Abstract
Autophagy is a membrane-trafficking mechanism that delivers cytoplasmic constituents into the lysosome/vacuole for bulk protein degradation. This mechanism is involved in the preservation of nutrients under starvation condition as well as the normal turnover of cytoplasmic component. Aberrant autophagy has been reported in several neurodegenerative disorders, hepatitis, and myopathies. Here, we generated conditional knockout mice of Atg7, an essential gene for autophagy in yeast. Atg7 was essential for ATG conjugation systems and autophagosome formation, amino acid supply in neonates, and starvation-induced bulk degradation of proteins and organelles in mice. Furthermore, Atg7 deficiency led to multiple cellular abnormalities, such as appearance of concentric membranous structure and deformed mitochondria, and accumulation of ubiquitin-positive aggregates. Our results indicate the important role of autophagy in starvation response and the quality control of proteins and organelles in quiescent cells.
Introduction
There are two major protein degradation pathways in eukaryotic cells: the proteasome and the lysosome. The proteasome is a self-compartmentalized protease complex with catalytic activities inside its central proteinaceous chamber (Baumeister et al., 1998). It plays crucial roles in selective degradation of not only short-lived regulatory proteins but also abnormal proteins that should be eliminated from the cells (Goldberg, 2003). In contrast, the lysosome is a vesicle that contains many hydrolases, which are separated from the cytosol by the limiting membrane. In this lysosomal pathway, degradation of plasma membrane proteins and extracellular proteins is mediated by endocytosis, whereas degradation of cytoplasmic components is achieved through several pathways: macroautophagy, microautophagy, and chaperone-mediated autophagy (Seglen and Bohley, 1992; Dunn, 1994; Klionsky and Emr, 2000; Massey et al., 2004).
Macroautophagy (hereafter referred to as autophagy) is the main route for sequestration of the cytoplasm into the lysosome. The initial step of autophagy is elongation of the isolation membrane. The isolation membrane initially enwraps cytoplasmic constituents such as organelles, and then its edges fuse with each other forming a double membrane structure called autophagosome. Finally, the outer membrane of the autophagosome fuses with the lysosome/vacuole and the sequestered cytoplasmic components are degraded by the lysosomal/vacuolar hydrolases, together with the inner membrane of the autophagosomes (Mizushima et al., 2002).
In mammals, autophagy is considered necessary for the turnover of cellular components, particularly in response to starvation or glucagons (Mortimore and Poso, 1987). Yeast deficient in autophagy rapidly die under nutrition-poor conditions (Tsukada and Ohsumi, 1993), suggesting its important roles in preservation of nutrient supply. Indeed, autophagy is necessary for survival in early neonatal starvation period in mice (Kuma et al., 2004). Furthermore, autophagy plays a role in cellular remodeling during differentiation and development of multicellular organisms, such as fly, worm, and slime mold (Levine and Klionsky, 2004), and cellular defense against invading streptococcus (Nakagawa et al., 2004). Plants deficient in autophagy show accelerated senescence (Hanaoka et al., 2002). In humans, autophagy has been implicated in several pathological conditions (Shintani and Klionsky, 2004); e.g., low levels of autophagy were described in some malignant tumors (Liang et al., 1999). In contrast, elevated levels of autophagosome formation were reported in other human pathologies such as neurodegenerative diseases, myopathies, and liver injury (Mizushima et al., 2002; Perlmutter, 2002), and autophagy is implicated in the execution of cell death (Xue et al., 1999; Bursch, 2001). However, the high level of autophagosome formation does not necessarily reflect enhanced protein degradation because the formation of autophagosomes is increased in Danon cardiomyopathy, which is characterized by defective lysosomal degradation (Nishino et al., 2000; Tanaka et al., 2000). Thus, it is not clear whether increased levels of autophagosome formation reflect the activation or defective protein degradation.
Although autophagy has been extensively studied, little was known about its molecular mechanism until the recent discovery of ATG genes in budding yeast (Tsukada and Ohsumi, 1993). Of the many ATG genes, seven uniquely compose two ubiquitin-like conjugation systems: ATG12 and ATG8 conjugation systems (Mizushima et al., 1998; Ichimura et al., 2000; Ohsumi, 2001). The ubiquitin-like protein Atg12p covalently attaches to Atg5p in a reaction similar to ubiquitination. In this process, Atg12p is activated by an E1-like enzyme, Atg7p (Tanida et al., 1999), and transferred to an E2-like enzyme, Atg10p (Shintani et al., 1999), and then finally conjugates to Atg5p. Atg8p, another ubiquitin-like protein, is unique among other ubiquitin-like molecules, as it conjugates to phosphatidyl-ethanolamine (Ichimura et al., 2000). Atg8p is activated by Atg7p, which is common to the Atg12 conjugation system, and is transferred to Atg3p, an E2-like enzyme (Ichimura et al., 2000). In mammals, there exist at least three Atg8 homologues that can all be activated by Atg7 (Tanida et al., 2001), GATE-16, GABARAP, and LC3 (Ohsumi, 2001), and they localize to the autophagosome (Kabeya et al., 2000, 2004).
Here, we generated conditional knockout mice of Atg7 and analyzed the roles of autophagy in neonates and adult liver. Autophagosome formation and starvation-induced degradation of proteins and organelles was impaired in Atg7-deficient mice and adult livers. We also found an important role for autophagy in constitutive turnover of cytoplasmic components, and its loss resulted in accumulation of abnormal organelles and ubiquitinated proteins. Our results suggest that autophagy is important for clearance of ubiquitin-positive aggregates.
Results
Generation of Atg7 conditional knockout mice
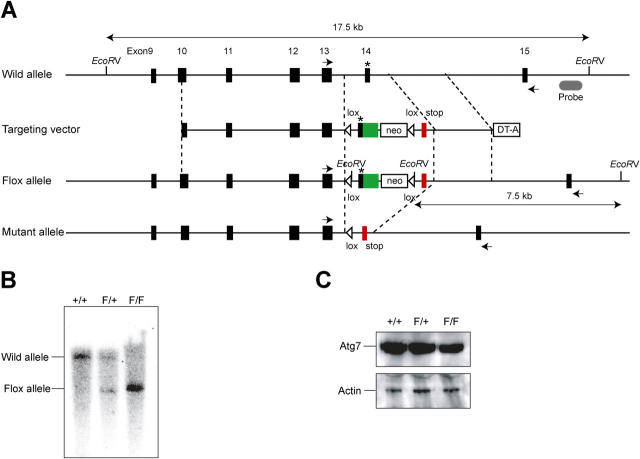
To investigate the physiological roles of autophagy in mammals, we generated Atg7 conditional knockout mice. Mouse Atg7 gene is encoded by 17 exons that span 216-kb long genomic DNA. The active site cysteine residue essential for activation of the substrates is encoded by exon 14 and the targeting vector is designed to conditionally disrupt this exon by Cre-loxP technology. The targeted exon 14 was modified so that it could express Atg7 even in the presence of neo-resistant gene cassette in intron 14 (Fig. 1 A). Mice homozygous for the Atg7 Flox allele (referred to as Atg7 F/F mice), which were expected to express intact Atg7, were born healthy and fertile without any noticeable pathological phenotypes. Fig. 1 B shows Southern blots of mice with the indicated genotypes. Immunoblot analysis revealed the presence of Atg7 protein in Atg7 F/F mouse embryonic fibroblasts (MEFs; Fig. 1 C), suggesting that Atg7 is efficiently expressed from the Atg7 Flox allele.
Figure 1.
Generation of Atg7F/F mice. (A) Schematic representation of the targeting vector and the targeted allele of Atg7 gene. The coding exons numbered in accordance with the initiation site as exon 1 are depicted by black boxes. Green and red boxes indicate Atg7 cDNA fragment (aa 1786–2097) and Atg7 cDNA fragment (aa 1669–1698) with stop codon, respectively. The open triangles denote loxP sequence. A probe for Southern blot analysis is shown as a gray ellipse. Arrows indicate the positions of PCR primers. The asterisk denotes the essential cysteine residue on exon 14. EcoRV, EcoRV sites; neo, neomycin-resistant gene cassette; DT-A, diphtheria toxin gene. (B) Southern blot analysis of genomic DNAs extracted from mice tails. Wild-type and Flox alleles are detected as 17.5- and 7.5-kb bands, respectively. (C) Immunoblot of Atg7 in MEFs. The lysates of MEFs of indicated genotypes were immunoblotted with Atg7 and actin.
The phenotype of Atg7-deficient mice
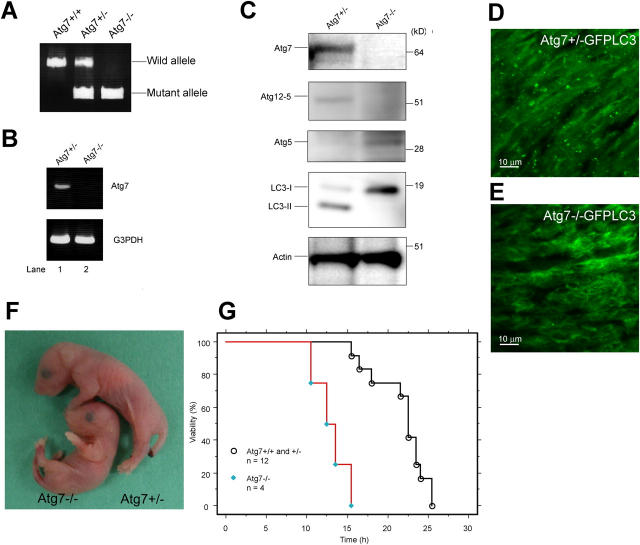
To examine the Atg7-deficient phenotype, we bred Atg7 F/F mice with a line of transgenic mice that express the Cre recombinase under the control of the Zp3 promoter in the oocyte (Lewandoski et al., 1997). The heterozygous mice (referred to as Atg7 +/−) were obtained from female Atg7 F/+:Zp3 mice. Atg7 +/− mice were born healthy and fertile without any noticeable pathological phenotypes for 1 yr. The Atg7 −/− mice, obtained by breeding Atg7 +/− mice, were born at Mendelian frequency (+/+: +/−: −/− = 21: 38: 19). The results of PCR genotyping are shown in Fig. 2 A. Neither Atg7 mRNA nor protein was detected in the homozygous mice (Fig. 2, B and C). We also tested the loss of Atg7 activity by examining the ATG conjugation systems in the neonate liver. A 56-kD protein, equivalent to Atg5–Atg12 conjugate, was detected by Atg5 antibody in the control Atg7 +/− but not Atg7 −/− liver (Fig. 2 C). In contrast, free Atg5 of 30 kD, which was faintly observed in the Atg7 +/− liver, increased in Atg7 −/− liver (Fig. 2 C). Mammalian Atg8p homologue LC3 has two forms (i.e., LC3-I and LC3-II; Kabeya et al., 2000). It is generally accepted that LC3-I is the free mature form whereas LC3-II is the lipidated form, in analogy to yeast Atg8p (Ichimura et al., 2000; Kabeya et al., 2000). Both forms were detected in Atg7 +/− liver whereas only the LC3-I form was detected and increased in Atg7 −/− liver (Fig. 2 C). When crossed with GFP-LC3 transgenic mice (Mizushima et al., 2004), the punctate structures representing autophagosomes were detected in Atg7 +/− but not in Atg7 −/− heart (Fig. 2, D and E). These results indicate that Atg7 is essential for ATG conjugation systems and autophagosome formation in mice.
Figure 2.
The phenotypes of Atg7-deficient mice. (A) PCR analysis of genomic DNA extracted from wild-type, Atg7 +/−, and Atg7 −/− mice tail. The amplified fragments derived from wild and mutant alleles are indicated. (B) Expression of Atg7 transcript. Atg7 transcript was detected by RT-PCR analysis. The region amplified was between exons 12 and 13. G3PDH cDNA was amplified as an internal control. (C) ATG conjugation systems in Atg7 −/− mice liver. The liver homogenate was centrifuged at 800 g for 10 min and the postnuclear supernatant (PNS) was immunoblotted with antibodies against Atg7, Atg5, LC3, and actin as a loading control. The bottom panel of Atg5 blotting is the long exposure of the top panel to detect free Atg5. Data shown are representative of three separate experiments. (D and E) Deficiency of autophagosome formation in Atg7 −/− heart. Atg7 +/− (D) and Atg7 −/− (E) mice expressing GFP-LC3 were obtained by caesarean delivery and analyzed by fluorescence microscopy. Representative results obtained from each neonatal heart at 3 h after caesarean delivery. (F) Morphology of Atg7 +/− and Atg7 −/− mice. (G) Kaplan-Meier curves of survival of newborn mice. Control and Atg7 −/− mice were delivered by caesarean section, and their survival was followed up to 26 h.
Although homozygous mice seemed normal at birth (Fig. 2 F) and had no apparent developmental defect by histological analyses (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1), the mean body weight of Atg7 −/− mice (0.983 ± 0.0763 g, ± SD, n = 9) was significantly lower than that of wild-type and heterozygote mice (1.20 ± 0.116 g, n = 29; P < 0.01), and Atg7 −/− mice died within 1 d after birth (n = 19). We considered that Atg7 −/− mice could survive in utero by virtue of the nutrients supplied through the placenta but could not survive when the supply terminates after birth, as recently reported (Kuma et al., 2004). We tested the survival time of Atg7 −/− neonates under starvation condition after caesarean delivery. Wild-type and heterozygous mice died at 21.7 ± 3.3 h after birth, whereas Atg7 −/− mice died at 13 ± 2.0 h (P < 0.01; Fig. 2 G). To further test whether the cause of earlier death correlates with lower nutrient supply, we measured amino acid concentrations in plasma at 10 h after caesarean delivery. Essential and branched-chain amino acid concentrations in the sera of Atg7 −/− mice were lower than those of wild-type mice (essential amino acids: 1.536 ± 0.087 vs. 1.291 ± 0.166 mmol/L, P < 0.05; branched-chain amino acids: 0.375 ± 0.038 vs. 0.268 ± 0.015 mmol/L, P < 0.01, respectively). The same results were also obtained using MEF cells from Atg7 −/− mice (unpublished data). Together, these results indicate that Atg7 is crucial for the recycling of amino acids in cells and survival of newborn mice under starvation condition.
Starvation response in adult mice liver
To delete Atg7 gene in the adult mice, we bred the Atg7 F/F mice with Mx1-Cre transgenic mice that express the Cre recombinase in response to interferon γ or its chemical inducer, polyinosinic acid–polycytidylic acid (pIpC). The Mx1-Cre transgenic mice can excise Flox allele completely in the liver and spleen and partially in the kidney and heart (Kuhn et al., 1995). Intraperitoneal injections of pIpC resulted in effective recombination of the Atg7 Flox allele in the liver and spleen (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1; and not depicted). No Atg7 transcript, protein, and activity were detected, similar to Atg7 −/− mice (Fig. S2). Next, we tested the autophagosome formation under fasting condition. 1-d fasting resulted in induction of typical autophagosomes in control Atg7 F/+:Mx1 mice (Fig. 3, A–D and I). In contrast, no such induction of autophagosome formation was noted in the liver of fasted Atg7 F/F:Mx1 mice (Fig. 3, E, F, and I). Although some autophagosome-like structures were occasionally observed both in fed and fasted mutant mice livers (Fig. 3, G and H), they tended to be smaller than those observed in fasted control liver and hardly contained large cytoplasmic organelles (compare with Fig. 3, C and D). The number of autophagosomes per hepatocyte was counted and the mean values are shown in Fig. 3 I. The mutant hepatocytes lacked typical glycogen area, in contrast to the fed hepatocytes (Fig. 3, A and E); however, well-developed glycogen granules (α granules) were observed between numerous smooth endoplasmic reticula (Fig. 3 E, inset). Immunofluorescent analysis also revealed the presence of many cup-shaped and ringlike structures representing autophagosomes in the control hepatocytes (Fig. 3, J and K). Although several LC3-positive dots were observed in the mutant hepatocytes, they were not induced in response to starvation and did not form cup-shaped and ringlike structures (Fig. 3, L and M).
Figure 3.
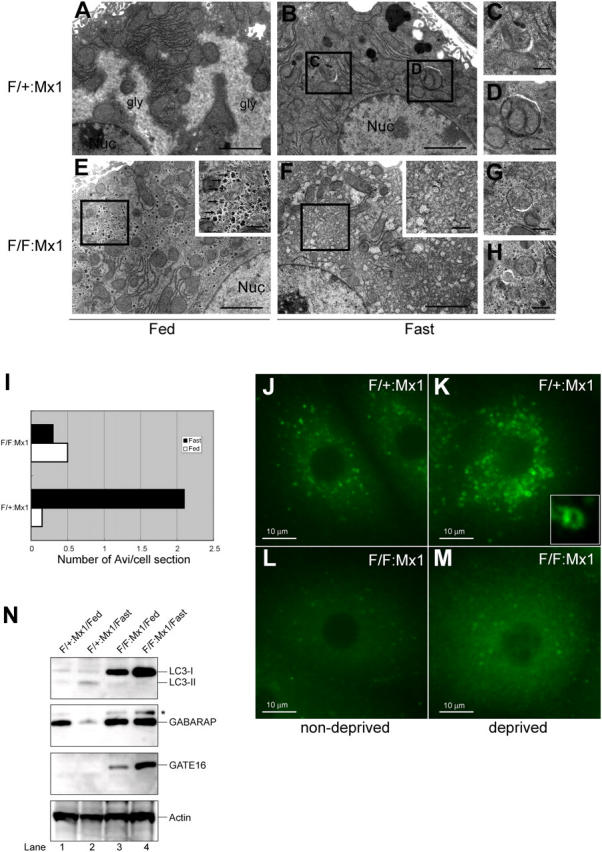
Impaired autophagosome formation in Atg7-deficient liver. (A–H) Electron micrographs of liver from Atg7 F/+:Mx1 (A–D) and Atg7 F/F:Mx1 (E–H) mice fed ad libitum (A and E) or fasted for 1 d (B and F). (C and D) Early stages of autophagic vacuoles observed in B are highlighted. (E and F) Autophagosome was not induced in mutant hepatocytes upon fasting. Insets show higher magnification views of glycogen granules. (G and H) Occasionally observed autophagosome-like structures in mutant hepatocytes. Bars: (A, B, E, and F) 5 μm; (C, D, G, and H) 0.5 μm. (I) Number of autophagosomes per hepatocyte (n = 20) in each genotype was counted and their averages are shown. (J–M) Immunofluorescent analysis of LC3 in primary cultured hepatocytes. Hepatocytes isolated from Atg7 F/+:Mx1 (J and K) and Atg7 F/F:Mx1 mice (L and M) were cultured in Williams' E (J and L) or Hanks' solution (K and M). Inset highlights the cup-like structure of LC3 observed in K. (N) Immunoblot analysis of Atg8 homologues in the liver. Atg7 F/+:Mx1 (lanes 1 and 2) and Atg7 F/F:Mx1 mice (lanes 3 and 4) were fed ad libitum (lanes 1 and 3) or fasted for 1 d (lanes 2 and 4), and then PNS fractions of liver were analyzed by immunoblotting with anti-LC3, GABARAP, GATE-16, and actin antibodies. Asterisk denotes a nonspecific band. Data shown are representative of three separate experiments.
Next, we examined the fasting response of LC3 and other homologues, GABARAP and GATE-16, by immunoblotting (Fig. 3 N). LC3 is known to be up-regulated and recruited to the autophagosome upon starvation and degraded in the lysosomes (Kabeya et al., 2000). Fasting slightly increased the modification of LC3 in heterozygous liver. In the mutant liver, no modification of LC3 was noted and LC3-I increased in response to fasting. These results suggest that LC3 is up-regulated, but its modification and degradation are impaired in mutant mice. The level of GABARAP did not change upon fasting in the mutant liver, whereas it decreased in the heterozygous liver, suggesting that GABARAP is not up-regulated but its degradation after modification is impaired in mutant liver. Although GATE-16 was hardly detected in the heterozygous liver under both fed and fasting conditions, it was clearly detected and increased upon fasting in the mutant liver. These results suggest that GATE-16 may be constitutively degraded even at fed condition in heterozygous mice and up-regulated in response to fasting under defective Atg7. The levels of all LC3 homologues were elevated even at fed condition in the mutant liver, suggesting their marked stabilization in autophagy-deficient condition. However, the possibility that their transcriptions are up-regulated at basal level due to Atg7 deficiency cannot be excluded. We sought to determine their localizations in the cells. However, our antibodies for these molecules were not applicable for immunofluorescent analyses, and those localizations remain to be clarified. In conclusion, all Atg8 homologues respond to fasting, although in a different manner, and their levels are affected by the absence of Atg7.
Atg7 is indispensable for fasting-induced degradation of cytosolic proteins and organelles in the mouse liver
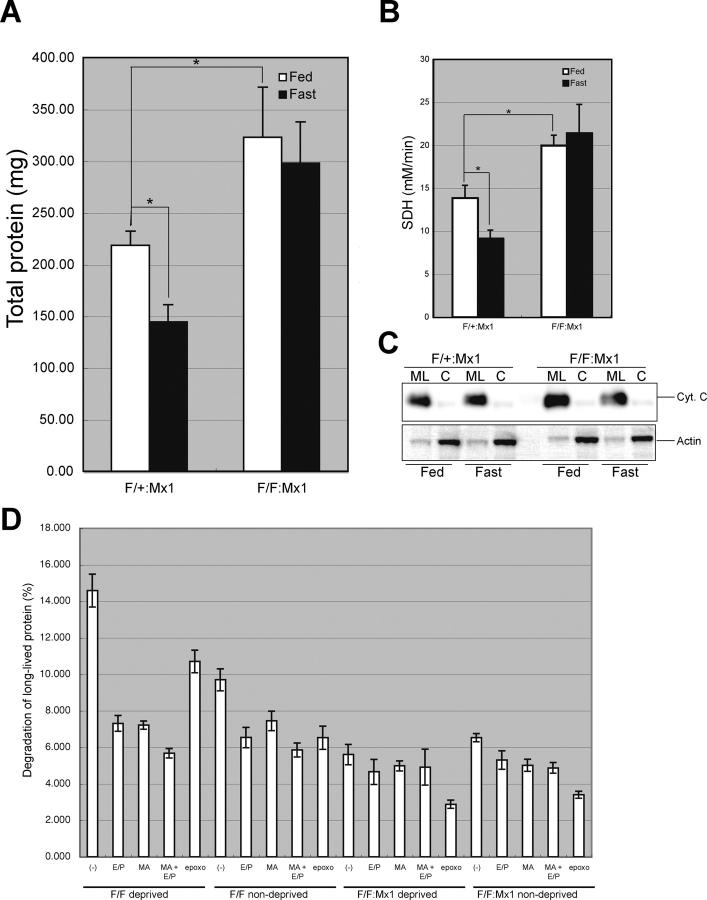
Given that autophagosome formation was impaired in Atg7-deficient liver, we next examined its effects on the bulk degradation of proteins and organelles under fasting condition. After 1-d fasting in control Atg7 F/+:Mx1 and mutant Atg7 F/F:Mx1 mice, the liver was dissected and the amount of total protein per whole liver was measured. The amount of total liver proteins decreased to ∼66% by 1-d fasting in the control liver (Fig. 4 A). In contrast, fasting did not significantly decrease the amount of total liver proteins in the mutant liver. Moreover, the amount of total proteins in the mutant liver was 1.5-fold that of control. These results indicate that the decrease of total proteins is dependent on Atg7 and autophagosome formation.
Figure 4.
Fasting response of Atg7-deficient liver. (A and B) Livers from Atg7 F/+:Mx1 and Atg7 F/F:Mx1 mice fed ad libitum (Fed) or fasted for 1 d (Fast) at 20 d after pIpC injection were dissected, and the amount of total protein (A) and SDH activity (B) per liver were measured. Data are mean ± SD values of five mice in each group; *, P < 0.01. (C) Cytochrome c levels in the cytosolic and mitochondria/lysosomal fractions of the liver at 20 d after injection. Equal amount of PNS fractions were centrifuged at 8,000 g for 10 min and the pellets were used as the mitochondrial/lysosomal fraction (ML). The supernatants were further centrifuged at 100,000 g for 1 h and the supernatant was used as the cytosolic fraction (C). Actin was blotted as control. Data shown are representative of two separate experiments. (D) Turnover of long-lived protein. Hepatocytes from Atg7 F/+:Mx1 and Atg7 F/F:Mx1 mice were isolated and labeled with [14C]leucine for 24 h, and degradation of long-lived protein in deprived or nondeprived condition was measured. Monomethylamine (MA) and/or E64d and pepstatin (E/P) or epoxomicin (epoxo) was added as indicated. Data are the mean ± SD of triplicate experiments.
We also examined whether or not fasting causes the degradation of cellular organelles such as mitochondria in the livers of mice. To quantify the amount of the mitochondria, we first measured the activity of mitochondrial enzyme succinate dehydrogenase (SDH) in total liver extracts. In the control livers, fasting was associated with a significant decrease of SDH activity, and the reduction was proportional with the decrease in the amount of total protein (Fig. 4 B). In contrast, fasting was not associated with any change in SDH activity in the mutant livers. Similar to total protein, the basal SDH activity in mutant liver was significantly higher than in control. The effect of fasting on the amount of the mitochondria was also assessed by immunoblot analysis of mitochondrial protein cytochrome c (Fig. 4 C). When equal amounts of proteins were loaded, the level of cytochrome c was equivalent in the two genotypes at either fed or fasting conditions, suggesting that the ratio of mitochondria versus total protein is not altered by fasting in both genotypes. Considering that the total protein amounts decreased by fasting in the control liver (Fig. 4 A), these results suggest that the mitochondria and cytoplasmic proteins are proportionally degraded upon fasting in heterozygous mice. However, such degradation is impaired in Atg7-deficient liver because the levels of both proteins and mitochondria are unchanged and kept at a higher level.
Next, we investigated the effect of autophagy deficiency on protein turnover. To quantify the turnover of long-lived protein, after each control and mutant hepatocytes had been labeled with [14C]leucine for 24 h and chased for 2 h, the release of TCA-soluble [14C]leucine was measured for 4 h. In control hepatocytes, nutrient deprivation significantly induced protein degradation, and such degradation was suppressed by the addition of lysosomal inhibitors such as monomethylamine and E64d and pepstatin (Fig. 4 D). The induced degradation was still observed in the presence of proteasome inhibitor epoxomicin, suggesting that such protein degradation is mediated in the lysosomal pathway rather than the proteasome (Fig. 4 D). In the mutant hepatocytes, degradation of long-lived protein was not induced by nutrient deprivation (Fig. 4 D), indicating that autophagy is the main route for lysosomal degradation under starvation condition. Consistent with these results, amino acid concentrations in starved mutant hepatocytes were lower than in control hepatocytes (unpublished data). Intriguingly, although lysosomal inhibitors inhibited protein degradation even at nondeprived condition in the control hepatocytes, such inhibition was not significant in the mutant hepatocytes (Fig. 4 D), indicating that significant amounts of proteins are constitutively degraded in the lysosome via autophagic pathway. Together, these results suggest that autophagy plays a significant role in turnover of long-lived protein.
Loss of Atg7 in the liver leads to hepatomegaly and accumulation of abnormal organelles in hepatic cells
We further chased the phenotypes of the mutant mice for up to 90 d after pIpC injection. Gross anatomy revealed severe enlargement of the liver, filling up most of the abdominal cavity (Fig. 5 A). Other major organs were normal histologically (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1). The mean liver weights of control and mutant mice at 90 d after pIpC injection were 1.39 ± 0.24 and 6.10 ± 2.06 g, respectively (n = 5 each). Histological analysis revealed disorganized hepatic lobules and cell swelling in the mutant liver (Fig. 5, D and E). No hepatocellular proliferation or regeneration was detected (unpublished data). Vacuolated hepatic cells were occasionally observed and those were associated with hepatic cell death, which is consistent with the leakage of alkaline phosphatase, aspartate aminotransferase, and alanine aminotransferase in the mutant mice sera (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1).
Figure 5.
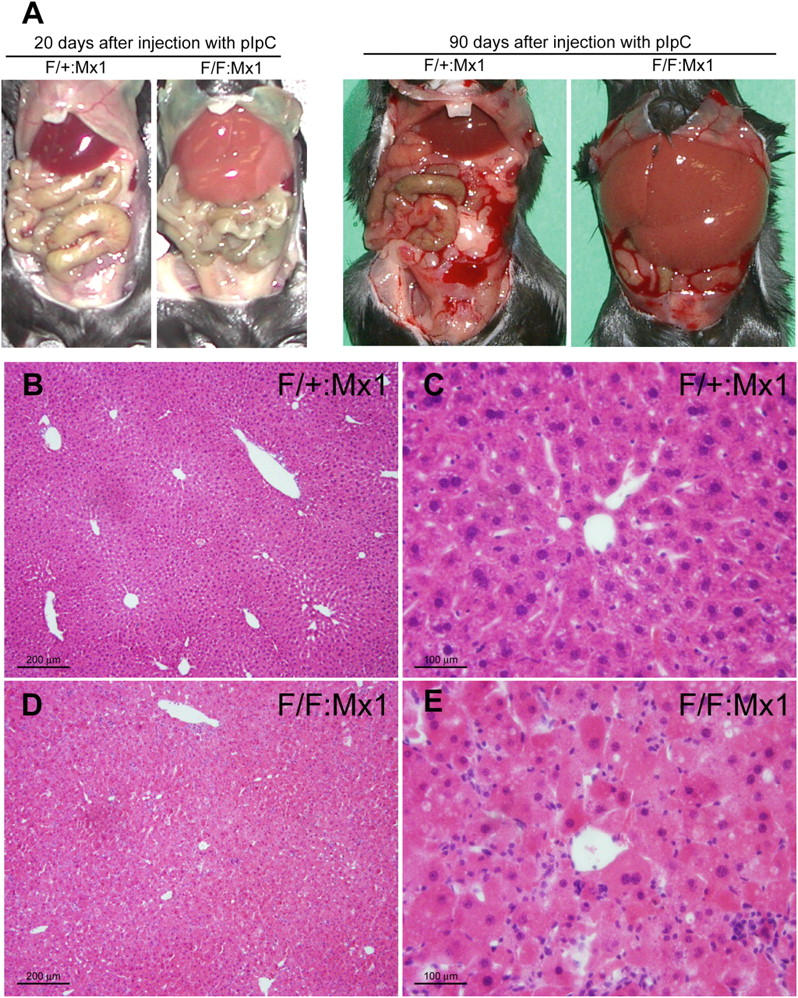
Atg7 deficiency in the liver causes hepatomegaly and hepatic cell swelling. (A) The gross anatomical views of representative mice at the indicated day after pIpC injection. (B–E) Histology of representative livers with Atg7 deficiency. Hematoxylin and eosin staining of Atg7 F/+:Mx1 (B and C) and Atg7 F/F:Mx1 (D and E) liver at 90 d after pIpC injection.
Although most hepatocytes were still alive in the mutant liver, ultrastructural analysis revealed the appearance of aberrant concentric membranous structures (Fig. 6, A and B), which were also observed as early as 20 d after pIpC-injected liver (not depicted). These structures surrounded various cytoplasmic constituents such as mitochondria, lipid droplets, and vesicular structures (Fig. 6 A). Their membranous elements were continuous with the rough ER (Fig. 6 B, arrowheads), and the corresponding structures were positive for calreticulin, an ER protein marker (not depicted), indicating that these structures originated from the rough ER. Accumulation of peroxisomes (Fig. 6 C) and deformed mitochondria (Fig. 6, C and D) was also observed in the mutant liver. These results suggest the important role of autophagy in turnover of organelles, and its defect results in accumulation of abnormal organelles.
Figure 6.
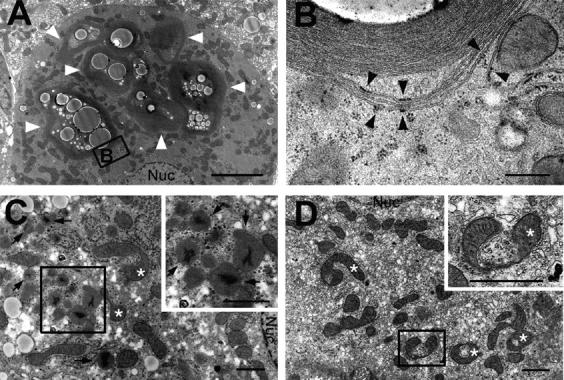
Electron micrographs of hepatic cells with Atg7 deficiency. (A and B) Note the presence of concentric membranous structures in the mutant cells (arrowheads). Higher magnification view (B) shows the membranous elements are continuous with the rough ER (arrowheads). (C and D) The mutant cells contained a high number of peroxisomes (arrows) and deformed mitochondria (asterisks). Insets show higher magnification views. Nuc, nucleus. Bars: (A) 5 μm; (B) 0.5 μm; (C and D) 1 μm.
Formation of ubiquitin-positive inclusions in Atg7-deficient hepatocytes
Autophagy has been implicated in not only organelle turnover but also in elimination of protein aggregates (Kopito, 2000). Protein aggregates are often ubiquitinated. In the next step, we immunostained the liver with an ubiquitin antibody to examine the presence of such aggregates. Several ubiquitin-positive particles of various sizes were detected in the Atg7 F/F:Mx1 but not in Atg7 F/+:Mx1 hepatic cells at both 10 and 90 d after pIpC injection (Fig. 7, A and B; and Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1). The immunoblots of the liver lysates revealed the accumulation of high-molecular mass polyubiquitinated proteins in the mutant liver (Fig. 7 G and Fig. S5), suggesting that the ubiquitin particles are aggregates of polyubiquitinated proteins. To further determine the localization of ubiquitin-positive dots, analysis of immunoelectron micrographs was performed. Numerous particles of colloidal gold, indicative of ubiquitin, were detected on lipid dropletlike structures, membranous structures, and amorphous substances in the cytoplasm (Fig. 7, C–F). Such signals were not observed in the wild-type liver (unpublished data).
Figure 7.
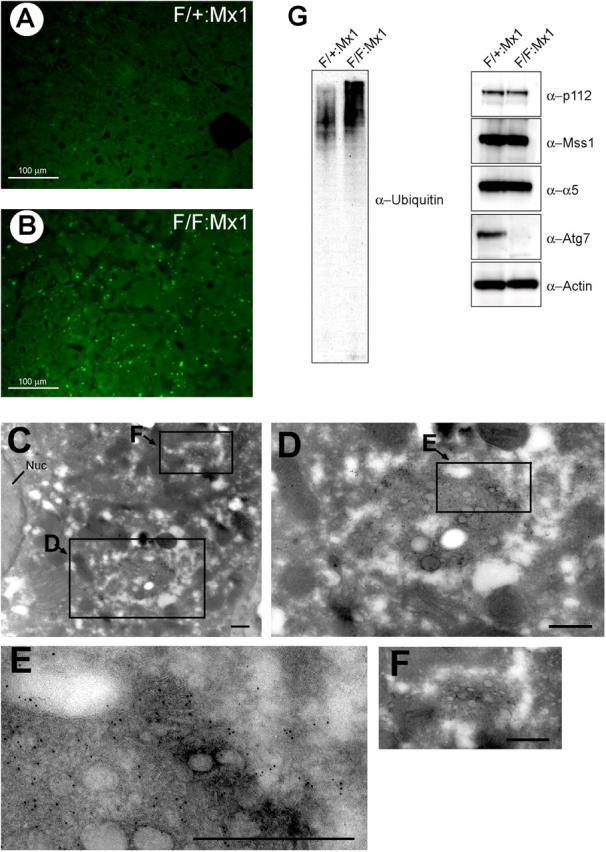
Accumulation of ubiquitin-positive aggregates in Atg7-deficient liver. (A and B) Immunofluorescent detection of ubiquitin in the Atg7 F/+:Mx1 (A) and Atg7 F/F:Mx1 (B) liver. (C–F) Immunoelectron micrographs of ubiquitin in a representative mutant liver. The high-magnification view shows ubiquitin particles near the lipid dropletlike structure (D–F). Bars, 0.5 μm. (G) Immunoblot analysis of the liver. PNS fractions of the liver at 90 d after injection were immunoblotted with the indicated antibodies. Data shown are representative of three separate experiments.
The accumulation of ubiquitin-positive inclusions in the cytoplasm prompted us to examine the effect of autophagy deficiency on proteasome function. Immunoblots of proteasome subunits (p112/Rpn2, Mss1/Rpt1, and α5) showed that their relative amounts were not affected in the mutant liver (Fig. 7 G). Furthermore, the activities of the proteasome, measured by Suc-LLVY-MCA as substrate, were also comparable between wild-type and mutant livers (unpublished data). These results indicate the accumulation of ubiquitin-positive aggregates in autophagy-deficient hepatocytes despite the apparently normal proteasome function.
Discussion
Autophagy is a bulk protein degradation pathway, which is conserved in eukaryotes, essential for the survival of unicellular organisms under nutrient-poor condition and for cellular remodeling of multicellular organisms (Mizushima et al., 2002; Levine and Klionsky, 2004). In the present study, we generated conditional knockout mice of Atg7 gene, which is an essential gene for autophagy in budding yeast, and analyzed its roles in mice.
In mammals, Atg7 was indeed essential for ATG12 conjugation, LC3 modification systems, and autophagosome formation (Fig. 2, Fig. 3, and Fig. S2). Immunofluorescent analyses revealed that LC3-positive dots appeared but did not form cup-shaped and ringlike structures in Atg7 F/F:Mx1 livers (Fig. 3). The LC3-I form is usually present at the S100 fraction and the LC3-II form at the P100 fraction (Kabeya et al., 2000). In the mutant liver, LC3-I was present in both S100 and P100 fractions (unpublished data), suggesting that the LC3-positive dots in the mutant hepatocytes are indeed the LC3-I form. These results suggest that LC3 may be recruited to the dot structures independent of the modification (Fig. 3). In mammals, LC3 has at least two homologues, GABARAP and GATE-16, which share common biochemical characteristics (Tanida et al., 2001) and localize to autophagosome in response to fasting (Kabeya et al., 2004). Indeed, the modification and levels of these molecules under fasting condition were affected in the mutant liver (Fig. 3 N). However, these LC3 homologues have been identified in a different biological pathway and may have diverse functions (Ohsumi, 2001). Thus, how their functions and localizations are affected in Atg7-deficient cells remains to be clarified.
Although Atg7 −/− mice were born at Mendelian ratio, and the major organs were almost normal histologically (Fig. S1), they had reduced body weight and died within 1 d after birth. Atg7 −/− mice had lower amino acid level and died earlier compared with wild type under nonsuckling condition after caesarean delivery (Fig. 2 G), suggesting that Atg7 is important for survival during the early neonatal starvation period, similar to recently reported Atg5 −/− mice phenotypes (Kuma et al., 2004). However, because suckling Atg7 −/− mice also died within 1 d after birth (unpublished data), the cause of death may not be only due to low level of amino acids. The reason for the reduced body size is also unclear and may be related to placental function or due to inefficient reutilization of nutrients during embryogenesis. It is of note that a lower level of autophagy occurs during embryogenesis (Mizushima et al., 2004) even when nutrients are supplied from the placenta. Furthermore, Atg7 null mice possess several ubiquitin-positive inclusions in some organs at the time of birth (unpublished data). This phenotype might be related to the earlier death of mutant. Further analysis of Atg7 −/− mice is required to unravel the roles of autophagy, and such analysis is currently under way by breeding the Atg7 F/F mice with several Cre-transgenic mice.
Starvation-induced autophagosomes appeared to sequester the cytoplasm randomly (Fig. 3). Consistent with this notion, the amount of mitochondria decreased in proportion with reduction in the amount of total protein (Fig. 4, A–C). These results suggest that mitochondria are degraded nonselectively under fasting condition. In Atg7-deficient liver, no autophagosome formation was noted and the degradation of proteins and organelles under fasting condition was largely impaired. These results suggest that the rapid reduction of proteins and organelles upon fasting is dependent on Atg7 and autophagosome formation.
Although autophagy can be induced by starvation, this pathway may take place even at feeding condition at basal level. This constitutive pathway may be important for turnover of organelles and cytoplasmic proteins. Indeed, the degradation of long-lived protein was inhibited in mutant hepatocytes irrespective of nutrient deprivation (Fig. 4 D), and multiple abnormalities of organelles (e.g., the presence of concentric membranous structure and accumulation of deformed mitochondria) were observed in Atg7-deficient hepatocytes (Fig. 6). Unexpectedly, the morphologically abnormal mitochondria appear to retain their function, as judged by the normal membrane potentials and the absence of cytochrome c leakage in the cytosol (unpublished data). In contrast to starvation-induced autophagy, whether or not constitutive autophagy eliminates abnormal and excess organelles in a degree of selectivity remains to be clarified.
Beclin 1, a human homologue of ATG6/VPS30 essential for autophagy in yeast, was recently identified as a tumor suppressor gene, and autophagy has been implicated in the regulation of cellular proliferation (Liang et al., 1999). Indeed, heterozygous disruption of mouse Beclin 1 led to enhanced tumorigenesis (Qu et al., 2003; Yue et al., 2003). Atg7 deficiency led to hepatomegaly (Fig. 5 A), suggesting that cell proliferation or malignant transformation might be induced in the Atg7-deficient cells. However, neither tumorigenesis nor enhanced cell proliferation was detected as tested by BrdU incorporation at 90 d after pIpC injection in the mutant liver compared with control mice (unpublished data). The hepatomegaly observed in the mutant mice was likely due to increased cellular volume rather than cell number, which is also supported by the swollen appearance of hepatocytes (Fig. 5, D and E).
In Atg7-deficient liver, we detected numerous ubiquitin-positive particles indicative of protein aggregates (Fig. 7 and Fig. S5). It has been reported that proteasome inhibition leads to aggregate formation. Conversely, the formation of protein aggregates inhibits the proteasome (Bence et al., 2001), resulting in a malignant cycle of aggregate formation and proteasome inhibition. In the mutant liver, failure of the proteasome was postulated; however, no impairment of proteasome function, in terms of its expression or peptidase activities, was noted (Fig. 7 G and not depicted). Our results suggest that the ubiquitinated proteins eventually aggregate even in the presence of functional proteasomes. Considering that such ubiquitinated aggregates must be difficult to unfold, and proteasomes need to unfold their substrate before degradation (Baumeister et al., 1998), it is likely that elimination of ubiquitin-positive aggregates in the cells is largely dependent on the autophagic process. Protein ubiquitination may also occur after protein aggregation. In either case, we propose the possibility that protein ubiquitination may serve as a signal to the autophagic process in addition to the proteasomes pathway. In this context, it is worth noting that sperm mitochondria are known to be ubiquitinated before degradation during fertilization (Sutovsky et al., 1999). It is now well established that ubiquitin regulates not only proteasomal degradation, but also lysosomal degradation. Thus, it is conceivable that ubiquitin could also regulate the autophagic pathway.
A growing number of disease-associated proteins have been found to accumulate in aggresome, including huntingtin, parkin, α-synuclein, and peripheral myelin protein 22 (Notterpek et al., 1999; Ciechanover and Brundin, 2003). The aggregation of these proteins is thought to be involved in the pathogenesis of Huntington's disease, Parkinson's disease, and peripheral neuropathies, respectively. Enhanced autophagosome formation is prevalent in most of these diseases (Mizushima et al., 2002), and autophagy has also been considered as a caspase-independent cell death pathway (Xue et al., 1999; Bursch, 2001). Our Atg7 mutant mice should be useful for examining the role of autophagy in the cell death pathway or in a cellular defense mechanism in the pathogenesis of these diseases.
Materials and methods
Generation of Atg7 F/F mice
The targeting vector was constructed by insertion of a loxP sequences within introns 13 and 14 of Atg7 gene. Exon 14 was fused to a cDNA fragment encoded by exons 15, 16, and 17 (aa 1786–2097) and polyA signal sequence was added after the stop codon. Neo resistant gene cassette (mc1-neo-pA) was ligated behind the polyA signal sequence followed by the second loxP sequence, splicing acceptor site, and exon 14 with stop codon preceding the active site. We electroporated the targeting vector into mouse TT2 ES cells, selected with 200 μg/ml G418 (GIBCO BRL), and then screened for homologous recombinants by PCR and Southern blot analyses. PCR primers were as follows: 5′-TGGCTGCTACTTCTGCAATGATGT-3′, 5′-GAAGGGACTGGCTGCTATTGGGCGAAGTGC-3′, and 5′-TTAGCACAGGGAACAGCGCTCATGG-3′. Southern blot analysis was performed by digestion of genomic DNA with EcoRV and hybridization with the probe shown in Fig. 1 A. Genotyping of mice by PCR was performed using the following two primers: 5′-TGGCTGCTACTTCTGCAATGATGT-3′ and 5′-CAGGACAGAGACCATCAGCTCCAC-3′. Progeny containing the Atg7 Flox allele were bred with Zp3-Cre and Mx1-Cre transgenic mice to produce Atg7 −/− and Atg7 F/F:Mx1 mice, respectively. With regard to Atg7 F/F:Mx1 mice, Cre expression in the liver was induced by i.p. injection of pIpC (Sigma-Aldrich). 300 μl pIpC solution (1 mg/ml in water) was injected three times at 48-h intervals. Mice were housed in specific pathogen-free facilities, and the experimental protocol was approved by the Ethics Review Committee for Animal Experimentation of the Tokyo Metropolitan Institute of Medical Science.
RT-PCR analysis
cDNA was synthesized from 5 μg of DNase I–treated total RNA using the SuperScript First-Strand Synthesis System (GIBCO BRL) and oligo (dT)12-18 primers. Specific primers for each gene were as follows: 5′-ATGCCAGGACACCCTGTGAACTTC-3′ and 5′-ACATCATTGCAGAAGTAGCAGCCA-3′ for Atg7, and 5′-GAGCTGAACGGGAAGCTCAC-3′ and 5′-ACCACCCTGTTGCTGTAGC-3′ for G3PDH.
Immunoblot analysis
The fractions were immunoblotted as described previously (Komatsu et al., 2001). The antibodies for Atg7 (Tanida et al., 1999) and Atg5 (Mizushima et al., 2001) were described previously. The antibodies for ubiquitin (DakoCytomation) and actin (MAB1501R; Chemicon International, Inc.) were purchased. The antibodies against LC3, GABARAP, and GATE-16 were raised in rabbits using their specific peptides as antigens. The antibodies against p112, Mss1, and α5 were provided by K.B. Hendil (August Krogh Institute, University of Copenhagen, Copenhagen, Denmark).
Caesarean delivery and measurement of amino acids
Newborns were delivered by caesarean section at 19.0 d postcoitus and placed in a humidified, thermostat-controlled chamber (30°C). Plasma was fixed in 3% sulphosalicylic acid. Amino acids in the supernatant from plasma samples were measured by an amino acid analyzer (L8500A; Hitachi).
Protein degradation assay
The assay was performed essentially as described previously (Gronostajski and Pardee, 1984). In brief, hepatocytes were plated at 5 × 104 cells/well in collagen-coated 24-well plates and cultured in Williams' E medium with 10% FCS (Williams' E/10% FCS) for 24 h. Cells were incubated with Williams' E/10% FCS containing 0.5 μCi/ml [14C]leucine for 24 h to label long-lived proteins. Cells were washed with Williams' E/10% FCS containing 2 mM of unlabeled leucine and incubated with the medium for 2 h to allow degradation of short-lived proteins and minimize the incorporation of labeled leucine, which was released by proteolysis into protein. The cells were then washed with PBS and incubated at 37°C with Krebs-Ringer bicarbonate medium and Williams' E/10% FCS in the presence or absence of protease inhibitors (5 mM monomethylamine, 10 μg/ml E64d and pepstatin, or 5 μM epoxomicin). After 4 h, aliquots of the medium were taken and a one-tenth volume of 100% trichloroacetic acid was added to each aliquot. The mixtures were centrifuged at 12,000 g for 5 min, and the acid-soluble radioactivity was determined using a liquid scintillation counter. At the end of the experiment, the cultures were washed twice with PBS, and 1 ml of cold trichloroacetic acid was added to fix the cell proteins. The fixed cell monolayers were washed with trichloroacetic acid and dissolved in 1 ml of 1 N NaOH at 37°C. Radioactivity in an aliquot of 1 N NaOH was determined by liquid scintillation counting. The percentage of protein degradation was calculated according to published procedures (Gronostajski and Pardee, 1984).
Histological examination
Tissues were dissected, fixed in 4% PFA, paraffin embedded, and sectioned. Sections were stained by Meyer's hematoxylin and eosin. For immunohistochemical analysis, sections were blocked in 5% normal goat serum in PBS containing 0.2% Triton X-100, and then incubated with antiubiquitin antibody (1B3; MBL International Corporation) and Alexa 488–labeled secondary antibody (Molecular Probes). Apoptotic cells were detected by TUNEL assay using Apoptag kit (Intergen Company) as described previously (Tateishi et al., 2001). For GFP-LC3 observations, tissues were fixed with 4% PFA, and the cryosections were imaged with a conventional fluorescence microscope. For LC3 staining, hepatocytes were fixed and stained with anti-LC3 antibody as described previously (Kabeya et al., 2000). All fluorescence images were obtained using a fluorescence microscope (model Q550FV; Leica) equipped with cooled charge-coupled device camera (model CTR MIC; Leica). Pictures were taken using Qfluoro software (Leica).
EM and immunoelectron microscopy
Livers were fixed by cardiac perfusion using 0.1 M phosphate buffer containing 2% PFA and 2% glutaraldehyde for conventional EM. They were post-fixed with 1% OsO4, embedded in Epon812, and sectioned. Immunoelectron microscopy was performed on cryothin sections as described previously (Waguri et al., 1995). In brief, livers were frozen in phosphate buffer with 2.3 M sucrose and 20% polyvinyl pyrrhoridon. Ultrathin sections were mounted on Formvar carbon-coated nickel grids, blocked with 1% BSA in PBS, and incubated with antiubiquitin antibody (1B3) and colloidal gold conjugated secondary antibody.
Other procedures
MEFs were prepared as described previously (Murata et al., 2001). Primary hepatocytes were prepared as described previously (Ueno et al., 1990). Cell starvation was conducted by incubating the cells in Hanks' balanced solution after three separate washes. The SDH activity was assayed as described previously (Ueno et al., 1990).
On line supplemental material
Fig. S1 shows the histological analyses of tissues from Atg7 +/− and Atg7 −/− mice at 1 d after birth. Fig. S2 shows the loss of Atg7 protein and activity in Atg7 F/F:Mx1 mouse liver. Fig. S3 shows the histological analyses of tissues from Atg7 F/+:Mx1 and Atg7 F/F:Mx1 mice. Fig. S4 shows the cell death in autophagy-deficient liver. Fig. S5 shows the accumulation of ubiquitin-positive inclusions at early stage of autophagy deficiency. Further comments on the data can be found in the legends. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200412022/DC1.
Acknowledgments
We thank T. Kaneko, T. Kouno, and K. Tatsumi for technical assistance. We also thank F. Kaji for his help in EM study; A. Kuma for technical guidance in caesarean delivery; K. Tateishi and H. Uozaki for discussion of liver pathology; and T. Fujimura and K. Murayama for amino acid measurements.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used in this paper: MEF, mouse embryonic fibroblast; pIpC, polyinosinic acid–polycytidylic acid; PNS, postnuclear supernatant; SDH, succinate dehydrogenase.
References
- Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 92:367–380. [DOI] [PubMed] [Google Scholar]
- Bence, N.F., R.M. Sampat, and R.R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 292:1552–1555. [DOI] [PubMed] [Google Scholar]
- Bursch, W. 2001. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ. 8:569–581. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A., and P. Brundin. 2003. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron. 40:427–446. [DOI] [PubMed] [Google Scholar]
- Dunn, W.A., Jr. 1994. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 4:139–143. [DOI] [PubMed] [Google Scholar]
- Goldberg, A.L. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature. 426:895–899. [DOI] [PubMed] [Google Scholar]
- Gronostajski, R.M., and A.B. Pardee. 1984. Protein degradation in 3T3 cells and tumorigenic transformed 3T3 cells. J. Cell. Physiol. 119:127–132. [DOI] [PubMed] [Google Scholar]
- Hanaoka, H., T. Noda, Y. Shirano, T. Kato, H. Hayashi, D. Shibata, S. Tabata, and Y. Ohsumi. 2002. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129:1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, Y., T. Kirisako, T. Takao, Y. Satomi, Y. Shimonishi, N. Ishihara, N. Mizushima, I. Tanida, E. Kominami, M. Ohsumi, et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492. [DOI] [PubMed] [Google Scholar]
- Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., N. Mizushima, A. Yamamoto, S. Oshitani-Okamoto, Y. Ohsumi, and T. Yoshimori. 2004. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117:2805–2812. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J., and S.D. Emr. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M., I. Tanida, T. Ueno, M. Ohsumi, Y. Ohsumi, and E. Kominami. 2001. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1-E2 complex formation. J. Biol. Chem. 276:9846–9854. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530. [DOI] [PubMed] [Google Scholar]
- Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. [DOI] [PubMed] [Google Scholar]
- Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036. [DOI] [PubMed] [Google Scholar]
- Levine, B., and D.J. Klionsky. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477. [DOI] [PubMed] [Google Scholar]
- Lewandoski, M., K.M. Wassarman, and G.R. Martin. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7:148–151. [DOI] [PubMed] [Google Scholar]
- Liang, X.H., S. Jackson, M. Seaman, K. Brown, B. Kempkes, H. Hibshoosh, and B. Levine. 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. [DOI] [PubMed] [Google Scholar]
- Massey, A., R. Kiffin, and A.M. Cuervo. 2004. Pathophysiology of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 36:2420–2434. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., T. Noda, T. Yoshimori, Y. Tanaka, T. Ishii, M.D. George, D.J. Klionsky, M. Ohsumi, and Y. Ohsumi. 1998. A protein conjugation system essential for autophagy. Nature. 395:395–398. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima, N., Y. Ohsumi, and T. Yoshimori. 2002. Autophagosome formation in mammalian cells. Cell Struct. Funct. 27:421–429. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., A. Yamamoto, M. Matsui, T. Yoshimori, and Y. Ohsumi. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell. 15:1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore, G.E., and A.R. Poso. 1987. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 7:539–564. [DOI] [PubMed] [Google Scholar]
- Murata, S., H. Udono, N. Tanahashi, N. Hamada, K. Watanabe, K. Adachi, T. Yamano, K. Yui, N. Kobayashi, M. Kasahara, et al. 2001. Immunoproteasome assembly and antigen presentation in mice lacking both PA28alpha and PA28beta. EMBO J. 20:5898–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, et al. 2004. Autophagy defends cells against invading group A Streptococcus. Science. 306:1037–1040. [DOI] [PubMed] [Google Scholar]
- Nishino, I., J. Fu, K. Tanji, T. Yamada, S. Shimojo, T. Koori, M. Mora, J.E. Riggs, S.J. Oh, Y. Koga, et al. 2000. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 406:906–910. [DOI] [PubMed] [Google Scholar]
- Notterpek, L., M.C. Ryan, A.R. Tobler, and E.M. Shooter. 1999. PMP22 accumulation in aggresomes: implications for CMT1A pathology. Neurobiol. Dis. 6:450–460. [DOI] [PubMed] [Google Scholar]
- Ohsumi, Y. 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2:211–216. [DOI] [PubMed] [Google Scholar]
- Perlmutter, D.H. 2002. Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J. Clin. Invest. 110:1579–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, X., J. Yu, G. Bhagat, N. Furuya, H. Hibshoosh, A. Troxel, J. Rosen, E.L. Eskelinen, N. Mizushima, Y. Ohsumi, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112:1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen, P.O., and P. Bohley. 1992. Autophagy and other vacuolar protein degradation mechanisms. Experientia. 48:158–172. [DOI] [PubMed] [Google Scholar]
- Shintani, T., and D.J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science. 306:990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., N. Mizushima, Y. Ogawa, A. Matsuura, T. Noda, and Y. Ohsumi. 1999. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 18:5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutovsky, P., R.D. Moreno, J. Ramalho-Santos, T. Dominko, C. Simerly, and G. Schatten. 1999. Ubiquitin tag for sperm mitochondria. Nature. 402:371–372. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., G. Guhde, A. Suter, E.L. Eskelinen, D. Hartmann, R. Lullmann-Rauch, P.M. Janssen, J. Blanz, K. von Figura, and P. Saftig. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 406:902–906. [DOI] [PubMed] [Google Scholar]
- Tanida, I., N. Mizushima, M. Kiyooka, M. Ohsumi, T. Ueno, Y. Ohsumi, and E. Kominami. 1999. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 10:1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida, I., E. Tanida-Miyake, T. Ueno, and E. Kominami. 2001. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 276:1701–1706. [DOI] [PubMed] [Google Scholar]
- Tateishi, K., M. Omata, K. Tanaka, and T. Chiba. 2001. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155:571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174. [DOI] [PubMed] [Google Scholar]
- Ueno, T., S. Watanabe, M. Hirose, T. Namihisa, and E. Kominami. 1990. Phalloidin-induced accumulation of myosin in rat hepatocytes is caused by suppression of autolysosome formation. Eur. J. Biochem. 190:63–69. [DOI] [PubMed] [Google Scholar]
- Waguri, S., N. Sato, T. Watanabe, K. Ishidoh, E. Kominami, K. Sato, and Y. Uchiyama. 1995. Cysteine proteinases in GH4C1 cells, a rat pituitary tumor cell line, are secreted by the constitutive and regulated secretory pathways. Eur. J. Cell Biol. 67:308–318. [PubMed] [Google Scholar]
- Xue, L., G.C. Fletcher, and A.M. Tolkovsky. 1999. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol. Cell. Neurosci. 14:180–198. [DOI] [PubMed] [Google Scholar]
- Yue, Z., S. Jin, C. Yang, A.J. Levine, and N. Heintz. 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 100:15077–15082. [DOI] [PMC free article] [PubMed] [Google Scholar]