Abstract
EB1 is a member of a conserved protein family that localizes to growing microtubule plus ends. EB1 proteins also recruit cell polarity and signaling molecules to microtubule tips. However, the mechanism by which EB1 recognizes cargo is unknown. Here, we have defined a repeat sequence in adenomatous polyposis coli (APC) that binds to EB1's COOH-terminal domain and identified a similar sequence in members of the microtubule actin cross-linking factor (MACF) family of spectraplakins. We show that MACFs directly bind EB1 and exhibit EB1-dependent plus end tracking in vivo. To understand how EB1 recognizes APC and MACFs, we solved the crystal structure of the EB1 COOH-terminal domain. The structure reveals a novel homodimeric fold comprised of a coiled coil and four-helix bundle motif. Mutational analysis reveals that the cargo binding site for MACFs maps to a cluster of conserved residues at the junction between the coiled coil and four-helix bundle. These results provide a structural understanding of how EB1 binds two regulators of microtubule-based cell polarity.
Introduction
The microtubule cytoskeleton, whose minus ends in most cells are located near the centrosome and plus ends extend toward the cell periphery, is involved in the long range transport of membrane organelles, mRNAs, protein complexes, and chromosomes (Karcher et al., 2002). To achieve these functions, microtubules serve as tracks for motor proteins that move along the microtubule surface. A second and more recently discovered type of intracellular transport is driven by the inherent polymerization of tubulin at the microtubule plus end (Desai and Mitchison, 1997). Several “plus end–tracking” proteins have been described that continually bind to the growing plus end of the microtubule and then dissociate, after a temporal delay, from the microtubule shaft. By tracking along the tips of microtubules, plus end–tracking proteins can be delivered to the cell cortex. Cortical polarity factors also can bind to plus end–tracking proteins, leading to the capture and selective stabilization of the microtubule and communication with the actin network (Korinek et al., 2000; L. Lee et al., 2000; Miller et al., 2000). Such interactions between microtubule plus ends and the cortex have been shown to be important for orienting the microtubule network, which can polarize the cell for migration or rotate the mitotic spindle during asymmetric cell divisions (L. Lee et al., 2000; Lu et al., 2001; Etienne-Manneville and Hall, 2003).
The two best characterized microtubule plus end binding proteins are Clip-170 and EB1, which are conserved from yeast to humans. The Clip-170 protein family is characterized by NH2-terminal microtubule-interacting Cap-Gly domains and a COOH-terminal region that recruits various cargos, including the dynein–dynactin complex (Lansbergen et al., 2004), and vesicles (Pierre et al., 1992). EB1 members have a similar bipartite composition: the NH2-terminal domain mediates microtubule plus end localization and a COOH-terminal cargo binding domain that captures cell polarity determinants. In Saccharomyces cerevisiae, an EB1 homologue, Bim1p, links microtubules to the cell cortex by interacting with the polarity determinant Kar9p (Korinek et al., 2000; L. Lee et al., 2000; Miller et al., 2000). In mammalian cells, the tumor suppressor adenomatous polyposis coli (APC) protein plays a functionally homologous role (Su et al., 1995; Lu et al., 2001). In addition to its role as a scaffold for the regulated degradation of β-catenin (Su et al., 1993), APC plays a role in microtubule-based cell polarity (Rubinfeld et al., 1993; Lu et al., 2001). EB1 was originally identified as an APC binding protein, and this interaction appears to be important for the later pathway (Su et al., 1995; Lu et al., 2001). Several signaling pathways establish polarity via EB1–APC recruitment including: (a) the sequential activation of CDC42, Par6-associated atypical PKC, and GSK-3β, which in turn localizes APC to the leading edge of astrocytes in a wounded monolayer (Etienne-Manneville and Hall, 2003; Kodama et al., 2003), (b) an NGF-induced axon growth pathway (Zhou et al., 2004), (c) the Rho-activated mDia recruitment of EB1 and APC in fibroblast cell migration (Wen et al., 2004), and (d) a microtubule actin cross-linking factor (MACF)–based recruitment of EB1 and APC to the muscle–tendon junction in Drosophila melanogaster (Subramanian et al., 2003). The EB1–APC interaction also is likely to be critical for tumorigenesis/metastasis of intestinal epithelial, probably through a loss of the normal polarity of the epithelia. The majority of APC mutations in human tumors involve deletion of its EB1 binding domain (Powell et al., 1992; Su et al., 1993).
Previous biochemical studies have shown that the COOH-terminal half of EB1 is sufficient for binding APC (Askham et al., 2000; Bu and Su, 2003). Likewise, in yeast, the corresponding region of Bim1p is used for Kar9p binding (Miller et al., 2000). Overexpression of the EB1 COOH-terminal domain also produced a dominant-negative phenotype in cell polarization (Wen et al., 2004; Zhou et al., 2004). Collectively, these studies establish EB1's COOH terminus as a cargo binding domain and a key determinant in search and capture processes. However, the structural mechanism by which EB1 recognizes its cargo is unknown. Here, we have identified a repeat motif in APC that binds EB1 and found a similar motif in the MACF family of spectraplakins, which allows these proteins to bind to EB1 and track along microtubule plus ends in vivo. We also solved the X-ray crystal structure of EB1's cargo binding domain and mapped the residues involved in MACF binding by alanine scanning mutagenesis. These results provide a structural understanding of how EB1 recruits cargo to the microtubule plus end.
Results
The EB1 COOH-terminal domain binds to a repeat motif in APC and MACFs
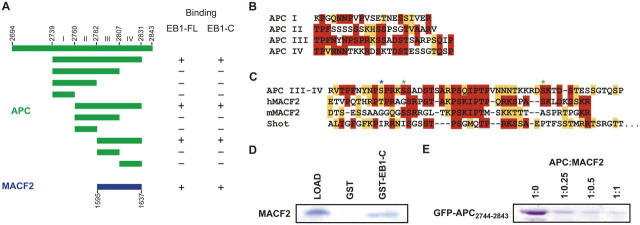
We first set out to define a minimal domain in human APC that interacts with the human EB1 COOH-terminal domain. Previous studies using a two-hybrid screen and in vitro binding revealed that the COOH-terminal 170 amino acids of APC can interact with EB1 (Su et al., 1995; Askham et al., 2000). To gain further insight on this APC region, we performed a detailed sequence analysis and identified four cryptic repeats of 21–25 amino acids (designated repeats I–IV), which are characterized by conserved prolines, serines, threonines, and positively charged residues (Fig. 1 B). The highest degree of homology was observed between repeats III and IV, which displayed 44% amino acid identity and 52% homology. To determine if all or a subset of these repeats constituted functional EB1 binding sites, we constructed several APC-GFP fusion constructs representing all contiguous combinations of the repeats and tested them for binding to a GST-EB1185-255 column. APC constructs containing repeats III and IV were retained on this column, but proteins lacking the repeat III–IV pair failed to bind, as did the single III or IV repeat proteins (Fig. 1 A). Mapping the EB1 binding site to repeats III and IV (APC2782-2831) corroborates data from Bu and Su (2003), who delineated an overlapping APC region for EB1 binding (APC2781-2820). The minimal construct defined by Bu and Su (2003) compromises the COOH-terminal 11 amino acids of repeat IV, which, although retaining EB1 binding, may result in decreased affinity. Additional support for EB1 binding to this region is derived from phosphorylation and phosphomimetic mutation studies of residues within repeats III and IV. In these studies, modulation of the CDC2 and PKA phosphorylation sites (Fig. 1 C) reduced or ablated EB1 binding (Askham et al., 2000; Nakamura et al., 2001; Bu and Su, 2003).
Figure 1.
Delineation of the EB1–APC and EB1–MACF2 interaction regions. (A) Summary of EB1 binding assays. APC constructs are delineated in green, MACF2 in blue. (B) Alignment of APC repeat regions I–IV. Identity is indicated in red, homology in orange. (C) Alignment of the EB1 binding motif of human APC, human MACF2, murine MACF2, and Drosophila Shot, with conservation colored as in B. Asterisks denote tentative PKA (green) and CDC2 (blue) phosphorylation sites. (D) Coomassie-stained polyacrylamide gel showing MACF21595-1637 retention on a GST-EB1185-255 column. (E) GFP-APC2744-2834 and MACF21596-1637 compete for binding to GST-EB1185-255. 10 μl GST-EB1185-255–bound glutathione Sepharose beads were preincubated with 0, 138, 225, and 550 nmol MACF21595-1637 for 30 min before incubation with 550 nmol GFP-APC2744-2842 for an additional hour. Resin was washed three times in batch with wash buffer, and the amount of bound GFP-APC2744-2843 was analyzed by Coomassie-stained SDS-PAGE.
We investigated whether or not any other proteins have repeat motifs homologous to those in APC that bind EB1. After performing a BLAST search (Altschul et al., 1990) using APC repeats III and IV, we identified a similar sequence (38% identity and 57% homology; Fig. 1 C) in the human and mouse spectraplakin family member MACF2 (Sun et al., 2001). Previous work has shown that ACF7 (a mouse MACF) binds to the plus ends of microtubules in mammalian tissue culture cells, but an interaction with EB1 was not investigated (Kodama et al., 2003). Like APC, MACFs are large, multidomain proteins characterized by tandem, NH2-terminal calponin homology domains that mediate binding to actin filaments, a plakin domain, a central rod-like region composed of dystrophin-like spectrin repeats that typically spans 1,000–4,000 residues, and a COOH-terminal region comprised of two EF-hand motifs, a GAS2/GAR2 domain that confers microtubule association (Sun et al., 2001), a series of quadruplet repeats typified by the sequence GSRA, and a final 50–100 residues of unknown function (Leung et al., 1999). The APC-like repeat motifs that we identified are found in this very COOH-terminal region of human MACF2, the murine MACF2, and the Drosophila MACF Shot (also known as Shortstop or Kakapo). Unlike APC, which has four repeat motifs, the MACFs only have two repeats that are more similar to APC repeats III and IV than to I and II.
We tested whether or not the dual MACF repeat is indeed capable of binding to EB1. We purified the human MACF2 repeat (MACF21595-1637) and tested its binding to GST fusions of full-length EB1 or its COOH-terminal (EB1185-255) domain prebound to glutathione-Sepharose. Both full-length GST-EB1 (not depicted) and GST-EB1185-255 retained the MACF21595-1637 peptide, whereas GST alone did not (Fig. 1 D). Thus, like APC, the MACF2 repeat motif is capable of specifically binding to the EB1 COOH-terminal domain in vitro. Additionally, competition experiments using GST-EB1185-255 prebound to glutathione-Sepharose, fixed levels of GFP-APC2744-2843, and increasing level of MACF21595-1637 revealed that MACF2 could compete with APC for binding to the EB1 COOH-terminal domain (Fig. 1 E).
DmEB1 associates with the Drosophila spectraplakin Shot
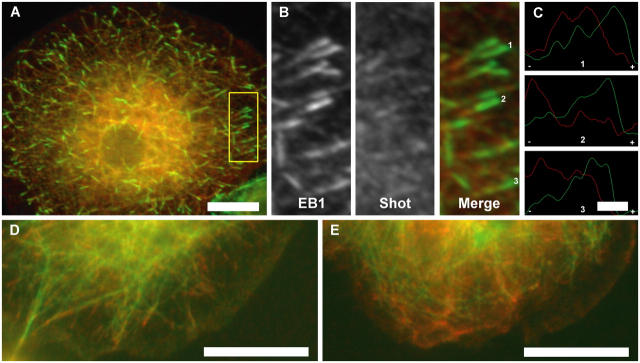
Next, we wished to determine if the MACF interaction with EB1 also occurs in vivo. We pursued this question using Drosophila S2 cells because DmEB1 can be very efficiently depleted in these cells by RNAi (Rogers et al., 2002). The solitary MACF homologue in Drosophila is Shot, which is expressed as many different isoforms. Shot was initially isolated in a screen for proteins involved in integrin-mediated adhesion and has subsequently been found to play a critical role during axon extension as a cytoskeletal cross-linker (Gregory and Brown, 1998; Lee and Kolodziej, 2002). Additional work demonstrated that Shot recruits DmEB1 and APC1 to the muscle–tendon junction to promote microtubule assembly, but it was not investigated whether the Shot–DmEB1 association was direct or indirect (Subramanian et al., 2003). In S2 cells, DmEB1 localizes to the plus ends of microtubules in a “comet-tail” pattern throughout the cell cycle as well as to centrosomes and spindle poles (Rogers et al., 2002). When cells were double stained for DmEB1 and Shot, we found that the distribution of the proteins overlapped but were not identical (Fig. 2, A–C). Shot antibodies labeled microtubules along their entire length but were enriched at their tips and also stained small punctae throughout the cell. However, close examination of the microtubules with double staining of Shot and DmEB1 revealed a terminal “cap” of DmEB1 (∼1 μm long) that lacked Shot followed by a region of overlap of both proteins in 106 out of 120 microtubules examined (Fig. 1 C). These results indicate that DmEB1 and Shot colocalize on microtubules but also maintain individual zones of localization, which is similar to observations made for EB1 and APC in mammalian cells (Juwana et al., 1999; Askham et al., 2000; Mimori-Kiyosue et al., 2000; Nakagawa et al., 2000; Barth et al., 2002).
Figure 2.
Shot is recruited to microtubule tips by DmEB1. (A) Colocalization of DmEB1 (green) and Shot (red) on microtubule tips in Drosophila S2 cells. (B) Enlarged view of the yellow boxed region outlined in A detailing microtubule tip localization. Individual channels and the resulting merged image are indicated. The distribution of DmEB1 (green) and Shot (red) on individual microtubule tips designated by numbers in B are represented in C normalized against individual maximum intensity along the microtubule axis with a minus and plus denoting the polarity of the microtubule tip. (D) Control S2 cells expressing GFP-tubulin (green) and immunostained for Shot (red); colocalization of Shot at microtubule tips can be seen. (E) After DmEB1 RNAi treatment, Shot (red) is diffusely localized and no longer stains at microtubules tips or along the length of the microtubule, as shown in this representative S2 cell expressing GFP-tubulin (green). 900 cells were analyzed in total. Bars: (A) 5 μm; (C) 1 μm; (D and E) 10 μm.
We expressed Shot (B isoform) fused to a COOH-terminal GFP and observed its localization in living S2 cells using spinning disk confocal microscopy. As predicted by immunofluorescence, Shot-GFP associated with microtubules, but exhibited two types of dynamic behaviors. On dynamic microtubules, Shot-GFP tracked along with growing microtubule plus ends at a velocity of 4.0 ± 0.8 μm/min (the rate of microtubule elongation in GFP-tubulin–expressing cells [Rogers et al., 2002]). This tip-associated Shot-GFP exhibited a comet-tail distribution, but unlike the short (∼2 μm) comet tails of EB1, the length of the microtubule decorated by Shot-GFP was often >5 μm (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200410114/DC1). Association of Shot-GFP with microtubule plus ends was also observed in mitosis, particularly on the halo of astral microtubules that emanated toward the cortex. A second population of microtubules associated with Shot-GFP along their length and were stable over extended (>20 min) periods of observation, exhibiting neither growth nor shrinkage typified by dynamic instability. Such highly stable microtubules were never observed in S2 cells expressing GFP-tubulin alone (unpublished data). Thus, overexpression of Shot-GFP compared with wild-type cells is likely to account for this suppression of microtubule dynamics. In support of this idea, when Shot-GFP was expressed at very high levels (2 d after transfection), exaggerated microtubule bundling and a complete suppression of microtubule dynamics was observed (Lee and Kolodziej, 2002; unpublished data).
We tested if EB1 was required for the plus end tracking of Shot-GFP by depleting EB1 (<1% of wild type) using RNAi (Rogers et al., 2002). RNAi-treated cells expressing GFP-tubulin under a constitutive actin promoter were double labeled for Shot and DmEB1 to identify unambiguously the DmEB1-depleted cells. Control cells showed colocalization of Shot at microtubule plus ends. In EB1 RNAi-treated cells, Shot never associated with microtubules (n = 900 cells), either at their tips or along their length (Fig. 2 E). Instead, Shot immunofluorescence staining revealed a punctuate pattern with no discernible microtubule association. In contrast, RNAi depletion of D-Clip-190 (another plus end–tracking protein) did not alter Shot's microtubule localization (unpublished data). These results indicate that association of Shot with microtubules requires DmEB1; however, albeit less likely, it cannot be ruled out that Shot mislocalization in DmEB1 RNAi-treated cells could be due to the relative undynamic nature of microtubules after DmEB1 depletion (Rogers et al., 2002).
The crystal structure of EB1's COOH-terminal cargo binding domain
To better understand the structural basis of cargo recognition by EB1, we crystallized its conserved COOH-terminal cargo binding domain (residues 185–255). Two crystal forms were obtained: P21 (two molecules per asymmetric unit, 2.0 Å resolution) and C2221 (one molecule per asymmetric unit, 1.8 Å resolution) (see Materials and methods). Phase information was obtained for both crystal forms by multiwavelength anomalous diffraction (MAD) using the selenomethionine (SeMet)-derivatized construct V243M, in which a methionine was incorporated at the homologous site of a methionine in the EB1 homologue RP1 (Fig. 3 D) to enhance MAD phasing power. The refinement of the structures yielded the following statistics: P21 crystal form, R = 21.9, Rfree = 25.1; C2221 crystal form, R = 22.1, Rfree = 25.9. Further details on the diffraction data, phasing, refinement, and model statistics are presented in Table I. Coordinates have been deposited in the Protein Data Bank under accession codes 1YIG (P21 crystal form) and 1YIB (C2221 crystal form).
Figure 3.
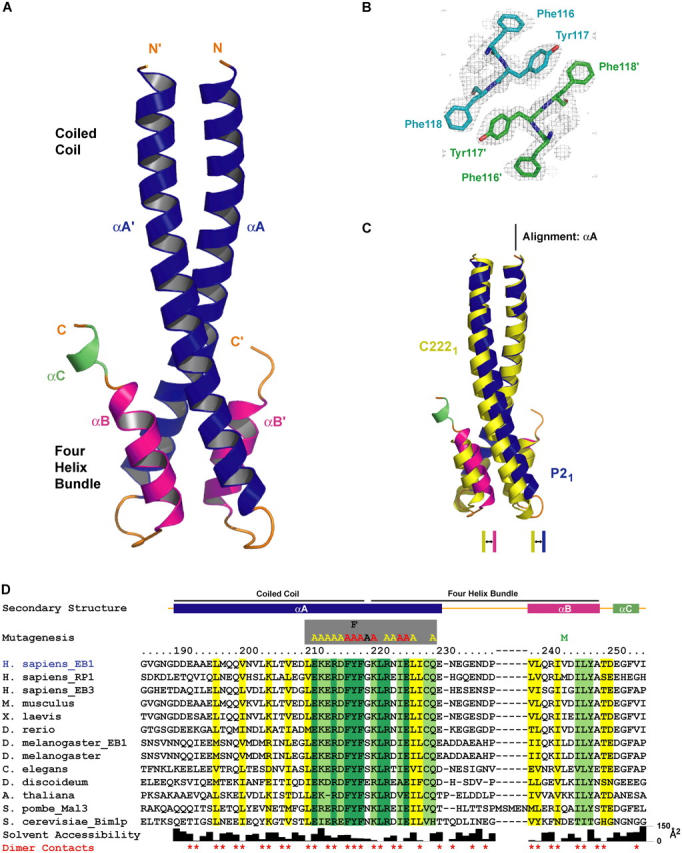
Crystal structure of the EB1 COOH-terminal domain. (A) Ribbons diagram of the homodimeric EB1 COOH-terminal domain (αA, blue; αB, pink; αC, green). (B) Experimental electron density map of the C2221 crystal form calculated to 2.0 Å, contoured at 1.0 σ, and superimposed over the conserved FYF′ motif located at the junction of the coiled coil–four-helix bundle junction. The view is directed down the two-fold axis of the coiled coil. (C) Comparison of the P21 and C2221 crystal structures after a least squares fit over αA. (D) Alignment of 13 EB1 family members from diverse species. Secondary structure is indicated in colors corresponding to those in A. The regions constituting the coiled coil and four-helix bundle are indicated. Invariance is indicated in dark green, 85% identity in light green, and 85% homology in yellow. Side chain solvent accessibility is indicated in the bar graph below. Residues involved in homodimer contacts are indicated with red asterisks. Residues mutated to alanine, phenylalanine, or methionine for binding studies and phasing purposes are indicated above the alignment.
Table I. Crystallographic data, phasing, and refinement.
| Beamline: Advanced Light Source 8.3.1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Space group: P21 | Cell dimensions: a=33.1 Å, b=37.3 Å, c=56.9 Å, α=90°, β=106.1°, γ=90° | Phasing Powerb | ||||||||
| Data Statistics: | dmin (Å) | Completeness (%) | I/σ | Rsym (%)a | +Friedel mate | −Friedel mate | ||||
| SeMet λ1 (0.97964 Å) | 2.00 | 92.8 (83.8) | 25.2 (6.4) | 3.4 (13.4) | 0.27 (0.17) | 0.81 (0.54) | ||||
| SeMet λ2 (0.97979 Å) | 2.00 | 93.6 (80.8) | 23.1 (2.4) | 3.8 (31.9) | 2.05 (0.71) | 2.05 (0.76) | ||||
| SeMet λ3 (1.12713 Å) | 2.00 | 82.3 (40.4) | 25.4 (6.1) | 3.0 (13.3) | Reference | 0.35 (0.17) | ||||
| Figure of meritc: | centrics: 0.81 (0.59) | acentrics: 0.57 (0.25) | ||||||||
| Refinement: | (50–2.0 Å) | |||||||||
| R Valued: | 21.9 (20.3) | |||||||||
| Rfree e: | 25.1 (23.3) | |||||||||
| Rmsd bond lengths (Å): | 0.006 | |||||||||
| Rmsd bond angles (°): | 0.92 | |||||||||
| Mean B factor (min/max): | 28.1 Å2 (7.9 Å2/65.7 Å2) | |||||||||
| Space group: C2221 | Cell dimensions: a=33.1 Å, b=108.3 Å, c=37.2 Å, α=90°, β=90°, γ=90° | Phasing Powerb | ||||||||
| Data Statistics: | dmin (Å) | Completeness (%) | I/σ | Rsym (%)a | +Friedel mate | −Friedel mate | ||||
| Native (1.12704 Å) | 1.80 | 97.2 (87.1) | 49.5 (11.2) | 2.9 (14.1) | ||||||
| SeMet λ1 (0.97973 Å) | 2.00 | 92.1 (88.6) | 20.9 (3.3) | 4.8 (25.7) | 1.74 (0.68) | 1.89 (0.74) | ||||
| SeMet λ2 (0.99987 Å) | 2.00 | 92.1 (83.9) | 22.8 (3.8) | 4.3 (21.3) | Reference | 0.29 (0.12) | ||||
| Figure of meritc: | centrics: 0.70 (0.43) | acentrics: 0.52 (0.20) | ||||||||
| Refinement: | (50–1.8 Å) | |||||||||
| R valued: | 22.1 (24.9) | |||||||||
| Rfree e: | 25.9 (28.8) | |||||||||
| Rmsd bond lengths (Å): | 0.004 | |||||||||
| Rmsd bond angles (°): | 0.82 | |||||||||
| Mean B factor (min/max): | 24.2 Å2 (12.0 Å2/58.5 Å2) | |||||||||
Values in parentheses are for the highest resolution shells.
Rsym = ShΣi|Ii(h)−,I(h)>|/ ΣhΣiIi(h), where I i(h) is the integrated intensity of the ith reflection with the Miller Index h, and <I(h)> is the average over Friedel and symmetry equivalents.
Mad phasing power is defined as [<|F D−F N | 2>/∫P(φ)(||F N Ie iφ+ΔF h |−F D | 2 )dφ] 1/2, where P(φ) is the experimental phase probability distribution. FN are structure factors at the designated reference wavelength and F D are structure factors of + or − Friedel mates at the other designated wavelength. ΔFh is the difference in heavy atom structure factors between two wavelengths.
Figure of merit is the weighted mean of the cosine of the deviation from αbest.
R value = Σ(|Fobs|−k|Fcalc|)/Σ|Fobs|.
Rfree is calculated using a 10% subset of the data that is removed randomly from the original data and excluded from refinement.
The crystal structure of the EB1 COOH-terminal domain reveals a unique homodimeric structure formed by a coiled coil that folds back at the COOH-terminal region to form a four-helix bundle (Fig. 3 A). A search of structural domains in the Protein Data Bank failed to identify a similar homodimeric fold. The homodimer engages both protomers in the P21 asymmetric unit, whereas a crystallographic two-fold in the C2221 form delineates the molecule's endogenous symmetry. Entirely helical in structure, the COOH-terminal domain is defined by helices αA and αA′ (where ′ refers to the homodimeric mate) that span residues 190–230, αB and αB′ that span residues 238–248, and a solitary helix αC that spans residues 251–254 found in only one protomer of the P21 crystal form. Helices αA and αA′ engage in a coiled coil over residues 190–218, burying conserved hydrophobic side chains and the aliphatic region of a conserved lysine (K204). No density was observed for residues 185–189 in either crystal form, thus delineating the start of the dimerization domain at residue 190. The remaining portion of αA and αA′ cooperate with αB and αB′ to form a four-helix bundle that contributes 1,540 Å2 to the total 3,340 Å2 of buried surface area formed in homodimerization. Superimposition of the P21 and C2221 crystal forms along αA reveals a plasticity in the relative twist of the coiled coil and four-helix bundle (Fig. 3 C). Differences in the organization of the loop bridging helices αA and αB are apparent, correlating with this region's low homology and exemplified by a five amino acid insert located within the corresponding region of Schizosaccharomyces pombe Mal3 (Fig. 3 D). The final helix, αC is ordered in one protomer of the P21 crystal form, most likely due to crystal packing contacts. Due to its disorder in the C2221 crystal form and in one protomer of the P21 crystal form, we suspect αC is flexible in solution, but due to its proximity to the MACF binding region described in the following section, it may transit to an ordered state upon cargo binding.
To elucidate tentative sites responsible for cargo interaction, we examined the spatial positions of conserved residues (defined by invariance, 85% identity and 85% homology) across 13 representative EB1 family members from yeast to vertebrates (Fig. 3 D). Under this classification system, conserved residues within the NH2-terminal region of the coiled coil (αA and αA′) are predominantly buried and constitute the hydrophobic core of the coiled coil (Figs. 3 D and 4 B). The highest near-continuous stretch of conservation occurs along the second half of αA, spanning residues 211–229 at the coiled coil–four-helix bundle junction (Fig. 3 D). Amidst this stretch of conservation are the invariant residues F216, Y217, and F218 (Fig. 3, B and D) that form symmetrical, stacked arrays of solvent-exposed hydrophobic side chains that flank each side of the coiled coil. The register of the coiled coil dictates that the spatial FYF′ motif be comprised of F216 and Y217 from one chain and F218′ from the homodimeric mate (Fig. 3 B). Additional conservation at this junction is provided by two solvent-exposed residues of the αB helix: I245 and Y247. This conserved surface is characterized by both a hydrophobic character and a net negative charge contributed by the acidic residues E211, E213, D215, and E225 (Fig. 4 D). The composite architecture of a conserved solvent-exposed region with hydrophobic and charged character implicated this zone as the tentative cargo binding site.
Figure 4.
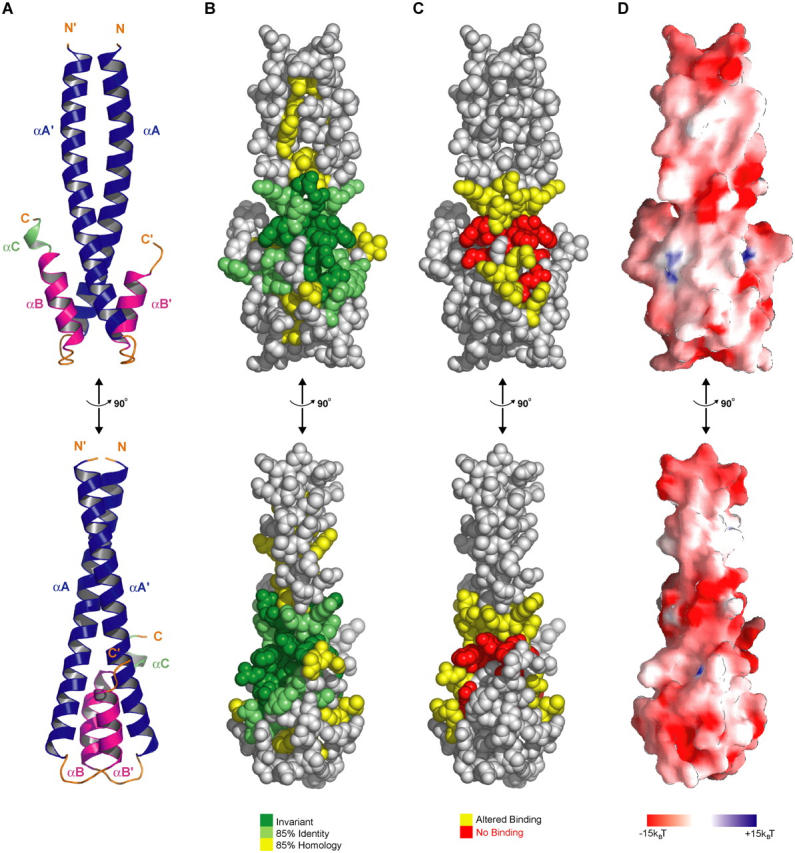
A conserved region at the coiled coil–four-helix bundle junction mediates cargo interactions. (A) Domain architecture of the EB1 heterodimer in ribbons format, with orientation maintained in B, C, and D. Bottom panel represents a 90° rotation about the y-axis. (B) CPK representation of conserved residues: dark green, invariant; light green, 85% identical; yellow, 85% homology (correlating to residues and colors used in Fig. 3 D). (C) CPK representation of mutations colored according to the effect these mutations had on binding to MACF21595-1637: yellow, altered binding; red, no binding. (D) Surface potential from −15 to +15 kBT (kB = Boltzmann's constant, T is the absolute temperature [K]) calculated in GRASP (Nicholls et al., 1991).
Conserved residues constitute the APC-MACF binding site
To identify the residues in EB1 that contact cargo, we developed a surface plasmon resonance binding assay. We prepared a construct of the MACF21595-1637 EB1 binding region with an NH2-terminal His-tag and a COOH-terminal motif (GGGLNDIFEAQKIEWHE) that becomes biotinylated at the lysine residue during expression in Escherichia coli (Beckett et al., 1999). MACF21595-1637 peptide was immobilized on a streptavidin sensor chip, increasing concentrations of NH2-terminally His-tagged full-length EB1 were injected, and binding was detected by the increased mass density at the sensor chip surface. Full-length EB1, rather than EB1185-255, was used to maximize the binding signal. Analysis revealed that the on-rate and maximal signal increased with increasing concentrations of EB1; however, a very slow off-rate was observed, which is most likely due to bivalent complex formations of two peptides with EB1, which retarded dissociation from the surface. The best fit of these curves was also achieved assuming a bivalent binding reaction (unpublished data). However, the very slow off-rate rate made it difficult to accurately determine kd1 and the equilibrium binding constant.
To test the individual binding contribution of residues within the 211–229 conserved span, we prepared single alanine mutations, with the exception of Y217 for which both alanine (Y217A) and phenylalanine (Y217F) mutants were generated. Mutant EB1 proteins were analyzed for MACF21595-1637 binding using the surface plasmon resonance assay at a fixed concentration of 2.5 μM and compared with the behavior of wild-type EB1 at the same concentration (Fig. 5 B). A spectrum of association rates was obtained for the various mutants and classified according to effect (altered association, yellow; no binding, red). Only one mutant of a nonconserved glycine (G219A) displayed wild-type association kinetics (Fig. 3 D). Several alanine mutations of charged/polar residues (E211A, K212A, E213A, R214A, D215A, R222A, N223A, and Q229A) displayed altered association. No binding was obtained for the F216A, Y217A, F218A, K220A, I224A, and E225A mutants. All mutants that showed no MACF2 binding were analyzed by gel filtration to determine if homodimerization had been compromised. All mutants eluted at the position of a homodimer at the concentrations used for the binding study (not depicted), except for I224A, which eluted at the position of a monomer (Fig. 5 C). I224 is the only residue mutated that is completely buried, situated at the core of the four-helix bundle. These results indicate that destabilization of the four-helix bundle compromises dimerization and that dimerization is essential for MACF2 binding.
Figure 5.
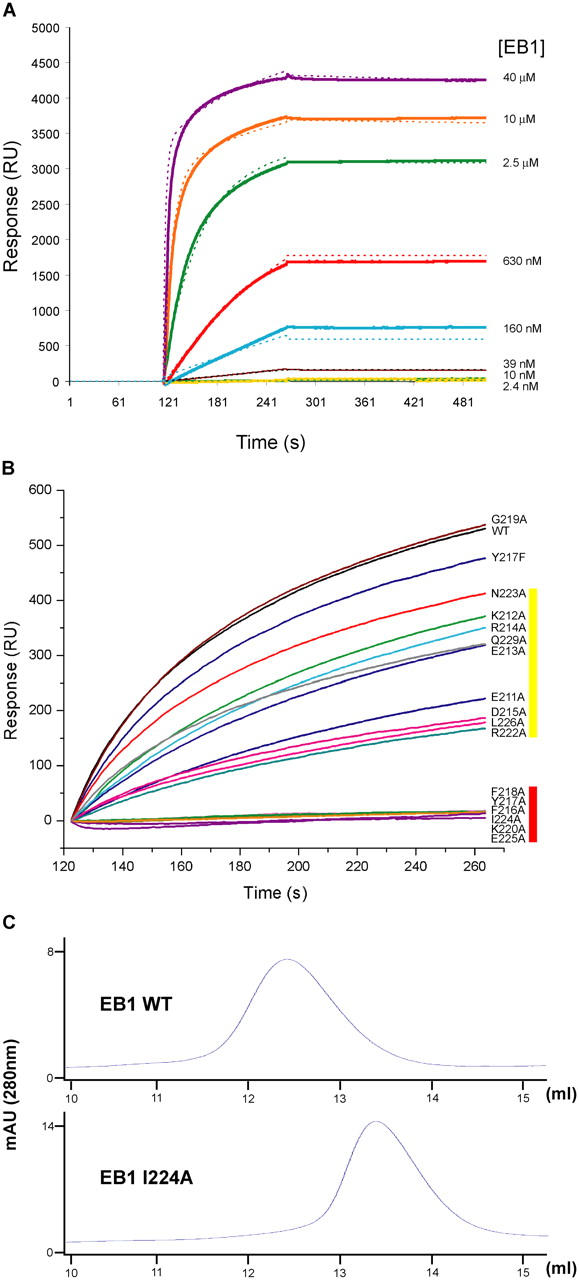
Mutations within the coiled coil–four-helix bundle junction abrogate binding to MACF21595-1637. (A) Surface plasmon resonance of EB1 (FL) at various concentrations (solid lines) on a sensor chip with immobilized MACF21595-1637. The resulting bivalent model is overlaid in dashed lines with colors corresponding to experimental EB1 concentrations. (B) Surface plasmon resonance of wild-type and mutant full-length EB1 constructs at 2.5 μM on a sensor chip with immobilized MACF21595-1637. Color coding at right designates the relative effect on the association of the EB1 mutants with surface-bound peptide. Yellow, altered binding; red, no binding. (C) Gel filtration analysis of EB1 full-length wild-type protein and the EB1 I224A mutant.
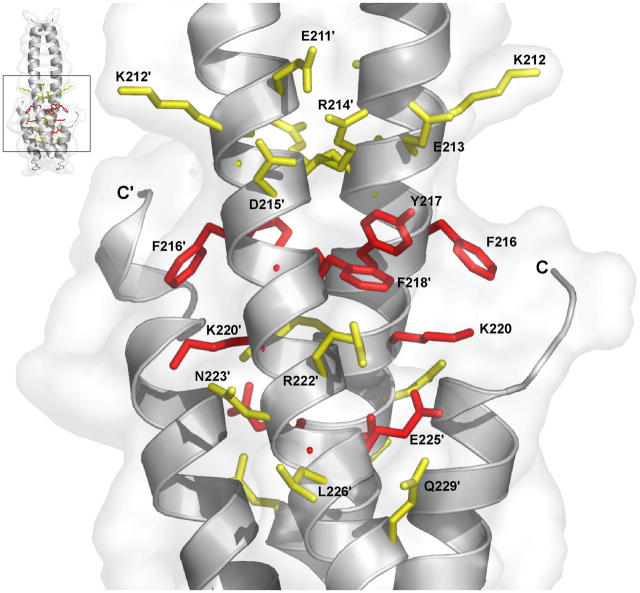
Mapping of mutant association results on the crystal structure reveals a cluster of key binding residues along helix αA, the dominant contributors to which are E211, D215, F216, Y217, F218, R222, E225, and L226 (Fig. 6). The FYF′ motif provides a dramatic contribution to the binding interaction as mutation of any of the three residues to alanine ablates binding. In contrast, the Y217F mutation produces only a modest decrease in binding. Thus, the tyrosine's hydrophobic Van der Waals contacts, rather than hydrogen bonding by the hydroxyl, makes a dominant contribution, either by contributing direct binding energy to the protein–protein interface or by positioning the spatially flanking phenylalanine residues 216 and 218′, which in turn confer crucial contacts for MACF2 association.
Figure 6.
Key hydrophobic and charged residues define a conserved MACF21595-1637 binding site. Residues mutated in the plasmon resonance study and their relative effect on the binding to the MACF21595-1637 peptide are shown, superimposed on a ribbons model of the COOH-terminal domain. A reference point is indicated in the top left. Residues are color coded according to the effect on MACF21595-1637 peptide binding: yellow, altered binding; red, no binding.
Discussion
In this work, we have characterized the binding of EB1 to APC and have identified a new EB1 binding partner, the spectraplakin MACF. We show using X-ray crystallography and structure-function studies that a repeat region shared by APC and MACF binds at an interface created by a four-helix bundle motif in the COOH-terminal domain of EB1. We discuss the implication of these findings for understanding how EB1 molecules attract cargo binding partners to microtubule tips.
Diverse cell polarity determinants share a common EB1 binding motif
We have elucidated a motif shared by mammalian APC and several mammalian spectraplakin MACFs. Although these two families bear no apparent sequence homology outside the identified EB1 binding site, they do share a similar composite domain architecture and cellular functions. Both proteins are extremely large (1,000–5,000 amino acids) due to repeated domains in their central regions (β-catenin and axin binding sites in APC [Su et al., 1993; Rubinfeld et al., 1993] and dystrophin-like spectrin repeats in MACFs MACFs [Leung et al., 1999]). Both proteins also are characterized by microtubule binding motifs (Smith et al., 1994; Deka et al., 1998; Sun et al., 2001) followed immediately after by the COOH-terminal EB1 recognition motif. In recent studies, both APC and the spectraplakins ACF7 and Shot have been shown to play roles in axon growth, cell polarity, and migration (S. Lee et al., 2000; Lu et al., 2001; Etienne-Manneville and Hall, 2003; Kodama et al., 2003; Subramanian et al., 2003; Wen et al., 2004; Zhou et al., 2004). In these studies, overexpression of the EB1 COOH-terminal domain resulted in a dominant-negative disruption of polarity signaling, indicative that an interaction with full-length EB1 is required for proper signal transduction/cytoskeletal stabilization by mammalian APC and spectraplakins. GSK-3β, a kinase implicated in cell polarity signal transduction pathways, also has been implicated in the regulation of APC and MACFs (Etienne-Manneville and Hall, 2003; Kodama et al., 2003; Zhou et al., 2004). It is possible that this kinase might, at least in part, control polarity by regulating EB1 interaction because in vitro studies have shown that phosphorylation of APC abrogates binding to EB1 (Askham et al., 2000; Nakamura et al., 2001; Bu and Su, 2003).
Interestingly, the two APC genes in Drosophila have the β-catenin/axin binding domains, but lack the COOH-terminal region in mammalian APC that contains the EB1 binding motif. To date, Drosophila APCs have not been observed tracking along microtubule tips, although DmAPC2 has been found to tether mitotic spindles to cortical actin (McCartney et al., 2001). Furthermore, the spectraplakin interaction with EB1 and microtubule tips appears to be widely conserved in all metazoans. Previous work has implicated the MACF Shot as an EB1-associated factor based on colocalization and coimmunoprecipitation, but a direct interaction was not shown (Subramanian et al., 2003). Here, we have definitively demonstrated that the in vivo association of Shot with the tips of microtubules is mediated by EB1. Thus, we speculate that the spectraplakins may have represented the original metazoan invention of a microtubule tracking, cell polarity factor and that vertebrates subsequently incorporated this feature into APC by duplication and gene integration. Consistent with this idea, the EB1 binding motifs are more similar between vertebrate MACF and APC than they are between MACFs from vertebrates and invertebrates.
In addition to the EB1 interaction, our localization studies suggest other features about the mechanism by which spectraplakins and APC interact with microtubules. Shot colocalizes only with the third of EB1 furthest from the microtubule plus end (Fig. 2 C). This pattern, which is not observed for other proteins such as RhoGEFs (Rogers et al., 2004), suggests that Shot does not coassemble with EB1 on the growing end of the microtubule but recognizes EB1 after a temporal delay, perhaps also suggesting some additional mechanism of regulation.
The EB1 COOH-terminal domain is a structural scaffold for dimerization and cargo binding
Our X-ray crystallographic studies show that the conserved EB1 COOH-terminal domain constitutes a unique coiled coil–four-helix bundle motif that confers both dimerization and cargo binding. Although EB1 and Bim1p were suggested to have a coiled coil domain based on sequence analysis (Rehberg and Graf, 2002), this is the first physical evidence that EB1 proteins are indeed dimers. Dimerization appears to rely not only on the coiled coil but also on the four-helix bundle motif, which provides nearly half of the buried surface area formed in dimerization. Consistent with this idea, destabilization of the four-helix bundle with the I224A mutation abolishes dimerization.
We also used alanine scanning mutagenesis, based on the crystal structure, to uncover a bivalent cargo binding site in the EB1 COOH-terminal domain. Both subunits of the homodimer appear to contribute residues to the cargo binding site, which is also supported by the finding that abrogating dimerization prevents cargo binding. Although the EB1 COOH-terminal domain is highly negatively charged, many of the key determinants in MACF2 binding are hydrophobic and form a discrete exposed hydrophobic pocket in the center of the domain. In comparison to the high conservation of EB1's COOH-terminal domain (from yeast to man), the repeat binding motifs found in APC and MACF2 are less well conserved, especially if one compares Drosophila to human. However, general features are the presence of several hydrophobic residues (mainly prolines) and a net positive charge. We speculate that these prolines and other hydrophobics dock into EB1's hydrophobic pocket, whereas the charged residues may augment the binding interaction by providing a peripheral gasket.
The conservation of solvent-exposed residues in EB1's COOH-terminal domain extends from vertebrates down to the yeast homologues Bim1 and Mal3. Kar9 is functionally homologous to APC and Shot in its general role of microtubule search and capture (Miller et al., 2000); however, Kar9 does not have a repeat motif architecture similar to APC/spectraplakins (Miller and Rose, 1998). A weak segment of homology with hydrophobic character was reported between regions of Kar9 and the EB1 binding motif (Bienz, 2001), but this site has not been experimentally confirmed. It will be interesting to compare and contrast the molecular basis for the EB1–APC interaction and the Bim1–Kar9 interaction to determine if a similar or unique set of determinants govern these binding interactions.
Our results also help to explain previous mutagenesis results of EB1 that were performed before the availability of a crystal structure. Wen et al. (2004) mutated conserved hydrophilic residues within aa 211–229 in EB1 and found that the mutations E211A/E213A, E211A/D215A, K220A/R222A, and E211A/E213A/K220A/R222A inhibited EB1–APC interaction in vitro and yielded supporting phenotypic behavior in vivo in an mDia-mediated cell polarity pathway. Our results predict the strongest effect for constructs containing the K220A mutation and milder effects for the one with the E213A mutation, which agrees with the observations by Wen et al. (2004). This correlation between EB1 mutants binding to MACF21595-1637 and APC also substantiates the likelihood of a similar binding architecture between these two classes of proteins and the EB1 COOH-terminal domain.
Implications of the EB1 COOH-terminal domain structure for other cargo binding interactions
Several EB1 family members exist in metazoan genomes (four in Drosophila [Rogers et al., 2002] and three in humans [Su and Qi, 2001]). Although the crystal structures reported in this paper describe a homodimeric structure, we cannot rule out the possibility that EB1 heterodimers exist and are used for diverse cargo recognition. The length of the coiled coil domains and the residues in the COOH-terminal domain that constitute the hydrophobic core and mediate dimer contacts are highly conserved across family members, especially within species (Fig. 3 D). Because each bivalent cargo binding site is formed from residues contributed by each of the EB1 molecules, heterodimers could confer complex, pair-wise cargo recognition motifs. Thus, the possibility of whether or not heterodimers of EB1 can form in vitro and within cells merits investigation.
The EB1 COOH-terminal domain also may contain cargo binding sites other than the one that we defined for APC and MACF at the coiled coil–four-helix bundle junction. Several proteins, including the dynein–dynactin complex (Valetti et al., 1999; Vaughan et al., 2002), Ncd (unpublished data; Ghoshima, G, personal communication), the microtubule-destabilizing kinesin Klp10A (Mennella et al., 2005), and RhoGEF2 (Rogers et al., 2004), have been reported to track along microtubule tips and associate with EB1. However, only in the cases of the p150Glued dynactin subunit (Askham et al., 2002; Tirnauer et al., 2002; Bu and Su, 2003; Wen et al., 2004) and Klp10A (Mennella et al., 2005) has a direct interaction with EB1 been shown. In the case of p150Glued, deletion studies have suggested that p150Glued and APC use overlapping yet unique binding sites on EB1's COOH-terminal domain (Askham et al., 2002; Bu and Su, 2003; Wen et al., 2004). The p150Glued binding site maps to EB1's coiled-coil–four-helix bundle but also requires EB1's COOH terminus (13 residues not included in our crystallization construct). Thus, it is possible that EB1 contains multiple cargo binding sites, some of which might be independent and others which might be mutually exclusive. The crystal structure reported here should provide a valuable tool for probing these future questions concerning EB1–cargo interactions.
Materials and methods
DNA manipulation, protein preparation, and immunoblots
Human cDNA encoding EB1 was obtained from the American Type Culture Collection and subcloned into the E. coli expression vectors pGEX-6P2 (Amersham Biosciences) and pET28a (Novagen). The EB1 conserved NH2-terminal domain (residues 12–133) and the conserved COOH-terminal domain (residues 185–255) were independently subcloned into pGEX-6P2. A COOH-terminal mutant, V243M, in EB1185-255 was created using the PCR sewing method and inserted into pGEX-6P2. Site-directed mutagenesis was conducted on the full-length EB1-pET28a construct using the Quickchange method (Stratagene) to create the point mutants described in Figs. 4–6. APC constructs (residues 2744–2843, 2694–2843, 2739–2831, 2739–2806, 2739–2781, 2739–2759, 2760–2831, 2760–2806, 2760–2781, 2782–2831, 2782–2806, and 2807–2831) and EB1 constructs (full length and residues 185–255) were subcloned into a modified pET15 (Novagen) vector to yield an NH2-terminal hepta-histidine tag followed by GFP and the specified region of APC. APC cDNA was provided by K. Kinzler and B. Vogelstein (Johns Hopkins University, Baltimore, MD). Residues 1595–1637 of KIAA0728 (human MACF2) were synthesized at the DNA level using optimal E. coli codon usage and subcloned into pGEX-6P2. A modified version of MACF21595-1637 (pGEX-6P2) was created using the PCR method to introduce a hexa-histidine tag NH2-terminal to MACF21595-1637 and a biotinylation tag (GGGLNDIFEAQKIEWHE; for in vivo biotinylation [Beckett et al., 1999]) at the COOH terminus.
All aforementioned constructs were transformed into BL21 (DE3) E. coli for recombinant expression (excluding a SeMet growth of the pGEX-6P2 EB1185-255 V243M mutant in which the construct was transformed into B834 E. coli and grown as previously described [Leahy et al., 1994]). Cultures were grown in 2× YT at 37°C to an optical density at 600 nm of 0.6, temperature was decreased to 20°C, and IPTG was added to a final concentration of 100 μM. Cultures were induced for 16 h, harvested, and frozen at −20°C. Bacterial pellets of pGEX-6P2 EB1 constructs were resuspended in lysis buffer (25 mM Tris, pH 8.0, 200 mM NaCl, and 0.1% β-mercaptoethanol [β-ME]) and lysed using a microfluidizer (Microfluidics, Inc.). 2 mM PMSF was added, and the lysate was clarified by centrifugation at 36,000 g for 45 min. Supernatant was incubated with glutathione-Sepharose beads for 30 min at 4°C with rotation. Resin was collected by centrifugation at 200 g for 10 min, loaded onto a column, washed with 100 column volumes of lysis buffer, and then protein was applied to the column to test for retention. Additional purification of GST-EB1185-255 and GST-EB1185-255 V243M proceeded as follows. GST fusion protein was step eluted using 5 column volumes of lysis buffer supplemented with 50 mM glutathione, pH 8.0. 30 μl of PreScission protease (Amersham Biosciences; 2 mg/ml stock) was added and incubated at 4°C for 12 h, which yielded near complete cleavage. Cleaved fusion protein was concentrated in a Centriprep YM-3 (Millipore) and diluted 100-fold into 25 mM Tris, pH 7.0, 0.1% β-ME, and loaded onto SP Sepharose Fast Flow resin (Amersham Biosciences) and eluted using a NaCl gradient of 0–1 M over 20 column volumes. Protein fractions were pooled, concentrated in a Centriprep YM-3, and exchanged into storage buffer (10 mM Tris, pH 7.0, 25 mM NaCl, and 0.1% β-ME). All buffers used for the EB1185-255 V243M SeMet purification also contained 5 mM methionine as a supplemental reducing agent. Additional purification of GST-H6-MACF21595-1637-Biotinylation Tag entailed PreScission protease cleavage, followed by binding to Ni2+-nitrilotriacetic acid resin, washing in 25 mM Tris, pH 8.0, 15 mM imidazole, 300 mM NaCl, 0.1% β-ME, and step elution using washing buffer supplemented with 285 mM imidazole. Eluted protein was concentrated and exchanged into 10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% β-ME using a Centriprep YM-3. All H7-GFP fusion proteins were purified as follows. Bacterial cell pellets were thawed and resuspended in lysis buffer (25 mM Tris, pH 8.0, 300 mM NaCl, 15 mM imidazole, and 0.1% β-ME), lysed in a microfluidizer, supplemented with 2 mM PMSF, and centrifuged at 36,000 g for 45 min at 4°C. Supernatant was incubated in batch with Ni2+-nitrilotriacetic acid resin for 30 min at 4°C. Resin was harvested by centrifugation at 200 g at 4°C, loaded into a column, and washed with 100 column volumes of lysis buffer. EB1 mutants were washed with 10 column volumes of lysis buffer supplemented with 15 mM imidazole, and step eluted with lysis buffer supplemented with 285 mM imidazole. All other H7-GFP fusion proteins were eluted using a gradient of 15–300 mM imidazole over 20 column volumes. Eluted H7-GFP fusion proteins were concentrated in a Centriprep YM-3, exchanged into 10 mM Tris, pH 8.0, 150 mM NaCl, and 0.1% β-ME, frozen in liquid nitrogen, and maintained at −80°C until used.
Drosophila tissue culture and immunofluorescence
Drosophila S2 cells were cultured, treated for RNAi, and processed for immunofluorescence as described previously (Rogers et al., 2002). Primary antibodies used for these studies were murine monoclonal anti-Shot (raised using the antigen Shot L(A) 1454–1909, which was prepared as a PreScission protease cleavage product from a GST fusion [Amersham Biosciences]), rabbit anti–Clip-190 (Lantz and Miller, 1998), and rabbit anti-DmEB1 (Rogers et al., 2002). Secondary antibodies (TMR anti-rabbit, Cy5 anti–mouse, Cy2 anti–rabbit, and Cy3 anti–guinea pig; Jackson ImmunoResearch Laboratories) were used at a dilution of 1:300.
Time-lapse observation of Shot-GFP in living cells was performed by cotransfection of S2 cells with pUAS-Shot (L) B-GFP (Lee and Kolodziej, 2002) and pMT-GAL4 (Rogers et al., 2004) using the Cellfectin transfection reagent (Invitrogen) following the manufacturer's protocols. GAL4-mediated Shot-GFP was induced with a 2-h induction using 1 μM copper sulfate and imaged using a spinning disk confocal microscope (Solamere Technology Group) mounted on an Axiovert 200 (Carl Zeiss MicroImaging, Inc.) equipped with an XR/Mega10 intensified charge coupled device camera (Stanford Photonics) driven by QED In Vivo software (QED Imaging).
Crystallization, data collection, data processing, model building, and refinement
The EB1 COOH-terminal domain (residues 185–255) was crystallized using the hanging drop vapor diffusion method at 20°C. 2 μl EB1 COOH-terminal domain at 15 mg/ml was added to 2 μl of mother liquor containing 22% PEG 200, 100 mM ammonium acetate, pH 4.6, and equilibrated over 1 ml of the mother liquor. Native crystals were crushed and used to streak-seed crystals of the EB1 COOH-terminal domain mutant V243M SeMet. The V243M SeMet protein crystallized in the space group P21 with two molecules in the asymmetric unit. Attempts to cocrystallize a complex of EB1185-255 with MACF21595-1637 using similar crystallization conditions yielded native and SeMet crystals of EB1 (without MACF21595-1637 present) in the space group C2221 with one molecule in the asymmetric unit. Crystals were transferred to Paratone-N and flash frozen in liquid nitrogen.
Diffraction data were measured at the Lawrence Berkeley National Laboratory Advanced Light Source beamline 8.3.1 using an ADSC 210 charge coupled device detector and maintained at 100°K using a nitrogen cryo stream. Data were processed and scaled using DENZO and SCALEPACK (Otwinowski, 1993). Heavy atom searches were performed using the CNS package (Brunger et al., 1998). Two selenium sites were found in each crystal form using an automated Patterson heavy atom search method and peak (P21 crystal form) or inflection (C2221 crystal form) anomalous wavelength data. The additional two selenium sites for the P21 crystal form were found using an anomalous difference Fourier map and an experimental density modified map that used phases from the two selenium sites found in the initial automated search. The coordinates for these two additional selenium sites were refined using the CNS package and used for subsequent phase determination and map building. MAD phasing used the selenium coordinates and the low energy remote wavelength as reference. Phases were improved by solvent flipping and histogram matching. Model building was done with O (Jones et al., 1991). Refinement was preformed with the CNS package and monitored with a random 10% of the data used for cross-validation. Model refinement used the MLHL target function, torsion angle molecular dynamics simulated annealing, B-factor refinement, and rebuilding in O with σA-weighted difference Fourier maps. The C2221 crystal form structure refinement was extended to the high resolution of the native data set using the MLF target function. Figures were prepared with PyMOL (DeLano Scientific) and GRASP (Nicholls et al., 1991). Coordinates have been deposited in the Protein Data Bank under accession codes 1YIG (P21 crystal form) and 1YIB (C2221 crystal form).
Surface plasmon resonance studies
Surface plasmon resonance experiments were conducted using a Biacore 1000 and Streptavidin sensor chips (Biacore). Flow cells were pretreated with 2 × 1 min injections of 1 M NaCl, 50 mM NaOH, and 3 × 1 min injections of regeneration solution (0.25% SDS). Flow cells were normalized and one flow cell was injected with 50 μl of 10 μM H6-MACF21595-1637-Biotinylation Tag protein at a 5 μl/min flow rate to yield an increase of 600 response units. The second flow cell was used as a control surface over which identical analyte injections were performed in triplicate. All runs were conducted at a flow rate of 20 μl/min in running buffer (10 mM Tris, pH 8.0, 200 mM NaCl, 0.1% β-ME, and 0.05% Tween-20) at 25°C. Sensor chips were prestabilized for 2 min with running buffer. Analyte was injected for 2.5 min followed by 6 min of running buffer for the analysis of dissociation. The sensor chip was regenerated by injection of 0.25% SDS for 1 min followed by a 5.5-min stabilization of running buffer before the start of the next sensogram. For full-length EB1 analysis, eight different concentrations of H6-EB1 were injected onto the sensor chip in triplicate (2.4 nM, 10 nM, 39 nM, 160 nM, 630 nM, 2.5 μM, 10 μM, and 40 μM). Binding studies using wild-type and mutant EB1 proteins were conducted at an analyte concentration of 2.5 μM and conducted in triplicate. Data was processed using BIAevaluation 3.2 RC1 software. Control analyte runs were subtracted from corresponding analyte plus MACF2 peptide runs and calibrated for comparison via baseline adjustment before analyte injection.
Gel filtration assay
Full-length human EB1 and EB1 point mutants (F216A, Y217A, F218A, K220A, I224A, and E225A) were diluted in running buffer (25 mM Tris, pH 8.0, 200 mM NaCl, and 0.1% β-ME) to a final concentration of 32 μM. 100 μl of each protein was injected onto a Superdex200 Tricorn analytical gel filtration column (Amersham Biosciences) equilibrated in running buffer and analyzed at a flow rate of 0.3 ml/min, measuring the absorbance at 280 nm.
Online supplemental material
GFP-Shot in interphase S2 cells shows microtubule plus end localization (Video 1). Frames for Video 1 were acquired at a rate of one frame per 5 s. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200410114/DC1.
Acknowledgments
This work was facilitated by use of the Lawrence Berkeley National Laboratory Advanced Light Source beamline 8.3.1 and the generous support of the 8.3.1 staff: James Holton, George Meigs, and Jane Tanamachi. We thank John Sotir for help with data collection, Luke Rice for advice on MAD phasing, and Gohta Goshima for assistance with S2 cell work. APC cDNA was kindly provided by Ken Kinzler and Bert Vogelstein.
The work was supported by the Agouron Institute Paul B. Sigler Fellowship of the Helen Hay Whitney Foundation (to K.C. Slep), the Wellcome Trust (to H. Ohkura), and the National Institutes of Health (grant NIH38499 to R.D. Vale).
Sarah L. Elliott's present address is Dept. of Biological Sciences, Stanford University, Stanford, CA 94305.
Abbreviations used in this paper: APC, adenomatous polyposis coli; β-ME, β-mercaptoethanol; MACF, microtubule actin cross-linking factor; MAD, multiwavelength anomalous diffraction; SeMet, selenomethionine.
References
- Altschul, S.F., W. Gish, W. Miller, E.W. Myers, and D.J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Askham, J.M., P. Moncur, A.F. Markham, and E.E. Morrison. 2000. Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene. 19:1950–1958. [DOI] [PubMed] [Google Scholar]
- Askham, J.M., K.T. Vaughan, H.V. Goodson, and E.E. Morrison. 2002. Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell. 13:3627–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, A.I., K.A. Siemers, and W.J. Nelson. 2002. Dissecting interactions between EB1, microtubules and APC in cortical clusters at the plasma membrane. J. Cell Sci. 115:1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, D., E. Kovaleva, and P.J. Schatz. 1999. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8:921–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz, M. 2001. Spindles cotton on to junctions, APC and EB1. Nat. Cell Biol. 3:E67–E68. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., P.D. Adams, G.M. Clore, W.L. DeLano, P. Gros, R.W. Grosse-Kunstleve, J.S. Jiang, J. Kuszewski, M. Nilges, N.S. Pannu, et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921. [DOI] [PubMed] [Google Scholar]
- Bu, W., and L.K. Su. 2003. Characterization of functional domains of human EB1 family proteins. J. Biol. Chem. 278:49721–49731. [DOI] [PubMed] [Google Scholar]
- Deka, J., J. Kuhlmann, and O. Muller. 1998. A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur. J. Biochem. 253:591–597. [DOI] [PubMed] [Google Scholar]
- Desai, A., and T.J. Mitchison. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., and A. Hall. 2003. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 421:753–756. [DOI] [PubMed] [Google Scholar]
- Gregory, S.L., and N.H. Brown. 1998. kakapo, a gene required for adhesion between and within cell layers in Drosophila, encodes a large cytoskeletal linker protein related to plectin and dystrophin. J. Cell Biol. 143:1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., J.Y. Zou, S.W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 47:110-9. [DOI] [PubMed] [Google Scholar]
- Juwana, J.P., P. Henderikx, A. Mischo, A. Wadle, N. Fadle, K. Gerlach, J.W. Arends, H. Hoogenboom, M. Pfreundschuh, and C. Renner. 1999. EB/RP gene family encodes tubulin binding proteins. Int. J. Cancer. 81:275–284. [DOI] [PubMed] [Google Scholar]
- Karcher, R.L., S.W. Deacon, and V.I. Gelfand. 2002. Motor-cargo interactions: the key to transport specificity. Trends Cell Biol. 12:21–27. [DOI] [PubMed] [Google Scholar]
- Kodama, A., I. Karakesisoglou, E. Wong, A. Vaezi, and E. Fuchs. 2003. ACF7: an essential integrator of microtubule dynamics. Cell. 115:343–354. [DOI] [PubMed] [Google Scholar]
- Korinek, W.S., M.J. Copeland, A. Chaudhuri, and J. Chant. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 287:2257–2259. [DOI] [PubMed] [Google Scholar]
- Lansbergen, G., Y. Komarova, M. Modesti, C. Wyman, C.C. Hoogenraad, H.V. Goodson, R.P. Lemaitre, D.N. Drechsel, E. Van Munster, T.W. Gadella Jr., et al. 2004. Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J. Cell Biol. 166:1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantz, V.A., and K.G. Miller. 1998. A class VI unconventional myosin is associated with a homologue of a microtubule-binding protein, cytoplasmic linker protein-170, in neurons and at the posterior pole of Drosophila embryos. J. Cell Biol. 140:897–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy, D.J., H.P. Erickson, I. Aukhil, P. Joshi, and W.A. Hendrickson. 1994. Crystallization of a fragment of human fibronectin: introduction of methionine by site-directed mutagenesis to allow phasing via selenomethionine. Proteins. 19:48–54. [DOI] [PubMed] [Google Scholar]
- Lee, L., J.S. Tirnauer, J. Li, S.C. Schuyler, J.Y. Liu, and D. Pellman. 2000. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 287:2260–2262. [DOI] [PubMed] [Google Scholar]
- Lee, S., and P.A. Kolodziej. 2002. Short Stop provides an essential link between F-actin and microtubules during axon extension. Development. 129:1195–1204. [DOI] [PubMed] [Google Scholar]
- Lee, S., K.L. Harris, P.M. Whitington, and P.A. Kolodziej. 2000. short stop is allelic to kakapo, and encodes rod-like cytoskeletal-associated proteins required for axon extension. J. Neurosci. 20:1096–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, C.L., D. Sun, M. Zheng, D.R. Knowles, and R.K. Liem. 1999. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 147:1275–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B., F. Roegiers, L.Y. Jan, and Y.N. Jan. 2001. Adherens junctions inhibit asymmetric division in the Drosophila epithelium. Nature. 409:522–525. [DOI] [PubMed] [Google Scholar]
- McCartney, B.M., D.G. McEwen, E. Grevengoed, P. Maddox, A. Bejsovec, and M. Peifer. 2001. Drosophila APC2 and Armadillo participate in tethering mitotic spindles to cortical actin. Nat. Cell Biol. 3:933–938. [DOI] [PubMed] [Google Scholar]
- Mennella, V.C., G.C. Rogers, S.L. Rogers, D.W. Buster, R.D. Vale, and D.J. Sharp. 2005. Functionally distinct Kinesin13 family members cooperate to regulate microtubule dynamics during interphase. Nat. Cell Biol. In press. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., and M.D. Rose. 1998. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 140:377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., S.C. Cheng, and M.D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., N. Shiina, and S. Tsukita. 2000. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J. Cell Biol. 148:505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, H., K. Koyama, Y. Murata, M. Morito, T. Akiyama, and Y. Nakamura. 2000. EB3, a novel member of the EB1 family preferentially expressed in the central nervous system, binds to a CNS-specific APC homologue. Oncogene. 19:210–216. [DOI] [PubMed] [Google Scholar]
- Nakamura, M., X.Z. Zhou, and K.P. Lu. 2001. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr. Biol. 11:1062–1067. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., K.A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. 1993. Oscillation data reduction program. Data Collection and Processing. N.I.L. Sawyer and S. Bailey, editors. Science and Engineering Research Council Daresbury Laboratory/Daresbury, Cheshire, UK. 56–62.
- Pierre, P., J. Scheel, J.E. Rickard, and T.E. Kreis. 1992. CLIP-170 links endocytic vesicles to microtubules. Cell. 70:887–900. [DOI] [PubMed] [Google Scholar]
- Powell, S.M., N. Zilz, Y. Beazer-Barclay, T.M. Bryan, S.R. Hamilton, S.N. Thibodeau, B. Vogelstein, and K.W. Kinzler. 1992. APC mutations occur early during colorectal tumorigenesis. Nature. 359:235–237. [DOI] [PubMed] [Google Scholar]
- Rehberg, M., and R. Graf. 2002. Dictyostelium EB1 is a genuine centrosomal component required for proper spindle formation. Mol. Biol. Cell. 13:2301–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., G.C. Rogers, D.J. Sharp, and R.D. Vale. 2002. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, U. Hacker, C. Turck, and R.D. Vale. 2004. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr. Biol. 14:1827–1833. [DOI] [PubMed] [Google Scholar]
- Rubinfeld, B., B. Souza, I. Albert, O. Muller, S.H. Chamberlain, F.R. Masiarz, S. Munemitsu, and P. Polakis. 1993. Association of the APC gene product with beta-catenin. Science. 262:1731–1734. [DOI] [PubMed] [Google Scholar]
- Smith, K.J., D.B. Levy, P. Maupin, T.D. Pollard, B. Vogelstein, and K.W. Kinzler. 1994. Wild-type but not mutant APC associates with the microtubule cytoskeleton. Cancer Res. 54:3672–3675. [PubMed] [Google Scholar]
- Su, L.K., and Y. Qi. 2001. Characterization of human MAPRE genes and their proteins. Genomics. 71:142–149. [DOI] [PubMed] [Google Scholar]
- Su, L.K., B. Vogelstein, and K.W. Kinzler. 1993. Association of the APC tumor suppressor protein with catenins. Science. 262:1734–1737. [DOI] [PubMed] [Google Scholar]
- Su, L.K., M. Burrell, D.E. Hill, J. Gyuris, R. Brent, R. Wiltshire, J. Trent, B. Vogelstein, and K.W. Kinzler. 1995. APC binds to the novel protein EB1. Cancer Res. 55:2972–2977. [PubMed] [Google Scholar]
- Subramanian, A., A. Prokop, M. Yamamoto, K. Sugimura, T. Uemura, J. Betschinger, J.A. Knoblich, and T. Volk. 2003. Shortstop recruits EB1/APC1 and promotes microtubule assembly at the muscle-tendon junction. Curr. Biol. 13:1086–1095. [DOI] [PubMed] [Google Scholar]
- Sun, D., C.L. Leung, and R.K. Liem. 2001. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J. Cell Sci. 114:161–172. [DOI] [PubMed] [Google Scholar]
- Tirnauer, J.S., S. Grego, E.D. Salmon, and T.J. Mitchison. 2002. EB1-microtubule interactions in Xenopus egg extracts: role of EB1 in microtubule stabilization and mechanisms of targeting to microtubules. Mol. Biol. Cell. 13:3614–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti, C., D.M. Wetzel, M. Schrader, M.J. Hasbani, S.R. Gill, T.E. Kreis, and T.A. Schroer. 1999. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell. 10:4107–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, P.S., P. Miura, M. Henderson, B. Byrne, and K.T. Vaughan. 2002. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 158:305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Y., C.H. Eng, J. Schmoranzer, N. Cabrera-Poch, E.J. Morris, M. Chen, B.J. Wallar, A.S. Alberts, and G.G. Gundersen. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6:820–830. [DOI] [PubMed] [Google Scholar]
- Zhou, F.Q., J. Zhou, S. Dedhar, Y.H. Wu, and W.D. Snider. 2004. NGF-induced axon growth is mediated by localized inactivation of GSK-3beta and functions of the microtubule plus end binding protein APC. Neuron. 42:897–912. [DOI] [PubMed] [Google Scholar]