Abstract
The mechanism for forming linear microtubule (MT) arrays in cells such as neurons, polarized epithelial cells, and myotubes is not well understood. A simpler bipolar linear array is the fission yeast interphase MT bundle, which in its basic form contains two MTs that are bundled at their minus ends. Here, we characterize mto2p as a novel fission yeast protein required for MT nucleation from noncentrosomal γ-tubulin complexes (γ-TuCs). In interphase mto2Δ cells, MT nucleation was strongly inhibited, and MT bundling occurred infrequently and only when two MTs met by chance in the cytoplasm. In wild-type 2, we observed MT nucleation from γ-TuCs bound along the length of existing MTs. We propose a model on how these nucleation events can more efficiently drive the formation of bipolar MT bundles in interphase. Key to the model is our observation of selective antiparallel binding of MTs, which can both explain the generation and spatial separation of multiple bipolar bundles.
Introduction
Microtubules (MTs) have important roles in numerous cellular processes, but to perform these roles MTs have to become ordered into arrays. In many cells, it is the centrosome that nucleates and organizes a radial array of MTs. Additional MT-based functionality is however required and cells organize radial and linear MT arrays that are not connected to the centrosome. It is becoming clear that the MTs that make up these arrays originate from noncentrosomal nucleation sites as well as detach from centrosomes by severing or breakage mechanisms (Keating and Borisy, 1999; Canaday et al., 2000; Dammermann et al., 2003; Karsenti and Nedelec, 2004). Key to the function of MT arrays is the regulation of MT polarity, i.e., the location of the so-called plus and minus ends of MTs. Self-organization mechanisms, involving molecular motors, are known to transform random networks of free MTs into astral arrays in which MT minus ends are focused at the center (Surrey et al., 2001; Vorobjev et al., 2001). In general, less is known about the regulation of MT polarity in linear arrays, such as observed in epithelial cells, neurons, myotubes, and plant cells (Canaday et al., 2000; Dammermann et al., 2003). A detailed observation of regulatory mechanisms is often hindered by the complexity and density of MT networks inside living cells.
The rod-shaped fission yeast Schizosaccharomyces pombe organizes a relatively simple linear MT array in interphase independent of the spindle pole body (SPB), which is the yeast analogue of the centrosome (Hagan, 1998). This array comprises about four distinct MT bundles that align along the long axis of the cell (Drummond and Cross, 2000; Tran et al., 2001). Each bundle, in its simplest form, contains two antiparallel oriented MTs that overlap slightly at their minus ends. The plus ends point to the cell ends and occasionally undergo catastrophes, i.e., they switch to a state of depolymerization. Depolymerization is halted at the region of MT overlap near the cell middle and is followed by MT regrowth, hence the region of overlap has become known as interphase MT organizing center (iMTOC). The minus ends that are embedded within the iMTOCs are believed to have no dynamics. Interphase bundles share a common feature with radial MT arrays in higher eukaryotes because MT minus ends are brought together in both cases. Furthermore, we can look upon interphase bundles as a very simple bipolar MT array: two plus ends or poles are separated by a region of MT overlap. Investigation of interphase bundles may therefore provide mechanistic insight into the formation of other bipolar structures, such as the mitotic spindle.
Recent work has characterized the interphase arrays, but many questions remain to be answered. It was shown that iMTOCs can bind to the nuclear membrane (Tran et al., 2001) and contain the well-conserved γ-tubulin complex (γ-TuC; Vardy and Toda, 2000; Zimmerman et al., 2004) that nucleates MTs and caps MT minus ends (Wiese and Zheng, 2000). New iMTOCs are continuously generated during interphase (Sawin et al., 2004), but we do not understand the mechanism nor do we know how the number of bundles is controlled or how MT bundling activity is prevented from bundling all MTs into one big bundle.
Here we identify a novel fission yeast protein mto2p as a protein involved in iMTOC formation. Mto2p associates with cytosolic γ-TuCs and is essential for MT nucleation from non-SPB sites in interphase. By direct observation of MT nucleation from γ-TuCs in wild-type cells we revealed the sequence of events that generate a region of MT overlap. Key are: (a) MT nucleation from γ-TuCs bound along the length of existing MTs and (b) selective bundling of antiparallel MTs. These events can efficiently transform single MTs into bipolar structures. Cells that lacked mto2p had primarily a single MT bundle during interphase and we used this phenotype to further investigate the known role of MT bundles in cell morphogenesis (Hagan, 1998) and nuclear positioning (Tran et al., 2001).
Results
Mto2p localizes to MTOCs and MTs
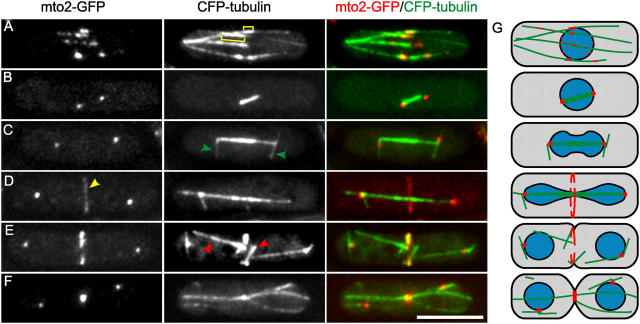
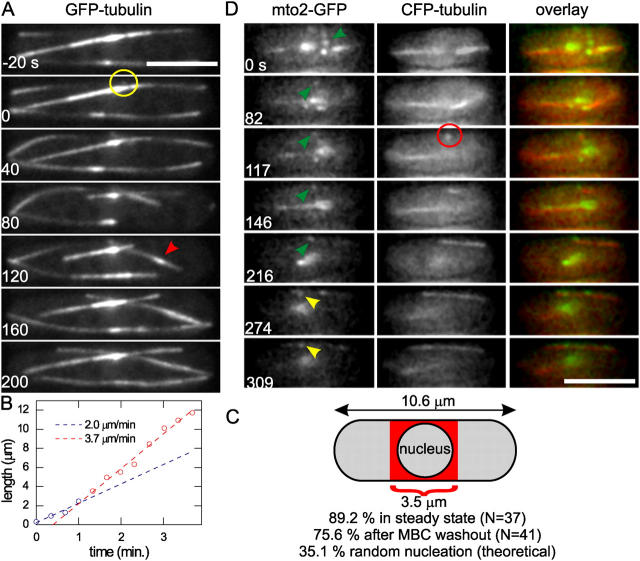
A part of the gene product of fission yeast ORF SPBC902.06, which we termed mto2 (MT Organizer 2), was previously shown to localize to the nucleus in a GFP-tagging screen (Ding et al., 2000). Because localization to the nuclear membrane is expected for iMTOC components, we further investigated this predicted 44.0-kD protein and analyzed its localization with respect to MTs at different stages of the cell cycle in cells expressing mto2-GFP and CFP-tubulin (Fig. 1, A–F; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1). Mto2-GFP localized primarily to the three known sites of MT nucleation (Hagan, 1998): iMTOCs near the middle of the cell (Fig. 1 A), the SPB during mitosis (Fig. 1, B–F), and the equatorial MTOCs (eMTOCs) at the actomyosin ring during the end of mitosis (Fig. 1, D–F). The localization of mto2-GFP is summarized in the diagram of Fig. 1 G. Because this localization was reminiscent of the localization described for the γ-TuC component alp4p (Spc97/hGCP2 family; Vardy and Toda, 2000; Zimmerman et al., 2004), we simultaneously expressed alp4-GFP and mto2-monomeric RFP (mRFP) and found that the localization of both proteins was nearly identical throughout the cell cycle (Fig. 2 A, for an interphase cell). Alp4-3HA also coimmunoprecipitated with mto2-GFP (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1) showing that mto2p associates with γ-TuCs. This association may exclude a subset of γ-TuCs at the SPB because the interphase SPB was rich in alp4-GFP but lacked mto2-mRFP (Fig. 2 A and Fig. S2 B).
Figure 1.
Localization of mto2-GFP to MTs. (A–G) Representative images of cells expressing mto2-GFP and CFP-tubulin (PT537). Imaging: projected deconvoluted wide-field images showing the complete cell depth. A delay of ∼20 s between the GFP and CFP image causes apparent nonperfect localization of mto2-GFP to MTs. (A) In interphase, mto2-GFP is localized primarily to the iMTOC regions of MT bundles that as a result of MT overlap are recognized by a brighter CFP signal (square brackets). (B–D) Mitotic spindles and astral MTs (green arrowheads) terminate at mto2-GFP rich SPBs. (D) Mto2-GFP is localized to the equatorial region (yellow arrowhead) before post-anaphase array MTs (red arrowheads) appear in E. (G) Cartoon of mto2-GFP localization and MT organization (Hagan, 1998). The nucleus is shown in blue. Bar, 5 μm.
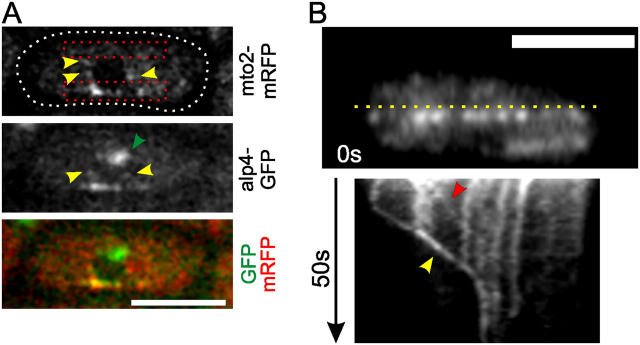
Figure 2.
Localization of mto2p–alp4p complexes in interphase. (A) Wild-type interphase cell expressing alp4-GFP and mto2-mRFP (PT568). The cell outline, from a separate DIC image, is shown as a dotted line. The SPB (green arrowhead) is visible in the alp4-GFP image but not in the mto2-mRFP image. Both proteins colocalize as linear arrays of dots along MTs (lower boxed region). The black circle in the mRFP image is a cross section through the nucleus. MTs are expected in the boxed regions because they are excluded from the nucleus and orient along the long axis of the cell. Some dots (yellow arrowheads) therefore suggest MT-independent colocalization along the nuclear rim. Imaging: two-color single plane confocal. 6.4-s time interval. (B) Kymograph of mto2-GFP fluorescence (PT300) along the line in the top panel. See text for notes on arrowheads. Imaging: single plane wide field (1-s time interval). Bars, 5 μm.
To examine the dynamics of mto2p–alp4p-containing γ-TuCs, we created kymographs of MT-associated mto2-GFP dots in interphase (Fig. 2 B). The predominantly vertical lines that run in parallel on the kymograph indicate that most of the dots were bound statically to an underlying MT bundle. Dots were seen along MTs throughout the cells, indicating that γ-TuC association with MTs is not limited to the iMTOC region. Using additional two color imaging we observed dots of mto2-GFP tracking tips of depolymerizing MTs (unpublished data). These events created diagonal lines in the kymographs that stop at the iMTOC where MTs are stabilized (Fig. 2 B, yellow arrowhead). Lastly, we occasionally observed the apparent motion of mto2-GFP dots along MTs (Fig. 2 B, red arrowhead). These events may correspond to dots bound to MTs that slide along each other within MT bundles. Two-color kymographs of alp4-GFP and mto2-mRFP showed that both proteins move as single entities within bundles (Fig. S2 B).
Mto2Δ cells have primarily a single MT bundle
To investigate the role of mto2p in MT organization we generated an mto2 deletion strain, which yielded viable but slightly bent cells (Fig. 3 A). In mto2Δ cells we observed alp4-GFP at the SPBs in both interphase and mitotic cells, but did not observe any non-SPB localization (Fig. 3 B and Fig. S3 B, available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1), suggesting that non-SPB γ-TuCs are absent. Consistent with this interpretation, we found important defects in the MT cytoskeleton of interphase mto2Δ cells but no defects in the mitotic spindle (Fig. S3 A). To observe MTs in relationship to the SPB, we performed time-lapse imaging of mto2Δ cells expressing GFP-tubulin and the SPB marker alp4-GFP (Fig. 3, C and D). The average number of MT bundles in mto2Δ cells (n = 1.3 ± 0.7 SD; Fig. 3 E) was significantly lower than in wild-type cells (3.6 ± 0.9). The bright medial section of these bundles indicated a region of MT overlap and was reminiscent to, but often longer than the iMTOC region in wild-type cells (Fig. 1 A). Similar to wild-type cells, alternating periods of MT growth from and shrinkage to the overlapped regions were observed in mto2Δ cells. Bundles in mto2Δ cells often contained more than two MTs (Fig. 3 C, first plane) and appeared to have more MT mass than wild-type bundles. The SPB in interphase mto2Δ cells was connected to an MT bundle in 49.7% of the cells (Fig. 3 C, n = 143). This connection was occasionally observed to break. Nonconnected SPBs (Fig. 3 D) had a low nucleation activity (0.0051 min−1; n = 5 observed nucleations), corresponding to approximately one event per interphase cycle. The SPB is therefore not the main source of MTs in interphase mto2Δ cells. The majority of bundles in mto2Δ cells (60.3%) was not connected to a SPB, suggesting that most bundles form independently of the SPB in mto2Δ cells. Nonconnected SPBs were not observed in wild-type cells (PT546; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1).
Figure 3.
MT and cell morphology defects in interphase mto2Δ cells. (A) DIC image of mto2Δ cells (PT541) grown in liquid to mid-log phase. Red and green arrowheads mark bent and wild-type looking cells. Bar, 5 μm. (B) Alp4-GFP localization in wild-type (top; PT544) and mto2Δ (bottom; PT538) interphase cells. Arrowheads point to the SPB and the boxed area indicates an iMTOC flanked by dots of alp4-GFP. Imaging: projected confocal planes showing the complete cell depth. Bar, 5 μm. (C and D) Time-lapse images of interphase mto2Δ cells expressing GFP-tubulin and alp4-GFP (PT541). Imaging: projected confocal planes showing the complete cell depth. Bars, 5 μm. (C) An MT-connected SPB (white arrowhead) oscillates in phase with the bright medial region of MT overlap (square bracket). Two MTs, which can splay apart (red arrowhead), are alternating between growth and shrinkage at the left end of this region. One MT is visible on the right. Short MTs with nearly constant length (arrow) are generated at cell ends during apparent MT breakage. Most of their apparent length change is caused by tumbling. 40-s time interval. (D) A non-SPB–connected MT bundle. 80-s time interval. (E) Distribution of MT bundles and single MTs in wild-type (n = 110 cells; PT546) and mto2Δ cells (n = 143 cells; PT541). (F and G) DIC images of tea1-GFP expressing mto2Δ cells (PT503) that recover from stationary phase. Time 0 corresponds to cell growth initiation. Bar, 2 μm. (H) Distribution of tea1-GFP at a cell tip. The far right image corresponds to the boxed area in G. The middle of the cell (red line) is judged from the full cell DIC image. Green dots indicate the centers of mass of tea1 patches. Imaging: overlaid DIC image and six collapsed wide-field image planes spaced 1 μm apart. Bar, 2 μm. (I) Tea1-GFP patch height (e.g., arrow in H) assayed as a function of time and averaged over the indicated number of cell ends (10, 7, and 10); SEM is plotted. Negative times (nongray area) corresponds to stationary cells.
Multiple interphase bundles are required for straight cell growth
The observed defects in interphase MT organization in mto2Δ cells are likely related to their bent phenotype (Hagan, 1998). A homogeneous deposition of the polarity marker tea1p (Behrens and Nurse, 2002) by multiple MT bundles is believed to be required for straight cell growth especially during the recovery of stationary, nongrowing, cells (Sawin and Snaith, 2004). Single MT bundles as observed in Fig. 3 (C and D) may pattern cell tips nonhomogeneously with tea1p, leading to bent growth. To test these ideas, we studied nutrient-starved stationary mto2Δ cells, which were short and in 50% of the cases had no visible bent phenotype. After being transferred to an agar pad with nutrients, cells initiated new growth on one or two sides within about 1 h. Growth of cell ends of initially straight cells occurred in 41% of the observed cases (n = 27 cell ends) in line with the main cell body (Fig. 3 F, and left side of the cell in Fig. 3 G), whereas the remainder of cell ends grew bent (Fig. 3 G, right side of the cell). We quantified the position of tea1p patches at cell ends during growth initiation in mto2Δ cells expressing tea1-GFP (Fig. 3 H, green dots). At cell ends that subsequently grew bent, these patches were on average located 0.72 μm above the cell middle at the moment of growth initiation (Fig. 3 I). This distance was only 0.22 μm for cell ends of straight-growing mto2Δ cells and 0.20 μm for cell ends of wild-type cells. The latter all grew straight after recovery. The distribution of tea1p therefore positively correlated with the position of future cell growth. The correlation increased during the 30 min before initiation of cell growth, indicating that tea1p patches were motile (Fig. 3 H) and were not inherited from growth zones that were active before starvation. The analysis shows that mto2Δ cells can still grow straight if by chance tea1p is positioned homogeneously.
Nuclear positioning defects in mto2Δ cells
Disorganized interphase MT arrays have been linked to nuclear positioning defects (Sawin et al., 2004; Zimmerman et al., 2004) and therefore septation defects (Chang and Nurse, 1996; Tran et al., 2000). Forces, generated by interactions between growing interphase MTs and the cell wall, were hypothesized to push the nucleus toward the middle in wild-type cells (Tran et al., 2001). Deformations of the nuclear membrane at sites of MT-membrane connections were observed in response to MT growth, but it is not known whether these forces are large enough to move the nucleus. To investigate nuclear positioning, we imaged the nuclear membrane in interphase mto2Δ cells that express the nuclear pore marker nup107-GFP and the SPB marker alp4-GFP (Fig. 4 A). Strong oscillations of the SPB, caused by growth of attached MTs (Fig. 3 C), generated rather large nuclear membrane extensions in comparison to wild-type cells (not depicted). Interestingly, the complete nucleus moved into the direction of these membrane extensions, showing that polymerization forces indeed displaced the nucleus. Large membrane extensions occurred in 60.9% of the cells (n = 87), but 11.3% of this subset showed no SPB oscillations. Extensions were therefore primarily mediated by MT attachments to the SPB, but also occurred by direct binding of MTs to the membrane. We tracked and projected the motion of the nucleus along the long axis of the cell for wild-type and mto2Δ cells (Fig. 4 B). SDs were calculated for positional traces that lasted 30–50 min. The distribution of SDs (Fig. 4 C) was narrow for wild-type cells (average SD is 0.21 ± 0.01 μm; ± SEM) but extended to both smaller and larger values for mto2Δ cells. Smaller values were primarily caused by mto2Δ cells that did not show large membrane extensions and likely had no MTs attached to the nucleus (Fig. 4 C, average value of this subset is 0.15 ± 0.01 μm). Larger motion was seen in mto2Δ cells that did show large membrane extensions (0.34 ± 0.03 μm). This comparison showed that nuclear oscillations in cells that had primarily a single MT bundle attached to the nucleus were larger than in wild-type cells, in which multiple bundles are attached. This observation is predicated by models of pushing-based positioning (Dogterom and Yurke, 1998; Tran et al., 2001). These show that multiple bundles generate partly counteracting forces that cause only small nuclear displacements but maintain a more precise central nuclear position.
Figure 4.

Strong nuclear oscillations in mto2Δ cells. (A) Mto2Δ cells expressing nup107-GFP and alp4-GFP (PT574). Red dots indicate the center of the nucleus. The SPB (white arrowhead) oscillates in response to polymerization of attached MTs. Imaging: five collapsed wide field image planes spaced 1 μm apart. 1-min time interval. Bar, 5 μm. (B) Nuclear displacements, projected along the long axis of the cell, in three nup107-GFP expressing cells. Shown are traces for a wild-type cell (green; PT53), an mto2Δ cell that showed large extensions of the nuclear membrane (solid red; PT510) and an mto2Δ cell without such extensions (hatched red; PT510). (C) SDs were calculated for many curves similar to B. Shown is the distribution of SDs for wild-type (green, n = 68 cells) and mto2Δ (red, n = 76) cells. The distribution is subdivided for mto2Δ cells that did (solid, n = 45) and did not show (hatched, n = 31) large membrane extensions.
Single MTs in mto2Δ cells exhibit treadmilling
In addition to dynamic MT bundles, we observed on average 1.5 ± 1.0 (±SD; Fig. 3 E) MTs with a near constant length in mto2Δ cells (Fig. 3 C). Individual MTs could be observed for up to 30 min with only several micrometers of length change and no catastrophes. These MTs were dim and lacked a bright central region, suggesting that they were single MTs. They often appeared when MT-bundles bent around cell ends and snapped (Fig. 3 C). These MTs disappeared by rare catastrophes or were seen to bind laterally to existing bundles. We reasoned that single MTs in mto2Δ cells may be treadmilling: a state of constant MT minus-end depolymerization balanced by plus-end polymerization (Shaw et al., 2003). Minus-end depolymerization in mto2Δ cells would be consistent with the proposed absence of non-SPB γ-TuCs because γ-TuCs normally cap minus ends (Wiese and Zheng, 2000). Several single MTs in mto2Δ cells showed speckles, i.e., regions of high intensity caused by the random incorporation of GFP-tubulin into the MT (Waterman-Storer et al., 1998). A kymograph analysis (Fig. 5 A) showed speckles moving from left to right, implicating growth on the left (MT plus end) and indeed depolymerization on the right (MT minus end). Using speckles as fiduciary marks we measured the growth velocity of plus ends (3.17 μm/min; Table I) and the shrinkage rate of minus ends (3.18 μm/min). The near equality explains the constant length of treadmilling MTs. Dimly labeled MTs (0.21 ± 0.47 SD per cell; Fig. 3 E), reminiscent of single MTs, were also observed in wild-type cells but were rare and did not show treadmilling.
Figure 5.
MT treadmilling and bundle formation in mto2Δ cells expressing alp4-GFP and GFP-tubulin (PT541). (A and B) Kymographs of the fluorescent patterns along indicated lines (line width = 0.5 μm) in top panels. Imaging: single plane time-lapse wide field (5-s time interval). (A) Kymograph of a treadmilling MT. (B) Kymograph of an MT bundle. The right side of a region of MT overlap (square bracket) is indicated in the kymograph by a dotted line. The nonconnected SPB (arrow) generates a weak vertical streak that should be neglected. The arrowhead indicates a catastrophe that reaches the overlapped region. Re-growth occurs after an apparent shortening of the overlapped region, which becomes less bright on the right. (C) Bundling of two treadmilling MTs (marked 1 and 2). The arrowhead points to a region of MT overlap. Imaging: projected confocal planes showing the complete cell depth. Bars, 5 μm.
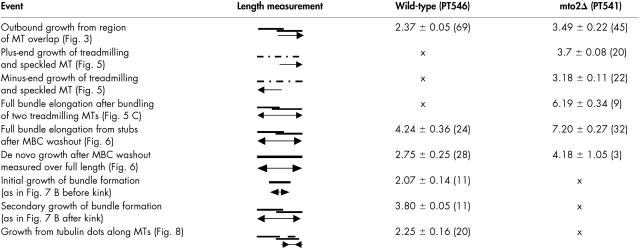
Table I.
MT growth velocities (μm/min) in interphase wild-type and mto2Δ cells.
MT growth velocities for indicated events in wild-type (PT546) and mto2Δ (PT541) cells expressing GFP-tubulin and alp4-GFP. Listed are: averages, standard errors, and the parenthesized number of events. All events were assayed in steady state except for the MBC experiments.
A kymograph analysis of an MT bundle in an mto2Δ cell is shown in Fig. 5 B. The bright medial region of the bundle oscillates from right to left and back in response to MT pushing forces. Speckles are visible on the MT, whose plus end grows out to the right of the medial bright region. These speckles do not move relative to the bright region, indicating that depolymerization of MT minus ends is prevented at regions of MT overlap. Similarly, we found that outbound growth from regions of MT overlap occurred in mto2Δ cell at 3.49 μm/min (Table I), in agreement with the plus-end growth velocity of treadmilling MTs. In rare cases, we observed the sudden bundling and elongation of two treadmilling MTs that met in the cytoplasm (nine events during 54 h of observation). Fig. 5 C shows a dramatic example: two treadmilling MTs align, and the formed bundle suddenly elongates at a full-length velocity of 7.3 μm/min during the first 40s. Later, this region behaves as a normal bundle in which catastrophes are followed by regrowth from the central region. In the other observed events, bundling was often more transient and MT separation was observed after initial bundling. Nonetheless, all nine events showed initial growth velocities after bundling between 4.0 and 8.0 μm/min (average 6.19 μm/min; Table I), which given the plus-end polymerization rate (3.17 μm/min; Table I), can only be achieved if both MTs in the bundle are stabilized at their minus end and grow in opposing directions. iMTOC-like regions can thus self-assemble during rare antiparallel encounters of two MTs. The MT minus ends in these overlapped regions are stabilized even in the absence of γ-TuCs. Not all bundles in interphase mto2Δ cells are formed by aligning treadmilling MTs because bundles sometimes appeared at the end of mitosis as remnants from spindle or astral MTs (Fig. S3 A).
Nucleation of new MTs upon MBC washout is mto2p dependent
To investigate the stability of overlapped regions in MT bundles, we treated cells with the MT depolymerizing drug methyl-benzidazole-carbamate (MBC). As described previously (Tran et al., 2001), MBC addition caused MTs in wild-type cells to depolymerize to short stubs, corresponding to iMTOCs (Fig. 6 A). Stubs maintained a nuclear-bound median position in the cell. MT depolymerization in mto2Δ cells yielded significantly longer stubs (Fig. 6 B), in agreement with the longer regions of MT overlap in unperturbed mto2Δ bundles (Fig. 3 C). The average number of stubs per cell after 10 min of MBC treatment in both wild-type (2.9 ± 0.7 SD; n = 11 cells) and mto2Δ cells (1.09 ± 0.67; n = 34 cells) was in reasonable agreement with the number of bundles shown in Fig. 3 E. Regions of MT overlap in mto2Δ and wild-type cells were thus similarly resistant to MBC treatment. Treadmilling MTs in mto2Δ cells quickly disassembled during MBC treatment confirming they are single MTs with no stabilized medial region (Fig. 6 B).
Figure 6.

Mto2p-dependent MT nucleation after MBC washout. Imaging: projected confocal planes showing the complete cell depth. (A) Wild-type cell expressing alp4-GFP and GFP-tubulin (PT546). After the addition of 25 μM MBC, at t0, MTs depolymerize to four short stubs (marked in red); one of them is connected to the SPB visualized by alp4-GFP. Two stubs disappear during the prolonged 10-min MBC incubation. After MBC washout, at t1, MT regrowth occurs from stubs (red arrowheads) and five new sites. (B) mto2Δ cell expressing alp4-GFP and GFP-tubulin (PT541). The bundle hardly shortens after MBC treatment, indicating a long stabilized region. Treadmilling MTs (red) first disappear but later reappear from MT breakage events (green arrowhead). Yellow arrowheads mark the SPB. Bar, 5 μm.
MT stubs in wild-type and mto2Δ cells immediately resumed growth after MBC washout at both sides (Fig. 6); velocities measured over the full length of the bundles were approximately two times higher as the velocities measured earlier for single sided growth from bundles (Table I). Immediately after washout we observed in wild-type cells a massive nucleation of new MTs that did not originate from stubs (3.7 ± 2.0 SD per cell) but later regrouped to form normal interphase bundles (Fig. 6 A). Their average initial growth velocity (2.75 μm/min measured over the first 24 s after washout; Table I) was close to single sided growth from bundles (2.37 μm/min) suggesting that new MT bundles first grew as single MTs. Growth of new MTs after MBC treatment in mto2Δ cells was very rare (three observed events corresponding to 0.15 ± 0.44 per cell), in agreement with an absence of non-SPB γ-TuCs. Growth occurred at 4.18 μm/min and instead of being single MTs, which should treadmill, these MTs may have originated from small, mobile, and hard to detect MT stubs.
MT nucleation of single MTs during steady state
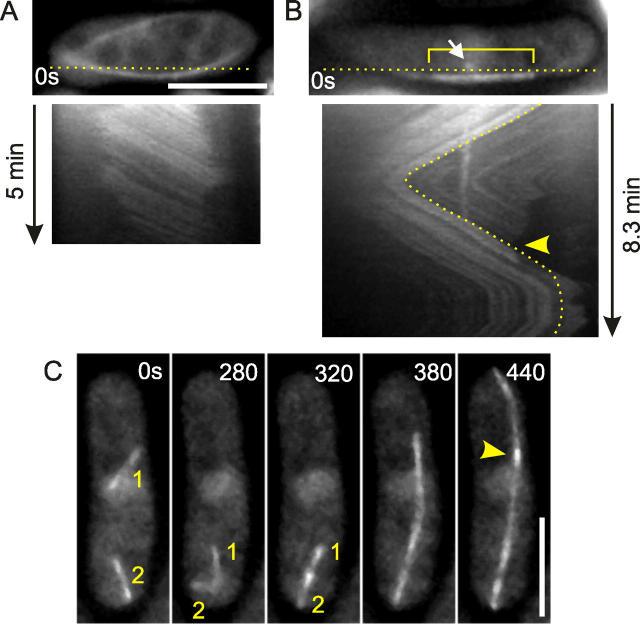
The appearance of novel MT bundles was previously reported in unperturbed interphase cells (Sawin et al., 2004). We reasoned that these bundles may originate from the apparent single MTs that were observed in steady-state wild-type cells (Fig. 3 E). These MTs appeared in two distinct manners: (1) short MT fragments separated from existing MT bundles during catastrophes (Fig. S4 B, available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1) or appeared near iMTOC regions shortly after catastrophes; and (2) MTs appeared independently of other MTs (Fig. 7 A). Rates of observations were 0.054 min−1 (31 class A events in 579 min of total cell observation time) and 0.064 min−1 (37 class B events). MTs were initially faint and a bright region of MT overlap sometimes became visible at a time scale of 1 min (Fig. 7 A). We measured the length increase after initial appearance for class B MTs and often (11 out of 37 events) observed a kink in the growth curve after ∼1 min (Fig. 7 B). The average elongation rate after kinks was about two times higher as before kinks (3.80 μm/min vs. 2.07 μm/min; Table I), suggesting that bundles indeed start as single MTs. The transformation to bundles occurred at a rate of at least 0.16 min−1 (11 kink events in 68.5 min of length measurements on 37 class B MTs), but apparently did not involve bundling to a separate second MT as observed earlier in mto2Δ cells (Fig. 5 C). A catastrophe on 17 out of the 37 new MTs did not lead to a complete disassembly of the MT (as in six other cases), but was rescued suggesting that in total 17 MTs made the switch to a bundle. The fate of the remainder 14 MTs could not be observed because these MTs moved close to an existing bundle and often seemed to fuse with them. Class A MTs behaved similarly (unpublished data).
Figure 7.
Formation of MT bundles in steady-state interphase cells. (A) Wild-type cell expressing alp4-GFP and GFP-tubulin (PT546). A dot of GFP-tubulin appears at t = 0 s around the cell center (yellow circle) and grows out as an MT bundle. A bright region appears at t = 120 s (red arrowhead). Imaging: projected confocal planes showing only the planes in which the nucleated MT is visible. (B) Length of the nucleated MT in A as a function of time. Growth is initially slow (blue dots) but then speeds up (red dots). Separate linear fits to the blue and red dots are shown. (C) Spatial map of MT nucleations. The percentage of nucleation events that occurred within the medial 3.5 μm (one third of the average cell length of 10.6 ± 0.3 μm; ±SEM, n = 48 cells) is indicated for PT546 cells during steady state and after MBC washout (Fig. 6). A theoretical value for random nucleation was calculated for a 10.6-μm cell (3.5-μm diam and spherical tips) with a nucleus (2.5-μm diam) whose volume is excluded for MT nucleation. (D) Wild-type cell (PT537) expressing mto2-GFP and CFP-tubulin. The nucleation of an MT (red circle) colocalizes with a dot of mto2-GFP (green arrowheads) that moved around in the cell middle. Initially the mto2-GFP dot remains at the left MT end (t = 117–216), during which time the MT growth velocity equals 2.0 μm/min. Later, more mto2-GFP dots appear along the MT, and the left end is free of mto2-GFP (t = 216–309 s). MT growth to the left occurs at 1.6 μm/min, with respect to a static mto2-GFP dot (yellow arrowheads). Imaging: single plane two-color wide field (5.8-s time interval). Bars, 5 μm.
We spatially mapped the appearance of new MTs (Fig. 7 C) and found that 89.2% of MT nucleations (class B MTs only) occurred within the middle 33% of cell length. This percentage was similar, though slightly lower (75.6%), for nucleation events after MBC treatment in wild-type cells. These values were expected to be 35.1% for random nucleation throughout the cytoplasm (excluding the volume of the nucleus, see note in Fig. 7 C). Nucleation was thus highly constrained to the region of the cell that contained the nucleus. Earlier, the protein mto1p (also known as mod20p/mbo1p) was shown to colocalize with alp4p at the nuclear membrane of cold treated and fixed cells, and short MTs were observed to emanate from cytoplasmic mto1p dots (likely containing alp4p) in fixed cells (Sawin et al., 2004). Similarly, we observed faint dots of mto2-GFP encircling the interphase nucleus (Fig. S1) and colocalization of mto2-mRFP and alp4-GFP near the nucleus (Fig. 2 A). To test whether these represent nuclear-bound γ-TuCs that nucleate single MTs, we imaged cells expressing mto2-GFP and CFP-tubulin. Fig. 7 D shows the nucleation of a new MT at a dot of mto2-GFP, whose medial location in the cell suggested an association with the nuclear membrane. MT growth occurred initially (between 117 and 216 s) at a single MT rate and during this time the mto2-GFP dot remained associated with one end. Based on earlier work on γ-TuCs (Wiese and Zheng, 2000), we may assume that this association is with the minus end. Single MTs are thus nucleated by nuclear γ-TuCs that cap their minus ends. These γ-TuCs require mto2p, because the nucleation rate of new MTs in mto2Δ cells was at least 100 times lower as in wild-type cells (0.0017 min−1 vs. 0.16 min−1; one possible new MT, excluding MT breakage events at cell tips, was observed during 581 min).
Mto2-dependent MT nucleation along existing MTs
The MT bundle depicted in Fig. 7 D initially grew only to the right, but later (>216 s) also to the left. This switch coincided with the appearance of additional mto2-GFP dots along the initially single MT. One of these dots might have nucleated a secondary MT in a direction opposing initial growth, thereby forming a bundle. MT nucleation was observed along existing MT bundles in wild-type cells (Fig. 8, A and B). In these cells, dots of GFP fluorescence occasionally moved along MT bundles. The fluorescence intensity at the dot location was about twice that of the underlying MT, suggesting they were very short single MTs. In agreement, these dots were observed to elongate at an average velocity of 2.25 μm/min while sliding (Table I). In both Fig. 8 (A and B), sliding motion stopped or slowed down when the iMTOC region of the underlying MT bundle was reached. We observed 52 inbound (with respect to the iMTOC) sliding events and no MTs that sled toward MT plus ends (outbound). The average velocity of these sliding events, analyzed before motion ceased, was 5.50 ± 0.25 μm/min (± SEM). Additionally we observed four MTs that were nucleated but grew without sliding. The nucleation rate of new MTs along bundles was 0.10 min−1 per bundle (56 events in almost 10 h of observation time spread over 32 cells, in which on average one bundle was in focus), which compares well to the rate at which single MTs are transformed to bundles (0.16 min−1/MT; Fig. 7). This strongly suggests that bundle formation occurs by nucleation of a second MT along the length of a single MT. In addition, nucleation along MTs likely caused the appearance of class A MTs (0.054 min−1/cell or 0.015 min−1/bundle), which separated from bundles after they were nucleated along MTs (Fig. S4 B). Note that all rates may be under estimated because we are likely to miss events.
Figure 8.

MT nucleation along existing MT bundles. (A and B) Wild-type interphase cells (PT546). Imaging: single plane time-lapse wide field (5-s time interval). (A) The boxed area is shown at successive time intervals. A region of MT overlap (yellow arrowhead), or iMTOC, becomes small and invisible from the fifth image. GFP fluorescence appears (red arrowhead) and moves at a speed of 8.1 μm/min toward the iMTOC region, where motion stops. (B) Kymograph of a similar event. MT growth occurs (green arrowhead) while the MT moves to the iMTOC region (red winding line). Initial rapid motion (blue line) slows down as the new MT grows. A bundle catastrophe (yellow arrowhead) is followed by a catastrophe of the new MT, starting on the left (red arrowhead). (C) Nucleation in cells expressing alp4-GFP and mRFP-tubulin (PT574). Initially, two MTs grow out on the left and one on the right side of a bright iMTOC (yellow bracket). One left MTs has a catastrophe (green arrowhead), after which two dots of alp4-GFP demarcate the region of MT overlap. An MT is nucleated (white arrowhead) from a dim alp4-GFP dot (red arrowhead and red outlined area) that moves toward the iMTOC. The dot associates with the left side of the nucleated MT (yellow arrowhead). Imaging: single plane two-color confocal (6.4-s time interval). Bars, 5 μm.
Double color imaging revealed that both mto2p and alp4p were associated with one end of nucleated MTs along MT bundles (Fig. 8 C and Fig. S4 A). Based on earlier work on γ-TuCs (Wiese and Zheng, 2000), we assumed that this was the minus end. In both Fig. 8 C and Fig. S4 A, the minus end of new MTs therefore points away from the iMTOC region of the underlying bundle. This indicates an antiparallel association of the nucleated MT with the underlying MT because minus ends are embedded in iMTOCs. The same orientation was found for eight additional events observed with alp4-GFP and mRFP-tubulin, but parallel orientations were not observed. Also the catastrophe observed in Fig. 8 B (red arrowhead), likely corresponds to a plus end, which would make the MT antiparallel after the initial nucleation.
Observations in mto2Δ cells showed that mto2p is required for active nucleation along MTs and the transformation of single MTs to bundles; treadmilling MTs (single MTs) were not observed to form bundles (apart from bundling with existing MTs) within a total observation time of 104 min. 17 transitions were expected if transitions would have occurred at wild-type rate (0.16 min−1).
Discussion
By characterizing a novel protein, mto2p, we have obtained new insights into MT nucleation from non-SPB sites in fission yeast. Simultaneous imaging of MT growth and alp4p–mto2p complexes identified active γ-TuCs along MTs and the nuclear membrane in interphase. This finding shows that nucleation occurs dispersed throughout cells and is not limited to discrete iMTOCs. Instead, iMTOCs turned out to be a dynamic structure that can be formed and regrouped during interphase. Mto2Δ cells had fewer MT bundles and as a consequence showed defects in cell morphology and nuclear positioning. We found no obvious homology between mto2p and proteins in other eukaryotes but we will discuss why the observed mechanism of MT bundling can be of general interest.
Observations in other organisms show that γ-TuCs along MTs and membranes may be conserved sites for nucleation of noncentrosomal MTs: MT nucleation from membrane associated γ-TuCs was observed for plant nuclei (Canaday et al., 2000), myotubes (Tassin et al., 1985), nuclear membrane fragments during spindle assembly in Drosophila spermatocytes (Rebollo et al., 2004), and Golgi membranes in mammalian cells (Chabin-Brion et al., 2001; Rios et al., 2004). Nucleation along MTs was recently reported in plant cells (Van Damme et al., 2004), in which γ-tubulin is found along MT arrays (Canaday et al., 2000; Drykova et al., 2003). Other findings show that MT nucleation may in general be more dispersed: in animal cells γ-tubulin is found along MTs in mitotic spindles (Lajoiemazenc et al., 1994) and punctuated γ-tubulin staining was observed along in vitro generated MT asters from egg extracts (Stearns and Kirschner, 1994). In mammalian cells, a majority of γ-TuCs does not associate with the centrosome but is cytosolic (Moudjou et al., 1996). New studies, furthermore, show that noncentrosomal MTs contribute to mitotic spindle formation, and aspects of MT nucleation need to be addressed (Tulu et al., 2003; Rebollo et al., 2004).
Our results show an association of mto2p with non-SPB γ-TuCs, which may be rather large and well-regulated protein complexes involving γ-tubulin, mto1p, the J-domain protein rsp1p, alp4p (hGCP2), alp6p (hGCP3), and alp16p (hGCP6; Vardy and Toda, 2000; Fujita et al., 2002; Sawin et al., 2004; Venkatram et al., 2004; Zimmerman et al., 2004). γ-Tubulin itself may have a role in binding γ-TuCs to MTs (Llanos et al., 1999), but another candidate is the centrosomin-related protein mto1p, which in a very recent study was found to localize all along MTs and the nuclear membrane when overexpressed (Samejima et al., 2005). Mto1p was furthermore shown to bind mto2p directly and associate with γ-TuCs in a strongly mto2p-dependent manner. Here, we found several clues on mto2p's function: In mto2Δ cells, non-SPB γ-TuCs were absent during interphase, but γ-TuCs at the SPB nucleated a normal mitotic spindle (Fig. S3 A). This excludes a function for mto2p in intranuclear γ-TuCs at the SPB. Furthermore, mto2-mRFP did not clearly localize to interphase SPBs but was at the SPB during mitosis. Therefore, a subset of γ-TuCs may shuttle between the SPB and the cytoplasm during the cell cycle. Mto2p may play a role in this process or alternatively may be more structurally involved in the assembly of non-SPB γ-TuCs.
In agreement with an absence of non-SPB γ-TuCs in mto2Δ cells, we observed the steady depolymerziation of MT minus ends in interphase. Fission yeast seems well suited for further studies on the regulation of free MT minus ends, which in general depolymerize in vivo (Dammermann et al., 2003). The deletion of mto2 also increased the polymerization rate of interphase MTs plus ends by 50% compared with wild-type cells (Table I) and caused MTs to curve around cell tips and break occasionally. A possible explanation lies in an increased concentration of free tubulin (Walker et al., 1988) caused by the decreased number of MTs in mto2Δ cells. However, contradicting this trend, we did not observe very fast polymerization in cells that almost completely lacked MTs (Fig. 5 C). Therefore, a structural change of MTs, related to their nucleation, may form an alternative explanation. All MTs in γ-TuC–deficient interphase cells may originate from γ-TuCs at the SPB, either from weak nucleation during interphase and mitosis (astral MTs) or as spindle MTs escaping from the nucleus (Sawin et al., 2004; Fig. S3 A). Such “centrosomally” derived MTs were shown to have less protofilaments compared with noncentrosomal MTs in developing wing epidermal Drosophila cells (Tucker et al., 1986).
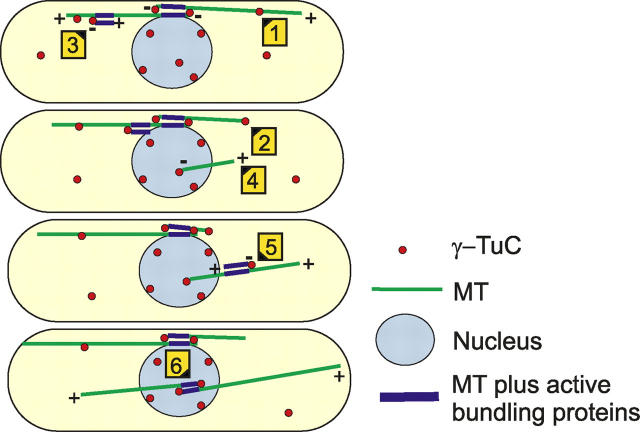
Based on our observations of MT nucleation, we propose a model for the generation of iMTOCs in fission yeast cells (Fig. 9): nucleation of single MTs from nuclear-membrane-associated γ-TuCs is followed by nucleation of secondary MTs from MT-bound γ-TuCs to form regions of bipolar MT overlap. Key to this model is the observation that iMTOC-like regions can self-assemble out of two MTs. In mto2Δ cells this occurs when two MTs by chance align in the cytoplasm and in wild-type cells the second MT is directly nucleated along the first MT. The latter mechanism is much more efficient for bundle formation and in fact increases the ratio between bundles and single MTs in cells by a factor of 20 (Fig. 3 E). In terms of MT stabilization and MBC resistance the iMTOCs in wild-type and mto2Δ cells are quite similar (Fig. 5 B and Fig. 6).
Figure 9.
Model for interphase MT bundle generation. A perinuclear and free cytoplasmic pool of γ-TuCs coexists and possibly exchanges. Cytoplasmic γ-TuCs bind to existing MTs (1) and move toward the nucleus during MT catastrophes (2) or are moved to the nucleus with an adhered nucleated MT (3). New MTs originate primarily from the perinuclear pool and first grow out as a single MT (4). Perinuclear nucleation may secure a nuclear attachment of the iMTOC that is under construction. A bipolar bundle is formed when a γ-TuCs adheres to a new MT and nucleates a second MT in an antiparallel manner (5). Two minus ends are moved towards each other by a mechanism that likely involves MT minus-end–directed motors and bundling proteins. A region of MT overlap is maintained that can bind to the nuclear membrane (6). In our model γ-TuCs demarcate regions of MT overlap as is observed (Fig. 8 C). The motion of small MTs along MT bundles largely explains the previously reported complex motion of MT associated proteins, like tip1p (CLIP-170) and mal3p (EB1; Busch and Brunner, 2004).
In our model, MTs selectively bind in an antiparallel orientation in accordance with observations (Fig. 5 C and Fig. 8 C). Parallel bundling was not observed and Fig. 3 C in fact shows that parallel MTs can splay apart within a single bundle. Antiparallel MTs were seen to slide along each other, apparently to bring their minus ends together and form an iMTOC. The observed velocity of MT sliding after MT nucleation from MT-associated γ-TuCs, suggests that molecular motor proteins transport MTs along each other. We speculate that a protein construct with two minus-end–directed motors may be responsible for sliding. If the heads of these two motors would be counter opposing these constructs could only bind and move antiparallel MTs. They could even force overlapping MTs to become antiparallel if the initial nucleation along the parent MTs would occur randomly oriented. Selective antiparallel MT associations were previously observed in vitro for the plus-end–directed motor protein MKLP-1 (Nislow et al., 1992), which has a role in spindle midzone assembly (Mishima et al., 2002). In our model, motors rupture the connection between a γ-TuC and parent MT after a new MT is nucleated. The γ-TuCs stay associated with nucleated MTs. This explains the apparent motility of some γ-TuCs along MTs (Fig. 2 B) and is mechanistically different from earlier reported dynein-dependent motility of MT nucleating material in animal cells, which can move without nucleating MTs (Vorobjev et al., 2001). After bringing minus ends together, the sliding motion should be balanced by proteins that bind statically along MTs to create a stable region of overlap. A candidate is the MT-bundling protein ase1p which associates with interdigitating MTs in the spindle midzone. Interestingly, fission yeast ase1p localizes to the iMTOC (Loïodice et al., 2005; Yamashita et al., 2005) and the mammalian homologue PRC1 binds molecular motors (Kurasawa et al., 2004; Zhu and Jiang, 2005). Ase1p may stabilize MT minus ends upon bundling in mto2Δ cells and rescue catastrophes of MT plus ends in iMTOCs.
Our model suggests that the number of MT bundles in cells may be regulated by a tubulin-dependent nucleation rate of γ-TuCs. For this we note the massive increase in MT nucleation after MBC washout in wild-type cells (32 events in 11 cells within 24 s after MBC washout corresponds to a nucleation rate of 7.3 min−1/cell vs. 0.064 min−1/cell during steady state), caused by a momentarily increased concentration of free tubulin upon MT disassembly. Similar effects were found after cold treatment (Sawin et al., 2004). In steady-state, γ-TuCs may be de-activated when there are too many MT bundles in the cell, and vise-versa. The accumulation of too many bundles may furthermore be regulated by the observed fusion of MT bundles during steady state (unpublished data), which could start near the nucleus. Only there MTs from two different bundles can bundle in an antiparallel manner. In this way, iMTOCs that move too close to each other on the nuclear membrane may be actively combined, making iMTOCs on average well dispersed over the nuclear membrane. After fusion, single MTs may get expelled from the bundle and be removed by catastrophes similar to class A MT appearance. MT contacts in iMTOCs with more than two MTs (Sagolla et al., 2003; Fig. 8 C) may be unstable because nonbundling parallel MT contacts become possible within the iMTOC.
Our analysis has shown how the generation of multiple and well-separated bipolar MT bundles in fission yeast is optimized by MT nucleation along MTs and selective bundling of antiparallel MTs. Similar mechanisms could be important for the generation of linear MT bundles in other cell types, e.g., the cortex of plant cells (Shaw et al., 2003; Van Damme et al., 2004), neurons and myotubes. Based on the existence of large pools of cytoplasmic γ-TuCs (Moudjou et al., 1996), we further hypothesize that the described mechanisms may act in more complex bipolar arrays such as mitotic spindles. Of particular interest are chromosomal MTs, which are important for the formation of robust bipolar spindles (Karsenti and Nedelec, 2004). They are randomly nucleated around mitotic chromosomes but are later bipolarly organized. MT-plus-end–directed motors would be required to overlap MT plus ends in this instance. The full spindle is more complex and MTs bundling occurs both parallel and antiparallel (Chakravarty et al., 2004). In general, the ability to discriminate between parallel and antiparallel may be key to forming morphological different MT networks, and regulation of bundling activity may give cells the ability to dynamically change MT structures. Further analysis of the fission yeast system should yield key proteins involved in bundling/sliding, and provide insight into the molecular mechanism of maintaining a stable region of MT overlap. This may prove to be instrumental to understanding aspects of MT organization in higher eukaryotic cells.
Materials and methods
Yeast strains and genetic methods
Standard media and genetic methods were used as described in the Nurse Lab Handbook (http://www.sanger.ac.uk/PostGenomics/S_pombe/links.shtml#pombe). Strains used are listed in Table S1. Mto2-GFP and mto2-deletion strains were constructed using a PCR-based knockout method (Bahler et al., 1998). For primers, see supplemental material.
COOH (pPT73) and NH2 (pPT76) termini nmt1-based mRFP tagging vectors were constructed by replacing GFP by mRFP (Campbell et al., 2002) in pSGP572 (NotI–SalI) and pSGP573 (XhoI–NotI; Siam et al., 2004). Mto2 (XhoI–NotI) and tub1 (NotI–SalI) were then subcloned to create mto2-mRFP (pPT79) and mRFP-tub1 (pPT77) vectors.
Cell preparations
All cells containing nmt-based plasmids were grown on EMM + 5 μg/ml thiamine agar plates for 1–2 d at 30°C. Non-plasmids cells were grown on YE5S. Cells were then grown overnight in a 2-ml shaking liquid culture of the same media at 25°C. Cells in mid-log phase were transferred to an agar pad for imaging (Tran et al., 2001). Exceptions to this procedure were made for cells containing mRFP and CFP plasmids for which pads were used that contained 50 μM n-propyl gallate (Fluka) to minimize photobleaching (Sagolla et al., 2003). Furthermore, for time-lapse imaging of CFP-tubulin (pRL71; Glynn et al., 2001) we transferred very fresh cells directly from plate to pad, which yielded a higher fluorescence signal. The mRFP-fluorescence signal in mid-log growing cells in liquid was very weak, possibly as a result of a delayed folding of mRFP (Campbell et al., 2002). To circumvent this problem, cells were overgrown for an additional day in liquid, which slowed down cell growth and increased the fluorescence signal. These cells were transferred to fresh medium 3 h before imaging. Thiamine suppression (5 μg/ml) was used for pPT79 but not for pPT80.
MT depolymerization and regrowth experiments were conducted on yeast cells that were bound to the bottom of a flow chamber (Browning et al., 2003; Zimmerman et al., 2004). To maximize binding we incubated cells with Con A (Sigma-Aldrich; 5 min in 5 mg/ml in EMM). Cells were washed (three times in EMM) and bound to a poly-l-lysine (P-1274; Sigma-Aldrich)–coated coverslip. To depolymerize MTs, 25 μg/ml MBC (Sigma-Aldrich) was flown into the chamber.
For cell growth initiation studies, we grew a liquid mid-log cell culture for an additional 36 h to reach stationary state. Cells were directly transferred to an agar pad for imaging.
Microscopy
Microscopy was performed using a wide-field microscope (model Eclipse TE2000; Nikon; equipped with 100×/1.45 NA Plan Apo objective, Nikon CY GFP and EN GFP HQ filter sets, and Hamamatsu ORCA-ER camera) or a spinning disk confocal scanner (Perkin Elmer combined with a Nikon eclipse E600, equipped with 100×/1.45 NA Plan Apo objective, electronically controlled filter wheels [Sutter Instruments] and Hamamatsu ORCAII-ERG camera). When noted, images were deconvoluted with Softworx (Applied Precision). In confocal mode, the 488- and 568-nm laser lines of an Argon/Krypton laser (Melles Griot) were used for excitation of GFP and mRFP in combination with a triple band pass dichroic mirror. Microscope control, image acquisition and image analysis were done using Metamorph software (Universal Imaging). All imaging was done at room temperature (close to 23°C).
For MT speckle analysis we used wide-field microscopy because of its large depth of field in comparison to confocal microscopy. This allowed for the observations of speckles on MTs that were not completely in focus over their full length. Especially in mto2Δ cells MTs buckled out of plane.
Data analysis
Velocities of MT growth and motion were obtained using linear least square fits to distance versus time data. Statistical means and errors (Table I) were obtained by weighting data with the total distance covered in individual events. Outbound growth of MTs from regions of MT overlap in wild-type and mto2Δ cells were analyzed by measuring distances between MT tips and bright central regions. Curved lines were used to measure lengths of buckled MTs. Plus-end polymerization and minus-end depolymerization rates of treadmilling MTs were obtained by measuring distances between MT tips and individual speckles. Only speckles were selected that were visible for at least six frames. Elongation of MTs bundles after MBC treatment was measured over the full length of the bundle (from plus end to plus end) between the first two frames taken after MBC washout (24-s time interval). Here, lengths were not measured from maximum projected images but in the full three-dimensional stacks. In this way, we minimized errors due to the rotation of small MTs in space. The same procedure was applied to the length of short MTs directly after their nucleation (Fig. 7 A). Sliding motion of nucleated MTs (Fig. 8) was analyzed with respect to the bright central region of the underlying MT bundle. In cases were MTs growth could be analyzed while sliding, we measured the velocity of the MT side that pointed away from the central region of the underlying bundle.
Tea1p patches at cell tips were quantified using ImageJ (http://rsb.info.nih.gov/ij/). All images were rotated such that the central cell axis was horizontal. A rolling ball–type background subtraction (radius 0.5 μm) was used to isolate the patch from background fluorescence. Additional background subtraction with a fixed value was applied to make every pixel outside the patch zero. We calculated the center of gravity over a 5- by 2.5-μm area that was centered on the cell tip and measured its vertical position relative to the cell middle (estimated from a simultaneously acquired differential interference contrast [DIC] image). The sign of this height was chosen positive in the direction of future cell growth (up or down) or was chosen such that it was positive at the moment of cell growth initiation for the case of straight cell growth.
Displacements of nuclei (Fig. 4) were quantified using Metamorph's track objects algorithm. As a reference image, a cropped image of a nucleus was selected at a time point at which no nuclear deformations were visible. Tracked positions in other frames largely corresponded to the main mass of the nucleus and were not skewed toward deformations.
Online supplemental material
Fig. S1 shows localization of mto2-GFP along the nuclear membrane in interphase cells. Fig. S2 shows immunoprecipitation of alp4-3HA with mto2-GFP and motion of alp4p–mto2p complexes along MTs. Fig. S3 displays defects in MT organization and alp4-GFP localization in mitotic mto2Δ cells. Fig. S4 A shows additional evidence for selective anti-parallel MT bundling after MT nucleation along existing MT bundles and Fig. S4 B shows the detachment of a class A MT from an existing bundle along which it was nucleated. Used cell strains are listed in Table S1 and a supplemental text lists DNA primers. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200410119/DC1.
Acknowledgments
We thank Ryan Cochran and Shenbagam Krishnan for constructing mRFP plasmids; Isabelle Loïodice and other members of the Tran lab for discussions and support; Duane Compton, David Drubin, and Doug Drummond for helpful discussions; Fred Chang (Columbia University, New York, New York), Roger Tsien (University of California, San Diego, San Diego, CA), Takashi Toda (The Cancer Research UK London Research Institute, London, UK), Iain Hagan (Paterson Institute for Cancer Research, Manchester, UK), and Susan Forsburg (USC, Los Angeles) for strains and reagents; Marileen Dogterom and Erfei Bi for a critical reading of the manuscript; and Nikon, Hamamatsu and Universal Imaging for microscopy support. M. Janson is supported by a fellowship of the Netherlands Organization for Scientific Research (NWO).
This work is supported by the ADF funds from the University of Pennsylvania to P.T. Tran.
Note added in proof. Mto2Δ cells largely lack post-anaphase array MTs (Fig. S3). A recent study (Venkatram et al., 2005. Mol. Biol. Cell. 10.1091/mbc.E04-12-1043) shows that mto2Δ cells consequently have a defective anchoring of the cytokinetic actin ring to the medial region of the cell.
Abbreviations used in this paper: MBC, methyl-benzidazole-carbamate; MT, microtubule; mRFP, monomeric RFP; MTOC, MT organizing center; DIC, differential interference contrast; SPB, spindle pole body; iMTOC, interphase MTOC; eMTOC, equatorial MTOC; γ-TuC, γ-tubulin complex.
References
- Bahler, J., J.Q. Wu, M.S. Longtine, N.G. Shah, A. McKenzie, A.B. Steever, A. Wach, P. Philippsen, and J.R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 14:943–951. [DOI] [PubMed] [Google Scholar]
- Behrens, R., and P. Nurse. 2002. Roles of fission yeast tea1p in the localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 157:783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning, H., D.D. Hackney, and P. Nurse. 2003. Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5:812–818. [DOI] [PubMed] [Google Scholar]
- Busch, K.E., and D. Brunner. 2004. The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14:548–559. [DOI] [PubMed] [Google Scholar]
- Campbell, R.E., O. Tour, A.E. Palmer, P.A. Steinbach, G.S. Baird, D.A. Zacharias, and R.Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaday, J., V. Stoppin-Mellet, J. Mutterer, A.M. Lambert, and A.C. Schmit. 2000. Higher plant cells: γ-tubulin and microtubule nucleation in the absence of centrosomes. Microsc. Res. Tech. 49:487–495. [DOI] [PubMed] [Google Scholar]
- Chabin-Brion, K., J. Marceiller, F. Perez, C. Settegrana, A. Drechou, G. Durand, and C. Pous. 2001. The Golgi complex is a microtubule-organizing organelle. Mol. Biol. Cell. 12:2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, A., L. Howard, and D.A. Compton. 2004. A mechanistic model for the organization of microtubule asters by motor and non-motor proteins in a mammalian mitotic extract. Mol. Biol. Cell. 15:2116–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, F., and P. Nurse. 1996. How fission yeast fission in the middle. Cell. 84:191–194. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., A. Desai, and K. Oegema. 2003. The minus end in sight. Curr. Biol. 13:R614–R624. [DOI] [PubMed] [Google Scholar]
- Ding, D.Q., Y. Tomita, A. Yamamoto, Y. Chikashige, T. Haraguchi, and Y. Hiraoka. 2000. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 5:169–190. [DOI] [PubMed] [Google Scholar]
- Dogterom, M., and B. Yurke. 1998. Microtubule dynamics and the positioning of microtubule organizing centers. Phys. Rev. Lett. 81:485–488. [Google Scholar]
- Drummond, D.R., and R.A. Cross. 2000. Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr. Biol. 10:766–775. [DOI] [PubMed] [Google Scholar]
- Drykova, D., V. Cenklova, V. Sulimenko, J. Volc, P. Draber, and P. Binarova. 2003. Plant γ-tubulin interacts with alpha beta-tubulin dimers and forms membrane-associated complexes. Plant Cell. 15:465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, A., L. Vardy, M.A. Garcia, and T. Toda. 2002. A fourth component of the fission yeast γ-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when γ-tubulin function is compromised. Mol. Biol. Cell. 13:2360–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn, J.M., R.J. Lustig, A. Berlin, and F. Chang. 2001. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr. Biol. 11:836–845. [DOI] [PubMed] [Google Scholar]
- Hagan, I.M. 1998. The fission yeast microtubule cytoskeleton. J. Cell Sci. 111:1603–1612. [DOI] [PubMed] [Google Scholar]
- Karsenti, E., and F. Nedelec. 2004. The mitotic spindle and actin tails. Biol. Cell. 96:237–240. [DOI] [PubMed] [Google Scholar]
- Keating, T.J., and G.G. Borisy. 1999. Centrosomal and non-centrosomal microtubules. Biol. Cell. 91:321–329. [PubMed] [Google Scholar]
- Kurasawa, Y., W.C. Earnshaw, Y. Mochizuki, N. Dohmae, and K. Todokoro. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23:3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoiemazenc, I., Y. Tollon, C. Detraves, M. Julian, A. Moisand, C. Guethhallonet, A. Debec, I. Sallespassador, A. Puget, H. Mazarguil, et al. 1994. Recruitment of antigenic γ-tubulin during mitosis in animal-cells—presence of γ-tubulin in the mitotic spindle. J. Cell Sci. 107:2825–2837. [DOI] [PubMed] [Google Scholar]
- Llanos, R., V. Chevrier, M. Ronjat, P. Meurer-Grob, P. Martinez, R. Frank, M. Bornens, R.H. Wade, J. Wehland, and D. Job. 1999. Tubulin binding sites on γ-tubulin: identification and molecular characterization. Biochemistry. 38:15712–15720. [DOI] [PubMed] [Google Scholar]
- Loïodice, I., J. Staub, T. Gangi Setty, N. Nguyen, A. Paoletti, and P.T. Tran. 2005. Ase1p organizes anti-parallel microtubule arrays during interphase and mitosis in fission yeast. Mol. Biol. Cell. 16:1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima, M., S. Kaitna, and M. Glotzer. 2002. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2:41–54. [DOI] [PubMed] [Google Scholar]
- Moudjou, M., N. Bordes, M. Paintrand, and M. Bornens. 1996. γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109:875–887. [DOI] [PubMed] [Google Scholar]
- Nislow, C., V.A. Lombillo, R. Kuriyama, and J.R. McIntosh. 1992. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 359:543–547. [DOI] [PubMed] [Google Scholar]
- Rebollo, E., S. Llamazares, J. Reina, and C. Gonzalez. 2004. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. Plos Biol. 2:54–64; 10.1371/journal.pbio.0020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios, R.M., A. Sanchis, A.M. Tassin, C. Fedriani, and M. Bornens. 2004. GMAP-210 recruits γ-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell. 118:323–335. [DOI] [PubMed] [Google Scholar]
- Sagolla, M.J., S. Uzawa, and W.Z. Cande. 2003. Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J. Cell Sci. 116:4891–4903. [DOI] [PubMed] [Google Scholar]
- Samejima, I., P.C. Lourenco, H.A. Snaith, and K.E. Sawin. 2005. Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol. Biol. Cell. 10.1091/mbc.E04-11-1003. [DOI] [PMC free article] [PubMed]
- Sawin, K.E., P.C.C. Lourenco, and H.A. Snaith. 2004. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14:763–775. [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., and H.A. Snaith. 2004. Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J. Cell Sci. 117:689–700. [DOI] [PubMed] [Google Scholar]
- Shaw, S.L., R. Kamyar, and D.W. Ehrhardt. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 300:1715–1718. [DOI] [PubMed] [Google Scholar]
- Siam, R., W.P. Dolan, and S.L. Forsburg. 2004. Choosing and using Schizosaccharomyces pombe plasmids. Methods. 33:189–198. [DOI] [PubMed] [Google Scholar]
- Stearns, T., and M. Kirschner. 1994. In-vitro reconstitution of centrosome assembly and function—the central role of γ-tubulin. Cell. 76:623–637. [DOI] [PubMed] [Google Scholar]
- Surrey, T., F. Nedelec, S. Leibler, and E. Karsenti. 2001. Physical properties determining self-organization of motors and microtubules. Science. 292:1167–1171. [DOI] [PubMed] [Google Scholar]
- Tassin, A.M., B. Maro, and M. Bornens. 1985. Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, P.T., V. Doye, F. Chang, and S. Inoue. 2000. Microtubule-dependent nuclear positioning and nuclear-dependent septum positioning in the fission yeast Schizosaccharomyces pombe. Biol. Bull. 199:205–206. [DOI] [PubMed] [Google Scholar]
- Tran, P.T., L. Marsh, V. Doye, S. Inoue, and F. Chang. 2001. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 153:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, J.B., M.J. Milner, D.A. Currie, J.W. Muir, D.A. Forrest, and M.J. Spencer. 1986. Centrosomal microtubule-organizing centers and a switch in the control of protofilament number for cell surface-associated microtubules during Drosophila wing morphogenesis. Eur. J. Cell Biol. 41:279–289. [Google Scholar]
- Tulu, U.S., N.M. Rusan, and P. Wadsworth. 2003. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr. Biol. 13:1894–1899. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., K. Van Poucke, E. Boutant, C. Ritzenthaler, D. Inze, and D. Geelen. 2004. In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136:3956–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., and T. Toda. 2000. The fission yeast γ-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19:6098–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatram, S., J.J. Tasto, A. Feoktistova, J.L. Jennings, A.J. Link, and K.L. Gould. 2004. Identification and characterization of two novel proteins affecting fission yeast γ-tubulin complex function. Mol. Biol. Cell. 15:2287–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev, I., V. Malikov, and V. Rodionov. 2001. Self-organization of a radial microtubule array by dynein-dependent nucleation of microtubules. Proc. Natl. Acad. Sci. USA. 98:10160–10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.A., E.T. Obrien, N.K. Pryer, M.F. Soboeiro, W.A. Voter, H.P. Erickson, and E.D. Salmon. 1988. Dynamic instability of individual microtubules analyzed by video light-microscopy—rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., A. Desai, J.C. Bulinski, and E.D. Salmon. 1998. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8:1227–1230. [DOI] [PubMed] [Google Scholar]
- Wiese, C., and Y.X. Zheng. 2000. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2:358–364. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., M. Sato, A. Fujita, M. Yamamoto, and T. Toda. 2005. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol. Biol. Cell. 16:1378–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C.J., and W. Jiang. 2005. Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc. Natl. Acad. Sci. USA. 102:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, S., P.T. Tran, R.R. Daga, O. Niwa, and F. Chang. 2004. Rsp1p, a J domain protein required for disassembly and assembly of microtubule organizing centers during the fission yeast cell cycle. Dev. Cell. 6:497–509. [DOI] [PubMed] [Google Scholar]