Abstract
Wound healing of mammalian tissue is an essential process in the maintenance of body integrity. The general mechanism of wound healing usually studied in adult mammals is repair, in contrast to the regeneration seen in more primitive vertebrates. We recently have discovered that MRL/MpJ mice, unlike all other strains of mice tested, undergo rapid and complete wound closure that resembles regeneration. Specifically, through-and-through surgical ear hole wounds close without scarring in <4 weeks with normal gross and microanatomic architecture, including chondrogenesis. We also demonstrated that this healing is a heritable trait in inbred mice. In this study, we present results pertaining to its genetic control in progeny segregating for this phenotype. To identify the genetic loci that control the wound closure process, a genome-wide scan was performed on (MRL/MpJ-Faslpr × C57BL/6)F2 and backcross populations. In the primary screens of these populations, quantitative trait loci that control the extent of wound closure were detected on chromosomes 8, 12, and 15 and at two separate locations on chromosome 13. Evidence of further genetic control of healing was found on chromosome 7. All alleles that contribute to full wound closure are derived from the MRL/MpJ-Faslpr parent except for the quantitative trait locus on chromosome 8, which is derived from C57BL/6.
The biological response to tissue injury in higher organisms falls into two main categories: wound repair and regeneration (1). In amphibians, the form of wound healing seen is often epimorphic regeneration, where entire limbs can be reformed after amputation (1). In adult mammals, the form of healing seen most often is wound repair or tissue regeneration, accomplished by the replacement of mature cells through cell proliferation (2) or the replenishment of cells, but not organs, from immature stem cells (3–5). There are, however, several examples of epimorphic regeneration that exist in mammals. These include the replacement of antlers (6) and the closure of ear holes, originally described in the rabbit (7, 8), where a through-and-through hole placed in the ear is healed to completely normal tissue.
Ear hole closure in the rabbit is considered to be the result of regeneration and not wound repair because there is the replacement of multiple tissues and there is perfect healing (9). Unlike wound repair, contracture of skin cannot occur in this model because ear skin is connected tightly to the cartilage and there is no wound bed caused by the through-and-through nature of this wound. In addition, epithelial “crawling” and, hence, the normal sequence of repair processes cannot occur or is diminished greatly across the hole because there can be no provisional matrix formed by fibrin clots, fibroblasts, extracellular matrix, and granulation tissue. What does occur in this ear hole model is the formation of a wound blastema resembling that seen in amphibian wounds (9), leading to closure of the hole and regrowth of cartilage, phenomena not generally seen elsewhere in mammals.
The ability of the mouse strain MRL/MpJ to heal a through-and-through ear hole wound like the rabbit has been described recently (10). The method used to reveal this phenotype, ear hole punching, is a standard technique for identifying mice by number in the animal colony and usually has lifelong use; in our experience, no other mouse strain tested has the capability of healing this identification mark. MRL/MpJ-Faslpr mice, better known for their inherited susceptibility to autoimmune disorders (11–14), are also capable of completely closing 2.1-mm surgical holes in their ears within 30 days whereas all other mice tested showed no closure when treated in the same way. Moreover, the type of healing seen in MRL/MpJ mice is reminiscent of that of the classical model of regeneration reported in rabbits. What is remarkable in both rabbits and MRL/MpJ mice is that their ear hole closures not only display full healing but also show the recovery of normal architecture, collagen structure, angiogenesis, the appearance of hair follicles and sebaceous glands and cartilage, and the lack of scarring (10). In many ways, the closure of ear holes resembles what is seen during mammalian development and neonatal wounding more than it resembles adult wound healing.
To determine the genetic elements involved in wound closure, the two parental phenotypes must be significantly different, the mode of inheritance in the F1 hybrid must be clear, and the segregating progeny must be discriminated easily for this trait. These conditions have been met well by investigations of wound healing in the two inbred mouse strains we have used. In the present study, DNA-based microsatellite markers were screened on the parental strains, MRL/MpJ-Faslpr (superhealer) and C57BL/6 (normal, poor healer), and those that exhibited polymorphism were used to analyze F2 and backcross progeny derived from matings of these two strains. The healing capacity of progeny animals was tested after the placement of standard (2.1-mm) holes in the ears, and the size of the hole remaining at day 30 was measured and used as a quantitative trait for analysis with respect to the inheritance of the microsatellite markers. We report our initial results that loci on chromosomes 7, 8, 12, 13, and 15 play a role in the regeneration process.
MATERIALS AND METHODS
Mice.
The parental strain MRL/MpJ-Faslpr (hereafter referred to as MRL/lpr) was obtained from The Jackson Laboratory, and the C57BL/6 parental strain was acquired from Taconic Farms. Both parents were bred and maintained under standard conditions at The Wistar Institute (Philadelphia, PA). F1, F2, and MRL/lpr backcross populations were generated to conduct the genetic studies. The female parent used for generating the F2 and backcross animals was MRL/lpr. Alleles derived from the C57BL/6 parent are designated B, and the MRL/lpr-derived alleles are designated S in this report.
Phenotyping.
At 6 weeks of age all mice were ear-punched, resulting in a 2.1-mm diameter hole through the center of both ears. Measurements of the hole diameter were taken at day 14 and at endpoint day 30. An average between the hole diameter of the right and left ear then was used for any quantitative trait analysis. At this time, animals were killed, and livers were frozen at −70°C.
Genetic Analysis.
Genomic DNA was prepared from the liver of each animal in the (MRL/lpr × C57BL/6) × (MRL/lpr × C57BL/6) F2 population. These preparations were performed by homogenization of the frozen tissue followed by an overnight proteinase K (100 mg/ml) digestion. Samples were treated with two phenol:chloroform extractions and one final chloroform extraction. Finally, genomic DNA was purified by an overnight dialysis against Tris·EDTA buffer. PCR primers, purchased from Research Genetics (Huntsville, AL), were used to perform a genome-wide scan of the mouse. Amplification was conducted by using Boehringer Mannheim reagents with the following concentrations: 1× PCR buffer, 0.375 mM dNTPs, 0.5 units/μl of Taq polymerase, 0.165 μM of each primer, and 160 ngm/20 μl of genomic DNA. Cycling conditions include a 1 min at 95°C denaturing, 35 to 50 cycles of 1 min at 94°C, 1 min 30 sec at 55°C, 2 min 10 sec at 72°C, and a 6 min final extension at 72°C. PCR products were resolved by using 3% Metaphor agarose (FMC) and were visualized through ethidium bromide staining. This method was followed for the majority of polymorphic markers. In the case of small base pair differences, PCR amplification was carried out by using [32P]ATP labeled forward primers as described (15). Radiolabeled PCR products then were size fractionated on 6% denaturing polyacrylamide gels.
Statistics.
Genotype data was organized and analyzed through the use of map manager qt (16). For quantitative trait analysis, critical threshold values for significance of linkage were determined by the permutation test, which was based on a regression model developed by Churchill and Doerge (17, 18). The values for the additive model of inheritance were calculated in terms of a likelihood ratio statistic (LRS). The threshold in the F2 under assumptions of the additive model for suggestive linkage is LRS > 3.3 and for significant linkage is LRS > 10.7. The dominant, free, and recessive models also were tested and did not show a significant difference (i.e., they were <18-fold different) in resultant P values. The additive regression model was used because it is the simplest model and because it is consistent with the mode of inheritance of the quantitative trait loci (QTL) determined in this study. It also should be noted that the use of the additive regression model does not assume the pattern of inheritance to be strictly additive, and at present the degree of additivity and dominance has not been determined. The threshold values in the backcross are 3.7 and 11.8, respectively. Loci were named as healing QTL if they independently attained significance in either cross or a suggestive level of significance in one cross, confirmed in the other (P < 0.05). Microsatellite markers were evaluated individually for linkage to the healing phenotype, based on the threshold values. In addition, a mean healing score for markers closely linked to each healing QTL was calculated independently of other loci by using ANOVA, using a Bonferroni/Dunn post hoc test for making the three possible pairwise comparisons in the F2, resulting in single-locus genotypic values (see Table 3) (22).
Table 3.
Single locus genotypic values for heal1 to heal5
| D8Mit211 (heal1) | D13Mit115 (heal2) | |||||
|---|---|---|---|---|---|---|
| Genotype | Average | SD | P* | Average | SD | P |
| b/b | 0.73 | 0.27 | 0.0319 | 0.96 | 0.25 | 0.0904 |
| b/s | 0.84 | 0.27 | 0.1214 | 0.84 | 0.29 | 0.0567 |
| s/s | 0.95 | 0.28 | 0.0013*** | 0.72 | 0.26 | 0.0024*** |
| D13Mit129 (heal3) | D15Mit244 (heal4) | |||||
| Average | SD | P | Average | SD | P | |
| b/b | 0.96 | 0.28 | 0.2156 | 0.95 | 0.19 | 0.0485 |
| b/s | 0.84 | 0.22 | 0.141** | 0.86 | 0.30 | 0.0677 |
| s/s | 0.74 | 0.34 | 0.0022*** | 0.70 | 0.27 | 0.0013*** |
| D12Mit132 (heal5) | ||||||
| Average | SD | P | ||||
| b/s | 0.51 | 0.40 | 0.0005*** | |||
| s/s | 0.15 | 0.32 | ||||
Averages and SDs for wound diameters at day 30 are given for mice sorted by the genotype of markers near each healing QTL.
P values in the F2 are listed vertically for b/b vs. b/s, s/s vs. b/s, or b/b vs. s/s; in the backcross, P values are for the heterozygote b/s vs. the homozygote s/s. Genotype values are significant at α = 0.05 (**) or α = 0.01 (***) in post hoc analyses.
RESULTS
The Pattern of Inheritance of the Wound Healing Trait.
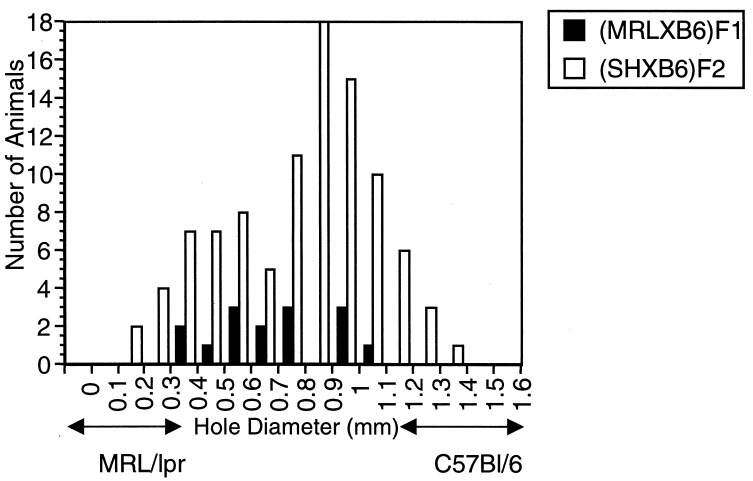
All MRL mice quickly and effectively close the wounds in their ears; C57BL/6 (B6) mice are unable to completely close their wounds (10). The F1 has an intermediate phenotype, with considerable variability (Fig. 1). Like the F1, the (MRL/lpr × B6)F2 population demonstrates a bell-shaped curve of healing diameters (Fig. 1). The backcross (BC1) population to MRL/lpr [(MRL/lpr × B6)F1 × MRL/lpr] displays a curve skewed to MRL/lpr-type healing whereas, in the backcross population to B6 [(MRL/lpr × B6)F1 × B6], the progeny show a mean displaced to B6-type (i.e., poor) healing (10).
Figure 1.
Histogram of wound closure in (MRL/lpr × C57BL/6)F1 and F2 intercross populations.
The healing profiles of each of the populations used in this study are given in Table 1. The distribution of the variance in each of these populations compared with those of the parental and F1 mice were used to give a rough estimate of the number of unlinked genes that contribute to this quantitative trait (19, 20). From this calculation, it is likely that, at minimum, three to four unlinked loci (QTLs) have an impact on the healing trait in this strain combination.
Table 1.
Residual wound size
| Parental strains and hybrid | Backcross and intercross progeny | ||
|---|---|---|---|
| C57BL/6 | 1.10 ± 0.27 | F1 × MRL/lpr | 0.35 ± 0.27 |
| MRL//lpr | 0.04 ± 0.06 | F1 × B6 | 0.95 ± 0.25 |
| (MRL × B6)F1 | 0.73 ± 0.22 | (MRL × B6)F2 | 0.83 ± 0.28 |
Numbers are the diameter of holes in millimeters ± SD on day 28–30. F1, (MRL × C57BL/6)F1 hybrid.
Mapping of Simple Sequence Length Polymorphism Markers and Significant Threshold Values.
A total of 436 simple sequence length polymorphism markers were tested for potential polymorphisms between the two strains of mice. Ninety-two markers detected allelic variants in the parental strains and therefore were used for genotyping of the segregating populations. Markers were chosen based on their location in the genome in an attempt to make a linkage map with an even spacing of 20 centimorgans between markers to generate a complete genome-wide scan (21). In regions in which linkage to the healing trait was detected, the density of markers was increased to obtain a more accurate genetic dissection in the area of interest. Overall, genomic coverage reached ≈97.7 percent across the 19 autosomes. Surprisingly, despite the distant genetic relationship between these two inbred strains (11, 12), no polymorphisms were found on the X chromosome (0/18 primers tested; data not shown).
The 92 polymorphic microsatellite markers then were used to map the wound healing/regeneration trait in 101 mice from the (MRL/lpr × C57BL/6)F2 intercross. For assessment of the probability of genetic linkage, critical values were calculated from this database by using the permutation test (17) (see Materials and Methods). The order of all markers in this linkage analysis was consistent with the order predicted by the available genomic maps (Whitehead Institute, Massachusetts Institute of Technology, and Mouse Genome Database (MGD), The Jackson Laboratory: http://www.informatics.jax.org).
Mapping of Quantitative Trait Loci Linked to the Healing Phenotype in the F2 Population.
Table 2 shows all of the microsatellite markers that were positive for linkage to the healing phenotype in two crosses, and Table 3 lists the healing scores for markers associated with wound closure. In the F2 cross, QTLs that contribute to the healing phenotype and are derived from MRL/lpr exist in three primary support intervals. Two of these QTLs are located on individual sites on chromosome 13 and are designated heal2 and heal3. These QTLs are located as follows (with their corresponding microsatellite marker): heal2, on proximal chromosome 13, near D13Mit115 (P = 0.0019) and heal3, at a more distal location near D13Mit129 (P = 0.0010). One of the two QTLs on chromosome 13 (heal3) has achieved significant likelihood of linkage to the healing trait whereas the other (heal2) is suggestive, though confirmed in a second cross (see below). Multiple markers near these loci show a suggestive level of significance, including, for heal2, D13Mit135 and D13Mit116 and, for heal3, D13Mit53, D13Mit151, D13Mit144, and D13Mit107 (Table 2). D13Nds1 and D13Mit191 both achieved highly suggestive LRS values, but the assignment of a QTL in this region is provisional at the present time because the 95% confidence intervals for these markers and the two flanking QTLs overlap (not shown). Nevertheless, there are distinct breaks between these QTLs in the level of significance for their linkage to healing with no deviation from the predicted order of these markers. A second region that contains a QTL with significant linkage to the healing phenotype was detected on chromosome 15, in the region of marker D15Mit244 (P = 0.0011). Other microsatellite markers mapping to this location that meet the criteria for suggestive linkage to wound healing include D15Mit172 and D15Mit14. We have designated this QTL heal4.
Table 2.
Location of heal QTL as determined in the intercross (F2) and the backcross (BC1) progeny
| Mouse Genome Database, centimorgans | Primers | F2 LRS* | F2 P value | BC1 LRS† | BC1 P value | QTL |
|---|---|---|---|---|---|---|
| 71.5 | D1Mit288 | 6.1 | 0.0130 | ns | ||
| 105 | D2Mit148 | nd | 4.8 | 0.0279 | ||
| 55.6 | D4Nds2 4.2 | 4.2 | 0.0400 | ns | ||
| 77.5 | D4Mit127 | 5.5 | 0.0190 | ns | ||
| 52.4 | D7Mit220 | ns | 10.2‡ | 0.0014 | ||
| 21 | D8Mit191 | ns | 6.4 | 0.0115 | ||
| 33 | D8Mit132 | 5.5 | 0.0188 | ns | ||
| 37 | D8Mit249 | 7.3 | 0.0069 | ns | ||
| 49 | D8Mit211 | 10.7 | 0.0011 | ns | heal1‡ | |
| 56 | D8Mit166 | 7.3 | 0.0068 | nd | ||
| 34 | D12Mit4 | ns | 7.1 | 0.0077 | ||
| 52 | D12Mit233 | 6.1 | 0.0137 | 9.4 | 0.0022 | |
| 52 | D12Mit132 | nd | 10.9 | 0.0009 | heal5ठ| |
| 9 | D13Mitl35 | 9.4 | 0.0022 | 4.8 | 0.0290 | |
| 11 | D13Mit115 | 9.7 | 0.0019 | 5.0 | 0.0261 | heal2ठ|
| 13 | D13Mitll6 | 8.7 | 0.0030 | 5.1 | 0.0230 | |
| 30 | D13Mit245 | 7.5 | 0.0335 | nd | ||
| 32 | D13Nds1 | 10.2‡ | 0.0014 | ns | ||
| 44 | D13Mit126 | 7.4 | 0.0065 | ns | ||
| 44 | D13Mit191 | 10.2‡ | 0.0014 | ns | ||
| 47 | D13Mit29 | 6.5 | 0.0101 | nd | ||
| 48 | D13Mit107 | 7.4 | 0.0064 | ns | ||
| 49 | D13Mit144 | 6.9 | 0.0088 | nd | ||
| 60 | D13Mit129 | 10.8 | 0.0010 | 8.3 | 0.0040 | heal3ठ|
| 62 | D13Mit53 | 10.5 | 0.0012 | 7.1 | 0.0077 | |
| 71 | D13Mit151 | 8.2 | 0.0042 | 4.6 | 0.0318 | |
| 54.5 | D15Mit171 | 6.6 | 0.0104 | nd | ||
| 55.6 | D15Mit242 | 7.1 | 0.0079 | nd | ||
| 57.8 | D15Mit172 | 7.3 | 0.0070 | nd | ||
| 56.8 | D15Mit244 | 10.7 | 0.0011 | nd | heal4‡ | |
| 56.8 | D15Mit14 | 8.5 | 0.0035 | ns |
Underlining indicates the LRS values that were used for assigning heal QTL. ns, not significant; nd, not determined.
Threshold LRS for significant linkage = 10.7 and for suggestive linkage = 3.3.
Threshold LRS for significant linkage = 11.8 and for suggestive linkage = 3.7.
See text for discussion.
Confirmed in second cross.
In addition to the QTL on chromosome 13 and chromosome 15, which have MRL/lpr-derived healing alleles, a B6-derived healing QTL was mapped to chromosome 8, near the marker D8Mit211, with a significant LRS value (10.7; P = 0.0011). Other markers in this location that showed linkage were D8Mit132, D8Mit166, and D8Mit249. This locus was the first QTL identified and was designated heal1.
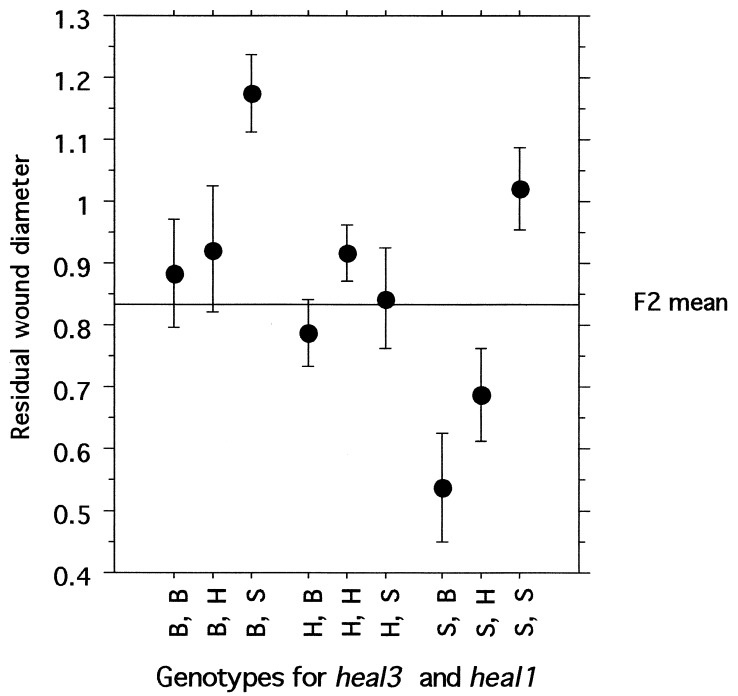
The contribution of each heal locus to the process of wound closure, as expressed by the single-locus genotypic values (22) (Table 3), generally fit an additive mode of inheritance, with the heterozygote healing score approximately halfway between the scores of the two homozygotes. One exception is the heal3 QTL, which may be recessive (i.e., heal3s/s homozygotes show significantly better wound closure than either heterozygotes or heal3b/b homozygotes). In addition, the heal3 QTL appears to interact with heal1 to give the most completely healed ear holes (ANOVA, P = 0.017). The average residual wound in heal1b/b homozygotes is 0.73 + 0.27; however, in animals that are both heal1b/b and heal3s/s, residual wound size is 0.53 + 0.30 (Fig. 2). Conversely, in mice homozygous for heal1s/s but also homozygous for heal3b/b, the residual wound size is 1.2 + 0.22. Other pairs of heal QTLs also show these largely additive interactions but do not attain statistical significance.
Figure 2.
Additive effects of heal1 and heal3 on wound closure. Average residual wound diameters are plotted for each genotype, with the results grouped by D13Mit129 and D8Mit211. The mean of all F2 mice ± 1 SEM is depicted as a horizontal line in this graph. B, H, and S designate mice homozygous for the healb/b allele from C57BL/6, heterozygous for the healb/s alleles from C57BL/6 and MRL/lpr, or homozygous for the heals/s allele from MRL/lpr, respectively.
Mapping of Quantitative Trait Loci Associated with the Healing Phenotype in the Backcross.
To confirm the linkage assignments seen in the F2, we conducted a small backcross study (42 mice) by using (MRL/lpr × B6)F1 females and MRL/lpr males as parents. The F1 between MRL/lpr and B6 has an intermediate wound closure phenotype, and all progeny in this cross were expected to show intermediate to good healing. In fact, this was largely the case, although several mice in the backcross had poor wound healing. An analysis of this cross showed linkage to healing at locations that coincided with the supported intervals of association seen in the F2 for both of the two QTLs on chromosome 13 (heal2 and heal3). These QTLs were near markers D13Mit115 (LRS value = 5.0, P = 0.0261) and D13Mit129 (LRS value = 8.3, P = 0.0040), respectively (Table 2). In addition, linkage to the healing phenotype also was detected on chromosome 12 (heal5) at marker D12Mit132 with a LRS value of 10.9 (P = 0.0009) and at the closely linked marker, D12Mit233 (LRS = 9.4, P = 0.0022). This linkage was supported by suggestive linkage in the F2. The single locus genotypic value for residual wound diameter for the heal5 (D12Mit132) QTL also is given in Table 3. Finally, a locus showing a highly suggestive LRS value of 10.2 (P = 0.0014) was found on chromosome 7 near marker D7Mit220.
DISCUSSION
The MRL mouse model of wound healing and regeneration is amenable to genetic mapping by using genome exclusion methods (15). A complete genome scan was carried out to map the QTL that control healing in MRL/lpr-derived F2 and backcross progeny. Our results revealed several major features of this model system. First, there are at least five unlinked genes that can contribute to the healing phenotype in two different types of genetic crosses. Three of the QTL independently attained significance in the F2 (heal1, heal3, and heal4) and one was confirmed in the backcross (heal5). The fifth QTL linked to healing achieved significance in the backcross. Second, the wound-healing trait is quantitative, and the pattern of inheritance of most QTLs was largely additive, with intermediate phenotypes that span a linear range between the healer and the nonhealer parents. Third, the separate QTL can interact with one another in an additive fashion to effect more complete wound closure.
The Mouse Genome Database and associated literature were searched for potential candidates near the heal QTL (Table 4). The first locus, heal1, located on chromosome 8, is derived from C57BL/6 and is one of the strongest QTL of the five (P = 0.0011). In the absence of contributing genes from MRL, C57BL/6 mice clearly cannot accomplish complete wound closure with heal1 alone. It has been shown that alleles that contribute to a trait from a parental strain that does not display that trait are not uncommon (23). Thus, it is not surprising that heal1 shows a strong additive effect with MRL loci in the F2 (see Fig. 2). Candidate genes for heal1 (Table 4) in the strongest-supported interval include the guanine nucleotide binding protein α 0, Gnao, an α subunit of a heterotrimeric G-protein that interacts with an activated G protein coupled receptor, preceding downstream signaling (24). It is interesting to speculate that the basis of the additive interaction of heal1 with heal3 is an interaction between the gene products encoded by Gnao and the G protein coupled receptor 18 found in the heal3 interval.
Table 4.
Candidate genes in genomic intervals containing QTLs
| OTL | Mouse Genome Database, centimorgans | Candidate genes in interval |
|---|---|---|
| heal1 | 33 | Comp, cartilage oligomeric matrix protein |
| 39 | pdw, proportional dwarf | |
| 42 | Os, oligosyndactylism | |
| 46 | Gna0 | |
| 51.5 to 67 | Cadherin family | |
| heal2 | 7 | Nid, nidogen |
| 8 | Gli3, GLI-Kruppel family member GL13 | |
| 10 | Amph, amphiphysin | |
| 10 | Inhba, inhibin beta-A | |
| 10 | Rasl1, Ras-like, family 1 | |
| heal3 | 32 | Msx2, hox8 |
| 32.5 | Fgfr4, fibroblast growth factor receptor | |
| 33 | mes, mesenchymal dysplasia | |
| 36 | Tgfbi, transforming growth factor induced | |
| 44 | Cspg2, chondroitin sulfate proteoglycan | |
| 45 | Rasa, ras p21 GTPase activating protein | |
| 56 | Gpcrl8, G-protein coupled receptor 18 | |
| 62 | Itga 1,2, integrin alpha 2 (Cd49b) | |
| heal4 | 51.6 | Pdgfec, platelet derived growth factor |
| 56.8 | Col2a1, procollagen, type 11, alpha 1 | |
| 56.8 | Ela1, elastase 1 | |
| 57 | Emb, embigin | |
| 57.1 | Hoxc, homeo box C cluster | |
| 57.1 | Rarg, retinoic acid receptor, gamma | |
| 57.5 | Dhh, desert hedgehog homolog | |
| 58.7 | Krt2, keratin gene complex 2 | |
| 60 | Itga5, integrin alpha 5 | |
| 61.1 | Itgb7, integrin beta 7 | |
| 63 | Glycam 1 adhesion molecule | |
| heal5 | 40 | Fos, FBJ osteosarcoma oncogene |
| 41 | Tgfb3, transforming growth factor, beta | |
| 44.6 | Chx10, C elegans ceh-10 homeo domain con | |
| 45 | Pgf, placental growth factor |
Though the method used for determining QTL cannot separate multiple loci in the same linkage group, we have observed highly suggestive LRS values for two regions of chromosome 13 in addition to the two QTLs identified (Table 2). That these may be unique QTLs is supported by the fact that they are separable from each other in phenotype congenics that have been generated (data not shown). One of these regions is located on chromosome 13, near marker D13nds1. Msx2 (also known as Hox8) is found in this interval and is expressed in regenerating amphibian tissue (25) as well as regrowing fingertips in neonatal mice (26). In this regard, we have evidence that Msx2 expression is different in healing ear tissue between MRL and B6 mice (S. Samulewicz, X.-M.Z., and E.H.-K., unpublished data ). This difference could be caused by a polymorphism in the Msx2 gene itself or by the indirect effect of a fibroblast growth factor signaling difference (27) mediated through the fibroblast growth factor receptor, FGFR4 (28), which also is encoded by a candidate gene located in this interval.
Heal4 is found on chromosome 15 near marker D15Mit244. The chromosome 15 QTL is strongly associated with the wound closure trait (P = 0.0011) and is located in an area rich in candidate genes, including the gene encoding retinoic acid receptor gamma (Rarg), members of the keratin family that influence differentiation in the epidermis, as well as developmental genes known as homeobox and wnt genes. The retinoic acid pathway is known for its role in regeneration in amphibians (29–31). Furthermore, the gamma subtype of the RAR displays preferential expression over the α and β subtypes in skin and cartilage tissues (32), which is the site where the MRL/lpr healing trait is evaluated in the present study.
Finally, this set of genes does not include the fas gene, H-2, or any other gene known to play a role in the autoimmune profile of MRL/lpr mice. Several lines of evidence support this. First, previous findings showed that the MRL/MpJ mice heal similarly to MRL/MpJ-Faslpr mice (10). Secondly, intervals containing wound-healing genes in our crosses showed no overlap with those from another report on the genetic analysis of MRL/lpr autoimmune phenotypes (33). Third, we tested the lymph node cell number, a parameter associated with lymphoproliferation, of each of 101 F2 mice in a regression comparison with their healing phenotype, and no association was found with the healing trait (r2 = 0.0002, P = 0.89) (E.P.B., L.D.C., and E.H.-K., unpublished data).
The MRL/lpr mouse strain originally was selected for its large size (11, 12); it subsequently was found to have a major defect in immune regulation caused by a retrotransposon insertion into the second intron of the fas gene (34, 35). This mouse exhibits immunological defects closely mimicking those of the human disease systemic lupus erythematosus and other lymphoproliferative disorders (13, 14). In the present study, however, none of the five healing QTLs nor the highly suggestive regions identified displays linkage to the fas gene or other genes proposed thus far to control the other autoimmune phenotypes seen in this strain of mouse.
Acknowledgments
We thank Sarah Schuler for her assistance and James Handelman for his enthusiastic support. This work was supported generously by the G. Harold and Leila Y. Mathers Charitable Foundation.
ABBREVIATIONS
- LRS
likelihood ratio statistic
- QTL
quantitative trait locus
References
- 1. Gross J. Wound Repair Regeneration. 1996;4:190–202. doi: 10.1046/j.1524-475X.1996.40205.x. [DOI] [PubMed] [Google Scholar]
- 2.Michalopoulos G K, DeFrances M C. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 3.Spangrude G J, Heimfeld S, Weisman I L. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 4.Potten C S, Morris R J. In: Stem Cells. Lord B I, Dexter T M, editors. Cambridge: Company of Biologists; 1988. pp. 45–62. [Google Scholar]
- 5.Clark R A F. The Molecular and Cellular Biology of Wound Repair. New York: Plenum; 1996. pp. 3–35. [Google Scholar]
- 6.Goss R J. Clin Orthop. 1970;69:227–238. [PubMed] [Google Scholar]
- 7.Joseph J, Dyson M. Brit J Surg. 1966;53:372–380. doi: 10.1002/bjs.1800530415. [DOI] [PubMed] [Google Scholar]
- 8.Goss R J, Grimes L N. J Morphol. 1975;146:533–542. doi: 10.1002/jmor.1051460408. [DOI] [PubMed] [Google Scholar]
- 9.Stocum D L. Differentiation. 1984;27:13–28. doi: 10.1111/j.1432-0436.1984.tb01403.x. [DOI] [PubMed] [Google Scholar]
- 10.Clark L D, Clark R K, Heber-Katz E. Clin Immunol Immunopathol. 1998;88:35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- 11.Murphy E D, Roths J B. In: Genetic Control of Autoimmune Disease. Rose N R, Bigazzi P E, Warner N L, editors. New York: Elsevier; 1979. pp. 207–220. [Google Scholar]
- 12.Murphy E D. In: Immunological Defects in Laboratory Animals. Gershwin M E, Merchant B, editors. Vol. 2. New York and London: Plenum; 1981. pp. 143–73. [Google Scholar]
- 13.Cohen P L, Eisenberg R A. Annu Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- 14.Theofilopoulos A N. In: The Molecular Pathology of Autoimmune Diseases. Bona C, Siminovitch K A, Zanetti M, Theofilopoulos A N, editors. Chur, Switzerland: Harwood; 1993. pp. 281–316. [Google Scholar]
- 15.Dietrich W F, Miller J C, Steen R G, Merchant M, Damron D, Nahf R, Gross A, Joyce D C, Wessel M, Dredge R D, et al. Nat Genet. 1994;7:220–245. doi: 10.1038/ng0694supp-220. [DOI] [PubMed] [Google Scholar]
- 16.Manly K F. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- 17.Doerge R W, Churchill G A. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churchill G A, Doerge R W. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright S. In: Quantitative Genetics. Reeve E C, Waddington C H, editors. London: Her Majesty’s Stationery Office; 1952. pp. 5–41. [Google Scholar]
- 20.Hartl D L. Basic Genetics. Boston: Jones and Bartlett; 1991. pp. 218–220. [Google Scholar]
- 21.Lander E S, Krugylak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 22.Cheveraud J M, Routman E J. Genetics. 1995;139:1455–1461. doi: 10.1093/genetics/139.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel W. Trends Genet. 1995;11:471–477. doi: 10.1016/s0168-9525(00)89155-6. [DOI] [PubMed] [Google Scholar]
- 24.Simon M I, Strathmann M P, Gautum N. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 25.Bryant S V, Gardinar D M. In: Metamorphosis and Regeneration: Keys to Tissue Regeneration. Stocum D, editor. Indianapolis: Indiana Univ. Press; 1997. p. 12. [Google Scholar]
- 26.Reginelli A D, Wang Y-Q, Sassoon D, Muneoka K. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- 27.Chuong C M, Widelitz R B, Ting-Berreth S, Jiang T X. J Invest Dermatol. 1996;107:639–646. doi: 10.1111/1523-1747.ep12584254. [DOI] [PubMed] [Google Scholar]
- 28.Ron D, Reich R, Chedid M, Lengel C, Cohen O E, Chan A M, Neufeld G, Miki T, Tronick S R. J Biol Chem. 1993;268:5388–5394. [PubMed] [Google Scholar]
- 29.Ragsdale C W, Gates P, Hill D S, Brockes J P. Mech Dev. 1992;40:99–112. doi: 10.1016/0925-4773(93)90091-b. [DOI] [PubMed] [Google Scholar]
- 30.Percrino L T, Lo D C, Brockes J P. Development. 1994;20:325–333. doi: 10.1242/dev.120.2.325. [DOI] [PubMed] [Google Scholar]
- 31.Chernoff E A G, Stocum D. Dev Growth Diff. 1995;37:133–147. doi: 10.1046/j.1440-169X.1995.t01-1-00002.x. [DOI] [PubMed] [Google Scholar]
- 32.Sucov H M, Evans R M. Mol Neurobiol. 1995;10:169–184. doi: 10.1007/BF02740674. [DOI] [PubMed] [Google Scholar]
- 33.Watson M L, Rao J K, Gilkeson G S, Ruiz P, Eicher E M, Pisetsky D S, Matsuzawa A, Rochelle J M, Seldin M F. J Exp Med. 1992;176:1645–1656. doi: 10.1084/jem.176.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah M, Foreman D M, Ferguson M W J. J Cell Sci. 1995;108:15–17. doi: 10.1242/jcs.108.3.985. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe-Fukunaga R, Brannan C, Copeland N G, Jenkins N A, Nagata S. Nature (London) 1992;356:314–316. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]