have been sequenced and described previously (Chureau et al., 2002). The BAC probes for Scurfy and Irak1 have been described previously and were BAC 196K10 (Brunkow et al., 2001) and BAC 228O4 (Reichwald et al., 2000), respectively. The BAC probe for Hprt was RPCI23 173F3 (GenBank/EMBL/DDBJ accession no. BX649621). All BACs were acquired from Research Genetics. α-Globin and L23mrp cosmid probes have been described previously (Gribnau et al., 2003). The Mecp2 probe was an 11-KB KpnI fragment covering part of the Mecp2 gene. The Smcx probe was a 15-KB XhoI fragment subcloned from BAC 330G24 (Tsuchiya et al., 2004). All probes were digoxygenin labeled by nick translation (Roche) or biotin labeled with a random prime labeling kit (Invitrogen), purified over G50 columns, precipitated, and resuspended in hybridization mix.
After hybridization, slides were washed in 2× SSC (5 min; 37°C), in 50% formamide, 2× SSC (3 × 10 min; 37°C), and in 0.1 M Tris, 0.15 M NaCl, and 0.05% Tween 20 (2 × 5 min; RT), and then incubated in 2 mg/ml BSA in 0.1 M Tris and 0.15 M NaCl, in a humidified chamber (30 min; RT). Detection was with subsequent incubation steps with anti-digoxygenin (Boehringer), anti–sheep (FITC; Jackson ImmunoResearch Laboratories; only when necessary), anti-BrdU (DakoCytomation), and anti–mouse (rhodamine red; Jackson ImmunoResearch Laboratories) antibodies in 0.1 M Tris, 0.15 M NaCl (30 min; RT). For double label DNA FISH, detection of biotin was with anti-biotin (Roche) and anti–mouse (rhodamine red; Jackson ImmunoResearch Laboratories) and detection of BrdU was with anti-BrdU (Abcam) and anti–rat (AMCA; Jackson ImmunoResearch Laboratories). Slides were washed twice in between each detection step with 0.1 M Tris, 0.15 M NaCl, and 0.05% Tween 20, mounted with Vectashield (Vector Laboratories), and stored at 4°C. Fluorescence was detected by epifluorescence/CCD. For replication timing coordination studies, more than 20 BrdU-positive cells with two SD signals were counted per cell line. Images were acquired using a microscope (model E800; Nikon) equipped with a 100× DIC H oil immersion lens with 1.4 NA. The camera was a Princeton Instruments, Inc. (model RTE/CCD 1317 = k/2) with a KAF-1400 1317 × 1035 chip (Kodak). As acquisition software, we used Openlab2.2 (Improvision).
RNA-DNA FISH analysis
RNA FISH analysis was performed as described previously (Panning and Jaenisch, 1996). Differentiated ES cells were grown on coverslips, extracted with cytoskeletal buffer, and fixed in 4% PFA in PBS. The Xist probe was a cDNA sequence (Wutz and Jaenisch, 2000), which was digoxygenin labeled by nick translation (Roche). After overnight hybridization, slides were washed in 2× SSC (5 min; 37°C), in 50% formamide, 2× SSC (3 × 10 min; 37°C), and fixed for 15 min in 4% PFA/PBS at RT. Slides were washed twice with PBS and dehydrated, and target sequences were denatured. DNA FISH was performed as described in the previous paragraph. The tet operator probe was a 500-bp fragment containing seven direct tet repeats that was biotin labeled with a random prime labeling kit (Invitrogen). Image acquisition was performed as described in the previous paragraph.
RT-PCR analysis
RNA was isolated with Trizol (Invitrogen), and 5 μg RNA was DNase treated and reverse transcribed with Superscript II (Invitrogen). Xist RNA was amplified with primer pair Xist-forward 5′-TTCCCATGTTTCTCCTGCAT-3′ and Xist-reverse 5′-GGAGACATGCAAAGGAAGGA-3′. These primers amplify a length polymorphism in exon 7 of Xist and amplification results in a 790-bp Mus musculus–specific product and an 820-bp Mus castaneus–specific product, which were resolved on a 1.5% agarose gel.
Acknowledgments
We thank Kathrin Plath for critically reading the manuscript and Barbara Panning, Andy Chess, Albrecht Sippel, and Alex Ensminger for stimulating discussions.
This work was conducted using the W.M. Keck biological imaging facility at the Whitehead Institute and was supported by a fellowship from the Human Frontier Science Program to J. Gribnau, a grant from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek to K. Monkhorst, and a grant from the National Institutes of Health/National Cancer Institute (5-R01-CA87869) to R. Jaenisch.
J. Gribnau and S. Luikenhuis contributed equally to this work.
J. Gribnau's and K. Monkhorst's present address is Dept. of Cell Biology, Erasmus MC, 3015 GE Rotterdam, Netherlands.
Abbreviations used in this paper: cas, castaneus; dpc, days past coitum; ES, embryonic stem; MEF, mouse embryonic fibroblast; SD, single-double; Xa, active X; Xce, X controlling element; Xi, inactive X; Xic, X inactivation center.
References
- Avner, P. and E. Heard. 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2:59–67. [DOI] [PubMed] [Google Scholar]
- Azuara, V., K.E. Brown, R.R. Williams, N. Webb, N. Dillon, R. Festenstein, V. Buckle, M. Merkenschlager, and A.G. Fisher. 2003. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 5:668–674. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N. 1998. The role of Xist in X-inactivation. Curr. Opin. Genet. Dev. 8:328–333. [DOI] [PubMed] [Google Scholar]
- Brockdorff, N., A. Ashworth, G.F. Kay, P. Cooper, S. Smith, V.M. McCabe, D.P. Norris, G.D. Penny, D. Patel, and S. Rastan. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 351:329–331. [DOI] [PubMed] [Google Scholar]
- Brown, C.J., A. Ballabio, J.L. Rupert, R.G. Lafreniere, M. Grompe, R. Tonlorenzi, and H.F. Willard. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 349:38–44. [DOI] [PubMed] [Google Scholar]
- Brunkow, M.E., E.W. Jeffery, K.A. Hjerrild, B. Paeper, L.B. Clark, S.A. Yasayko, J.E. Wilkinson, D. Galas, S.F. Ziegler, and F. Ramsdell. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 27:68–73. [DOI] [PubMed] [Google Scholar]
- Cerrato, F., W. Dean, K. Davies, K. Kagotani, K. Mitsuya, K. Okumura, A. Riccio, and W. Reik. 2003. Paternal imprints can be established on the maternal Igf2-H19 locus without altering replication timing of DNA. Hum. Mol. Genet. 12:3123–3132. [DOI] [PubMed] [Google Scholar]
- Chess, A., I. Simon, H. Cedar, and R. Axel. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. [DOI] [PubMed] [Google Scholar]
- Chureau, C., M. Prissette, A. Bourdet, V. Barbe, L. Cattolico, L. Jones, A. Eggen, P. Avner, and L. Duret. 2002. Comparative sequence analysis of the X-inactivation center region in mouse, human, and bovine. Genome Res. 12:894–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D.W., J.L. VandeBerg, G.B. Sharman, and W.E. Poole. 1971. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat. New Biol. 230:155–157. [DOI] [PubMed] [Google Scholar]
- Csankovszki, G., B. Panning, B. Bates, J.R. Pehrson, and R. Jaenisch. 1999. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 22:323–324. [DOI] [PubMed] [Google Scholar]
- Gribnau, J., K. Hochedlinger, K. Hata, E. Li, and R. Jaenisch. 2003. Asynchronous replication timing of imprinted loci is independent of DNA methylation, but consistent with differential subnuclear localization. Genes Dev. 17:759–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis, A.K., M. Gertsenstein, M. Ikawa, M. Okabe, and A. Nagy. 1998. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 76:79–90. [DOI] [PubMed] [Google Scholar]
- Hansen, R.S., T.K. Canfield, A.D. Fjeld, and S.M. Gartler. 1996. Role of late replication timing in the silencing of X-linked genes. Hum. Mol. Genet. 5:1345–1353. [DOI] [PubMed] [Google Scholar]
- Kitsberg, D., S. Selig, M. Brandeis, I. Simon, I. Keshet, D.J. Driscoll, R.D. Nicholls, and H. Cedar. 1993. Allele-specific replication timing of imprinted gene regions. Nature. 364:459–463. [DOI] [PubMed] [Google Scholar]
- Lee, J.T., and N. Lu. 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 99:47–57. [DOI] [PubMed] [Google Scholar]
- Lingenfelter, P.A., D.A. Adler, D. Poslinski, S. Thomas, R.W. Elliott, V.M. Chapman, and C.M. Disteche. 1998. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 18:212–213. [DOI] [PubMed] [Google Scholar]
- Luikenhuis, S., A. Wutz, and R. Jaenisch. 2001. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21:8512–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon, M.F. 1996. X-chromosome inactivation. Pinpointing the centre. Nature. 379:116–117. [DOI] [PubMed] [Google Scholar]
- Lyon, M.F., A.G. Searle, C.E. Ford, and S. Ohno. 1964. A mouse translocation suppressing sex-linked variegation. Cytogenetics. 15:306–323. [DOI] [PubMed] [Google Scholar]
- Marahrens, Y., B. Panning, J. Dausman, W. Strauss, and R. Jaenisch. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11:156–166. [DOI] [PubMed] [Google Scholar]
- Marahrens, Y., J. Loring, and R. Jaenisch. 1998. Role of the Xist gene in X chromosome choosing. Cell. 92:657–664. [DOI] [PubMed] [Google Scholar]
- McMahon, A., and M. Monk. 1983. X-chromosome activity in female mouse embryos heterozygous for Pgk-1 and Searle's translocation, T(X; 16) 16H. Genet. Res. 41:69–83. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky, R., N. Singh, T. Tenzen, M. Goldmit, C. Gabay, S. Elizur, P. Qi, B.E. Reubinoff, A. Chess, H. Cedar, and Y. Bergman. 2001. Asynchronous replication and allelic exclusion in the immune system. Nature. 414:221–225. [DOI] [PubMed] [Google Scholar]
- Nesterova, T.B., C.M. Johnston, R. Appanah, A.E. Newall, J. Godwin, M. Alexiou, and N. Brockdorff. 2003. Skewing X chromosome choice by modulating sense transcription across the Xist locus. Genes Dev. 17:2177–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newall, A.E., S. Duthie, E. Formstone, T. Nesterova, M. Alexiou, C. Johnston, M.L. Caparros, and N. Brockdorff. 2001. Primary non-random X inactivation associated with disruption of Xist promoter regulation. Hum. Mol. Genet. 10:581–589. [DOI] [PubMed] [Google Scholar]
- O'Neill, L.P., T.E. Randall, J. Lavender, H.T. Spotswood, J.T. Lee, and B.M. Turner. 2003. X-linked genes in female embryonic stem cells carry an epigenetic mark prior to the onset of X inactivation. Hum. Mol. Genet. 12:1783–1790. [DOI] [PubMed] [Google Scholar]
- Ogawa, Y., and J.T. Lee. 2003. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell. 11:731–743. [DOI] [PubMed] [Google Scholar]
- Panning, B., and R. Jaenisch. 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10:1991–2002. [DOI] [PubMed] [Google Scholar]
- Penny, G.D., G.F. Kay, S.A. Sheardown, S. Rastan, and N. Brockdorff. 1996. Requirement for Xist in X chromosome inactivation. Nature. 379:131–137. [DOI] [PubMed] [Google Scholar]
- Rastan, S. 1983. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos—location of the inactivation centre. J. Embryol. Exp. Morphol. 78:1–22. [PubMed] [Google Scholar]
- Rastan, S., and E.J. Robertson. 1985. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morphol. 90:379–388. [PubMed] [Google Scholar]
- Reichwald, K., J. Thiesen, T. Wiehe, J. Weitzel, W.A. Poustka, A. Rosenthal, M. Platzer, W.H. Stratling, and P. Kioschis. 2000. Comparative sequence analysis of the MECP2-locus in human and mouse reveals new transcribed regions. Mamm. Genome. 11:182–190. [DOI] [PubMed] [Google Scholar]
- Rougeulle, C., P. Navarro, and P. Avner. 2003. Promoter-restricted H3 Lys 4 di-methylation is an epigenetic mark for monoallelic expression. Hum. Mol. Genet. 12:3343–3348. [DOI] [PubMed] [Google Scholar]
- Sado, T., Z. Wang, H. Sasaki, and E. Li. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development. 128:1275–1286. [DOI] [PubMed] [Google Scholar]
- Selig, S., K. Okumura, D.C. Ward, and H. Cedar. 1992. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11:1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva, J., W. Mak, I. Zvetkova, R. Appanah, T.B. Nesterova, Z. Webster, A.H. Peters, T. Jenuwein, A.P. Otte, and N. Brockdorff. 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 4:481–495. [DOI] [PubMed] [Google Scholar]
- Simmler, M.C., B.M. Cattanach, C. Rasberry, C. Rougeulle, and P. Avner. 1993. Mapping the murine Xce locus with (CA)n repeats. Mamm. Genome. 4:523–530. [DOI] [PubMed] [Google Scholar]
- Simon, I., T. Tenzen, B.E. Reubinoff, D. Hillman, J.R. McCarrey, and H. Cedar. 1999. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 401:929–932. [DOI] [PubMed] [Google Scholar]
- Singh, N., F.A. Ebrahimi, A.A. Gimelbrant, A.W. Ensminger, M.R. Tackett, P. Qi, J. Gribnau, and A. Chess. 2003. Coordination of the random asynchronous replication of autosomal loci. Nat. Genet. 33:339–341. [DOI] [PubMed] [Google Scholar]
- Takagi, N. 1974. Differentiation of X chromosomes in early female mouse embryos. Exp. Cell Res. 86:127–135. [DOI] [PubMed] [Google Scholar]
- Takagi, N., and M. Sasaki. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 256:640–642. [DOI] [PubMed] [Google Scholar]
- Takagi, N., and K. Abe. 1990. Detrimental effects of two active X chromosomes on early mouse development. Development. 109:189–201. [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K.D., J.M. Greally, Y. Yi, K.P. Noel, J.P. Truong, and C.M. Disteche. 2004. Comparative sequence and x-inactivation analyses of a domain of escape in human xp11.2 and the conserved segment in mouse. Genome Res. 14:1275–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz, A., and R. Jaenisch. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 5:695–705. [DOI] [PubMed] [Google Scholar]
- Xiong, Z., W. Tsark, J. Singer-Sam, and A.D. Riggs. 1998. Differential replication timing of X-linked genes measured by a novel method using single-nucleotide primer extension. Nucleic Acids Res. 26:684–686. [DOI] [PMC free article] [PubMed] [Google Scholar]