Abstract
Here, we study an insect hnRNP M protein, referred to as Hrp59. Hrp59 is relatively abundant, has a modular domain organization containing three RNA-binding domains, is dynamically recruited to transcribed genes, and binds to premRNA cotranscriptionally. Using the Balbiani ring system of Chironomus, we show that Hrp59 accompanies the mRNA from the gene to the nuclear envelope, and is released from the mRNA at the nuclear pore. The association of Hrp59 with transcribed genes is not proportional to the amount of synthesized RNA, and in vivo Hrp59 binds preferentially to a subset of mRNAs, including its own mRNA. By coimmunoprecipitation of Hrp59–RNA complexes and microarray hybridization against Drosophila whole-genome arrays, we identify the preferred mRNA targets of Hrp59 in vivo and show that Hrp59 is required for the expression of these target mRNAs. We also show that Hrp59 binds preferentially to exonic splicing enhancers and our results provide new insights into the role of hnRNP M in splicing regulation.
Introduction
In eukaryotic cells, premessenger RNAs bind to a large number of proteins to form premessenger RNP complexes (premRNPs). The proteins that bind premRNA and that are not stable components of other RNA complexes, such as snRNP complexes, are collectively termed hnRNP proteins (for review see Dreyfuss et al., 2002). In mammalian cells, ∼20 abundant proteins, designated A1 to U, have been identified as major constituents of the premRNPs. Although the hnRNP proteins were initially regarded as RNA packaging proteins, a more detailed study of their functions has revealed that individual hnRNP proteins have specific functions in the expression of specific genes. For instance, human hnRNP A1 plays a role in splice site selection both in vitro and in vivo (Mayeda and Krainer, 1992; Caceres et al., 1994), hnRNP K binds DNA and has been implicated in transcription regulation (for review see Bomsztyk et al., 2004), and hnRNP E regulates the stability of cytoplasmic globin mRNA (Kiledjian et al., 1995). HnRNP proteins thus appear to be involved in every step of gene expression, as they influence the interactions between the premRNAs and other cell components involved in mRNA function.
HnRNP proteins are evolutionarily conserved. In Drosophila melanogaster, several families of hnRNP proteins have been identified and they are very similar to mammalian hnRNP proteins (Haynes et al., 1990; Amero et al., 1991; Matunis et al., 1992; Hovemann et al., 2000). HnRNP proteins have also been studied in the dipteran Chironomus tentans due to the opportunities that the Balbiani ring (BR) system offers for in situ studies of gene expression (for reviews see Wieslander, 1994; Daneholt, 2001; Kiesler and Visa, 2004). In C. tentans, two large puffs called BR1 and BR2 can be identified in the polytene chromosomes of the salivary gland cells. The premRNPs synthesized in the BR1 and BR2 puffs, referred to as BR RNP particles, can be unambiguously identified in the transmission EM, and their synthesis, processing and transport can be directly visualized (for review see Daneholt, 2001). The BR system has been used to study the behavior of hnRNP proteins in relation to nucleo-cytoplasmic transport (Visa et al., 1996) and to analyze the intranuclear movement of premRNPs (Singh et al., 1999; Miralles et al., 2000).
We have previously identified Hrp65 as an hnRNP protein of C. tentans with two classical RNA-binding domains of the RRM type (Miralles et al., 2000). Three Hrp65 isoforms are generated by alternative splicing from a single premRNA (Miralles and Visa, 2001). In the salivary glands of C. tentans, Hrp65 is associated with specific loci in the polytene chromosomes, and it is also located in nonchromatin nucleoplasmic fibers known as connecting fibers (CFs). The CFs interact with premRNP particles in the nucleoplasm, and this suggests that Hrp65 is involved in mRNA biogenesis at the posttranscriptional level (Miralles et al., 2000). Moreover, recent studies have shown that the Hrp65-2 isoform plays a role in the transcription of class II genes (Percipalle et al., 2003). Hrp65 is evolutionarily conserved, and two of its homologues in mammals, PSF and p54nrb/NonO, have been implicated in both premRNA processing and transcription regulation (for review see Shav-Tal and Zipori, 2002). Using Hrp65 as a bait in a yeast two-hybrid screen, we have identified a novel hnRNP protein of C. tentans that we refer to as Hrp59. We show here that Hrp59 is an hnRNP protein of the hnRNP M type that binds cotranscriptionally to premRNA and accompanies the transcripts to the nuclear pore. We also show that Hrp59 binds preferentially to a subset of transcripts. Taking advantage of the tools available for genome-wide studies in Drosophila, and thanks to the evolutionary conservation of Hrp59, we have been able to identify the preferred RNA targets of Hrp59 in vivo by protein–RNA coimmunoprecipitation and whole-genome array hybridization. Interestingly, Hrp59 binds preferentially to a purine-rich sequence motif previously identified as an exonic splicing enhancer, and this observation provides new insights into the proposed role of hnRNP M in splicing regulation.
In spite of the importance of hnRNP proteins for mRNA biogenesis, the understanding of their functions in vivo has been constrained by the lack of methods for systematically identifying their natural RNA targets. The combination of RNA–protein coimmunoprecipitation and microarray hybridization that we have used for the study of Hrp59 can be applied to other hnRNP proteins, and will contribute to elucidating hnRNP protein function.
Results
Hrp59 is a novel hnRNP M/Myef-2–like protein of C. tentans that interacts with Hrp65
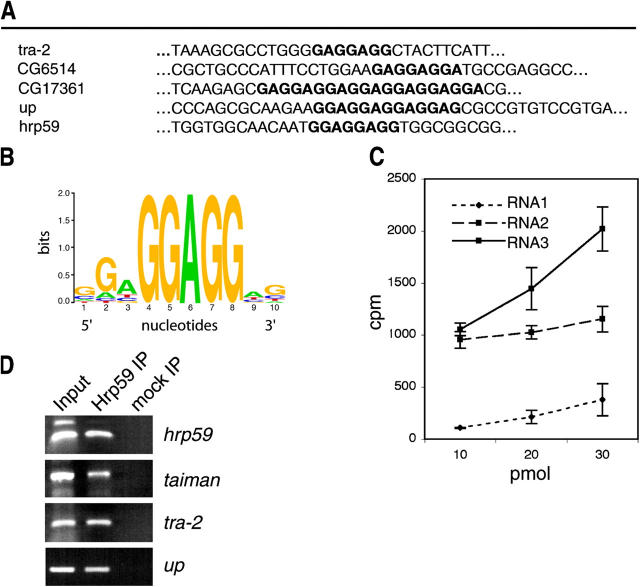
The coding sequence of Hrp65-1 was used as a bait to screen a C. tentans cDNA library using a yeast two-hybrid system (Kiesler et al., 2003). Out of 54 isolated positive clones, five independent clones encoded a novel protein of C. tentans here designed “Ct-Hrp59”, or “Hrp59” for simplicity (Fig. 1 A). No other RNA-binding proteins were found among the positive clones, except for Hrp65 itself (Kiesler et al., 2003), which suggests that the interaction between Hrp65 and Hrp59 in the yeast two-hybrid system is not mediated by RNA. Moreover, binding of Hrp65 to the Hrp59 gene product was further confirmed in vitro using recombinant proteins (Fig. 1 B).
Figure 1.
Interaction of Hrp65 and Hrp59. (A) Yeast two-hybrid analysis. Yeast cells were transformed with a plasmid encoding Hrp59 fused to the GAL4 activation domain (pAD-Hrp59) together with either a plasmid encoding Hrp65-1 fused to the GAL4 DNA-binding domain (pDBD-Hrp65) or with negative control plasmids encoding the GAL4 DBD fused to p53 (pDBD-p53), or alone (pDBD). As a positive control, cells were transformed with a plasmid encoding the complete GAL4 factor (pGAL4). Protein–protein interactions were detected by growing the cells on double selective −His −Ade medium. (B) In vitro binding assay. [35S]methionine-labeled Hrp59 and CAT proteins were made by in vitro translation in rabbit reticulocyte lysate, and each protein was incubated with recombinant His-tagged Hrp65 captured on Ni-NTA agarose (Ni-Hrp65), or with control Ni-NTA–agarose beads (Ni-NTA), in RIPA buffer. The bound proteins were eluted, resolved on a 10% SDS-PAGE gel and autoradiographed. The [35S]Hrp59 and [35S]CAT proteins were loaded as inputs.
Sequence analysis revealed that the isolated Hrp59 cDNA encodes a 5′-end truncated ORF followed by ∼500 bp of 3′ untranslated region (GenBank/EMBL/DDBJ accession no. AJ785003). The predicted protein encoded in the Hrp59 ORF is 527 aa long, has a predicted pI of 9.2, and contains three RNA-recognition motifs (RRMs), as illustrated in Fig. 2 A.
Figure 2.

The primary structure of Hrp59. (A) Schematic representation of C. tentans Hrp59. (B) The amino acid sequences of C. tentans Hrp59 (GenBank/EMBL/DDBJ accession no. AJ785003), Drosophila CG9373 and human hnRNP M and Myef-2 were compared and aligned using ClustalW and BoxShade at http://workbench.sdsc.edu.
BLAST searches in the genomes of D. melanogaster and Anopheles gambiae revealed the existence of one putative Hrp59 orthologue in each species: CG9373 in Drosophila (48% identity) and EAA01814 in A. gambiae (54% identity). Comparisons with the D. melanogaster and A. gambiae sequences (Fig. 2 B) suggested that ∼15 aa are missing from the NH2 terminus of the isolated Ct-Hrp59 cDNA.
The BLAST searches also revealed that Hrp59 is similar to two related proteins in mammals, Myef-2 (NP057216) and hnRNP M (NP112480). HnRNP M is one of the major hnRNP protein groups in human cells (Datar et al., 1993), was found associated with early spliceosomes (Kafasla et al., 2002) and was implicated in the regulation of premRNA splicing under stress conditions (Gattoni et al., 1996). Myef-2 is a single-stranded DNA-binding protein involved in transcription repression (Haas et al., 1995; Muralidharan et al., 1997). Myef-2 and hnRNP M are 43% identical to each other and display the same modular domain organization characterized by three RRMs and by the presence of multiple GM-repeats in the region located between RRM2 and RRM3 (Fig. 2 B). The GM-repeats were not found in the Hrp59 sequence.
The Hrp59 orthologue in D. melanogaster, CG9373, is the only D. melanogaster protein with high similarity to the mammalian hnRNP M and Myef-2 proteins. The same is true for the product of the A. gambie EAA01814 gene. These observations indicate that the mammalian hnRNP M/Myef-2 family has only one member in insects, and they suggest that Hrp59 is the orthologue of hnRNP M/Myef-2 in C. tentans.
In summary, we have identified Hrp59 as the orthologue of hnRNP M/Myef-2 in C. tentans, and we have shown that Hrp59 interacts specifically with Hrp65 in the yeast two-hybrid system and in vitro.
In vivo Hrp59 and Hrp65 interact both directly and via RNA
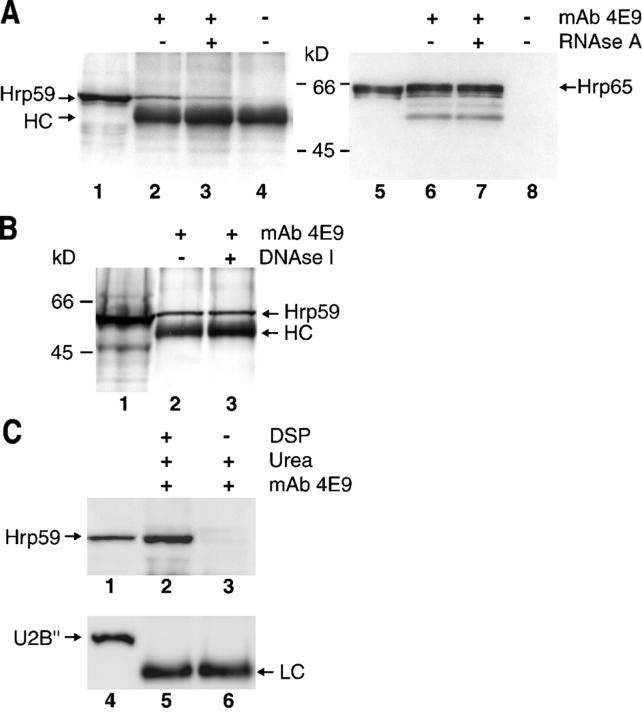
Hrp65-containing complexes were isolated by immunoprecipitation (IP) from nuclear extracts of C. tentans using the anti-Hrp65 mAb 4E9 (Miralles et al., 2000). Detection of the coimmunoprecipitated proteins by Western blotting using anti-Hrp59 antibodies showed the presence of Hrp59 in the Hrp65-containing complexes (Fig. 3 A, lane 2).
Figure 3.
In vivo interaction of Hrp59 and Hrp65. (A) RNA-mediated association of Hrp59 and Hrp65. Protein complexes were immunoprecipitated from native nuclear extracts of C. tentans tissue culture cells using mAb 4E9 against Hrp65. Mock IPs were performed in parallel in the absence of primary antibody (lanes 4 and 8). The immunoprecipitated material bound to Sepharose was incubated with or without RNase A and washed in order to remove RNase-released material. Proteins still bound after RNase digestion were analyzed by SDS-PAGE and Western blotting with antibodies against either Hrp59 (lanes 1–4) or Hrp65 (lanes 5–8). Nuclear extract was loaded as input (lanes 1 and 5). (B) Native coIP followed by DNase I digestion. The experiment was performed as in A and the bound proteins were incubated with (lane 3) or without (lane 2) DNase I and washed in order to remove DNase-released material. Nuclear extract was loaded as input (lane 1). (C) Direct interaction of Hrp59 and Hrp65: coimmunoprecipitation after in vivo cross-linking. C. tentans tissue culture cells were treated with (lanes 2 and 5) or without (lanes 3 and 6) DSP. Nuclear extracts were prepared in 8 M urea and used for IP with mAb 4E9 against hrp65 (lanes 2 and 3, and 5 and 6). Bound proteins were analyzed by SDS-PAGE and Western blotting with antibodies against either Hrp59 (lanes 1–3) or U2B′′ as a negative control (lanes 4–6). Nuclear extract from DSP-treated cells was loaded as input (lanes 1 and 4). HC and LC indicate cross-reactivity of the secondary antibodies used for Western blotting with the heavy and light chains, respectively, of the rabbit anti–mouse immunoglobulin used for IP.
Protein complexes isolated by IP with the anti-Hrp65 antibody were treated with RNase A to investigate whether the association between Hrp59 and Hrp65 was mediated by RNA. The amount of coimmunoprecipitated Hrp59 was significantly reduced after RNase digestion (Fig. 3 A, compare lanes 2 and 3), whereas the amount of bound Hrp65 was not affected (Fig. 3 A, lanes 6 and 7). By quantifying the relative intensities of Hrp59 and Hrp65 in IP complexes treated with or without RNase in three independent experiments, we estimated that ∼50% of the association between Hrp59 and Hrp65 was RNase-sensitive. Instead, digestion with DNase I before elution did not affect the amount of coimmunoprecipitated Hrp59 (Fig. 3 B). These results show that Hrp59 and Hrp65 are partially associated through binding to a common RNA substrate.
To investigate whether Hrp59 and Hrp65 interact directly with each other, we performed in vivo cross-linking experiments followed by IP. For this purpose, we treated C. tentans tissue culture cells with dithiobis-succinimidyl propionate (DSP), a cell-permeable, reversible short range (11 Å) cross-linker (Wang and Richards, 1974). Nuclear extracts were then prepared in the presence of urea to disrupt noncross-linked protein–protein interactions, and used in IP experiments with the anti-Hrp65 antibody. Hrp59 was coimmunoprecipitated with Hrp65 from cross-linked nuclear extracts (Fig. 3 C, lane 2), whereas the strong denaturing conditions of the experiment prevented any binding of Hrp59 to Hrp65 in noncross-linked nuclear extracts (Fig. 3 C, lane 3). To ascertain the specificity of the cross-linking experiment, we used an antibody against the U2B′′ snRNP protein as a negative control and found that U2B′′ was not present in IP complexes from cross-linked nuclear extracts (Fig. 3 C). To conclude, we have shown that Hrp59 and Hrp65 are associated in vivo, and that this association is partly due to a direct protein–protein interaction and partly mediated by RNA.
Hrp59 and Hrp65 colocalize on polytene chromosomes
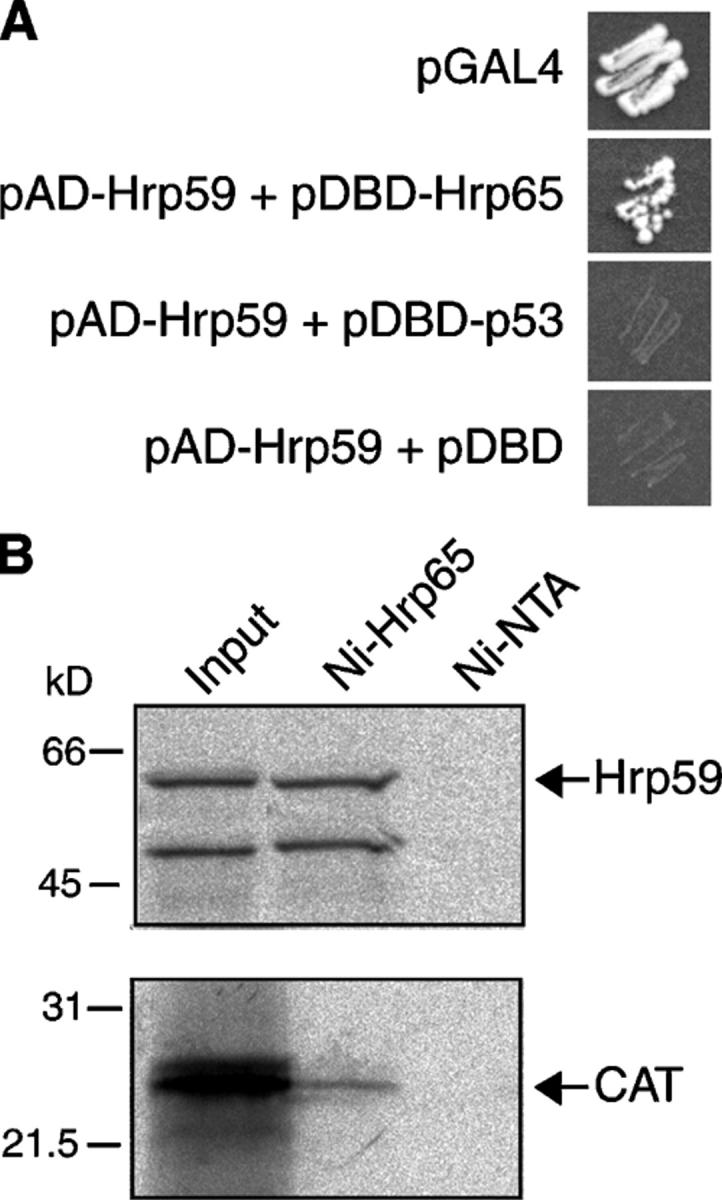
We examined the distribution of Hrp59 in polytene chromosomes by immunofluorescent staining. Hrp59 is associated with many bands in the chromosomes and is enriched in a subset of specific loci (Fig. 4 A, long arrow). Double labeling of Hrp59 and Hrp65 revealed that the two proteins are colocalized to a high extent on all chromosomes (Fig. 4 A). In some bands, a very evident colocalization could be observed in terms of both location and relative intensity. In other bands, differences were observed in the relative abundance of the two proteins. Only a few bands were stained with only one of the two antibodies (Fig. 4 A, short arrows).
Figure 4.
Distribution of Hrp59 and Hrp65 in the polytene chromosomes. (A) Squash preparations of C. tentans polytene chromosomes were double labeled with antibodies against Hrp59 (green) and Hrp65 (red). The BR1 and BR2 puffs in chromosome IV are indicated by arrowheads. The large arrow points to a band with intense labeling for both Hrp59 and Hrp65. The short arrows point to bands labeled by only one of the two antibodies. Note that the distributions of Hrp59 and Hrp65 are similar. (B) Polytene chromosome double labeled with antibodies against Hrp59 (green) and Hrp45 (red). The arrowheads point to the BR1 and BR2 puffs. Hrp45 and Hrp59 show different labeling patterns. (C) Transcriptionally active loci were visualized by BrUTP incorporation (red). The glands were coimmunostained with anti-Hrp59 antibody (green). C, cytoplasm; N, nucleolus; NP, nucleoplasm; PC, polytene chromosome. (D) The distributions of Hrp59 (green) and Hrp65 (red) in the polytene chromosomes were analyzed after heat shock. Note the redistribution of both proteins to the heat-shock locus at 5C (compare chromosome IV in A and D). (E) Tissue culture cells of C. tentans, either nontreated (22°C) or heat shocked (37°C), were double stained with anti-Hrp59 (green) and anti-Hrp65 (red) antibodies. All images correspond to optical sections with a thickness of ∼1 μm. Bars: (A, B, and D) ∼10 μm; (C) 20 μm; (E) 5 μm.
The distribution of Hrp59 and Hrp65 in the polytene chromosomes does not resemble those of other major hnRNP proteins, as shown by comparisons of the Hrp59 distribution to those of Hrp45 (Fig. 4 B) and Hrp36 (not depicted), two abundant hnRNP proteins of C. tentans (Alzhanova-Ericsson et al., 1996; Visa et al., 1996). For instance, the large BR puffs on chromosome IV (Fig. 4 B, arrowheads) were strongly stained by the anti-Hrp45 and anti-Hrp36 antibodies, but were less stained by the anti-Hrp59 antibody, whereas Hrp59 was enriched in other bands on the same chromosome that showed low levels of Hrp45.
In summary, there is a high degree of colocalization between Hrp59 and Hrp65, and the pattern of distribution of these two proteins is specific and different from that of other hnRNP proteins such as Hrp36 and Hrp45.
Hrp59 is recruited cotranscriptionally to the nascent premRNA
To determine whether the chromosomal bands labeled with antibodies against Hrp59 and Hrp65 correspond to sites of active transcription, newly synthesized RNA was labeled by incorporation of bromo-UTP (BrUTP) and the location of Hrp59 was detected by immunofluorescence. An almost complete overlap was observed in the two channels (Fig. 4 C), which indicated that the Hrp59-positive loci correspond to sites of active transcription. However, we also observed that the intensity of Hrp59 labeling does not parallel the level of BrUTP incorporation. For instance, Hrp59 is not enriched, but present, in the very actively transcribing BR genes. Several explanations can be envisioned to explain the lack of correlation between the intensities of the Hrp59 and BrUTP stainings. Differential accessibility effects cannot be totally ruled out, but immunostainings with antibodies against other hnRNP proteins, such as Hrp23 and Hrp36, closely correlate with BrUTP patterns (unpublished data), which suggests that in general antibodies against hnRNP proteins do not suffer from restricted accessibility. Thus, the lack of correlation between Hrp59 and BrUTP stainings is most likely due to preferential association of Hrp59 to specific loci.
We next asked whether Hrp59 and Hrp65 are dynamically redistributed in response to changes in gene expression, and we analyzed the distribution of the two proteins after heat shock. This resulted in repressed activity of the BR genes and in the recruitment of Hrp59 and Hrp65 to major heat-shock puffs (Sass, 1995), such as the 5C locus on chromosome IV (Fig. 4 D). Accumulation of Hrp59 and Hrp65 at specific sites upon heat-shock induction was also observed in a diploid cell line of C. tentans (Fig. 4 E).
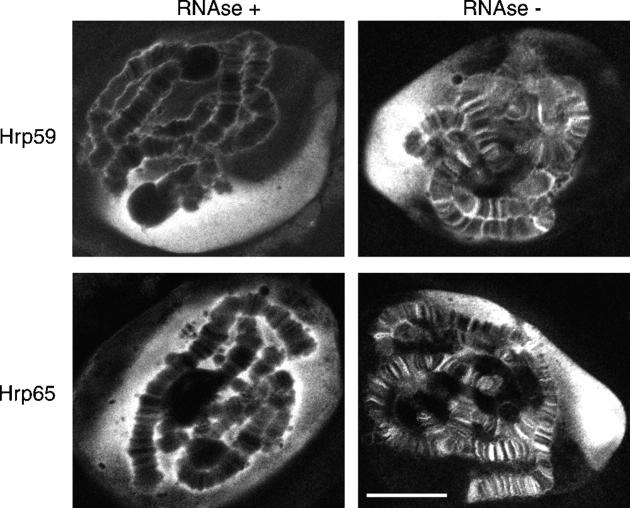
To investigate whether the binding of Hrp59 and Hrp65 to the transcribed loci was to RNA or to DNA, salivary glands were mildly permeabilized and digested with RNase A before fixation and immunostaining. The intensity of the immunofluorescent bands in the chromosomes was drastically reduced by the RNase A digestion (Fig. 5 A), which indicates that the association of Hrp59 and Hrp65 with the chromosomes is mostly mediated by RNA. Interestingly, no significant difference was observed in the intensity of the nucleoplasmic labeling, suggesting that the association of the two proteins with the nucleoplasm is independent of RNA.
Figure 5.
RNA-dependent distribution of Hrp59 and Hrp65. Salivary glands were permeabilized and treated with or without RNase A before fixation and immunostaining with antibodies against either Hrp59 or Hrp65. Note that the labeling of the chromosomes is drastically reduced after RNase digestion, both for Hrp59 and Hrp65, whereas the nucleoplasmic labeling is RNase resistant. The images are optical sections with a thickness of ∼1 μm. Bar, 20 μm.
Together, our results show that Hrp59 and Hrp65 bind to nascent premRNA in a large number of loci. These two proteins show a marked preference for certain genes, and this preference is not quantitatively related to the transcriptional activity.
Hrp59 remains associated with the mRNA from the gene to the nuclear pore
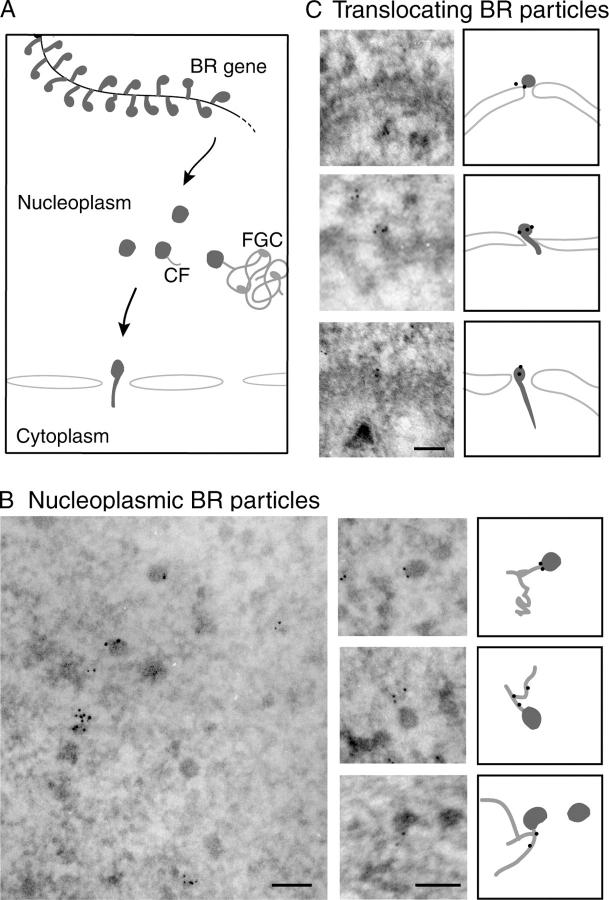
We performed immuno-EM experiments to determine whether Hrp59 remains associated with the premRNA after transcription. We labeled salivary gland cryosections with the anti-Hrp59 antibody followed by a gold-conjugated secondary antibody, and we analyzed the association of immuno-gold markers with BR RNP particles in the nucleoplasm of the salivary gland cells (Fig. 6). We found that a significant fraction of the nucleoplasmic BR particles (21%) was labeled by the anti-Hrp59 antibody (Fig. 6 B). We also found some labeling on BR RNP particles in transit through the nuclear pores, and in all cases the gold markers were located at the nuclear side of the translocating BR RNP particle (Fig. 6 C). This suggests that Hrp59 is released from the RNP during nucleo-cytoplasmic translocation. In summary, the immuno-EM analysis shows that Hrp59 remains bound to the BR RNP particles after transcription termination and accompanies the BR RNP to the nuclear pores.
Figure 6.
Hrp59 accompanies the BR RNP particles to the nuclear pore. (A) Schematic representation of BR premRNP particles showing successive stages of synthesis and assembly of the premRNP at the BR gene, transport through the nucleoplasm including transient interactions with CFs and fibrogranular clusters (FGCs), and translocation through the nuclear pores. (B) Immuno-EM analysis of Hrp59 in the nucleoplasm of the salivary gland cells. Cryosections of salivary gland were labeled with anti-Hrp59 and with a gold-conjugated secondary antibody. The figure shows examples of immunolabeling on BR particles as well as on CFs (arrows). Interpretations of the images are provided next to each micrograph. (C) Immuno-EM analysis of Hrp59 in BR particles in translocation through the NPC. Bars, 100 nm.
The immuno-EM analysis also revealed that only 5% of the nucleoplasmic gold markers were associated with BR RNP particles. Specific labeling was observed on the CFs adjacent to BR RNP particles (Fig. 6 B), as well as on fibrillar structures likely to represent the fibrogranular clusters previously described in the nucleoplasm of the salivary gland cells (Miralles et al., 2000). The observation that a fraction of Hrp59 remains associated with the nucleoplasm after RNase digestion (Fig. 5) is in agreement with its location at nonchromatin structures in the nucleoplasm.
Hrp59 binds preferentially to a subset of mRNAs
The specific distribution of Hrp59 in the polytene chromosomes (Fig. 4 A), and the observation that recruitment of Hrp59 to the transcribed loci is not proportional to the amount of synthesized RNA (Fig. 4 C), suggest that in vivo Hrp59 binds preferentially to a subset of transcripts. To identify the mRNAs associated with Hrp59 in vivo we took advantage of the evolutionary conservation of Hrp59 and we used the tools available for genome-wide studies in Drosophila. The anti-Hrp59 antibody cross-reacts against the Drosophila Hrp59 orthologue (Fig. 7 A) and was used to immunoprecipitate Hrp59-containing RNP complexes from Drosophila S2 cells. The coimmunoprecipitated RNA was purified and hybridized to high density oligonucleotide microarrays covering >13,500 Drosophila genes. Four chips were probed with the following RNA preparations: RNA from two independent Hrp59 IPs (IP arrays), RNA from a mock IP performed in the absence of primary antibody (mock array), and total RNA purified from the same extract used for IP (input array). The signal intensities of each transcript in the IP arrays were compared to the mock and to the input arrays (four comparisons in total), and the transcripts showing stronger signals in the IP arrays than in the mock and input arrays in all four comparisons were selected for further study. We then averaged the fold increase of the two independent IPs and selected those genes for which the increase was greater than a factor of two, i.e., at least twofold average enrichment in relation to both the input and the mock arrays. This procedure resulted in the identification of the most represented target RNAs (Table I). Among them, we found the mRNA coding for Drosophila Hrp59 itself, indicating the Hrp59 binds to its own mRNA. Interestingly, tra and tra-2, two mRNAs that code for regulators of alternative splicing (Forch and Valcarcel, 2003), were also significantly enriched.
Figure 7.
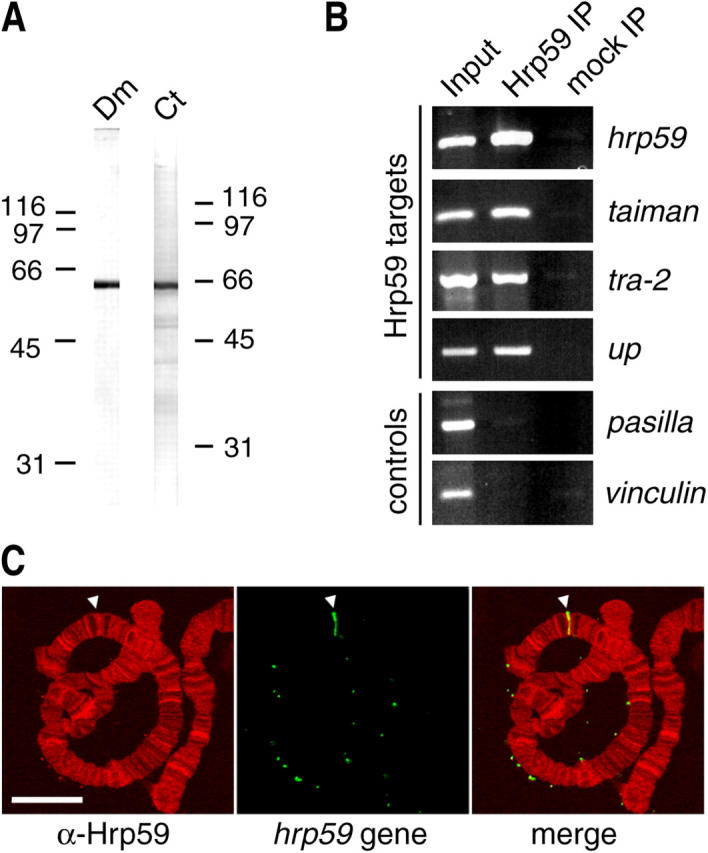
Coimmunoprecipitation of Hrp59-bound mRNA. (A) Western blot showing the specificity of the anti-Hrp59 antibody in D. melanogaster (Dm) and C. tentans (Ct). The mobility of molecular mass standards is indicated (kD). (B) Extracts were prepared from D. melanogaster S2 cells and the Hrp59-bound RNAs were coimmunoprecipitated with anti-Hrp59 antibody and protein A Sepharose (Hrp59 IP). Negative control IPs were performed in parallel in the absence of antibody (mock IP). RNA was purified from Hrp59 and mock IPs, and the coIP of transcripts corresponding to hrp59, taiman, tra-2, and up was analyzed by RT-PCR. Two additional transcripts not enriched in the Hrp59-IP but abundant in the total RNA, pasilla and vinculin, were also analyzed in parallel as negative controls. Input RNA was purified from 1% of the original extract used for IP, and analyzed in parallel. (C) Control experiment showing association of Hrp59 with the hrp59 locus in C. tentans chromosomes. Polytene chromosome squashes were hybridized with the hrp59 probe (green) and immunolabeled with anti-Hrp59 antibodies. The hrp59 locus (arrowheads) is labeled by the anti-Hrp59 antibody. Bar, 10 μm.
Table I. Drosophila RNAs coimmunoprecipitated with Hrp59.
| Hrp59 vs. input RNAa | Hrp59 vs. mock IPa | Gene | Description | [ga][atc]ggagg[atc][gct]b |
|---|---|---|---|---|
| 3.8 | 4.6 | CG9373 c | RNA-binding protein | ••••• |
| 3.7 | 5.6 | CG8301 | Transcription regulator, Zn finger C2H2 | ••••• |
| 3.7 | 2 | TpnC25D | Ca ion–binding protein, ligand binding or carrier | •• |
| 2.6 | 2.4 | Lsd1 | Chaperone, lipid storage | •••• |
| 2.6 | 2 | CG11417 | Unknown function | •••••• |
| 2.5 | 2.4 | tra | mRNA splicing, sex determination | |
| 2.5 | 2.2 | CG18854 | Inositol-triphosphate-3-kinase activity | • |
| 2.3 | 4.6 | up | Up/tropomyosin binding | •••••••••••••••• |
| 2.3 | 3.6 | wun2 | Phosphatidate phosphatase activity | |
| 2.3 | 2.7 | PP2A-B | Protein phosphatase 2A regulatory B subunit | •••• |
| 2.3 | 2.6 | taiman | Border cell migration, regulation of transcription | •••••••○○○○○○○○○○○○○ |
| 2.3 | 2.1 | tra-2 | mRNA splicing, sex determination | • |
| 2.3 | 2.1 | PtP4E | Protein tyrosine phosphatase 4E | •• |
| 2.3 | 2 | mod(mdg4) | RNA polymerase II transcription factor | • |
| 2.2 | 2.5 | chiffon | DNA binding domain, regulation of replication | ••••••• |
| 2.2 | 2.3 | CG3689 | mRNA cleavage | • |
| 2.2 | 2.1 | CG17361 | Nucleic acid binding, Zinc finger C2H2 | •• |
| 2.1 | 3.6 | vvl | RNA polymerase II transcription factor | •••• |
| 2.1 | 2.3 | CG3635 | Triacylglycerol lipase-like enzyme | • |
| 2.1 | 2.1 | CG5315 | Hormone-mediated signaling | • |
| 2.1 | 2 | CG6388 | tRNA modification | •• |
| 2 | 2.3 | CG14896 | Unknown function | •• |
| 2 | 2.3 | CG7255 | Cationic amino acid transporter | ••• |
The table includes RNAs from two independent experiments with at least two-fold average enrichment in relation to both the input and the mock IP.
No. of matches in each gene. Closed and open circles indicate the occurrence of the GGAGG-containing motif in exons and introns, respectively.
hrp59.
Two types of control experiments were performed to assess the specificity of our results. First, the enrichment of selected transcripts in the Hrp59-IP was analyzed by RT-PCR. As expected, the analyzed transcripts were detected in the Hrp59-IP, but not in the mock IP (Fig. 7 B). Second, the hrp59 gene in the polytene chromosomes of C. tentans was located by in situ hybridization, and the presence of the Hrp59 protein at the hrp59 locus was visualized by immunofluorescence (Fig. 7 C). This result confirms that Hrp59 binds to its own transcript cotranscriptionally.
Hrp59 is required for proper expression of its target mRNAs
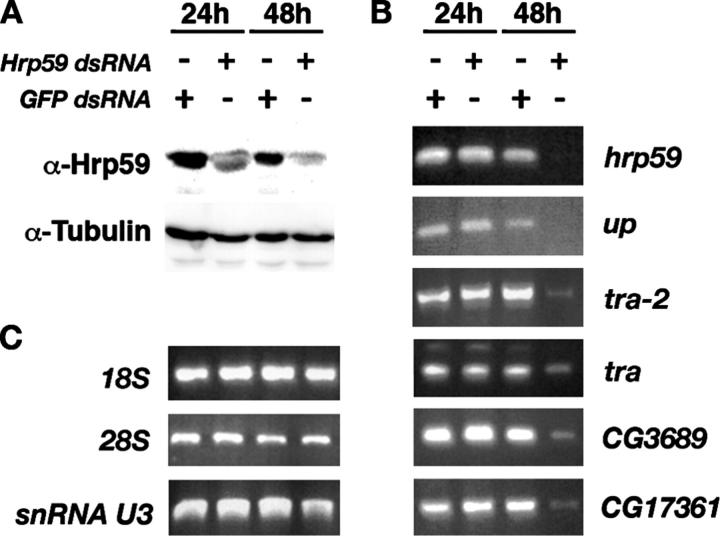
Next we asked whether the binding of Hrp59 to the target mRNAs identified above was of functional importance for their expression. We depleted Hrp59 in S2 cells by RNA interference (RNAi) and analyzed the levels of some of the Hrp59 target RNAs by RT-PCR. As shown in Fig. 8 A, the levels of Hrp59 protein were strongly reduced 48 h after administration of dsRNA corresponding to the hrp59 gene. The depletion was specific as judged by the constant levels of tubulin observed throughout the experiment (Fig. 8 A), and by the observation that administration of an unrelated dsRNA complementary to GFP did not affect the expression of hrp59 (Fig. 8 B, top). The expression of hrp59, up, tra-2, tra, CG3689, and CG17361 was analyzed by RT-PCR at 24 and 48 h. As shown in Fig. 8 B, all the analyzed targets showed specific reduction of RNA levels at 48 h. To assess the specificity of the observed effects, three unrelated RNAs were analyzed in parallel and were found unchanged (Fig. 8 C).
Figure 8.
Effect of Hrp59 depletion on the expression of Hrp59 target transcripts. S2 cells were treated with either Hrp59 dsRNA or GFP dsRNA (control) as indicated. (A) The effect of the RNAi treatments on the expression of Hrp59 was analyzed by Western blotting with anti-Hrp59 antibody 24 and 48 h after dsRNA administration. Staining with an anti-tubulin antibody served as a loading control. (B) Total RNA was isolated 24 and 48 h after addition of the dsRNAs. The levels of expression of hrp59, up, tra-2, tra, CG3689, and CG17361 were analyzed by RT-PCR. In all cases, the RNA levels were found to be drastically reduced at 48 h after dsRNA treatment. The effect was specific for Hrp59 as administration of GFP-dsRNA did not change the expression of the analyzed targets. (C) The levels of expression of three unrelated transcripts were also analyzed in parallel to assess the specificity of the RNAi treatment.
Although we have not yet analyzed all the Hrp59 targets identified in the microarray experiment, the reproducible effects observed for the five targets analyzed suggest that binding of Hrp59 to its target mRNAs is required for their correct expression.
Identification of common sequence patterns among the Hrp59 targets reveals an exonic purine-rich motif
A search for conserved sequence motifs among the Hrp59 targets was performed using the Teiresias software, a two-phase combinatorial algorithm for general purpose pattern discovery (Rigoutsos and Floratos, 1998). The Teiresias alignment of the sequences of the 23 most enriched transcripts revealed a common purine-rich motif present in exons and characterized by a GGAGG core (Fig. 9, A and B). The occurrence of this motif, defined as [gc][ga][atc]ggagg[atc][gct], is significantly higher among the Hrp59 targets than in the total Drosophila transcriptome (P < 0.005) or in RNAs enriched in the mock-IP (P < 0.01), as shown in the statistical analysis available as Online supplemental material at http://www.jcb.org/cgi/content/full/jcb.200407173/DC1.
Figure 9.
Hrp59 binds preferentially to an exonic purine-rich motif. (A) Examples of common motifs found in exons of Hrp59 targets. Sequences of the Drosophila genes tra2, CG6514, CG17361, up, and hrp59 are aligned and matches to the purine-rich motif are shown bold. (B) Sequence logo displaying the base frequencies over a 10-bp sequence obtained from alignment of the Hrp59 targets shown Table I. The y axis indicates the sequence conservation in each position expressed in bits, being two bits the maximum possible sequence conservation per site (Crooks et al., 2004). The figure was made using the sequence logo software at http://weblogo.berkeley.edu/. (C) Relative binding affinities of Hrp59 to different ribonucleotides. Hrp59 protein isolated from S2 cells by IP was incubated with increasing amounts of radioactively labeled oligonucleotides, and the bound radioactivity was measured by liquid scintillation. RNA1 (UA)10, RNA2 N20, RNA3: N5GAGGAGGNGN6. Error bars represent SDs from three independent experiments. RNA3 displays the highest affinity for Hrp59. (D) Total RNA was purified from either Hrp59 IP or mock IP, and the presence of intron-containing transcripts in the immunoprecipitated material was analyzed for hrp59, taiman, tra-2, and up by RT-PCR. Input RNA was purified from 1% of the original extract used for IP, and analyzed in parallel.
We next asked whether Hrp59 shows any preference for the identified purine-rich motif, and we performed protein–RNA binding assays in vitro. Hrp59 was isolated by IP under denaturing conditions in order to disrupt protein–protein and RNA–protein interactions, and the isolated Hrp59 immobilized on Sepharose beads was incubated with 32P-labeled 20-mer oligoribonucleotides containing either a random sequence (RNA1), a repeated AU motif (RNA2) or the purine-rich motif (RNA3). The amount of bound oligoribonucleotide was measured by liquid scintillation. As shown in Fig. 9 C, Hrp59 showed higher affinity for the purine-rich motif (RNA3) than for the random oligonucleotide (RNA2) or for the AU oligonucleotide (RNA1).
Interestingly, the purine-rich sequence identified above as a binding site for Hrp59 has been described as an exonic splicing enhancer (ESE; see Discussion). This raised the possibility that Hrp59 is involved in premRNA splicing regulation. If this is the case, Hrp59 should bind to its targets before intron removal. To check whether this is the case, we tested the ability of the anti-Hrp59 antibody to coimmunoprecipitate intron-containing transcripts. RNAs coimmunoprecipitated with the anti-Hrp59 antibody were reverse transcribed and the presence of specific intron-containing transcripts was analyzed by PCR using primer pairs designed to detect exclusively intron-containing transcripts. The tra, taiman, up and hrp59 transcripts were tested, and in all cases the intron-containing transcripts could be detected specifically in the Hrp59-IP (Fig. 9 D). Moreover, the RT-PCR reactions shown in Fig. 7 B were performed using primers that cover the exon-exon junctions and amplify spliced transcripts exclusively. Therefore, we can conclude that Hrp59 binds to the intron-containing premRNAs and remains bound to the transcripts after splicing.
Discussion
We have identified and characterized Hrp59, an insect hnRNP M protein. Hrp59 has many features in common with typical hnRNP proteins: it is a relatively abundant protein, has a modular domain organization containing three RRM domains, is dynamically recruited to transcribed genes, and binds to premRNA cotranscriptionally. Hrp59 accompanies the mRNA from the gene to the nuclear envelope, and is released from the mRNA at the nuclear pore complex (NPC), as shown by immuno-EM studies in C. tentans. The association of Hrp59 with transcribed genes is not quantitatively dependent on the amount of synthesized RNA, and in vivo Hrp59 binds preferentially to a subset of transcripts. Using a genome-wide approach in D. melanogaster, we have identified the preferred RNA targets of Hrp59, and we have shown that Hrp59 is required for the normal expression of these target transcripts. Moreover, we have shown that Hrp59 binds preferentially to exonic splicing enhancer sequences.
Hrp59 binds to the RNA during transcription and is released at the NPC
The hnRNP M proteins are evolutionarily conserved from yeast to human (Datar et al., 1993; Kafasla et al., 2000, 2002). However, little was known about their cellular distribution and about their association with premRNAs in vivo. Here, we have addressed these questions in C. tentans. Using immuno-EM, we have found that the insect orthologue of hnRNP M, Hrp59, binds to the premRNP cotranscriptionally and remains bound to BR RNP particles in the nucleoplasm. In agreement with this observation, we have shown by IP and RT-PCR that Hrp59 is bound to both intron-containing and spliced transcripts, which indicates that Hrp59 remains associated with the mRNA after splicing.
Hrp59 is also associated with BR RNP particles at the NPC. Interestingly, only the nuclear side of the translocating BR particles was labeled in immuno-EM studies, which indicates that either the epitopes recognized by the antibody become concealed during translocation or, more likely, Hrp59 leaves the BR RNP during translocation through the NPC. We conclude that the insect counterpart of mammalian hnRNP M accompanies the RNP to the nuclear pore and is released during nucleo-cytoplasmic translocation. HnRNP M is a target for SUMO (small ubiquitin-related modifier) modification (Vassileva and Matunis, 2004; Vertegaal et al., 2004), and a model has been proposed in which reversible SUMO modification of hnRNP M at the NPC facilitates remodeling of mRNP complexes during translocation through the nuclear pore, thus facilitating the transport of the mRNPs to the cytoplasm (Vassileva and Matunis, 2004). This model is based on the fact that Nup358, a nucleoporin located at the cytoplasmic filaments of the NPC, is a SUMO E3 ligase and enhances the SUMO modification of hnRNP M in vitro, and on the observation that SUMO modification alters the interactions between hnRNP proteins and nucleic acids (Vassileva and Matunis, 2004). Our present observation that Hrp59 is indeed associated with mature mRNP complexes during nucleo-cytoplasmic transport is consistent with the model proposed by Vassileva and Matunis (2004).
RNA coimmunoprecipitation and hnRNP protein function
Different experimental approaches have been used to determine the RNA-binding specificity of hnRNP proteins in vitro, but studies of hnRNP protein function in vivo have been difficult due to the lack of methods to identify their endogenous RNA targets. The combination of RNA–protein coimmunoprecipitation with genome-wide analysis of the immunoprecipitated transcripts allows the identification of the RNA targets for specific RNA-binding proteins. Such an approach, referred to as ribonomics by Tenenbaum et al. (2002), has been successfully applied to study the specificity of mRNA export factors in yeast (Hieronymus and Silver, 2003). We have followed a similar strategy to determine the specificity of Hrp59 in vivo. Due to the lack of genome-wide tools in Chironomus, we have taken advantage of the tools available in Drosophila and we have identified transcripts associated with Hrp59 in Drosophila S2 cells.
We applied scaling factors in the microarray experiments based on the average intensities of the whole chips according to standard Affymetrix procedures. This provides a reliable identification of the most enriched transcripts, but results in overestimation of the intensity values for the mock IP and, for this reason, our present study cannot provide a comprehensive description of all the endogenous targets for Hrp59. New scaling algorithms based on external calibrators are under development that will make it possible to estimate the total number of Hrp59 targets and to describe the complete transcriptome bound to Hrp59.
Hrp59 binding to exonic splicing enhancers
The analysis of the preferred Hrp59 targets led to the identification of a purine-rich sequence motif recognized by Hrp59. This purine-rich motif resembles a common ESE of the (GAR)n type (for review see Blencowe, 2000). Consensus sequences at the intron-exon boundaries of premRNAs are not sufficient for splice-site selection, and ESEs contribute to the definition of splice-sites by binding SR proteins that promote prespliceosome assembly (Shen et al., 2004). SR proteins contain RNA-binding domains of the RRM type, and the RRMs target specific SR proteins to particular ESE sequences (for review see Black, 2003).
The enhancer function of purine-rich motifs has been best documented in mammals. However, several observations suggest that in insects Hrp59 binds to purine-rich sequences that may be involved in splicing regulation. First, the Hrp59 motif is located mostly in exons. Second, an ESE similar to the Hrp59 target motif has been identified in the avian troponin T mRNA, where this sequence is required for the regulation of exon 5 splicing (Xu et al., 1993), and the Drosophila orthologue of troponin T, up, is one of the targets identified in our RNA coimmunoprecipitation study. Interestingly, several copies of the Hrp59 motif are located in the exon 5 of the Drosophila up mRNA. Third, the Hrp59 target sequence is also identified as a ESE by ESEfinder, a web-based resource for identification of putative ESEs responsive to several human SR proteins (Cartegni et al., 2003). Moreover, ESEfinder identifies the Hrp59 motif as a binding site for SF2/ASF2, an SR protein involved in splicing regulation known to bind (GAR)n sequences. Fourth, BLAST analyses show that one of the RRMs of Hrp59, RRM3, is very similar to the RRM of human ASF/SF2. In fact, the RRM3 of Hrp59 is as similar to the ASF/SF2 RRM as it is to the RRM3 of human hnRNP M.
We have shown that Hrp59 is required for expression of its target mRNAs. Although we have not directly addressed whether this requirement is at the level of transcription, splicing, export or stability, the observations reported above suggest that Hrp59 competes with ASF/SF2 for binding to the purine-rich ESE and plays a role in splicing maybe by preventing prespliceosome assembly.
Materials and methods
Animals and cell culture
C. tentans were raised under the conditions described by Lezzi et al. (1981). Fourth instar larvae were used for the experiments. For heat shock treatment, larvae were kept in water at 37°C for 90 min C. tentans tissue culture cells were cultivated at 22°C as described by Wyss (1982) and heat shock was for 90 min at 37°C.
Antibodies
The mAbs 3G10, 2E4 and 4E9 against Hrp36, Hrp45 and Hrp65 have been previously characterized by Kiseleva et al. (1997), Wurtz et al. (1996) and Miralles et al. (2000). The rabbit anti-Hrp59 antibody was raised and characterized by Falk et al. (2003), who referred to it as Y38 antibody. This antibody was used either as serum or affinity purified on recombinant Hrp59. The anti-U2B antibody was purchased from Research Diagnostics Inc.
Yeast two-hybrid
The complete coding sequence of Hrp65-1 (DBD-Hrp65) was used to screen a cDNA library of C. tentans tissue culture cells as described previously (Kiesler et al., 2003). Upon isolation from positive yeast clones, the library plasmid pAD-Hrp59 was cotransformed into AH109 yeast cells (CLONTECH Laboratories, Inc.) together with plasmids pBDGAL4-Hrp65 (Kiesler et al., 2003), pBDGAL4 or p53 control plasmid (Stratagene), respectively. Transformants were grown on selective medium to monitor the expression of the HIS3 and ADE2 reporter genes.
In vitro protein–protein binding
T7-Hrp65-2-His (Kiesler et al., 2003) was expressed in E. coli Bl21(DE3) cells and purified on Ni-NTA agarose (QIAGEN). The rabbit reticulocyte TnT system (Promega) was used for the in vitro synthesis of [35S]methionine-labeled Hrp59 and chloramphenicol acetyltransferase (CAT) proteins. Ni-NTA agarose containing purified Hrp65 (or Ni-NTA agarose without bound protein) was washed in RIPA buffer (1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.1 mg/ml PMSF in PBS) and incubated with 35S-labeled Hrp59 or with 35S-labeled CAT in RIPA buffer containing 50 mM imidazole. After extensive washing in RIPA buffer containing 50 mM imidazole, proteins were eluted, separated by 10% SDS-PAGE and analyzed by autoradiography.
Preparation of nuclear extracts from C. tentans tissue culture cells
C. tentans tissue culture cells were homogenized in PBS containing 0.2% NP-40 using a glass homogenizer. The homogenate was centrifuged at 2,000 g for 10 min at 4°C. For native IP experiments, the pellet containing the nuclei was resuspended in PBS, whereas for IP experiments using cross-linked cells the pellet was resuspended in PBS containing 8 M urea. Nuclei were sonicated three times for 4–5 s each time, and centrifuged as above. The resulting supernatant was the soluble nuclear extract, and it was supplemented with NP-40 to a final concentration of 0.1% and used as the input for IP experiments. All buffers contained 0.1 mg/ml PMSF.
IP of native protein complexes for analysis of protein–protein interactions
The monoclonal antibody 4E9 against hrp65 was added to nuclear extracts prepared from C. tentans tissue culture cells, and the specimen was incubated for 90 min at 4°C with gentle rotation. Rabbit anti–mouse IgG immunoglobulins covalently coupled to protein G–Sepharose beads (Zymed Laboratories) were added, and the incubation was continued for an additional 90 min at 4°C. The beads were washed four times with PBS containing 0.1% NP-40, once with PBS, and the proteins were eluted with 0.5% SDS at RT. The eluted proteins were precipitated with acetone and analyzed by Western blotting.
To test RNA-mediated interactions, the beads were incubated with antibodies and Sepharose beads as above, washed twice in PBS containing 0.1% NP-40, and incubated in 0.1 mg/ml of RNase A in PBS, or in PBS alone, for 40 min at 4°C followed by a 10-min incubation at RT. The beads were then washed and eluted as above.
To test DNA-mediated interactions, an equivalent amount of beads were incubated with 40 U/ml rDNase I (Ambion) in DNase I buffer (10 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2), or in PBS, for 30 min at 37°C. The beads were then washed three times with PBS containing 5 mM EDTA and 0.1% NP-40 and once with PBS, and the proteins were eluted as above.
IP of proteins after in vivo cross-linking
Proteins in C. tentans tissue culture cells were cross-linked for 20 min at RT with DSP (Sigma-Aldrich) dissolved in DMSO, or with DMSO alone, in standard culture medium to a final concentration of 1.5 mM. Nuclear extracts were prepared in 8 M urea as described above, diluted 10 times with PBS containing 0.2% NP-40 and 1 mM PMSF, and immediately incubated with mAb 4E9 for 90 min at 4°C, followed by a 90-min incubation upon addition of protein G agarose (Zymed Laboratories).
Immunostaining of C. tentans chromosome squashes
Salivary glands were dissected, fixed in 3.7% formaldehyde/45% acetic acid/PBS for 10 min and squashed in a drop of 45% acetic acid. The slides were frozen on dry ice, the coverslips pried off, and the preparations were further fixed with 3.7% formaldehyde in PBS for 15 min. The slides were washed with PBSG (PBS containing 0.1 M glycine) for 10 min, and blocked with a solution of 3% BSA in PBSG. The slides were incubated with either mAB 4E9 against Hrp65 or mAb 2E4 against Hrp45 for 2 h, followed by incubation with Texas red– and FITC-conjugated secondary antibodies (ICN/Cappel).
Immunofluorescent labeling of salivary glands
Salivary glands were fixed in 3.7% formaldehyde in PBS for 15 min, washed in PBS, and permeabilized for 9 min in 0.2% SDS in PBS. After washes in PBS, the glands were incubated with primary antibodies diluted in 0.3% BSA in PBS. Immunolabelings were visualized using Texas red– and FITC-conjugated secondary antibodies (ICN/Cappel).
Labeling of transcription sites by BrUTP incorporation
Salivary glands were dissected, washed in TBS and incubated in glycerol buffer (20 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.5 mM PMSF, 0.5 mM EGTA, 25% glycerol) for 3 min. The glands were then preextracted by a 3-min incubation in 0.05% Triton X-100 in glycerol buffer, washed in transcription buffer (50 mM Tris-HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 25% glycerol) and incubated for 20 min with transcription buffer supplemented with 0.5 mM each of ribonucleotides ATP, CTP and GTP, and 0.2 mM of BrUTP. Glands were subsequently washed for 3 min in 0.5% Triton X-100/TBS, 5 min in TBS, and fixed and coimmunostained with an mAb against bromo-uridine (Roche) and the polyclonal anti-Hrp59, as described above for immunofluorescent labeling of salivary gland cells.
RNase treatment of C. tentans salivary glands
Salivary glands were dissected and preextracted as for BrUTP incorporation, washed in TBS and incubated for 30 min with 0.1 mg/ml DNase-free RNase A (Sigma-Aldrich) in TBS, or with TBS alone. The glands were then fixed and immunostained using antibodies against either Hrp65 or Hrp59 as described for immunofluorescent labeling of salivary gland cells.
Acquisition of confocal images
Images were taken with a laser scanning microscope (model LSM 510; Carl Zeiss MicroImaging, Inc.) equiped with PlanApochromat objectives 40×/1.0 oil and 63×/1.4 oil, using immersion oil Immersol 518F (Carl Zeiss MicroImaging, Inc.). The optical sections are ∼1-μm thick.
Immuno-EM
IEM was performed using cryosectioned salivary glands as described by Kiesler et al. (2002). Salivary glands were fixed for 20–25 min in 4% PFA and 0.1% glutaraldehyde, and cryoprotected with 2.3 M sucrose. Secondary antibodies were conjugated to 6-nm gold particles (Jackson ImmunoResearch Laboratories). After immunolabeling, the sections were stained with 2% aqueous uranyl acetate and embedded in polyvinyl alcohol (Sigma-Aldrich). The specimens were examined and photographed in a electron microscope (model CEM 902; Carl Zeiss MicroImaging, Inc.) at 80 kV.
RNA coimmunoprecipitation and microarray hybridization
Drosophila S2 cells were homogenized in lysis buffer containing PBS, 2 mM MgCl2, 30 mM EDTA, 0.2% NP-40, 1 U/μl RNase OUT (Invitrogen), and supplemented with Complete Protease Inhibitor Cocktail (Roche). The homogenate was sonicated and centrifuged at 10,000 g for 15 min at 4°C. The supernatant was precleared using protein A–Sepharose beads (Sigma-Aldrich) and used for IP as indicated above for native protein complexes, using affinity-purified anti-Hrp59 antibody and protein A Sepharose (Sigma-Aldrich). Total RNA and the coimmunoprecipitated RNAs were purified using an RNeasy Mini Kit (QIAGEN) and analyzed with the Agilent Bioanalyzer using the RNA 6000 Nano LabChip Kit. Antisense cRNAs (targets) were generated using the Superscript Choice system (Invitrogen) and the ENZO BioArray HighYield RNA Transcript Labeling Kit (Enzo Inc.), and hybridized onto GeneChip Drosophila Genome Arrays (Affymetrix). The chips were hybridized using the Affymetrix Gene chip Fluidic Station 400 and analyzed with the Hewlett Packard Gene Array Scanner.
Microarray data analysis
The GCOS and DMT software from Affymetrix were used for data analysis. All files were user defined normalized to 1 and the target intensity signal set to 100. The Hrp59-IP RNA was compared with the mock-IP RNA. The data was filtered for probe sets either absent in the mock IP or increased in the hrp59-IP RNA with a factor of increase that was greater than two. This data was further filtered by comparing Hrp59-IP RNA to the input RNA, selecting for probe sets present and increased in hrp59-IP RNA with a factor of increase greater than two. Finally, we paired the two hrp-59 IP arrays and averaged the signal log ratios. Given the small number of experiments performed, no statistical criteria could be applied to determine the optimal increase factor to be used in the comparisons and a cutoff 2 was chosen arbitrarily. However, the statistical analysis of the frequency of the purine-rich motif (Supplement 2, available at http://www.jcb.org/cgi/content/full/jcb.200407173/DC1) indicated that setting the cutoff at 2 resulted in a group of targets with significantly higher representation of the purine-rich motif, compared with mock-IP and to the total Drosophila transcriptome.
Protein–RNA binding assay
The following oligoribonucleotides (Ambion) were used: RNA-1 was N20, RNA-2 was (UA)10, and RNA-3 was N4GAGGAGGNGN5. The oligonucleotides were 5′ end labeled with γ-[32P]ATP using T4 polynucleotide kinase (Ambion) and purified using G-25 Sephadex columns (Roche). For the RNA binding assay, 10, 20, and 30 pmol of radiolabeled RNA oligonucleotide were incubated at RT for 1 h with 30 μl Hrp-59–bound beads in a total of 200 μl RNA-binding buffer containing PBS, 100 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1% glycerol, 0.1% Triton X-100, 0.3 mg/ml tRNA, and 1 unit/μl RNase OUT (Invitrogen). The beads were then washed three times in RNA-binding buffer containing 0.5% Triton X-100. After the final wash the pelleted beads were incubated in 1% SDS in PBS at 65°C for 10 min to elute the bound RNA. The eluted RNA was quantified by liquid scintillation.
RT-PCR
Coimmunoprecipitated RNA was isolated as described above, and used as a template for the generation of single-stranded cDNA using a hexanucleotide mix (Roche) as primer and Superscript reverse transcriptase (Invitrogen). Different primer sets were designed to amplify 150–300-bp fragments for selected transcripts. For detection of unspliced transcripts, one primer was placed on exonic sequences and the other primer on intronic sequences. PCR amplification was performed using Taq Polymerase (Invitrogen). The reaction conditions were slightly different for the different primer combinations, but in all cases the Hrp59 and mock IPs were run in parallel under the same conditions. The sequences of the PCR primers are provided in Supplement 1, available at http://www.jcb.org/cgi/content/full/jcb.200407173/DC1.
In situ hybridization on polytene chromosome squashes
The hrp59 cDNA was digoxigenin labeled by nick translation and used as a probe. Salivary gland polytene chromosomes were hybridized as described by Miralles and Visa (2001). Detection of the hybridization signal was performed with a FITC-conjugated anti-DIG antibody (Roche) and an anti-FITC conjugated to Alexa-188 (Molecular Probes).
RNA interference in Drosophila S2 cells
For RNAi studies, dsRNA was produced with the Ambion MEGAscript RNAi kit. A 560 bp fragment corresponding to the second exon of the hrp59 gene was amplified by PCR from Drosophila genomic DNA. T7 promotor sequences were added at both ends of the initial PCR product in a second PCR reaction. For control experiments, GFP T7-tailed DNA was produced. RNAi treatment of Drosophila S2 cells was performed as described in Clemens et al. (2000). In brief, 20 nM of dsRNA was added to 106 cells in 1-ml serum-free Schneider's Drosophila medium. After 1 h, 2 ml of Schneider's medium containing 15% FBS were added, and the cells were cultured for 24 or 48 h before analysis. Total RNA was isolated from cells with RNAsy Mini Kit (QIAGEN) and reverse transcribed following standard procedures.
Online supplemental material
The sequences of the PCR primers used in this study are provided in Supplement 1. Supplement 2 presents the statistical analysis of the frequency of the purine-rich motif in the Hrp59 targets. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200407173/DC1.
Acknowledgments
We thank M. Tibäck and I. Granell for technical assistance, B. Daneholt for antibodies against Hrp45 and Hpr36, G. Farrants for correcting the English of the manuscript, and J. Liden (Affymetrix Core Facility at Novum, Karolinska Institute) for the microarray experiments. We are grateful to the Wallenberg Consortium North for supporting the Affymetrix core facility at NOVUM. We thank L. Wieslander for critical reading of the manuscript and B. Blencowe for helpful discussions.
Our research is financed by the Swedish Research Council and the Åke Wiberg Foundation. M. Hase is recipient of a stipend from the Carl Trygger Foundation.
E. Kiesler and M. Hase have contributed equally to this work.
Abbreviations used in this paper: BR, Balbiani ring; BrUTP, bromo-UTP; CAT, chloramphenicol acetyltransferase; CF, connecting fiber; DSP, dithiobis-succinimidyl propionate; ESE, exonic splicing enhancer; IP, immunoprecipitation; NPC, nuclear pore complex; premRNP, premessenger RNP complex; RRM, RNA-recognition motif.
References
- Alzhanova-Ericsson, A.T., X. Sun, N. Visa, E. Kiseleva, T. Wurtz, and B. Daneholt. 1996. A protein of the SR family of splicing factors binds extensively to exonic Balbiani ring pre-mRNA and accompanies the RNA from the gene to the nuclear pore. Genes Dev. 10:2881–2893. [DOI] [PubMed] [Google Scholar]
- Amero, S.A., S.C. Elgin, and A.L. Beyer. 1991. A unique zinc finger protein is associated preferentially with active ecdysone-responsive loci in Drosophila. Genes Dev. 5:188–200. [DOI] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291–336. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106–110. [DOI] [PubMed] [Google Scholar]
- Bomsztyk, K., O. Denisenko, and J. Ostrowski. 2004. hnRNP K: one protein multiple processes. Bioessays. 26:629–638. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F., S. Stamm, D.M. Helfman, and A.R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 265:1706–1709. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., J. Wang, Z. Zhu, M.Q. Zhang, and A.R. Krainer. 2003. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31:3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, J.C., C.A. Worby, N. Simonson-Leff, M. Muda, T. Maehama, B.A. Hemmings, and J.E. Dixon. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks, G.E., G. Hon, J.M. Chandonia, and S.E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneholt, B. 2001. Assembly and transport of a premessenger RNP particle. Proc. Natl. Acad. Sci. USA. 98:7012–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datar, K.V., G. Dreyfuss, and M.S. Swanson. 1993. The human hnRNP M proteins: identification of a methionine/arginine-rich repeat motif in ribonucleoproteins. Nucleic Acids Res. 21:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss, G., V.N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195–205. [DOI] [PubMed] [Google Scholar]
- Falk, R., C. Agaton, E. Kiesler, S. Jin, L. Wieslander, N. Visa, S. Hober, and S. Stahl. 2003. An improved dual-expression concept, generating high-quality antibodies for proteomics research. Biotechnol. Appl. Biochem. 38:231–239. [DOI] [PubMed] [Google Scholar]
- Forch, P., and J. Valcarcel. 2003. Splicing regulation in Drosophila sex determination. Prog. Mol. Subcell. Biol. 31:127–151. [DOI] [PubMed] [Google Scholar]
- Gattoni, R., D. Mahe, P. Mahl, N. Fischer, M.G. Mattei, J. Stevenin, and J.P. Fuchs. 1996. The human hnRNP-M proteins: structure and relation with early heat shock-induced splicing arrest and chromosome mapping. Nucleic Acids Res. 24:2535–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, S., A. Steplewski, L.D. Siracusa, S. Amini, and K. Khalili. 1995. Identification of a sequence-specific single-stranded DNA binding protein that suppresses transcription of the mouse myelin basic protein gene. J. Biol. Chem. 270:12503–12510. [DOI] [PubMed] [Google Scholar]
- Haynes, S.R., G. Raychaudhuri, D. Johnson, S. Amero, and A.L. Beyer. 1990. The Drosophila Hrb loci: a family of hnRNA binding proteins. Mol. Biol. Rep. 14:93–94. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., and P.A. Silver. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33:155–161. [DOI] [PubMed] [Google Scholar]
- Hovemann, B.T., I. Reim, S. Werner, S. Katz, and H. Saumweber. 2000. The protein Hrb57A of Drosophila melanogaster closely related to hnRNP K from vertebrates is present at sites active in transcription and coprecipitates with four RNA-binding proteins. Gene. 245:127–137. [DOI] [PubMed] [Google Scholar]
- Kafasla, P., M. Patrinou-Georgoula, and A. Guialis. 2000. The 72/74-kDa polypeptides of the 70-110 S large heterogeneous nuclear ribonucleoprotein complex (LH-nRNP) represent a discrete subset of the hnRNP M protein family. Biochem. J. 350(Pt 2):495–503. [PMC free article] [PubMed] [Google Scholar]
- Kafasla, P., M. Patrinou-Georgoula, J.D. Lewis, and A. Guialis. 2002. Association of the 72/74-kDa proteins, members of the heterogeneous nuclear ribonucleoprotein M group, with the pre-mRNA at early stages of spliceosome assembly. Biochem. J. 363:793–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesler, E., and N. Visa. 2004. Intranuclear pre-mRNA trafficking in an insect model system. Prog. Mol. Subcell. Biol. 35:99–118. [DOI] [PubMed] [Google Scholar]
- Kiesler, E., F. Miralles, and N. Visa. 2002. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 12:859–862. [DOI] [PubMed] [Google Scholar]
- Kiesler, E., F. Miralles, A.K. Ostlund Farrants, and N. Visa. 2003. The Hrp65 self-interaction is mediated by an evolutionarily conserved domain and is required for nuclear import of Hrp65 isoforms that lack a nuclear localization signal. J. Cell Sci. 116:3949–3956. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M., X. Wang, and S.A. Liebhaber. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 14:4357–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva, E., N. Visa, T. Wurtz, and B. Daneholt. 1997. Immunocytochemical evidence for a stepwise assembly of Balbiani ring premessenger ribonucleoprotein particles. Eur. J. Cell Biol. 74:407–416. [PubMed] [Google Scholar]
- Lezzi, M., B. Meyer, and R. Mahr. 1981. Heat shock phenomena in Chironomus tentans I. In vivo effects of heat, overheat, and quenching on salivary chromosome puffing. Chromosoma. 83:327–339. [DOI] [PubMed] [Google Scholar]
- Matunis, E.L., M.J. Matunis, and G. Dreyfuss. 1992. Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol. 116:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., and A.R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 68:365–375. [DOI] [PubMed] [Google Scholar]
- Miralles, F., and N. Visa. 2001. Molecular characterization of Ct-hrp65: identification of two novel isoforms originated by alternative splicing. Exp. Cell Res. 264:284–295. [DOI] [PubMed] [Google Scholar]
- Miralles, F., L.G. Ofverstedt, N. Sabri, Y. Aissouni, U. Hellman, U. Skoglund, and N. Visa. 2000. Electron tomography reveals posttranscriptional binding of pre-mRNPs to specific fibers in the nucleoplasm. J. Cell Biol. 148:271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan, V., A. Tretiakova, A. Steplewski, S. Haas, S. Amini, E. Johnson, and K. Khalili. 1997. Evidence for inhibition of MyEF-2 binding to MBP promoter by MEF-1/Pur alpha. J. Cell. Biochem. 66:524–531. [DOI] [PubMed] [Google Scholar]
- Percipalle, P., N. Fomproix, K. Kylberg, F. Miralles, B. Bjorkroth, B. Daneholt, and N. Visa. 2003. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc. Natl. Acad. Sci. USA. 100:6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoutsos, I., and A. Floratos. 1998. Combinatorial pattern discovery in biological sequences: The TEIRESIAS algorithm. Bioinformatics. 14:55–67. [DOI] [PubMed] [Google Scholar]
- Sass, H. 1995. Transcription of heat shock gene loci versus non-heat shock loci in Chironomus polytene chromosomes: evidence for heat-induced formation of novel putative ribonucleoprotein particles (hsRNPs) in the major heat shock puffs. Chromosoma. 103:528–538. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., and D. Zipori. 2002. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 531:109–114. [DOI] [PubMed] [Google Scholar]
- Shen, H., J.L. Kan, and M.R. Green. 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol. Cell. 13:367–376. [DOI] [PubMed] [Google Scholar]
- Singh, O.P., B. Bjorkroth, S. Masich, L. Wieslander, and B. Daneholt. 1999. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp. Cell Res. 251:135–146. [DOI] [PubMed] [Google Scholar]
- Tenenbaum, S.A., P.J. Lager, C.C. Carson, and J.D. Keene. 2002. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 26:191–198. [DOI] [PubMed] [Google Scholar]
- Vassileva, M.T., and M.J. Matunis. 2004. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol. Cell. Biol. 24:3623–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal, A.C., S.C. Ogg, E. Jaffray, M.S. Rodriguez, R.T. Hay, J.S. Andersen, M. Mann, and A.I. Lamond. 2004. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 279:33791–33798. [DOI] [PubMed] [Google Scholar]
- Visa, N., A.T. Alzhanova-Ericsson, X. Sun, E. Kiseleva, B. Bjorkroth, T. Wurtz, and B. Daneholt. 1996. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 84:253–264. [DOI] [PubMed] [Google Scholar]
- Wang, K., and F.M. Richards. 1974. An approach to nearest neighbor analysis of membrane proteins. Application to the human erythrocyte membrane of a method employing cleavable crosslinkages. J. Biol. Chem. 249:8005–8018. [PubMed] [Google Scholar]
- Wieslander, L. 1994. The Balbiani ring multigene family: coding repetitive sequences and evolution of a tissue-specific cell function. Prog. Nucleic Acid Res. Mol. Biol. 48:275–313. [DOI] [PubMed] [Google Scholar]
- Wurtz, T., E. Kiseleva, G. Nacheva, A. Alzhanova-Ericcson, A. Rosen, and B. Daneholt. 1996. Identification of two RNA-binding proteins in Balbiani ring premessenger ribonucleoprotein granules and presence of these proteins in specific subsets of heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 16:1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss, C. 1982. Chironomus tentans epithelial cell lines sensitive to ecdysteroids, juvenile hormone, insulin and heat shock. Exp. Cell Res. 139:309–319. [DOI] [PubMed] [Google Scholar]
- Xu, R., J. Teng, and T.A. Cooper. 1993. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol. Cell. Biol. 13:3660–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]