Abstract
Importin-β (Impβ) is a major transport receptor for Ran-dependent import of nuclear cargo. Impβ can bind cargo directly or through an adaptor such as Importin-α (Impα). Factors involved in nuclear transport have been well studied, but systems analysis can offer further insight into regulatory mechanisms. We used computer simulation and real-time assays in intact cells to examine Impα–β-mediated import. The model reflects experimentally determined rates for cargo import and correctly predicts that import is limited principally by Impα and Ran, but is also sensitive to NTF2. The model predicts that CAS is not limiting for the initial rate of cargo import and, surprisingly, that increased concentrations of Impβ and the exchange factor, RCC1, actually inhibit rather than stimulate import. These unexpected predictions were all validated experimentally. The model revealed that inhibition by RCC1 is caused by sequestration of nuclear Ran. Inhibition by Impβ results from depletion nuclear RanGTP, and, in support of this mechanism, expression of mRFP-Ran reversed the inhibition.
Introduction
The transport of proteins in and out of the nucleus is a major cellular process (Görlich and Kutay, 1999; Macara, 2001; Weis, 2002). Transport not only localizes proteins destined for the nucleus or cytoplasm, but also functions as a key step in signal transduction pathways and in the regulation of cell cycle progression. The nuclear pore complex is a large multi-protein assembly that penetrates the nuclear envelope and connects the cytoplasm to the nucleus (Fahrenkrog and Aebi, 2003; Suntharalingam and Wente, 2003). Although small proteins <20–40 kD can move through the NPC by passive diffusion, larger proteins require soluble transport receptors called karyopherins (Chook and Blobel, 2001). Importin-β (Impβ) is one of the predominant karyopherins that drive import. Although a small number of cargo proteins may bind Impβ directly, most cargoes require the adaptor protein importin-α (Impα).
Impα contains a COOH-terminal region that binds directly to proteins containing an NLS sequence (Conti et al., 1998; Herold et al., 1998). However, Impα also contains an NH2-terminal auto-inhibitory domain that blocks the NLS-binding site (Kobe, 1999). Binding of Impβ to Impα relieves this auto-inhibitory blockade and allows Impα to bind cargo proteins with high affinity (Fanara et al., 2000).
Translocation of the Impα–Impβ–cargo complex through the NPC is thought to be mediated by weak hydrophobic interactions between Impβ and nucleoporins (Bayliss et al., 2000). On the nuclear side of the NPC, RanGTP dislodges Impβ from the complex (Görlich et al., 1996). The Impα–cargo heterodimer then dissociates, assisted by the high affinity binding of CAS to NLS-free Impα. CAS is a karyopherin that serves to export Impα in association with RanGTP (Kutay et al., 1997). Both the RanGTP–Impβ and RanGTP–CAS–Impα complexes translocate back to the cytoplasm where RanGAP (assisted by RanBP1 and Impα) hydrolyzes the RanGTP to RanGDP (Bischoff and Görlich, 1997; Floer et al., 1997; Petersen et al., 2000). The export complexes disassociate and the transport receptors are recycled for another round of import (Fig. 1).
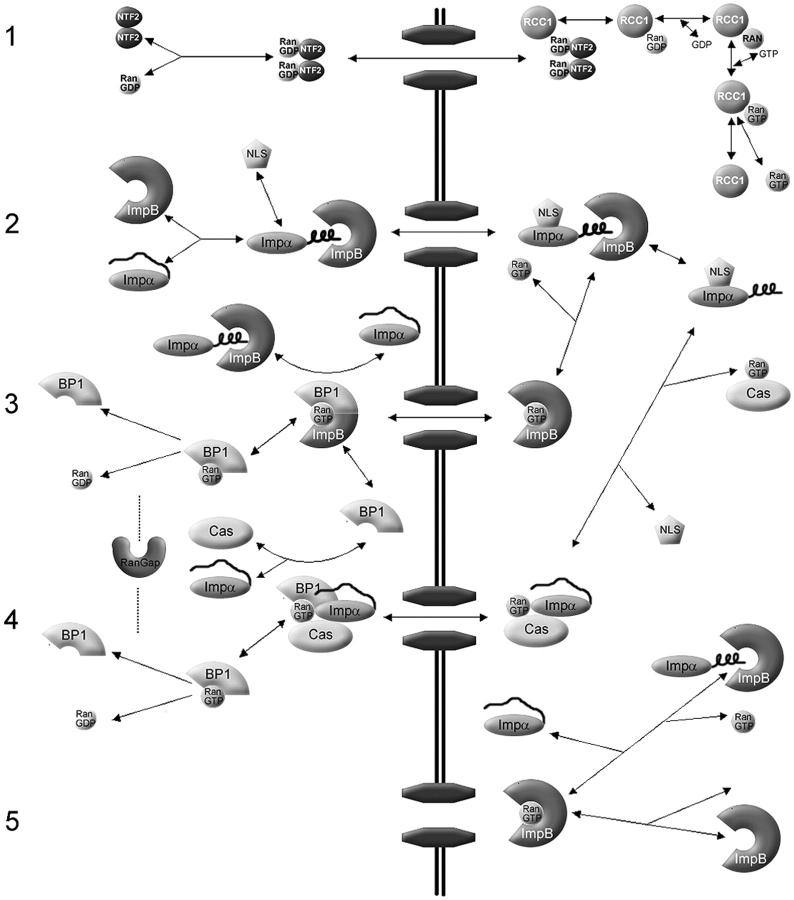
Figure 1.
Overview of the Impα–β nuclear protein import model (adapted in part from Catimel et al., 2001). (1) The NTF2 homodimer imports Ran into the nucleus. In the model, Ran can also translocate the NPC without NTF2, but with a much lower permeability constant. In the nucleus, RCC1 acts in a four-step reversible reaction to convert RanGDP to RanGTP. (2) Impα contains an auto-inhibitory region that prevents binding to an NLS. Upon binding to Impβ, the NH2-terminal auto-inhibitory domain folds into an α-helix, allowing NLS cargo to bind to Impα. Translocation of Impα–Impβ–cargo through the NPC is a reversible reaction, represented by a permeability constant. On the nuclear side of the NPC, RanGTP acts to dissociate Impβ from the complex. (3) The Impα–cargo heterodimer dissociates, assisted by high affinity binding of CAS–RanGTP to Impα. The RanGTP–Impβ complex translocates back to the cytoplasm where RanGAP (assisted by RanBP1 and Impα) catalyzes the hydrolysis of RanGTP to RanGDP, resulting in dissociation of Ran from the Impβ. (4) RanGTP–Cas–Impα complexes translocate back to the cytoplasm. RanGAP–RanBP1 hydrolyzes RanGTP to RanGDP. The complexes disassociate and transport receptors are recycled for another round of import. (5) Other association–dissociation reactions that occur in the nucleus. Note that passive diffusion by Ran, NLS-cargo, and Impα, and translocation of empty receptors through the NPC, are not depicted. Reversible reactions are represented by double-headed arrows. Essentially irreversible reactions are shown by a single-headed arrow. For simplicity, reactions involving the generic receptors and endogenous cargo, which are incorporated in the computer model, are not depicted here. The complete schematic is in Fig. S1. Concentrations and kinetic constants for the all components of the model are provided in Tables I–III.
The gradient of RanGTP between the nucleus and cytoplasm provides the motive force to drive transport. This gradient is created by an asymmetric distribution of the nucleotide exchange factor RCC1 (also called RanGEF) and of RanGAP. RCC1 is a resident nuclear protein that promotes the exchange of RanGDP to RanGTP (Bischoff and Ponstingl, 1991a). Excluded from the nucleus, RanGAP acts to maintain Ran in the GDP-bound state in the cytoplasm (Fig. 1). The Ran gradient is highly dynamic, with RanGDP rapidly imported by transport factor NTF2 (Ribbeck et al., 1998; Smith et al., 1998).
Smith et al. (2002) conducted the first in silico analysis of Ran transport. A system of ordinary differential equations (ODEs) was created using the Virtual Cell software to describe reactions and fluxes of proteins involved in Ran Transport (Loew and Schaff, 2001; Smith et al., 2002). These proteins were set at known initial concentrations and allowed to come to steady state. A jump in cytoplasmic Ran concentration was used to simulate the micro-injection of recombinant Ran. Sensitivity analysis showed that RCC1 dominated changes in steady-state Ran flux, whereas NTF2 and the permeability of NTF2–RanGDP had smaller effects. Increases in RanBP1, RanGAP, karyopherins, and the permeability of the RanGTP–karyopherin complex had no effect on Ran flux at steady state. A similar analysis to determine rate-limiting components of the initial rate of Ran import after cytoplasmic injection showed that NTF2 was limiting, but that the initial rate of import was insensitive to RCC1.
To compare simulation results with Ran flux in live cells, a fluorescently tagged Ran (RanFl) was injected into BHK21 cells and imaged using confocal microscopy. Time-lapse images were used to quantify nuclear accumulation and import rate of the Ran. Co-injected NTF2 produced positive changes in the initial rate of import and steady-state N/C ratio of Ran, as predicted by the model. RanGAP produced no changes, which also is consistent with the model. RCC1 depletion was tested using tsBN2 cells. These cells possess a temperature-sensitive RCC1 mutation, so that at 39.5°C the mutant RCC1 is rapidly degraded (Uchida et al., 1990). Temperature-shifted tsBN2 cells showed reduced nuclear accumulation of Ran but only small affects on initial rate of Ran import, which is consistent with the model.
Görlich and colleagues (Görlich et al., 2003) developed a model to study how the nuclear–cytoplasmic Ran gradient couples to nuclear import through Impβ. The authors modified several kinetic parameters in their simulation of the Ran GTPase system. Most significantly, the Michaelis-Menten model of RCC1 used by Smith et al. (2002) was replaced by a four-step reversible series of reactions for GTP–GDP exchange, based on a detailed in vitro kinetic analysis by Klebe et al. (1995b). The kcat/Km for RanGAP was also adjusted upward to reflect enzyme activity at 37°C. This simulation predicted that decreases in nuclear RCC1 would reduce RanGTP accumulation, but that RCC1 is not limiting for the system. Conversely, the simulation suggested that RanGAP is limiting. Lastly, nuclear accumulation of RanGTP was shown to be heavily dependent on the GDP/GTP ratio used in the model. The model also predicted a close relationship between the RanGTP gradient and the nuclear/cytoplasmic ratio of cargo concentration at steady state.
In summary, the Smith and Görlich kinetic models show how the RanGTP gradient is established and used to drive a nuclear cargo gradient, and that even a simple system of interacting proteins can generate behavior that would difficult to predict using a static model.
In the current study, we extend the RanGTPase model to show how the Ran gradient couples to nuclear cargo import through the Impα–Impβ system. Using a compartmental model, we show how the system of known biochemical reactions involved in Impα–Impβ import can successfully recapitulate nuclear import seen in vivo. Predictions from the model were tested in vivo by monitoring nuclear import in whole cells. Unexpectedly, the model predicted that increasing the concentrations of RCC1 or Impβ would inhibit rather than stimulate the initial rate of cargo import, and these effects were validated experimentally.
Results
We constructed a compartmental model of Impα–Impβ-mediated nuclear transport using the program Jarnac, a biochemical simulation package for Windows (Sauro et al., 2003). An overview of the model is shown in Fig. 1 and is described in Materials and methods. A more detailed schematic is provided in the Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200409024/DC1), together with a script file of the model than can be run in Jarnac (http://64.17.162.114/downloads/ISetup.exe).
Quantitative models of nuclear protein import are necessarily complex, because of the large number of factors involved in a single transport cycle. Moreover, there are multiple pathways for nuclear import and export, all of which use some factors such as Ran and NTF2 in common, but which use different karyopherins (Görlich and Kutay, 1999; Chook and Blobel, 2001; Macara, 2001). The concentrations and even the identities of all the cargoes for each of these karyopherins remain mostly unknown. However, the kinetics of the Ran cycle and of individual components of the Impα–β pathway have been well studied. Therefore, in our computer model we used experimentally determined parameters for Ran and Impα–β, and included terms for a generic karyopherin to account for other nuclear transport pathways. The same kinetic parameters were used for the generic karyopherin as for Impβ. This approach seems reasonable because karyopherins such as transportin bind Ran with an affinity similar to that of Impβ, while karyopherins that have a much lower affinity for Ran will not significantly perturb the system. Another complication is that there are probably hundreds of endogenous cargoes for Impα–β, most of which have not yet been characterized, and their cytoplasmic concentrations are unknown. Necessarily, therefore, we initially treated the concentration of endogenous cargo as a floating parameter. Endogenous cargo and labeled cargo reactions were modeled with identical but separate reaction pathways (Fig. S1) using the parameters listed in Tables I–III.
Table I. Reactant concentrations.
| Reactant
|
Concentration
|
Source
|
|---|---|---|
| μM | ||
| Ran | 5 |
Bischoff and Ponstingl, 1991b; Bischoff and Görlich, 1997 |
| RanBP1 | 2 | Bischoff et al., 1995 |
| RanGAP | 0.5 | Klebe et al., 1995a |
| RCC1 | 0.25 | Bischoff and Ponstingl, 1991b |
| NTF2 | 0.6 | Görlich et al., 2003 |
| Impα | 1 | Estimate |
| Impβ | 3 | Ribbeck et al., 1998 |
| CAS | 3 | Ribbeck et al., 1998 |
| Native β cargo | 1 | Estimate |
| Native α–β cargo | 10 | Estimate |
| GTP | 470 | Görlich et al., 2003 |
| GDP | 1.6 | Görlich et al., 2003 |
| Other transport receptors | 3.6 | Estimate |
The computer model was first run in the absence of any perturbation for a period of 1,000 s, to bring the system to steady state. A RanGTP gradient becomes established, and the transport factors distribute between the cytoplasmic and nuclear compartments (Table IV).
Table IV. Steady-state reactant concentrations.
| Reactant
|
Nucleus
|
Cytoplasm
|
|---|---|---|
| μM | μM | |
| Total RanGTP | 7.13564 | 0.49 |
| Free RanGTP | 1.23 | 0.0019 |
| Total Ran | 9.50 | 3.46 |
| Total Impα | 0.86 | 0.98 |
| Total Impβ | 4.095 | 2.43 |
| Total NTF2 | 0.36 | 0.67 |
| Total Cas | 2.55 | 2.52 |
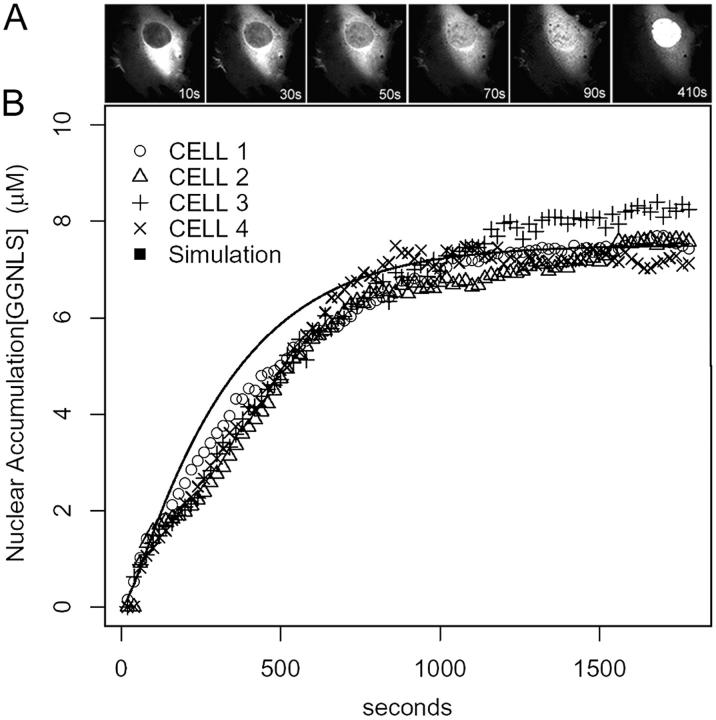
To simulate the micro-injection of labeled cargo, the cytoplasmic concentration of labeled cargo was stepped up from zero to some chosen initial value, and the simulation was run for an additional 500 s to track the import of this cargo into the nuclear compartment. Initial rates of cargo import were calculated from the rates of nuclear accumulation during the first 60 s after the addition of labeled cargo. To compare the results of the model with import in vivo, we injected GST-GFP-NLS (GGNLS) into the cytoplasm of whole cells and imaged nuclear import by confocal microscopy (Fig. 2 A). Both the initial rate and steady-state concentration of the virtual cargo compare well with the nuclear transport of GGNLS observed in live cells (Fig. 2 B).
Figure 2.
Nuclear import of GGNLS. (A) HeLA cells were cytoplasmically injected with 3 μM GGNLS. Cells were kept at 37°C in physiological saline for the duration of the experiment. Nuclear accumulation was quantified by time-lapse confocal microscopy. Images were taken every 20 s and pixel values within the nucleus were converted to concentration by the use of a standard series of known protein concentrations. (B) Results are overlaid with a simulation of identical conditions in the compartmental model.
Permeability terms
A key feature in any model of nucleo-cytoplasmic transport is the permeability of the karyopherins, cargo, and other components across the nuclear envelope. Rather than model the detailed structure of individual pores, one can take advantage of the fact that translocation between the nuclear and cytoplasmic compartments behaves to a first approximation like passive diffusion, and can therefore be modeled using a simple permeability term (Table III). The permeabilities for some components, such as NTF2, have been determined previously, using reconstituted transport in digitonin-treated cells (Ribbeck and Görlich, 2001). We have also measured permeabilities both using digitonin-treated cells and micro-injection into intact cells to measure permeabilities for Impα, Impβ, and GST-GFP through the NPC (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200409024/DC1). Impβ, labeled with Oregon green isothiocyanate (OGI), gave a permeability of 0.4 s−1 in whole cells, compared with 0.95 s−1 reported in a previous study in digitonin-permeabilized cells (Görlich et al., 2003). The leakage of free RanGTP from the nucleus to the cytoplasm was represented using a permeability of 0.03 s−1, taken from the previously measured permeability for RanGDP (Ribbeck and Görlich, 2001).
Table III. NPC permeabilities.
| Reactant | Permeability factor sec− 1 | Source |
|---|---|---|
| Ran | 0.03 | Görlich et al., 2003 |
| NTF2 | 1 | Görlich et al., 2003 |
| NTF2-Ran | 0.5 | Görlich et al., 2003 |
| Impβ | 0.4 | Experimentally measured |
| Impα | 0.03 | Experimentally measured |
| Impα–Impβ | 0.25 | Estimate |
| Impα–Impβ–GGNLS | 0.2 | Estimate |
| GGNLS | 0.0005 | Experimentally measured |
| CAS | 0.3 | Estimate |
| CAS-RanGTP-Impα | 0.2 | Estimate |
| RanGTP-Impβ | 0.07 | Estimate |
| Impβ-cargo | 0.25 | Estimate |
One important limitation of the model is that it does not include terms for competition between different transport pathways at the level of permeability across the nuclear envelope. Thus, permeabilities measured in vitro (using digitonin-treated cells) are likely to be rather higher than permeabilities measured for the same proteins in intact cells where the proteins are competing with other endogenous proteins for access to the pores. However, we know that the NPC has a high capacity and the import rate is, to a first approximation, linear with concentration over the concentration range of GGNLS used in this study. Competition is unlikely, therefore, to be a major source of error in the modeling. Moreover, the sensitivity analysis (see below) suggests that the cargo import rate is not limited by the permeabilities of many of the transport components in the model.
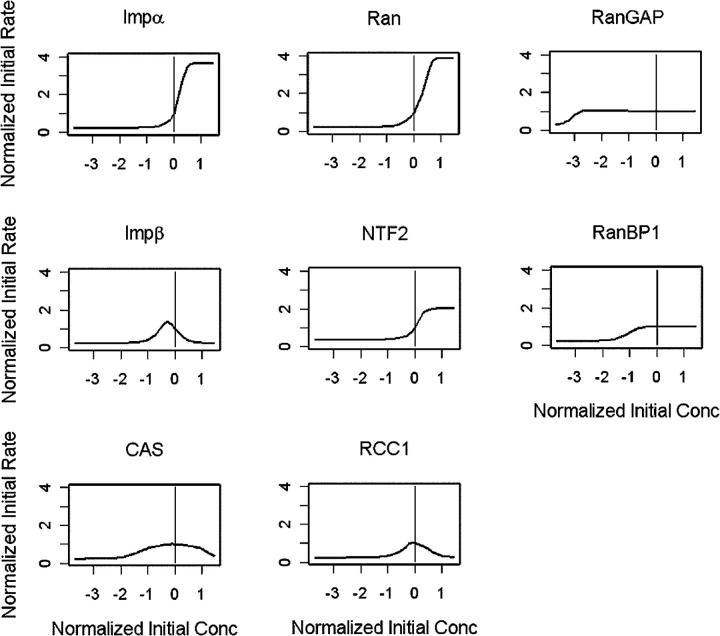
Sensitivity analysis
To identify possible rate-limiting steps in nuclear protein import, we performed a global sensitivity analysis on reactant concentrations in the model (Fig. 3). Reactant concentrations were varied individually over a five log scale from 0.0001 to 10 times the initial concentration. After allowing the system to come to steady state for 1,000 s, the cytoplasmic injection of GGNLS was simulated. Normalized initial rates of GGNLS import were then plotted as a function of initial concentration.
Figure 3.
Global sensitivity analysis of the computer model. Individual parameters in the model were varied over a five log scale, and initial rates of nuclear accumulation were calculated for each value. Graphs are normalized so that (0,1) indicates the original value used in the model.
These simulations showed that GGNLS import responds most dynamically to the Impα and Ran concentrations, and is limited by the estimated cellular concentration of these factors. The slope of the curve for Impα is higher than that for Ran, suggesting that transport is more highly responsive to small deviations from the basal concentration of Impα than of Ran. Surprisingly, however, import is predicted to be relatively insensitive to the cellular concentration of CAS, which recycles Impα back to the cytoplasm.
The initial import rate is also highly sensitive to the concentration of NTF2, which mediates the import of Ran into the nucleus, although the dynamic capacity is not so large as for Impα or Ran. Perturbations in the concentration of NTF2 will alter the import rate because the RanGTP gradient across the nuclear envelope is coupled to the rate at which RanGDP is recycled back into the nucleus.
The sensitivity analysis showed that RanGAP is not limiting for cargo import, and that the concentration of its cofactor, RanBP1, can be reduced at least 10-fold before cargo import is affected. Cytoplasmic RanBP1 forms a RanBP1–Impβ–RanGTP complex but then is immediately released in the next reaction step when RanGAP triggers GTP hydrolysis, and the complex disassociates (Fig. 1). This rapid recycling of RanBP1 allows it to be effective at submicromolar levels. By contrast, the nucleotide exchange factor for Ran, RCC1, shows an unexpectedly complex relationship to cargo import, in that either a reduction or an increase in the cellular RCC1 concentration relative to the endogenous level strongly inhibits import. A reduction in RCC1 reduces the concentration of free nuclear RanGTP, which leads to a nuclear accumulation of Impβ–Impα–cargo complexes that cannot dissociate, and of Impα which cannot be exported back to the cytoplasm by CAS. But why would an increase in RCC1 also inhibit cargo? The explanation lies in the four-step kinetic model for nucleotide exchange, described by Wittinghofer and colleagues (Klebe et al., 1995b). In this model, RCC1 has a significant affinity for Ran, and high concentrations of the exchange factor sequester Ran in the nucleus as an RCC1–Ran complex. The free concentration of RanGTP available to dissociate import complexes and export Impα is therefore reduced, with a consequent inhibition of cargo import.
Another unexpected outcome of the sensitivity analysis was the inhibitory effect of high Impβ concentrations. Intuitively, one might have predicted that increasing the amount of the transport receptor for GGNLS cargo would stimulate import. However, the receptor and Ran levels are finely balanced in the model and, if in excess, the Impβ translocates into the nucleus in an unloaded state, then sequesters free RanGTP, and delivers it to the cytoplasm where it is hydrolyzed. This process is called futile cycling, and it depletes the nucleus of RanGTP, thereby inhibiting cargo import. A similar, but less dramatic effect is seen at high concentrations of CAS, which also reduce import (Fig. 3).
We also performed a sensitivity analysis of the NPC permeability constants for various transport components (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200409024/DC1), which predicts that the system is fairly robust to perturbations in many of these constants. Thus, the predictions of the computer simulations are not dependent on their exact magnitudes. This is important, because we do not have experimental data on the permeabilities of all the components of the model.
Experimental approach
Experimental tests of this type of global sensitivity analysis are complicated by the need to change the endogenous concentration of each factor by a known amount, before the injection of the fluorescent import cargo. Ectopic expression from a plasmid varies from cell to cell, and cannot easily be measured until after the experiment has been completed. Micro-injection is also highly variable from cell to cell, because the injected volume is very sensitive to back pressure and needle diameter. Therefore, we chose in most cases to use co-injection of the GGNLS cargo with known concentrations of recombinant factors. Although the amount of injected cargo and factor will vary from cell to cell, the actual concentrations can be calculated from the total GFP fluorescence intensity within each cell. One advantage of computational modeling is the ease with which effects on transport rate can be calculated and compared with experimental data even when there are two variables for each data point (the cargo concentration and the factor concentration).
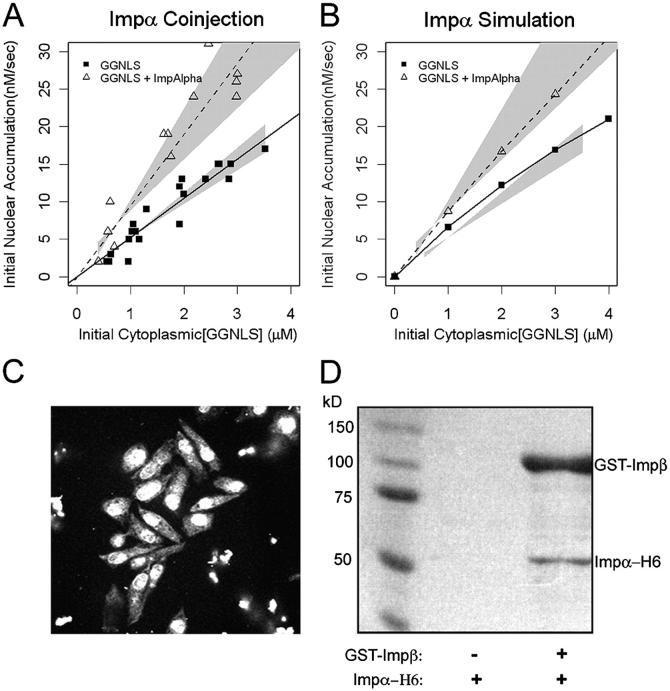
Impα is a limiting reactant for nuclear import
To test the predictions of the computer model for nuclear import, we performed co-injection experiments using 20 μM GGNLS as the labeled cargo. The GGNLS was injected together with recombinant transport factors into the cytoplasm of HeLa cells that were maintained at 37°C. The total amount of injected cargo and its initial rate of import were then quantified as described in Materials and methods. The variability in the injected volume from cell to cell generates a range of initial cytoplasmic concentrations that can be plotted against the initial rates. To provide a direct comparison with the model, we generated a series of simulations that directly mirrored the conditions used in the co-injection experiments, over the same range of initial cargo and factor concentrations.
The initial cytoplasmic concentration of cargo varied from ∼0.5 to 4 μM, and the initial import rate was linear with concentration over this range (Fig. 4 A), showing that, as predicted by our model, the Impα–β transport system is operating well below maximum capacity in HeLa cells. The concentration at half-maximal import rate (K0.5) for cargo in silico was calculated to be ∼12 μM.
Figure 4.
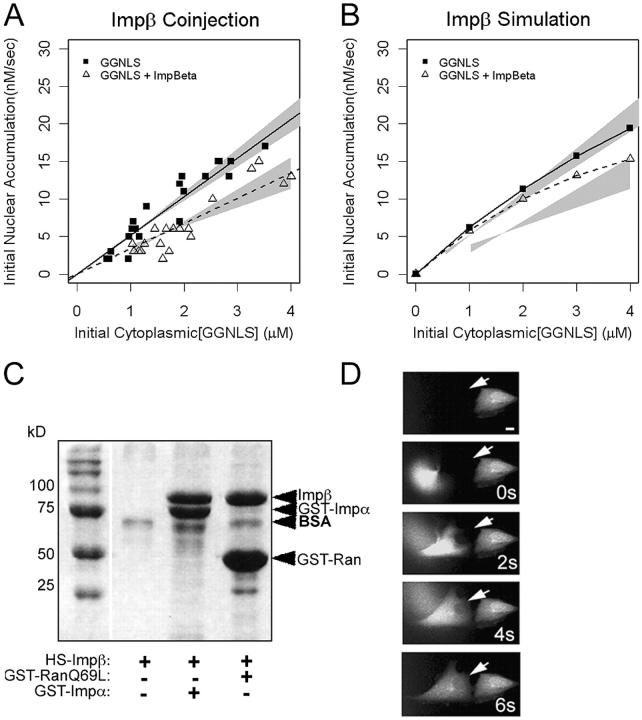
Effect of Impα-His6 on GGNLS nuclear import. (A) 20 μM GGNLS was microinjected with 5 μM Impα into the cytoplasm of HeLa cells maintained at 37°C and nuclear accumulation was recorded every 20 s. The initial concentration was calculated from the point at which nuclear and cytoplasmic GGNLS concentrations were equal, by comparison of mean pixel intensity in the cell with those of a series of protein standards. Initial rates were quantified as the slope of the import rate for the first 30–60 s of import. Data were plotted versus initial GGNLS concentration and fitted by linear regression analysis. Lines show the best fit to the data and gray areas indicate the 99% confidence limits of the slopes. Co-injection of Impα-His6 significantly increased the initial rate over baseline by two-way ANOVA (P = 0.00028). (B) The computer model was used to generate import data over the same range of GGNLS and Impα concentrations. Gray areas are taken from A. (C) Nuclear import of Impα. To verify the functionality of the His-tagged Impα the protein (1 mg/ml) was labeled with OGI and added to digitonin-permeabilized cells, which were then imaged by confocal microscopy. (D) Impα binds to Impβ. As a further test of functionality, 40 μg Impα-His6 was incubated with beads to which either 20 μg GST or 20 μg GST-Impβ had been attached. After extensive washing, bound proteins were analyzed by SDS-PAGE and stained with Coomassie blue.
We first tested the effect of co-injecting recombinant Impα. The model predicted that this factor is rate-limiting for cargo import (Fig. 4 B). To ensure that the injected factor was functional, we used a permeabilized cell assay. A portion of the protein was fluorescently labeled and incubated with Impβ, Ran, NTF2, RanBP1, and RanGAP. The label rapidly accumulated in the nuclei of permeabilized cells (Fig. 4 C). The Impα was also able to bind to GST-Impβ attached to glutathione–Sepharose beads (Fig. 4 D). An unlabeled portion of the same Impα preparation was then co-injected into intact HeLa cells with the GGNLS cargo. As shown in Fig. 4 A, the Impα induced a significant increase in the initial rate of cargo import, similar in magnitude to that predicted by the computer model.
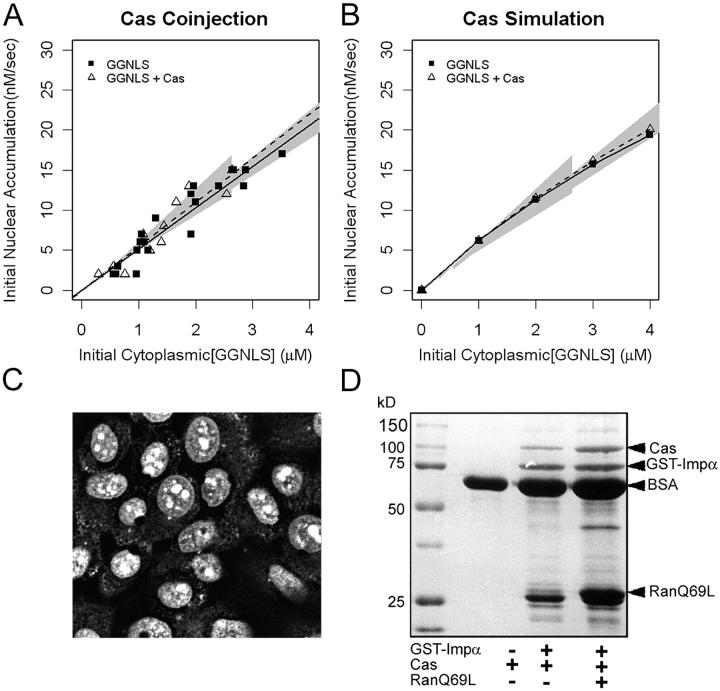
Import is insensitive to elevated CAS concentration
During each transport cycle, Impα needs to be recycled back to the cytoplasm. It is exported in a complex with RanGTP and CAS. Therefore, we expected that increasing the concentration of CAS in the cell would increase the availability of cytoplasmic Impα and therefore increase the cargo import rate. However, the model predicts that CAS is not limiting for import (Fig. 3 and Fig. 5 B). We addressed this possibility using recombinant CAS. To test the functionality of the CAS, we used a permeabilized cell assay, and observed that the recombinant CAS could accumulate in the nuclei of HeLa cells (Fig. 5 C). Second, we conducted binding assays between CAS, Impα, and RanGTP, using a constitutively active mutant of Ran (RanQ69L) (Fig. 5 D). The recombinant CAS bound Impα and showed increased binding in the presence of RanGTP. However, when the CAS was co-injected with GGNLS into intact HeLa cells, it had no detectable effect on import. These data show that CAS is not limiting for the initial rate of α–β cargo import into the nucleus.
Figure 5.
Effect of CAS on nuclear accumulation for GGNLS. (A) 20 μM GGNLS was microinjected with 5 μM recombinant CAS-His6 into the cytoplasm of HeLa cells and initial rates of nuclear import were measured and analyzed as in Fig. 4. (B) Initial rates were calculated from the computer model over the same range of GGNLS/CAS concentrations as in A. (C) To test the functionality of the recombinant CAS, the protein (1.3 mg/ml) was labeled as in Fig. 4 and incubated with digitonin-permeabilized cells. (D) Binding of CAS to Impα. 54 μg GST-Impα was attached to beads and incubated with 3 μg CAS ± 9 μg Ran(Q69)L-His6. After extensive washing the bound proteins were separated by SDS-PAGE and stained with Coomassie blue.
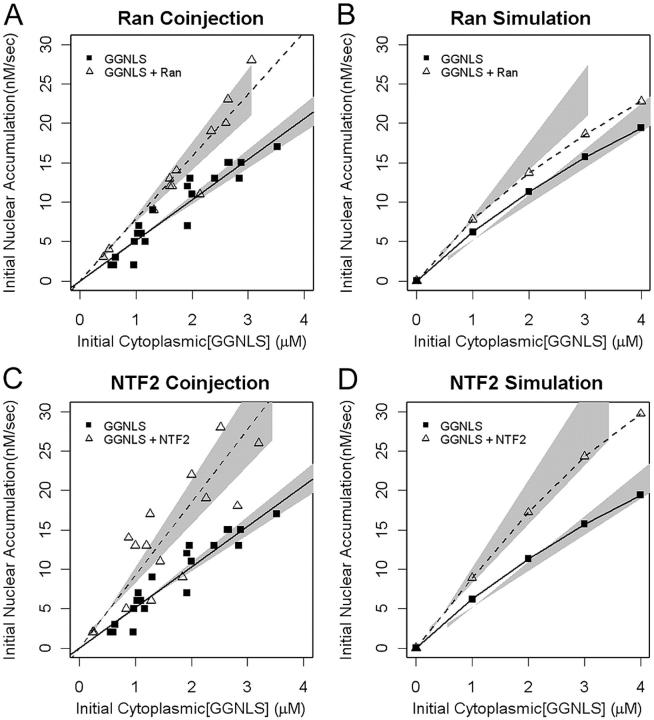
Factors in the RanGTPase system are limiting for nuclear import
Cargo import via Impα–β depends on the nucleo-cytoplasmic gradient not only for the recycling of Impα, but also for the release of cargo from the Impα–β complex. We have shown previously that NTF2 is limiting for Ran import, and we therefore expected that both NTF2 and Ran might be limiting for NLS cargo import. Indeed, the computer model supports this idea (Figs. 3 and 6). To test these predictions we co-injected GGNLS with either wild type His6-tagged Ran or with NTF2. In both cases, a significant increase in the initial rate of cargo import was observed (Fig. 6, A and C). The effect of Ran co-injection was slightly larger, experimentally, than predicted from the model. This difference arises in part because we make the assumption (because of scatter in the data) that the initial rate varies linearly with GGNLS concentration, over the experimental range used, whereas in the model the rate curve is hyperbolic. Additionally, secondary effects, such as competition for access to the nuclear pores, were not included in the model.
Figure 6.
Effects of Ran and NTF2 on GGNLS import. Import assays were performed and analyzed as in Fig. 4, using recombinant Ran-His6 (A) or NTF2 (C). Co-injection of either Ran or NTF2 significantly increased the initial rate over baseline (P = 0.00016 for Ran; P = 0.000069 for NTF2). The functionality of these proteins has been described previously. Simulations were performed over the same ranges of concentration as in the experiments (B and D).
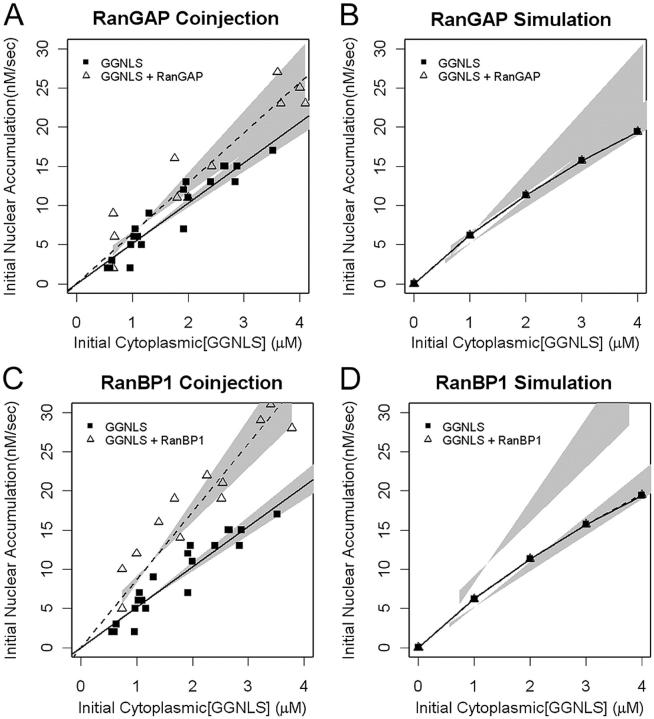
We next tested other components of the Ran system that are required for maintenance of a RanGTP gradient (Fig. 7). The Ran gradient has been predicted to be sensitive to the concentration of extra-nuclear RanGAP (Ribbeck and Görlich, 2001). However, our cargo import model shows that RanGAP is not limiting for cargo import in intact cells at 37°C (Fig. 3 and Fig. 7 B). Experimental assays confirmed this (Fig. 7 A): although co-injection of active RanGAP appeared to slightly increase import, this effect falls below the level of statistical significance (P > 0.05).
Figure 7.
Effects of RanGAP and RanBP1 on GGNLS import. Import assays were performed and analyzed as in Fig. 4, using recombinant GST-RanGAP (A) or GST-RanBP1 (C). The functionality of these proteins has been described previously. Simulations were performed over the same ranges of concentration as in the experiments (B and D). Co-injection of RanBP1 significantly increased the initial rate over baseline (P = 0.0022) but the effect of RanGAP fell under the threshold of significance (P = 0.067).
Next, we tested the effect of the RanGAP coactivator, RanBP1, on the system. The computer model predicted that RanBP1 is not limiting, but the co-injection of recombinant RanBP1 reproducibly increased the initial rate of cargo import (Fig. 7, C and D). Why is this? The global sensitivity analysis performed on the model shows that RanBP1 acts catalytically to stimulate RanGAP, and is needed at only substoichiometric levels to maintain an optimal transport rate. Large increases in RanBP1 concentration in the model have little effect on import compared with the baseline level (Fig. 3). The unexpected disparity between the simulation and experimental results, together with the observation that elevated RanGAP does not significantly increase import, suggests that the role of RanBP1 may not be completely understood.
Increased nuclear RCC1 inhibits cargo import
One unexpected prediction of the computer simulations was that the endogenous RCC1 concentration is finely balanced to provide the maximum RanGTP gradient. A reduction in the amount of RCC1 in the nucleus will of course reduce the production of RanGTP, but an increase in the amount of RCC1 also is predicted to reduce the free RanGTP concentration, through the formation of an RCC1–RanGTP complex.
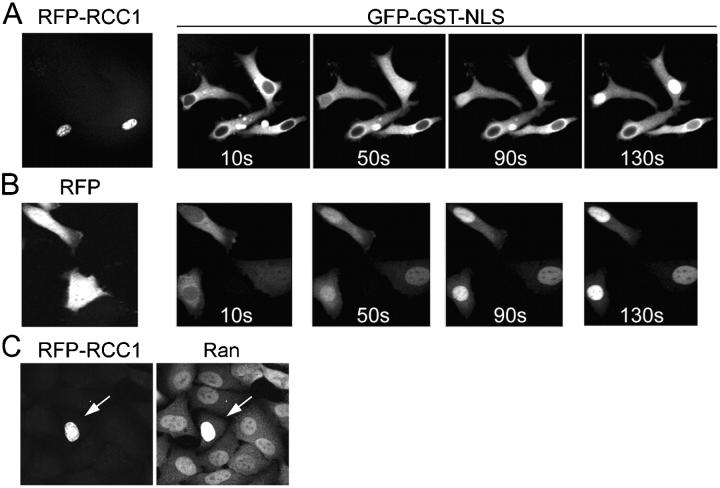
A small decrease in nuclear RanGTP in response to elevated RCC1 was proposed previously (Görlich et al., 2003), but this decrease was not predicted to alter the dynamic capacity of the system for Ran-driven transport, and no analysis of transport rates was performed. We found that cytoplasmic co-injection of recombinant RCC1 with GGNLS in equimolar ratio did inhibit import (unpublished data) but this result could arise because the RCC1 generates RanGTP in the cytoplasm, which would disassociate import complexes, and it might compete with the GGNLS for binding to Impα (Nemergut and Macara, 2000). Therefore, we tested the effect of RCC1 by transfection of HeLa cells with an mRFP-RCC1 construct, and then compared the import rates for injected GGNLS in cells that expressed the RFP fusion protein versus those that did not. As shown in Fig. 8 A, GGNLS import was dramatically inhibited in those cells that express mRFP-RCC1, as compared with adjacent cells that were injected 10–20 s earlier and that do not express the fusion protein. Cells transfected with mRFP alone showed no inhibition of import (Fig. 8 B), confirming that the effect is dependent on RCC1 expression and is not a nonspecific response to the mRFP.
Figure 8.
Effects of RCC1 on GGNLS import. HeLa cells were transiently transfected with pK-mRFP-RCC1 (A) or as a control with pK-RFP (B), and after 18 h were injected with GGNLS. Cells that visibly expressed the RCC1 and those that did not were both injected. Images show nuclear accumulation of GGNLS over time. (C) Cells expressing RFP-RCC1 accumulate more Ran in the nucleus. Cells were transfected as in A, then fixed and stained for endogenous Ran. RFP-RCC1 concentration was estimated to be 10–20 μM, based on standards using recombinant RFP protein (not depicted). Arrow indicates cell expressing RFP-RCC1
RCC1 possesses an NH2-terminal NLS that can bind Impα. It is conceivable, therefore, that elevated RCC1 levels could inhibit cargo import by sequestering Impα in the nucleus. This explanation is perhaps unlikely, because free Impα has a very low affinity for cargo (Fanara et al., 2000), and the total nuclear concentration of NLSs is probably high. Nonetheless, we tested this possibility by staining RFP-RCC1 transfected cells for endogenous Ran. Sequestration of Impα in the nucleus would not be expected to alter nuclear Ran, whereas sequestration of Ran by the RCC1 would be predicted to cause an increase in the nuclear/cytoplasmic ratio of Ran. This outcome is seen in Fig. 8 C.
Excess Impβ suppresses the initial rate of nuclear import
Counter-intuitively, the model predicts that the co-injection of Impβ with cargo suppresses rather than stimulates the initial rate of GGNLS import (Fig. 3 and Fig. 9 B). Why does this surprising effect occur? The simulation shows that excess Impβ undergoes futile cycling—that is, it translocates into the nucleus without cargo, where it sequesters free RanGTP. Without an available pool of free RanGTP, the Impα–Impβ–cargo heteromer cannot dissociate, so the cargo is not released, and will not accumulate in the nucleus.
Figure 9.
Effects of Impβ on GGNLS import. (A) Import was measured and analyzed as in Fig. 4, using recombinant His6-Impβ. Co-injection of His6-Impβ significantly decreased the initial rate compared with baseline (P = 0.046). (B) Simulations were calculated as in Fig. 4. (C) Binding of His6-Impβ to GST-Impα and GST-Ran(Q69L). 54 μg GST-Impα and 21 μg GST-Ran(Q69L) were attached to beads and incubated with 60 μg His6-Impβ. After washing, the bound proteins were separated by SDS-PAGE and stained with Coomassie blue. (D) To test the functionality of the Impβ, the protein (15 mg/ml) was labeled as in Fig. 4, then injected into HeLa cells. Nuclear accumulation of the protein was monitored over time. Arrows indicate cell injected at time 0.
Experimental observations support the prediction of the model. As shown in Fig. 9 A, co-injection of recombinant Impβ suppressed the initial rate of GGNLS import. However, fragments of Impβ that lack the Ran-binding domain can inhibit transport by binding with high affinity to the nuclear pore complex. Thus, proteolysis of the Impβ might produce inhibitory contaminants that inadvertently produce the result predicted by the model. To test the functionality of the recombinant Impβ, we performed both in vitro and in vivo tests. We first checked that the majority of the purified Impβ could bind GST-Impα and GST-Ran(Q69L) (Fig. 9 C). Second, we injected fluorescently labeled Impβ into the cytoplasm of live cells (Fig. 9 D). The rapid nuclear accumulation of Impβ in whole cells supports the conclusion that the Impβ is functional and should not inhibit nuclear transport by binding irreversibly to the pores.
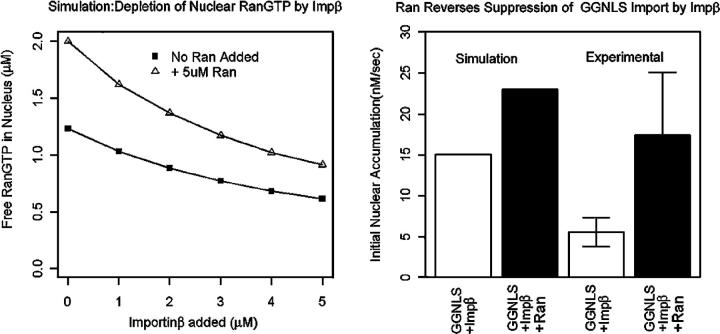
The underlying mechanism for the inhibition of cargo import by increased Impβ is the sequestration of nuclear RanGTP, and the model predicts that addition of exogenous Ran can reverse this effect (Fig. 10 A). The additional Ran accumulates in the nucleus in the GTP-bound form, replenishing the free RanGTP pool that has been reduced by the excess Impβ, and hence stimulating import (Fig. 10 B). To test this prediction, we first transfected HeLa cells with a fusion protein of Ran and RFP (mRFP-Ran). We then co-injected GGNLS and Impβ into those cells that showed nuclear accumulation of mRFP-Ran. Compared with nontransfected cells from the same population, the mRFP-Ran cells showed significant reduction of the Impβ effect on initial rate (Fig. 10 B). This result strongly argues that the observed effect of Impβ on import rates is not a consequence simply of contamination by inhibitory fragments of Impβ, and also validates the computer model.
Figure 10.
The inhibition of GGNLS import by elevated Imp is caused by depletion of nuclear RanGTP. (Left) Simulation results show that excess Impβ deletes free RanGTP in the nucleus. Addition of Ran increases the total pool of free RanGTP in the nucleus and offsets the addition of Impβ. (Right) HeLA cells were transfected with pK-mRFP-Ran. After 12 h, cells were cytoplasmically microinjected with 20 μM GGNLS + 5 μM Impβ. Initial rates of GGNLS import were then quantified for both transfected cells that showed nuclear accumulation of RFP-Ran and nontransfected cells in the same population. Both experimental groups were measured over a range of initial GGNLS concentrations between 2 and 4 μM, and show a significant difference in mean initial rates (P < 0.002). Simulation results using 1 μM Impβ, 5 μM Ran, and 4 μM GGNLS are displayed next to experimental results.
Discussion
Computer models are relevant to cell biology only to the extent that they can generate testable predictions, and provide new insights into cellular processes. They can suggest unexpected relationships between steps in a process, or expose flaws in our understanding by failing to correctly simulate an experimental outcome. We believe that our model of nuclear protein import satisfies both of these requirements. The model nicely describes the general features of Impα–Impβ-mediated import, in terms of rates and the requirement for a Ran-gradient but, more importantly, it made several unanticipated predictions about the response of the system to changes in the concentrations of certain factors. Moreover, it fails to account for certain experimental observations, which illuminates our ignorance of the functions of some of these factors. Specifically, it did not predict the observed increase in cargo import in response to an elevation in RanBP1 concentration. This failure suggests that RanBP1 performs roles not included in the model. We know that RanBP1 can form a ternary complex with RanGDP and Impβ, for which no function has yet been ascribed, and this interaction was not included in the model (Chi et al., 1997; Plafker and Macara, 2002). However, such a complex would likely reduce rather than increase the rate of cargo import, by inhibiting RanGDP import by NTF2, and therefore reducing the RanGTP gradient. Additionally, RanBP1 can shuttle in and out of the nucleus, but this process would again reduce rather than increase cargo import, by depleting RanGTP from the nucleus (Plafker and Macara, 2000a). We speculate that RanBP1 might either facilitate the dissociation of transporters from the NPC, or alter the permeability of the NPC to transporters. These processes were not included in the model, but might be expected to increase transport rates. However, we have not detected any accumulation of RanBP1 at the nuclear envelope in intact cells (unpublished data). Finally, RanBP1 is known to form a complex with Mog1 in the presence of Ran (Steggarda and Paschal, 2000). Mog1 is necessary for efficient nuclear import, although its function remains unknown, and elevated RanBP1 might in some way alter Mog1 activity (Baker et al., 2001).
The model was successful in making two unexpected and accurate predictions about the behavior of the nuclear transport system: that elevations in the cellular concentrations of either impβ or RCC1 would inhibit rather than stimulate cargo import. The inhibitory effect of Impβ demonstrates that futile cycling of transport receptors across the nuclear envelope can significantly deplete the Ran gradient. The levels of Impβ and of similar karyopherins must therefore be carefully regulated to reduce the steady-state concentration of free receptors in the cell. We have suggested previously that transport cofactors such as RanBP3 and NPAP60 might also function to reduce futile cycling (Lindsay et al., 2001; Nemergut et al., 2002). We have not included these cofactors in the current model, but expect that they might reduce the inhibitory effect of high Impβ levels.
The effect of RCC1 is of interest because it is a consequence of the kinetic model for exchange that was used in the transport model. In our earlier study, virtual cell simulations of Ran transport, we used simple Michaelis-Menten kinetics that did not produce the same outcome (Smith et al., 2002). These results therefore suggest that the four-step in vitro kinetic analysis of exchange by RCC1 provides an accurate reflection of the in vivo kinetics. Our model also predicts that RanGAP is not rate-limiting, and this was validated experimentally.
We believe that our model provides a foundation on which to test other concepts in nuclear transport. For example, it is not apparent why the cell uses the Impα adaptor to bind cargo, rather than allowing all cargo to interact directly with the Impβ karyopherin. One speculation is that the coupling of cargo import to the export of Impα, mediated by Cas–RanGTP, might drive the creation of a higher nuclear–cytoplasmic cargo gradient. The potential effects of competition between karyopherins for binding to the same cargo, or to the NPC, are also complex. In the future we will test these ideas both experimentally and in silico, using a modified transport model.
Materials and methods
Computer model
We first constructed a compartmental model of the Impα–Impβ import system using the program Jarnac, a biochemical simulation package for Windows (Sauro et al., 2003). Biochemical reactions were localized to the cytoplasm or nucleus in an idealized cell. Flux rates between compartments were modeled as the product of a permeability factor and the concentration difference of a reactant between the nucleus and cytoplasm. The reactions were converted internally by Jarnac to a series of coupled ODEs. Jarnac was then used to solve the ODEs based on a set of initial values. Protein concentrations and reactions rates were taken where possible from published reports (Tables I and II). Permeability constants for Impβ, Impα, and GST-GFP were determined experimentally as described in Tables I–III. The binding of Impα to Impβ as well as Impα–Impβ to the cargo were modeled as two-step reversible reactions (Catimel et al., 2001). We also adopted GTP and GDP concentrations and a four-step reversible reaction for nucleotide exchange by RCC1 (Klebe et al., 1995b). Concentrations of endogenous cargo proteins carried by Impα–Impβ or Impβ alone were treated as floating parameters. To account for the reduction of the Ran gradient by transport receptors other than Impβ, we included a reaction pathway for a “generic” karyopherin. This pathway used reactions rates identical to those used for the Impβ. Evidence exists in yeast (Gilchrist et al., 2002) that RanGTP and CAS act cooperatively to actively displace Impα from its cargo. Therefore, we modeled this reaction as a single-step displacement.
Table II. Rate constants.
| Interaction | kon μM− 1 sec− 1 | koff sec− 1 | Reference |
|---|---|---|---|
| RanGTP + Impβ | 9.6E4 | 4.8E-6 | Bischoff and Görlich, 1997 |
| RanGDP + NTF2 | 1E7 | 1 | Chaillan-Huntington et al., 2000 |
| RanGDP + RCC1 | 7.4E7 | 55 | Klebe et al., 1995b |
| RCC1Ran + GTP | 6E5 | 19 | Klebe et al., 1995b |
| RCC1Ran + GDP | 1.1E7 | 21 | Klebe et al., 1995b |
| RCC1 + RanGTP | 1E8 | 55 | Klebe et al., 1995b |
| Impα + Impβ | (kon1)4.9E5 (kon2)7,900 |
(koff11).017 (koff2).00025 |
Catimel et al., 2001 |
| Impα–Impβ + NLS | (kon1)1.5E5 (kon2)6,400 |
(koff11).075 (koff2).0048 |
Catimel et al., 2001 |
| Impα–Impβ–NLS + RanGTP | 2E4 | 4.8E-6 | Estimate |
| Cas + RanGTP | 1E4 | 4.8E-3 | Estimate |
| Impα–NLS + RanGTP–Cas | 1E5 | 1E-4 | Estimate from kD (Kutay et al., 1997) |
| Impβ + cargo | (kon1)4.9E5 (kon2)7,900 |
(koff1)0.017 (koff2).00025 |
Catimel et al., 2001 |
| Impβ–cargo + RanGTP | 2E4 | 4.8E-6 | Estimate |
| RanGTP–CAS–Impα + RanBP1 | 3E5 | 0.0004 | Estimate from Villa Braslavsky et al. (2000) |
| RanGTP–Impβ + RanBP1 | 3E5 | 0.0004 | Kuhlmann et al., 1997 |
| GTP hydrolysis reaction | kcat (Sec − 1) | Km (μM) | Reference |
| RanGTP–CAS–Impα–RanBP1 + RanGAP | 2 | 0.1 | Estimate based on Görlich et al. (2003) |
| RanGTP–RanBP1–Impβ + RanGAP | 2 | 0.1 | Estimate based on Görlich et al. (2003) |
| RanGTP–RanBP1 + RanGAP | 21.2 | 0.1 | Görlich et al., 2003 |
After construction of the model, parameter optimization was conducted using a genetic algorithm implemented in the Jarnac scripting language (Bäck and Schwefel, 1993).
Constructs, cell culture, and transfections
The ORFs for human Ran and RCC1 were subcloned into a mammalian vector (pKmRFP1) to generate mRFP-Ran and mRFP-RCC1 fusion proteins (T. Chen, University of Virginia). The mRFP1 was provided by R. Tsien (University of California, San Diego, La Jolla, CA; Campbell et al., 2002) and recombinant Cas by M. Yamada (University of Virginia). HeLa cells were transfected with the mRFP vectors using Effectene (QIAGEN) according to the manufacturer's instructions. The medium was changed to Ringer's solution 12–24 h after transfection, and the cells were imaged using confocal microscopy.
Immunofluorescence
HeLa cells were fixed and immunostained with anti-Ran and anti-Impα mAbs (Signal Transduction Laboratories) as described previously (Smith et al., 1998).
Protein binding assays
Proteins were purified as described previously (Plafker and Macara, 2000b; Brownawell and Macara, 2002; Smith et al., 2002). Recombinant Impβ was tested for its ability to bind both RanGTP and Impα. γ-Bind–Sepharose beads were first blocked with 3% BSA. GST-Impα and Impβ were then added in binding buffer and incubated for 1 h at 4°C (20 mM Tris, pH 7.4, 2 mM MgCl2, 200 mM NaCl, 0.1% BSA, 0.1% Tween, 2 mM DTT). The beads were washed with binding buffer lacking BSA. Proteins were eluted with Laemmli sample buffer, separated by SDS-PAGE and visualized by Coomassie brilliant blue staining. Binding of Impβ to the constitutively active Ran mutant, RanGTP(Q69L), and of CAS to GST–Impα and Ran(Q69L), were tested in a similar manner.
Microinjection
HeLa cells were cultured in Bioptek Delta-T dishes. The medium was exchanged to a physiological saline (25 mM Hepes, pH 7.2, 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM glucose, 1 mM KH2PO4, 1 mg/ml BSA) before microinjection. A fusion protein of GGNLS was used as a cargo to quantify nuclear import. The GGNLS was diluted with microinjection buffer to a final concentration of 20 μM. Co-injected proteins were added to give a final concentration of 5 μM in the injection mixture. Samples were centrifuged at 10,000 g at 4°C for 5 min before injection.
Confocal microscopy
A confocal microscope (model Meta LSM510; Carl Zeiss MicroImaging, Inc.) was used to collect time-lapse images. Cells were imaged using a 20× objective lens (NA = 0.65) at a zoom of 1.0. Detector gain was kept beneath 550 to maintain linearity of pixel intensity to concentration. Cells were kept at 37°C and the medium was changed every 15 min to avoid changes in concentration caused by evaporation. Cells were injected using Femtotips on an Eppendorf Injectman NI 2. After cytoplasmic injection, time-lapse images were taken every 20 s to quantify nuclear import for GGNLS. Average nuclear pixel intensity was related to protein concentration by the use of a standard dilution series of known concentration. Initial rates were calculated as the linear regression of the first three data points of nuclear accumulation. The significance of differences between control and experimental rates over a range of GGNLS concentrations was computed by a two-way ANOVA, using the R statistical package (http://cran.r-project.org/). Differences were assumed to be significant for P < 0.05. To quantify permeability factors, Impβ and Impα were first labeled with OGI. After cytoplasmic injection, images were acquired every second. The slope of initial nuclear concentration versus initial rate provided the permeability factor for each protein.
Online supplemental material
Computer model. A complete schematic of the computer model is provided in Fig. S1. A script file containing the model is also included and can be run using the program Jarnac, which can be downloaded from http://64.17.162.114/downloads/ISetup.exe.
Permeability assays. Fig. S2 shows import rate data for recombinant OGI-labeled Impα and Impβ, and for GST-GFP.
Sensitivity analysis of permeabilities. Fig. S3 shows the effects of varying permeabilities for various components of the model on the initial rate of GGNLS import. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200409024/DC1.
Acknowledgments
We thank Roger Tsien for the mRFP1 plasmid, Masami Yamada for recombinant CAS, and Ting Chen for the pK-mRFP-RCC1 and pK-mRFP-Ran plasmids. Thanks also to Leslie Loew, Boris Slepchenko, and Jim Schaff of the Virtual Cell Group at the University of Connecticut for useful early discussions concerning compartmental simulation.
This work was supported by grants GM50526 and GM064437 from the National Institutes of Health and Department of Health and Human Services.
Abbreviations used in this paper: GGNLS, GST-GFP-NLS; Impα, importin-α; Impβ, importin-β; NPC, nuclear pore complex; ODE, ordinary differential equation; OGI, Oregon green isothiocyanate.
References
- Bäck, T., and H.-P. Schwefel. 1993. An overview of evolutionary algorithms for parameter optimization. Evol. Comput. 1:1–23. [DOI] [PubMed] [Google Scholar]
- Baker, R.P., M.T. Harreman, J.F. Eccleston, A.H. Corbett, and M. Stewart. 2001. Interaction between Ran and Mog1 is required for efficient nuclear protein import. J. Biol. Chem. 276:41255–41262. [DOI] [PubMed] [Google Scholar]
- Bayliss, R., T. Littlewood, and M. Stewart. 2000. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 102:99–108. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., and D. Görlich. 1997. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 419:249–254. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., H. Krebber, E. Smirnova, W. Dong, and H. Ponstingl. 1995. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 14:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, F.R., and H. Ponstingl. 1991. a. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 354:80–82. [DOI] [PubMed] [Google Scholar]
- Bischoff, F.R., and H. Ponstingl. 1991. b. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc. Natl. Acad. Sci. USA. 88:10830–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell, A.M., and I.G. Macara. 2002. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 156:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel, B., T. Teh, M.R. Fontes, I.G. Jennings, D.A. Jans, G.J. Howlett, E.C. Nice, and B. Kobe. 2001. Biophysical characterization of interactions involving importin-α during nuclear import. J. Biol. Chem. 276:34189–34198. [DOI] [PubMed] [Google Scholar]
- Chaillan-Huntington, C., C.V. Braslavsky, J. Kuhlmann, and M. Stewart. 2000. Dissecting the interactions between NTF2, RanGDP, and the nucleoporin XFXFG repeats. J. Biol. Chem. 275:5874–5879. [DOI] [PubMed] [Google Scholar]
- Chi, N.C., E.J.H. Adam, and S.A. Adam. 1997. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J. Biol. Chem. 272:6818–6822. [DOI] [PubMed] [Google Scholar]
- Chook, Y.M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703–715. [DOI] [PubMed] [Google Scholar]
- Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 94:193–204. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog, B., and U. Aebi. 2003. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 4:757–766. [DOI] [PubMed] [Google Scholar]
- Fanara, P., M.R. Hodel, A.H. Corbett, and A.E. Hodel. 2000. Quantitative analysis of nuclear localization signal (NLS)-importin alpha interaction through fluorescence depolarization. Evidence for auto-inhibitory regulation of NLS binding. J. Biol. Chem. 275:21218–21223. [DOI] [PubMed] [Google Scholar]
- Floer, M., G. Blobel, and M. Rexach. 1997. Disassembly of RanGTP karyopherinb complex, an intermediate in nuclear protein import. J. Biol. Chem. 272:19538–19546. [DOI] [PubMed] [Google Scholar]
- Gilchrist, D., B. Mykytka, and M. Rexach. 2002. Accelerating the rate of disassembly of karyopherin-cargo complexes. J. Biol. Chem. 277:18161–18172. [DOI] [PubMed] [Google Scholar]
- Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607–660. [DOI] [PubMed] [Google Scholar]
- Görlich, D., N. Pante, U. Kutay, U. Aebi, and F.R. Bischoff. 1996. Identification of different roles for RanGDP and RanGTP In nuclear protein import. EMBO J. 15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich, D., M.J. Seewald, and K. Ribbeck. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, A., R. Truant, H. Wiegand, and B.R. Cullen. 1998. Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe, C., F.R. Bischoff, H. Ponstingl, and A. Wittinghofer. 1995. a. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 34:639–647. [DOI] [PubMed] [Google Scholar]
- Klebe, C., H. Prinz, A. Wittinghofer, and R.S. Goody. 1995. b. The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry. 34:12543–12552. [DOI] [PubMed] [Google Scholar]
- Kobe, B. 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6:388–397. [DOI] [PubMed] [Google Scholar]
- Kuhlmann, J., I. Macara, and A. Wittinghofer. 1997. Dynamic and equilibrium studies on the interaction of ran with its effector, RanBP1. Biochemistry. 36:12027–12035. [DOI] [PubMed] [Google Scholar]
- Kutay, U., F.R. Bischoff, S. Kostka, R. Kraft, and D. Görlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 90:1061–1071. [DOI] [PubMed] [Google Scholar]
- Lindsay, M., J. Holaska, B. Paschal, and I.G. Macara. 2001. RanBP3 is a cofactor for Crm1-mediated nuclear protein export. J. Cell Biol. 153:1391–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew, L.M., and J.C. Schaff. 2001. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 19:401–406. [DOI] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut, M.E., M.E. Lindsay, A.M. Brownawell, and I.G. Macara. 2002. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J. Biol. Chem. 277:17385–17388. [DOI] [PubMed] [Google Scholar]
- Nemergut, M.E., and I.G. Macara. 2000. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, C., N. Orem, J. Trueheart, J.W. Thorner, and I.G. Macara. 2000. Random mutagenesis and functional analysis of the Ran-binding protein, RanBP1. J. Biol. Chem. 275:4081–4091. [DOI] [PubMed] [Google Scholar]
- Plafker, K., and I.G. Macara. 2000. a. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol. Cell. Biol. 20:3510–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker, K.S., and I.G. Macara. 2002. Fluorescence resonance energy transfer biosensors that detect Ran conformational changes and a Ran.GDP:importin-{beta}:RanBP1 complex in vitro and in intact cells. J. Biol. Chem. 277:30121–30127. [DOI] [PubMed] [Google Scholar]
- Plafker, S.M., and I.G. Macara. 2000. b. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., G. Lipowsky, H.M. Kent, M. Stewart, and D. Görlich. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauro, H.M., M. Hucka, A. Finney, C. Wellock, H. Bolouri, J. Doyle, and H. Kitano. 2003. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. OMICS. 7:355–372. [DOI] [PubMed] [Google Scholar]
- Smith, A., A. Brownawell, and I.G. Macara. 1998. Nuclear import of Ran:GDP is mediated by NTF2. Curr. Biol. 8:1403–1406. [DOI] [PubMed] [Google Scholar]
- Smith, A.E., B.M. Slepchenko, J.C. Schaff, L.M. Loew, and I.G. Macara. 2002. Systems analysis of Ran transport. Science. 295:488–491. [DOI] [PubMed] [Google Scholar]
- Steggarda, S.M., and B.M. Paschal. 2000. The mammalian Mog1 protein is a guanine nucleotide release factor for Ran. J. Biol. Chem. 275:23175–23180. [DOI] [PubMed] [Google Scholar]
- Suntharalingam, M., and S.R. Wente. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function. Dev. Cell. 4:775–789. [DOI] [PubMed] [Google Scholar]
- Uchida, S., T. Sekiguchi, H. Nishitani, K. Miyauchi, M. Ohtsubo, and T. Nishimoto. 1990. Premature chromosome condensation is induced by a point mutation in the hamster RCC1 gene. Mol. Cell. Biol. 10:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa Braslavsky, C.I., C. Nowak, D. Görlich, A. Wittinghofer, and J. Kuhlmann. 2000. Different structural and kinetic requirements for the interaction of Ran with the Ran-binding domains from RanBP2 and importin-beta. Biochemistry. 39:11629–11639. [DOI] [PubMed] [Google Scholar]
- Weis, K. 2002. Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol. 14:328–335. [DOI] [PubMed] [Google Scholar]