Abstract
The promyelocytic leukemia (PML) protein is aggregated into nuclear bodies that are associated with diverse nuclear processes. Here, we report that the distance between a locus and its nearest PML body correlates with the transcriptional activity and gene density around the locus. Genes on the active X chromosome are more significantly associated with PML bodies than their silenced homologues on the inactive X chromosome. We also found that a histone-encoding gene cluster, which is transcribed only in S-phase, is more strongly associated with PML bodies in S-phase than in G0/G1 phase of the cell cycle. However, visualization of specific RNA transcripts for several genes showed that PML bodies were not themselves sites of transcription for these genes. Furthermore, knock-down of PML bodies by RNA interference did not preferentially change the expression of genes closely associated with PML bodies. We propose that PML bodies form in nuclear compartments of high transcriptional activity, but they do not directly regulate transcription of genes in these compartments.
Keywords: PML nuclear body; major histocompatibility complex; transcription; gene density; nuclear organization
Introduction
Our understanding of the structural and functional compartmentalization of the mammalian interphase nucleus has been refined over the last few decades. It is now known that the nucleus contains chromosome territories and subnuclear domains that are involved in DNA replication, transcription, and RNA processing (Cremer and Cremer, 2001). In each chromosome territory, genes are arranged functionally, with certain active genes and clusters located on the external surfaces (Dietzel et al., 1999; Volpi et al., 2000). The positions of chromosome territories within the nucleus appear to be related to gene density, with gene-rich territories tending to be placed more centrally (Croft et al., 1999). Analysis of individual loci within the nucleus has revealed that certain silent genes associate with centromeric heterochromatin in cycling lymphocytes (Brown et al., 2001), whereas other genes associate with nuclear structures such as SC-35 domains or coiled bodies (Frey and Matera, 1995; Smith et al., 1999).
One of the most prominent nuclear structures is the promyelocytic leukemia (PML) nuclear body, also known as PML oncogenic domain, nuclear domain 10, or Kremer body. There are usually between 5 and 30 PML bodies per nucleus, depending on the cell type and the stage of the cell cycle (Koken et al., 1995). The PML protein, which is a major and essential constituent of these bodies (Dyck et al., 1994; Weis et al., 1994), is a member of a protein family characterized by an RBCC motif (consisting of RING finger, B-box, and α-helical coiled-coil domains; Jensen et al., 2001). This motif allows the protein to interact with other proteins as well as to homodimerize. In acute promyelocytic leukemia (APL), the PML protein forms a fusion protein with retinoic acid receptor-α, resulting in the redistribution of PML nuclear bodies into microspeckles (de The et al., 1991; Goddard et al., 1991; Kakizuka et al., 1991).
PML bodies recruit a variety of other proteins, including Sp100, SUMO-1, Daxx, pRB, p53, and BLM, suggesting roles in DNA replication and repair, cell cycle control, and apoptosis (Seeler and Dejean, 1999; Ruggero et al., 2000). PML bodies are also implicated in gene transcription by their recruitment of the histone acetyltransferase, cAMP-response element binding protein (CBP), and RNA polymerase II (LaMorte et al., 1998; von Mikecz et al., 2000; Boisvert et al., 2001), and the accumulation of nascent RNA at their periphery (Boisvert et al., 2000). A role as a nuclear depot, coordinating the accumulation and release of regulatory proteins in response to external stimuli, has also been proposed (Negorev and Maul, 2001).
Using a novel statistical method, we recently showed that PML bodies associate in primary human fibroblast nuclei with the major histocompatibility complex (MHC), a gene-rich region of chromosome 6 (Shiels et al., 2001). The closest association was with the MHC class II region, which includes the transporter associated with antigen processing/large multifunctional protease (TAP/LMP) locus containing the LMP2/PSMB9, LMP7/PSMB8, TAP1, and TAP2 genes involved in MHC class I antigen presentation. In contrast, the gene-poor 6p24 region and the chromosome 6 centromere showed no association with PML bodies. These data showed for the first time that PML bodies associate with a specific genomic region. We have now extended this analysis to multiple gene-rich and gene-poor regions on other chromosomes in order to determine whether there is a spatial organization of PML bodies relative to particular regions of the genome. We find a significant correlation between association of PML bodies with genomic regions and the transcriptional activity and gene density within these regions. However, we also find that PML bodies do not serve as obligate transcription sites for associated genes tested, nor are basal transcription levels of these genes altered by knock-down of PML protein.
Results
Measurement of locus–PML spatial proximity
We examined associations of PML bodies with loci from chromosomes 1, 6, 7, 9, 14, 16, 17, 18, and 19 in comparison with the TAP/LMP locus in the MHC class II region (Fig. 1; Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200305142/DC1). The TAP/LMP locus was chosen as a reference, as we previously found that it was closely associated with PML bodies (Shiels et al., 2001). For each locus, three scores of association were made: the percentage of loci in contact with a PML body, the percentage of loci within 1 mm of the nearest PML body, and the mean minimum distance (mmd) of the loci to the nearest PML body (the last score was used for a statistical comparison with the TAP/LMP locus using paired t tests). As expected, and in agreement with others (Roix et al., 2003), we found that all three scores correlated closely with each other (Fig. S1). However, we also found that the differences in mmd association of each locus with PML bodies persisted after scores for alleles which were in direct contact with PML bodies were removed from the dataset (Fig. S1). This implies either that the association of loci with PML bodies does not require direct contact, or that loci that are statistically closer have a higher chance of contact of a PML body due to their dynamic nature (Chubb et al., 2002). We also found that all centromeres, with the exception of the chromosome 9 centromere, were among the least associated with PML bodies, using mmd measurements. However, due to the large FISH signals of centromeres within the nucleus, a high proportion of signals from centromeres are in contact with PML bodies. Therefore, we propose that statistical measurements based on distances, corrected for PML–PML distances, are less prone to artifact than visual counting. Similar distance measurements to measure proximity have also been used by others (Roix et al., 2003).
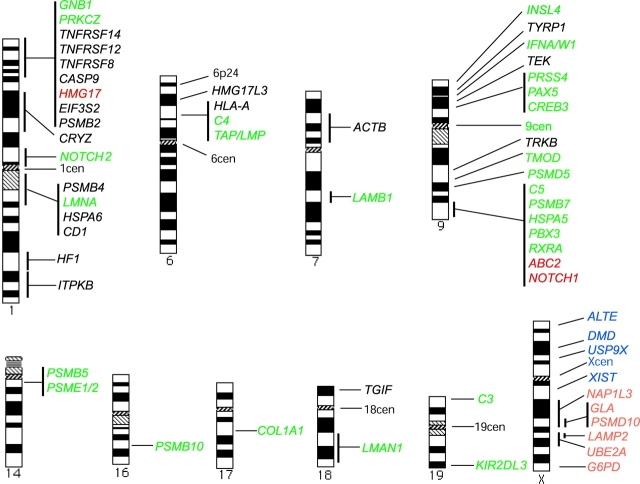
Figure 1.
Mapped locations of loci analyzed for association with PML bodies. Loci found to be more significantly associated with PML bodies than the TAP/LMP locus are shown in red. Loci that are as associated as the TAP/LMP locus are shown in green. Loci that are less significantly associated are shown in black. For the X chromosome, loci on the active and inactive homologues were compared. Loci that were more significantly associated with PML bodies on the active than the inactive chromosome are shown in orange. Loci that are equally associated with PML bodies on both X chromosomes are shown in blue.
PML bodies associate with regions of high transcriptional activity
Our initial observations suggested that regions of increased transcriptional activity were more likely to be closely associated with PML bodies. Results for chromosomes 1 and 9, each of which has regions of different gene densities and transcriptional activity, are shown in Fig. 2 as examples of our findings. Therefore, we analyzed the correlation statistically, using linear regression of the mean minimum locus–PML distances (mmd-locus) for 54 loci shown in Table S1 against their local transcriptional activity. For analysis of local transcriptional activity, expression levels were calculated in arbitrary units for all known genes within 2 centiRays around each locus using data from the human transcriptome map (Caron et al., 2001). The findings are represented in Fig. 3 A, and show a significant correlation between high local transcriptional activity and association with PML bodies. (R2 = 0.16, F = 9.69, P = 0.003). This significance persisted after weighting according to the variances. Furthermore, the strength of the association was increased by deleting the centromeric and outlying loci with >500 U (TAP/LMP, C4, PSMB10, GNB1, and PRKCZ), resulting in R2 = 0.38, F = 25.6, P < 0.00001 (Fig. 3 B).
Figure 2.
Comparison of locus–PML associations for genes on chromosomes 1 and 9 in relation to the local transcription activity, local gene density, and individual gene transcription. Local transcription activity was calculated for ± 1 centiRay around each locus, local gene density was calculated for ± 500 kb around each locus, and individual gene activity was determined relative to transcription of the β-actin gene. Locus–PML association is represented by mmd-locus in micrometers, with error bars showing 1 standard error.
Figure 3.
Linear regression graphs comparing locus–PML association with different parameters. A and B show the the local transcriptional activity, C shows individual gene transcription, and D shows the local gene density. For B, centromeric and outlying loci were excluded. The locus–PML association is represented by mmd-locus in micrometers. The local transcription activity, local gene density, and individual gene transcription are calculated as in Fig. 2. Each bullet represents a locus. Each graph displays the best line fit for the regression.
As the transcriptional activity of a region is dependent on both the local gene density and the transcriptional status of each gene, we wished to determine the relative contributions of both factors to the association. Regression analysis showed that there was no significant correlation between the transcription level of an individual gene (assessed by RT-PCR) and its association with PML bodies. The findings are represented in Fig. 3 C (R2 = 0.069, F = 3.86, P = 0.055). Local gene density was calculated within 1 megabase (Mb) of the gene tested (data from the National Cell Biology Institute and the Sanger Institute). Regions of 1 Mb were chosen, as it was found that genes separated by 500 kb can have statistically different associations with PML bodies (Fig. S2 A). Regression analysis showed a significant association of gene-rich regions with PML bodies compared with gene-poor regions (R2 = 0.096, F = 5.52, P = 0.02). The findings are represented in Fig. 3 D.
Therefore, our results showed that PML bodies associate with genomic regions of increased transcription, rather than with individual genes with high transcription levels. Conversely, genes such as NOTCH1 in regions of high activity, although not being highly expressed themselves, can have strong associations with PML bodies statistically. Within these regions of high local activity, PML bodies may associate with different genes at different times, again suggesting that the association of a region was not the effect of an individual gene (Fig. S2 B). Fig. S3 shows the regression analyses using loci in direct contact with PML bodies.
Association of PML bodies with loci on the active and inactive X chromosomes
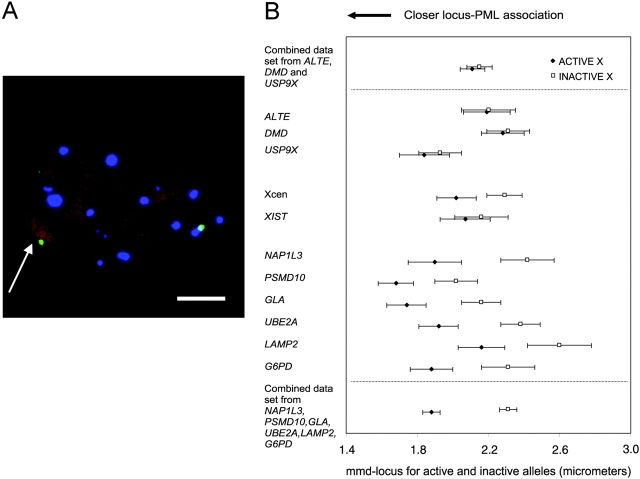
The active and inactive X chromosome territories have been shown to have similar volumes (Eils et al., 1996). When we compared the PML bodies around the active and inactive X chromosomes in a female fibroblast cell line, we found more PML bodies aggregating around the active X chromosome. This was confirmed using the paired t test (mean number of PML bodies around active X chromosome = 1.34, inactive X chromosome = 0.78, P = 0.0001; n = 51).
Specific loci along the active and inactive X chromosomes were then analyzed (Fig. 4). The homologues of six of eleven loci studied (G6PD, GLA, LAMP2, UBE2A, NAP1L3, and PSMD10) on the active X chromosome were found to be significantly more closely associated with PML bodies (P < 0.05) than those on the inactive X chromosome. In contrast, no significant difference was found in association with PML bodies between the ALTE, DMD, and USP9X loci in or around the pseudoautosomal regions on Xp (Carrel et al., 1999), or with the centromeres on these chromosomes. We also did not observe a more interior nuclear position of the Xp arm or the pseudoautosomal loci compared with the Xq arm or nonpseudoautosomal loci. Finally, the XIST genes on each X chromosome associated with PML bodies to a similar extent. The XIST gene, although only expressed from the inactive X chromosome (Brown et al., 1991), lies in a region not known to be a pseudoautosomal region on the Xq arm.
Figure 4.
Association of PML bodies with loci on the active and inactive X chromosomes. (A) WI-38 fibroblast cell nucleus showing the PSMD10 gene (green) on the X chromosome, PML bodies (blue), and propidium iodide counterstain (red). The inactive copy of the gene is distinguished by the Barr body (arrow). Bar, 5 μm. (B) Comparison of the locus–PML distances for loci along active and inactive X chromosomes. The locus–PML association is represented as the mmd-locus distances for the active and inactive copy of each locus, with error bars showing 1 standard error. Results for loci in and out of the pseudoautosomal regions are pooled and displayed at the top and bottom of the figure, respectively.
PML bodies have dynamic associations with cell cycle–regulated genes
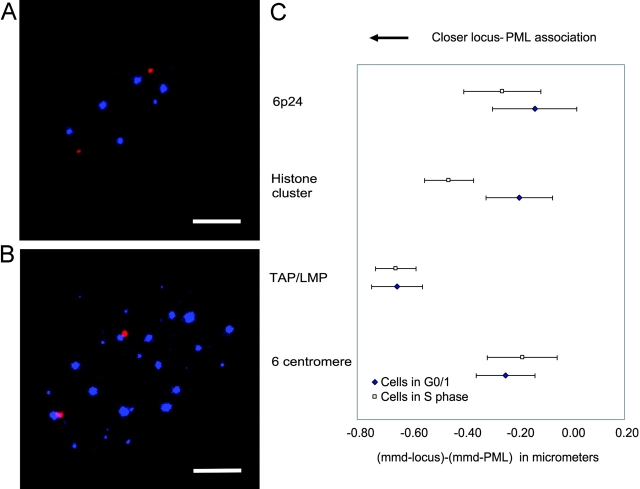
Previously, we showed that association of the TAP/LMP locus with PML bodies remains the same in the G0/G1 and S-phases of the cell cycle (Shiels et al., 2001). We have now extended these analyses to other regions of chromosome 6. These include the 6p22 region that contains genes encoding histone proteins, a large proportion of which are transcribed at high levels predominantly in S-phase (van der Meijden et al., 2002), as well as the chromosome 6 centromere and 6p24 region.
Cells in G0/G1 and S-phases were distinguished by incorporation of BrdU (Fig. 5, A and B). Cells in G2 detected by a double signal per allele were excluded. We found that cells in S-phase had more PML bodies, and hence a decrease in the PML–PML distances. Adjusting for this, no significant difference was found between these phases of the cell cycle for associations of PML bodies with the TAP/LMP, chromosome 6 centromere, and 6p24 loci. In contrast, the 6p22 region showed a significant increase in association in S-phase compared with G0/G1 phase (Fig. 5 C).
Figure 5.
Association of PML bodies with loci along chromosome 6, in cells in S-phase compared with cells in G0/G1 phase. (A and B) MRC5 cell nuclei showing the histone gene cluster (red) at 6p22, PML bodies (blue), and incorporated BrdU (green) [A, cell in non-S (G0/G1) phase; B, cell in S-phase]. Bar, 5 μm. (C) Locus–PML distances for different loci were compared between cells in S-phase versus G0/G1 phase. “(mmd-locus) − (mmd-PML)” on the x axis represents the distance to which a locus is further from or closer to the nearest PML body compared with the PML–PML distance, with error bars showing 1.4 standard errors. t tests were performed between the cells of G0/G1 and S-phases for the four loci. Only the histone cluster on 6p22 shows a significant difference in gene-PML association between cells in S- versus G0/G1 phase.
Proximity of a locus to PML bodies does not directly affect the transcription rate
Our work suggests that PML bodies associate with genomic regions that have high transcription, rather than with highly transcribed individual genes. To determine if PML bodies are directly involved in the transcription of the genes within these regions, we used RNA interference (RNAi) treatment on MRC5 cells, with small-interfering RNA (siRNA) as previously designed (Bruno et al., 2003). Using immunofluorescence, RNAi treatment resulted in 52% of cells expressing no PML bodies, with the remainder expressing approximately half the normal number of PML bodies (Fig. 6, A and B). With immunoblotting assays, we found the level of PML protein to be reduced by 60% (Fig. 6 C). We would expect this reduction of PML to lead to impaired transcription of genes that are normally closely associated with PML bodies. Semi-quantitative reverse-transcriptase PCR (RT-PCR) was initially performed for all genes in Table S1 on RNAi-treated cells compared with untreated and nonsilencing siRNA-treated cells. We found that the transcription levels of genes that are normally closely associated with PML bodies were not more substantially affected than those of genes that are less closely associated with PML bodies (Fig. S4 A). This measurement was confirmed using real-time PCR on 12 genes chosen to represent the different levels of association with PML bodies (Fig. 6 D).
Figure 6.
The effect of knock-down and formation of new PML bodies on transcription levels. (A and B) MRC5 cells showing typical PML body distribution in normal (A) and PML siRNA-treated (B) cells. (C) Immunoblot assay for PML and β-actin (ACTB) proteins from cell lysates in PML RNAi-treated MRC5 cells (a), nonsilencing RNAi-treated cells (b), and untreated cells (c). The PML protein in sample a was quantified by analysis software to be 40% of the intensity compared with sample b, and 55% compared with sample c. The β-actin protein in sample a was 97% compared with sample b, and 98% compared with sample c. (D) Graph showing the change in transcription levels of genes in cells treated with PML RNAi, measured by real-time PCR, as compared with cells treated with nonsilencing siRNA. Loci studied were arranged in increasing mmd-locus values. The vertical axis represents the ratio of transcription level of a gene in RNAi-treated cells to that of the control group (= 1), on a log2 scale. Therefore, a change above the line means an increase in expression of a gene, whereas a change below the line means a decrease in expression, compared with the control group. (E and F) NB4 cells showing the microspeckled pattern of PML protein (red) before ATRA treatment (E), and the formation of PML bodies (blue) after differentiation (F). F also shows four TAP/LMP loci (green) and 6p24 loci (red) in the treated cells (NB4 cells have a hypotetraploid karyotype). (G) A graph showing the change in transcription levels by real-time PCR of genes in NB4 cells after treatment with ATRA, compared with untreated cells. Horizontal and vertical axes are scaled as in D. Bars, 5 μm.
Previous papers showed that treatment of NB4 APL cells with all-trans retinoic acid (ATRA) results in the formation of discrete PML bodies, as well as a change in expression of multiple genes (Lee et al., 2002; Fig. 6, E and F). In our work, although more alterations in transcription levels were noted, we again found no differences between genes that are more closely associated with PML bodies and genes that are less closely associated (Fig. S4 B). Once again, real-time PCR analysis of a subset of these genes confirmed the initial results (Fig. 6 G). An inverse correlation was not observed between the effects of RNAi knock-down in MRC5 cells and ATRA treatment of NB4 cells, presumably due to the more complex changes in NB4 cells (Fig. S4 C).
However, we found that when NB4 cells were treated with ATRA, newly formed PML bodies were positioned statistically closer to the TAP/LMP locus than the 6p24 locus or the chromosome 6 centromere (mmd-TAP/LMP = 1.88 μm vs. mmd-6p24 = 2.41 μm, t score = 7.04, P < 0.01; mmd-TAP/LMP = 1.92 μm vs. mmd-6cen = 2.65 μm, t score = 8.18, P < 0.01). These experiments clearly show that PML bodies form preferentially in specific nuclear regions, in both ATRA-treated NB4 cells and fibroblasts. They also suggest that PML bodies form in the vicinity of (and in response to) local transcriptional activity, rather than being responsible for nearby activity.
PML bodies are not invariably sites of transcription
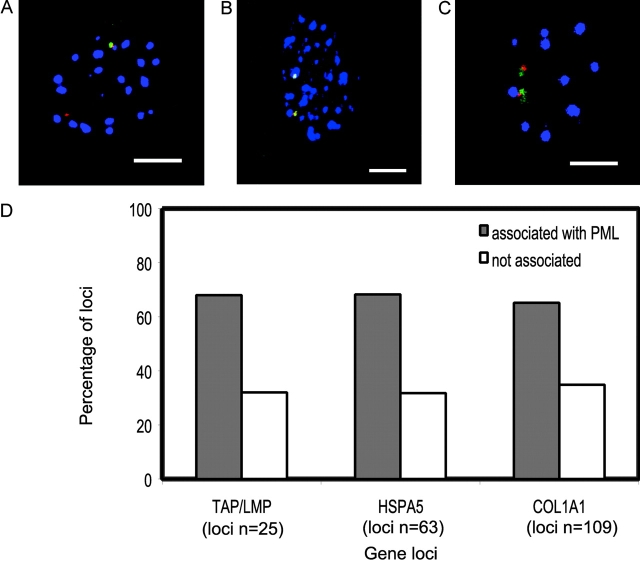
To determine whether PML bodies are themselves sites of transcription, we used RNA-FISH to visualize RNA transcripts for three loci: the TAP/LMP cluster (in which the coordinately expressed LMP2, LMP7, TAP1, and TAP2 were simultaneously detected), and the HSPA5 and COL1A1 genes (Fig. 7, A–C). To induce the expression and facilitate RNA-FISH, cells were either treated with IFNγ at 200 U/ml for detecting the TAP/LMP transcripts, or subjected to heat shock for the HSPA5 transcript. These genes were selected as they were among the most highly expressed in this paper after induction. We found that although the majority of RNA transcripts were adjacent to PML bodies, some transcripts were some distance away (Fig. 7 D). These findings suggest that neither PML bodies nor their immediate surroundings serve specifically as transcription foci, at least for these three loci.
Figure 7.
Association of RNA-FISH signals with PML bodies for the TAP/LMP, HSPA5, and COL1A1 loci. (A–C) MRC5 cells showing the RNA transcript signals (green), corresponding genomic loci (red), and PML bodies (blue) for the TAP/LMP (A), HSPA5 (B), and COL1A1 (C) loci. The TAP/LMP locus comprises the genes LMP2, LMP7, TAP1, and TAP2. (D) The proportion of RNA signals (in cells with detectable signals) that were in direct contact with a PML body for the three loci, compared with those not in direct contact. RNA signals tended to be larger than the corresponding DNA signals, suggesting the spreading of RNA transcripts from the gene location. RNA loci were considered in contact with a PML body if any part of the detectable signal was visually seen to be in contact. The percentages of RNA signals in contact with PML bodies were 68% (TAP/LMP), 68% (HSPA5), and 65% (COL1A1). For the same loci (with detectable RNA signals), the percentages of DNA signals in contact with PML bodies (which may reflect the site of transcription; not depicted) were 52% (TMP/LMP), 70% (HSPA5), and 39% (COL1A1). These scores were not directly comparable to the corresponding scores in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200305142/DC1) because for TAP/LMP and HSPA5, cells were treated such that the number of PML bodies were increased. Bars, 5 μm.
PML bodies associate with genes encoding proteins of diverse functions
Because the MHC contains a large number of genes involved in immune function and proteolysis, and there are several paralogous regions scattered around the genome, we wished to determine whether there is a preferential association of these types of genes with PML bodies. First, we reanalyzed the proteasome encoding genes on chromosomes 1, 9, 14, and 16. We found that PSMB2 and PSMB4 genes are statistically less associated using t test than the TAP/LMP locus (containing LMP2 and LMP7), whereas the other proteasome-encoding genes tested were comparable with the TAP/LMP locus (Table S1). Simultaneous visualization of the TAP/LMP, PSMB7, and PSMB10 loci showed that these loci do not preferentially associate with the same PML body (Fig. S5 A). Some of the proteasome-encoding genes (PSMB5, PSMB7, PSMB10, LMP2, LMP7, and PSME1/2) were noted to be IFN responsive. We examined the effect of 200 U/ml IFNγ on the TAP/LMP, PSMB7, and PSMB10 loci using the chromosome 6 centromere and a region on band 6p24 as controls, and found that none of the regions showed increased associations with PML bodies with IFNγ treatment (Table S2).
A second subset of genes examined were those paralogous to genes in the MHC. Of these, HMG17 (1p36), ABC2, and the adjacent NOTCH1 genes (9q34) were statistically more associated with PML bodies than the TAP/LMP locus, and were the most highly associated of all genes tested (Fig. S5 B). The NOTCH2 gene (1p11-13) was statistically as associated as the TAP/LMP locus, whereas the CD1 (1q21) and HSPA6 (1q12-21) genes were not associated.
Next, we examined a set of genes encoding structural proteins that had been tested elsewhere for association with SC-35 domains (Smith et al., 1999). The COL1A1, LMNA, and ACTB genes were reported to be associated with SC-35 domains, whereas LAMB1 was not. These findings were verified by our experiments (unpublished data). As PML bodies and SC-35 domains both lie in the interchromosomal space, the apparent association of a locus with a PML body may actually reflect its association with an SC-35 domain. Thus, we examined the association of these four genes with PML bodies, and found that although the COL1A1, LMNA, and LAMB1 genes associate with PML bodies to the same extent as the TAP/LMP locus, the ACTB gene was less significantly associated. We also studied these four loci, as well as the TAP/LMP locus, the chromosome 6 centromere, and the 6p24 region, with PML bodies and SC-35 domains labeled simultaneously, and found that these loci may be in contact with either PML bodies or SC-35 domains independently (Fig. S5 C).
These data suggest that PML bodies do not associate uniformly with particular sets of functionally related genes, nor are they limited in their association with only a single subset of genes. Furthermore, their association with certain genomic regions is unrelated to the proximity of these regions to SC-35 domains.
Discussion
The precise roles of PML bodies have remained elusive despite intensive research. In this work, we formally tested the hypothesis that PML bodies are located close to certain genomic regions and tested their function in transcriptional activity. By examining multiple genomic regions with different features, we found that PML bodies associate with genomic regions of high transcriptional activity, which is a function of both the gene density and the proportion of genes that are active. Single genes that are highly expressed do not alone show significant associations with PML bodies. The regression model showed that some loci were found outside the line fit, suggesting other factors may play a role in their association with PML bodies. For example, the chromosome 9 centromere showed a strong statistical association with PML bodies, possibly due to a region of high gene density and transcriptional activity located adjacent to the centromere on band 9p13. Alternatively, the heterochromatin in this centromere has been shown to have unique properties (Jolly et al., 2002). The organization of PML bodies in relation to transcriptionally active regions is further supported by the association of PML bodies with many genes on the active X chromosome compared with their homologues on the inactive X chromosome, and with replication-dependent histone genes on chromosome 6 in S-phase cells compared with those in G0/G1 phase cells. Newly formed PML bodies in NB4 cells after ATRA treatment also showed a preference for the TAP/LMP region compared with other loci along chromosome 6. Together, these findings show that PML bodies lie within or in close proximity to transcriptionally permissive nuclear compartments (Isogai and Tjian, 2003).
PML bodies have been linked with transcription in several reports. For example, RNA polymerase II transcription foci, visualized by fluorouridine, and nascent RNA have been observed in and around PML bodies (LaMorte et al., 1998; Boisvert et al., 2000; von Mikecz et al., 2000). PML bodies also recruit CBP, a transcriptional coactivator with histone acetylase activity (LaMorte et al., 1998; von Mikecz et al., 2000; Boisvert et al., 2001). However, PML bodies do not appear to be involved with all sites of transcriptional activity because only a minor proportion of transcription foci and regions of acetylated histones in the nucleus colocalize with PML bodies (Boisvert et al., 2000; Kiesslich et al., 2002).
In our experiments, certain loci were found to be associated more statistically with PML bodies, but this association was retained even when those loci that touch PML bodies are excluded from the calculations. This result may be explained by the fact that chromatin can move up to 2 mm (Chubb et al., 2002), and genes that are statistically closer might therefore associate intermittently with PML bodies more often. However, our RNA-FISH experiments with the loci TAP/LMP, HSPA5, and COL1A1 showed that these genes when being transcribed, or their transcripts, are not always adjacent or touching PML bodies, although the three loci themselves showed a high degree of association with PML bodies. This is similar to a previous report in which Cajal bodies associated with histone genes, but were not necessarily sites for transcription (Shopland et al., 2001). Therefore, we propose that a locus may form a statistically significant but indirect association with PML bodies as a result of its location within a nuclear compartment in which a PML body is likewise positioned.
The four genes in the TAP/LMP cluster are particularly interesting in this regard because they encode proteins that play a central role in the presentation of MHC class I molecules at the cell surface (van Endert, 1999). Expression of these genes was reported to be up-regulated after transfection of PML protein into mouse tumor cells with defective antigen presentation (Zheng et al., 1998). In our experiments using a human cell line, we found that PML bodies are not themselves sites of transcription for these genes, and that PML knock-down by RNAi does not diminish transcription of these genes. We have not yet explored the possibility that PML bodies are involved in post-transcriptional events, as proposed by others (Borden, 2002). However, in human cells, it has recently been shown that PML is not directly involved in the regulation of MHC class I antigen expression at the cell surface (Bruno et al., 2003), complementing our transcription analysis of the HLA-A and TAP/LMP genes.
These results also prompted us to examine whether PML bodies are associated with functional subsets of genes. Genes with immune function were of immediate interest in view of our previous demonstration of a spatial association between the MHC and PML bodies (Shiels et al., 2001), and other evidence linking PML bodies to immune function. For example, IFNγ treatment leads to increased expression of the PML gene and an increase in the number of PML bodies (Regad and Chelbi-Alix, 2001). IFNα increases the amount of nuclear DNA helicase II, nascent RNA transcripts, and CBP associating with PML bodies (Fuchsova et al., 2002; Kiesslich et al., 2002).
In our current experiments, significant associations with PML bodies were seen with many genes that have immune function and/or are IFN responsive. These include genes encoding the proteasome subunits LMP2, LMP7, PSMB10, PSME1, and PSME2, which are up-regulated by IFNγ, and the PSMB7, COL1A1, and LMNA genes, which are down-regulated by IFNγ (Boehm et al., 1997). Although these findings might suggest that PML bodies play a role in the transcription of such genes, no increased association of several of the IFN up-regulated genes with PML bodies was observed after IFNγ treatment. It should be noted that our statistical method takes into account the increase in the number of PML bodies in the nucleus, which is particularly important when assessing the effects of treatments that increase the number of PML bodies.
Many of the immune-related genes we tested lie in gene-rich regions and may be clustered to facilitate coordinated expression. Therefore, using our current statistical model, it is not possible to determine whether the association is due to the presence of functionally related genes rather than other factors such as gene density. It should also be noted that genes with nonimmune functions such as HMG17 (coding a chromosomal protein), ABC2 (transporter protein), and NOTCH1 (signaling receptor) show the most highly significant associations of all genes tested with PML bodies (Luciani et al., 1994; Artavanis-Tsakonas et al., 1995; Bustin et al., 1995). Thus, these data demonstrate that although PML bodies associate with a large group of immune-related genes, they are also strongly associated with other genes with disparate functions.
The central question to arise from these findings is whether PML bodies directly regulate transcription of genes with which they are spatially associated. Our RNAi experiments showed that knock-down of PML protein, leading to a reduction in the number of PML bodies, did not preferentially alter transcription levels of genes that were closer to PML bodies, or other subsets of genes such as those which are IFN responsive, mentioned earlier in this section. Regulation of transcription of specific genes by PML bodies has been previously reported to occur by the accumulation and release of transcriptional activators and repressors in a controlled manner (Vallian et al., 1998; Guo et al., 2000; Lehembre et al., 2001; Wu et al., 2002). However, these reports are not directly comparable to ours because they use transfection of exogenous PML protein and various transcription factors. Furthermore, they show that PML facilitates, but is not essential for, specific transcription pathways. Therefore, the role of PML bodies in transcription in the physiological setting has not been established. It may be that PML bodies only regulate transcription during overexpression or activation of a particular pathway, rather than basal transcription. Alternatively, the association of a genomic region with PML bodies may influence the expression of nearby genes that were not analyzed here by RT-PCR.
Previously, we showed that giant chromatin loops carrying the MHC region appear after IFN induction of MHC gene expression (Volpi et al., 2000). We now find that the positioning of signals at the painted chromosome territory surface or on large external chromatin loops does not correlate with increased PML association (Fig. S4, Table S3), despite PML bodies being found mostly in the interchromosomal space (Bridger et al., 1998). This result concurs with that of others (Williams et al., 2002). However, we noted that loci that had a higher incidence of external signals were also more likely to associate with PML bodies, although not necessarily at the same time. Both observations may be independently related to the transcriptional activity, as it has been shown that local gene density and transcriptional activity also affect the external localization of loci (Mahy et al., 2002). We excluded the possibility that the observed association of loci with PML bodies was due to their proximity to other nuclear inclusions such as Cajal bodies (Shiels et al., 2001) or SC-35 domains. It may be argued that PML bodies associate with specific loci due to their radial position in the interior of the cell nucleus. Our data on the pseudoautosomal region of the inactive X chromosome appear to suggest otherwise.
A role for PML bodies in transcription through the regulated recruitment and release of factors necessary for transcription has been mentioned above. A more general role for PML bodies as a nuclear depot containing transcription factors and other proteins that alter the nuclear activities in response to external stimuli, such as stress, has recently been proposed (Negorev and Maul, 2001). The recent discovery that proteasomes can accumulate in PML bodies suggests that degradation of excess proteins may occur at these sites as well (Mattson, 2000; Fabunmi et al., 2001). Therefore, we suggest that PML bodies are distributed so they function as depots for factors required for dense transcription foci in these regions, as shown in Fig. 8.
Figure 8.
Proposed model of PML body function and position in the nucleus. PML bodies aggregate at regions of highest density/levels of transcription activity. Transcribed genes that are in regions with low overall transcription activity are less likely to associate with PML bodies.
Further, we propose that this positioning of PML bodies is more predominant under normal cell growth. In specific circumstances, a proportion of PML bodies are also found aggregated at specific locations, such as at sites of stable transfected lac operator arrays after activation by the lac repressor protein (Tsukamoto et al., 2000). At sites of DNA damage, the BLM and MRE11 DNA repair proteins are recruited to the aggregated PML bodies (Bischof et al., 2001; Muto et al., 2001; Carbone et al., 2002). With these heterogeneous functions, individual PML bodies within a nucleus may have different constituent proteins and perform different functions, depending on the requirements of the nucleus (Bloch et al., 1999). However, we conclude that in interphase cells, PML bodies are distributed in a specific manner in proximity to nuclear domains of high transcriptional activity.
Materials and methods
Cell culture and treatment
MRC5 male and WI-38 female human primary fibroblasts were obtained from the American Type Culture Collection, and were cultured as monolayers in RPMI 1640 media supplemented with 10% FCS and 2 mM glutamine. NB4 (APL) cells were grown in suspension in the same medium. Heat shock of MRC5 cells was performed at 44°C for 1 h on a heating block. Recombinant human IFNγ (R&D Systems) was used at a concentration of 200 U/ml for 24 h to up-regulate transcription of MHC genes. Cells in S-phase were labeled by adding BrdU (Sigma-Aldrich) to the culture medium at a final concentration of 100 μM for 1 h immediately before fixation. Differentiation of NB4 cells was induced with 1 μM ATRA for 48 h. RNAi treatment on MRC5 cells was performed using 21-nt siRNA strands as described previously (Bruno et al., 2003). This oligonucleotide and a nonsilencing control siRNA oligonucleotide were obtained from QIAGEN. Transfection of siRNAs was performed using the TransMessenger™ reagent (QIAGEN) or the Oligofectamine® reagent (Invitrogen) 48–96 h before harvesting.
Antibodies and DNA probes
Antibodies used in this work were as follows: Rabbit polyclonal anti-PML (Borden et al., 1995) to detect PML nuclear bodies, mouse anti-BrdUTP (Boehringer) to detect BrdU incorporated into newly replicating DNA, and monoclonal mouse anti-splicing factor SC-35 (Sigma-Aldrich) to detect SC-35 domains. Secondary antibodies used were anti–mouse Alexa® 488, anti–rabbit Alexa® 546 (both from Molecular Probes, Inc.), and anti–rabbit cy5 (Amersham Biosciences).
The LMP/TAP locus in the MHC was detected using a pool of cosmids (HA14, U15, U10, and M4) containing the LMP2, LMP7, TAP1, and TAP2 genes (a gift from S. Beck, Sanger Institute, Cambridge, UK). A subsection of the 6p24 region was detected using pooled cosmids A9.5, B5.10, B10.10, B12.10, E11.2, and F1.6 (a gift from J. Ragoussis, University of Oxford, Oxford, UK). The histone H2-4 region in band 6p22 was detected using pooled cosmids A1037, H2317, and N1948 (a gift from J. Trowsdale, University of Cambridge, Cambridge, UK). Probes for loci on other chromosomes were bacterial artificial chromosomes and P1 artificial chromosomes derived from Dr. Pieter de Jong (Roswell Park Cancer Institute, Buffalo, NY) and provided by the Sanger Institute (listed in Table S4). The centromeres and whole-chromosome territories of chromosomes 1, 6, 9, 18 and 19, and X were detected using commercial biotinylated satellite probes and red or green whole-chromosome paints (Appligene Oncor). Xp and Xq arm probes (biotin and digoxigenin labeled, respectively) were obtained from Cambio. Because the satellite probe for chromosome 19 cross-hybridizes to chromosomes 1 and 6, it was hybridized together with the chromosome 19 paint.
Combined immunofluorescence and FISH (immuno-FISH)
Cells were grown, fixed, and permeabilized as described previously (Shiels et al., 2001). Cells were heat denatured on slides before hybridization of 300 ng denatured and biotin- and/or digoxigenin-labeled DNA probe at 37°C overnight, followed by standard formamide and SSC washes. DNA probes used are described in the previous section. Biotin-labeled DNA probes were detected using streptavidin-Alexa® 546 (Molecular Probes, Inc.) or streptavidin-Cy5 (Cambio), and digoxigenin-labeled DNA probes were detected using mouse and sheep mouse anti-digoxigenin-FITC or -Texas red (Sigma-Aldrich and Boehringer). Detection of nuclear proteins by immunofluorescence was performed simultaneously with the FISH detection steps. Female diploid fibroblast cells (WI-38) were counterstained in propidium iodide at 1 mg/ml. The inactive X chromosome was identified as a densely stained Barr body.
For RNA-FISH experiments, the biotinylated genomic probes for TAP/LMP, HSPA5, and COL1A1 were chosen at higher amounts of 600 ng per slide. Ribonuclease-free reagents were used where possible, and vanadyl ribonucleoside complex (New England Biolabs, Inc.) was added to the permeabilization and hybridization steps at a final concentration of 20 mM. The slide denaturation step was omitted, and detection of biotin was performed using the tyramide signal amplification system (PerkinElmer). Cells were then treated with ribonuclease A, heat denatured, and DNA-FISH (using digoxigenin-labeled probes for the same gene loci) and immunofluorescence were performed as above.
Microscopy and distance measurements
Image capture and analysis were as described previously (Shiels et al., 2001). Confocal image stacks were processed using the Image3D program (written by S.A. Islam). This produces three-dimensional coordinates of centroid positions for each locus and PML body, allowing the Euclidean distance between two foci to be calculated.
At least 30 nuclei were captured per experiment. For each locus, we calculated the distance to its nearest PML body, referred to as the minimum locus–PML distance. The mean minimum locus–PML distance (mmd-locus) for the two alleles was obtained for each nucleus, except for experiments involving the active/inactive X genes and locus position within chromosome territories where each allele was considered separately. In addition, for each PML body we calculated the minimum distance to its nearest PML body. The mean of the minimum PML–PML distances (mmd-PML) for all PML bodies was then calculated for each nucleus. Where three-color experiments were used to compare directly mmd-locus for two loci, the values of mmd-locus1 and mmd-locus2 were obtained as above. The percentage of minimum locus–PML distances that were <1 μm was also calculated. In addition, each cell was examined as a three-dimensional stack of images by two independent observers. Each locus was scored to be either physically touching a PML body, or not. Percentage of loci in contact with a PML body was calculated as the mean of the two observations.
Statistical analysis
To determine whether the associations of loci with PML bodies correlate with their transcriptional state, the gene density, or the transcriptional activity surrounding the loci, multiple linear regression analyses were performed. Where appropriate, weighted least squares were used with weights proportional to the inverse of the variance of the mmd-locus distance. Gene density was calculated for 500 kb on either side of the gene tested, using mean data from the National Cell Biology Institute gene mapping database (http://www.ncbi.nlm.nih.gov/) and the Sanger Institute (http://www.ensembl.org). Novel or predicted genes were included. The transcriptional level of a gene was determined by RT-PCR (see next section). Local transcriptional activity was calculated taking the expression data from the human transcriptome map (http://bioinfo.amc.uva.nl/HTMseq/controller). We summed the expression levels of genes within 1 centiRay on either side of the gene tested or the nearest available neighboring gene mapped, using the GB4 radiation hybrid map. As the MRC5 cell line is a fibroblast cell line derived from the lung, relative transcription units were derived for each gene from the averages of data from the normal lung and the skin fibroblast libraries. The distances (mmd-locus) for individual loci were analyzed against all three variables. Higher R2 and F indicated a stronger correlation, with P score values of <0.05 being considered a significant correlation.
For direct comparisons between the mmd for two loci, the t test was performed using two-sided P values, assuming the null hypothesis of equal means for the two loci. Where both loci were detected simultaneously using three-color experiments, paired t tests were used. For comparisons of mmd-locus between two different sets of cells, such as in a set of untreated cells versus treated cells, or cells in G0/G1 phase versus S-phase, the effect of treatment was then investigated using an unpaired t test on the two sets of mmd-locus values under the assumption of equal variance. Where the mmd-PML distances are statistically different in the two sets of cells, the comparison was made between the differences [(mmd-locus) − (mmd-PML)]. The difference in the association of PML bodies between two loci is said to be statistically significant if the P value is <0.05.
Total RNA isolation, RT-PCR, and real-time PCR
RNA was extracted from cells using TRIzol® reagent (GIBCO BRL) according to the manufacturer's protocol. 1 μg total RNA was then reverse transcribed using the First Strand cDNA Synthesis kit (Amersham Biosciences), using hexa-dNTP primer. cDNA made from 100 ng total RNA was mixed with 1 μM of each primer (see Table S5), and the appropriate volumes of the Taq PCR Master Mix (QIAGEN) and distilled water. The PCR reaction was performed for 30 cycles. PCR products were loaded onto 1.5% agarose gels labeled with ethidium bromide. The PCR product bands were scanned using UV light with LabWorks capture software (Ultra-Violet Products), which allowed the intensity of the bands to be quantified. Relative intensities were calculated using the intensity of the expressed gene β-actin (ACTB) as the reference (= 1). To compare expression in different cell populations (e.g., after RNAi or ATRA treatments), band intensities were first normalized on the ACTB gene for the corresponding cDNA sample. The relative intensities for the treated group and the control group were then calculated.
For real-time PCR, cDNA samples were prepared as above. Primers were redesigned (Table S5), and 300 nM of each primer pair was mixed with cDNA from 100 ng total RNA and the appropriate volumes of SYBR® green PCR Master Mix (Applied Biosystems) and distilled water. The PCR reaction was performed on a Sequence Detector System (ABI PRISM 7700; Applied Biosystems) for 40 cycles, with an annealing temperature of 60°C. Averages of calculated thresholds (Cts) were obtained for duplicated samples, and corrected against the ACTB reference. The difference in Cts between treated and control samples then allowed the relative expression of a gene to be quantitated as follows: ratio of treated to untreated = 2−[ (Ct-treated − Ct-ACTB) − (Ct-control − Ct-ACTB) ].
Protein electrophoresis and immunoblotting
Whole-cell lysates were obtained by adding 1 cell volume of 2× electrophoresis sample buffer (125 mM Tris, pH 6.8, 4% SDS, 10% glycerol, 0.006% bromophenol blue, and 2% DTT) to MRC5 cells treated with RNAi and controls. The lysate was boiled further for 5 min, and equal volumes were separated on 4–20% SDS-PAGE gels. Proteins were transferred to nitrocellulose, and PML and β-actin protein bands were detected with mouse monoclonals anti-PML (PG-M3; Santa Cruz Biotechnology, Inc.) and anti-β-actin (Sigma-Aldrich) antibodies, using a WesternBreeze kit (Invitrogen). Protein bands were quantitated using LabWorks software (Ultra-Violet Products).
Online supplemental material
Fig. S1 shows the relationship between loci–PML distance measurements and visual counts of loci in direct contact with PML bodies. Fig. S2 is representative of cells from immuno-FISH experiments used to detect two loci mapped in close proximity to each other, and their association with PML bodies. Fig. S3 correlates the proportion of genes in direct contact with PML bodies, with their local transcriptional activity, gene density, and individual gene transcription levels. Fig. S4 shows the effect of knock-down and formation of new PML bodies on the transcription levels of all genes analyzed by RT-PCR. Fig. S5 represents cells from immuno-FISH experiments in which PML bodies and loci from subsets of similar genes are detected. Fig. S6 illustrates the relationship between locus–PML associations and the position of loci within their chromosome territories.
Table S1 shows the detailed results of the locus–PML association for all autosomal loci analyzed, together with their local transcriptional activity, gene density, and individual gene transcription levels. Table S2 shows the effect of IFNγ on the associations of various loci along chromosome 6 with PML bodies. Table S3 compares the direct contact of loci and PML bodies with the location of the locus signals relative to the chromosome territory. Table S4 lists the bacterial/P-1 artificial chromosome probes used in the immuno-FISH experiments and the primers used in RT-PCR. Table S5 lists the primers used for genes analyzed using real-time PCR. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200305142/DC1.
Acknowledgments
We thank Stephan Beck, Jiannis Ragoussis, and John Trowsdale for genomic probes. We also thank Ian Tomlinson, Alistair Newall, Petros Takousis, and Emanuel Volpi for helpful discussions.
This work was supported by Cancer Research UK.
The online version of this article includes supplemental material.
Abbreviations used in this paper: APL, acute promyelocytic leukemia; ATRA, all-trans retinoic acid; CBP, cAMP-response element binding protein; MHC, major histocompatibility complex; mmd, mean minimum distance; PML, promyelocytic leukemia; RNAi, RNA interference; SiRNA, small-interfering RNA; TAP/LMP, transporter associated with antigen processing/large multifunctional protease.
References
- Artavanis-Tsakonas, S., K. Matsuno, and M.E. Fortini. 1995. Notch signaling. Science. 268:225–232. [DOI] [PubMed] [Google Scholar]
- Bischof, O., S.H. Kim, J. Irving, S. Beresten, N.A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, D.B., J.D. Chiche, D. Orth, S.M. de la Monte, A. Rosenzweig, and K.D. Bloch. 1999. Structural and functional heterogeneity of nuclear bodies. Mol. Cell. Biol. 19:4423–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, U., T. Klamp, M. Groot, and J.C. Howard. 1997. Cellular responses to interferon-γ. Annu. Rev. Immunol. 15:749–795. [DOI] [PubMed] [Google Scholar]
- Boisvert, F.M., M.J. Hendzel, and D.P. Bazett-Jones. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert, F.M., M.J. Kruhlak, A.K. Box, M.J. Hendzel, and D.P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K.L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden, K.L., M.N. Boddy, J. Lally, N.J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P.S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger, J.M., H. Herrmann, C. Munkel, and P. Lichter. 1998. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J. Cell Sci. 111:1241–1253. [DOI] [PubMed] [Google Scholar]
- Brown, C.J., A. Ballabio, J.L. Rupert, R.G. Lafreniere, M. Grompe, R. Tonlorenzi, and H.F. Willard. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 349:38–44. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., S. Amoils, J.M. Horn, V.J. Buckle, D.R. Higgs, M. Merkenschlager, and A.G. Fisher. 2001. Expression of α- and β-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606. [DOI] [PubMed] [Google Scholar]
- Bruno, S., F. Ghiotto, F. Fais, M. Fagioli, L. Luzi, P.G. Pelicci, C.E. Grossi, and E. Ciccone. 2003. The PML gene is not involved in the regulation of MHC class I expression in human cell lines. Blood. 101:3514–3519. [DOI] [PubMed] [Google Scholar]
- Bustin, M., L. Trieschmann, and Y.V. Postnikov. 1995. The HMG-14/-17 chromosomal protein family: architectural elements that enhance transcription from chromatin templates. Semin. Cell Biol. 6:247–255. [DOI] [PubMed] [Google Scholar]
- Carbone, R., M. Pearson, S. Minucci, and P.G. Pelicci. 2002. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 21:1633–1640. [DOI] [PubMed] [Google Scholar]
- Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M.C. Hermus, R. van Asperen, K. Boon, P.A. Voute, et al. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 291:1289–1292. [DOI] [PubMed] [Google Scholar]
- Carrel, L., A.A. Cottle, K.C. Goglin, and H.F. Willard. 1999. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl. Acad. Sci. USA. 96:14440–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb, J.R., S. Boyle, P. Perry, and W.A. Bickmore. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439–445. [DOI] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The, H., C. Lavau, A. Marchio, C. Chomienne, L. Degos, and A. Dejean. 1991. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 66:675–684. [DOI] [PubMed] [Google Scholar]
- Dietzel, S., K. Schiebel, G. Little, P. Edelmann, G.A. Rappold, R. Eils, C. Cremer, and T. Cremer. 1999. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp. Cell Res. 252:363–375. [DOI] [PubMed] [Google Scholar]
- Dyck, J.A., G.G. Maul, W.H. Miller, Jr., J.D. Chen, A. Kakizuka, and R.M. Evans. 1994. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 76:333–343. [DOI] [PubMed] [Google Scholar]
- Eils, R., S. Dietzel, E. Bertin, E. Schrock, M.R. Speicher, T. Ried, M. Robert-Nicoud, C. Cremer, and T. Cremer. 1996. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 135:1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi, R.P., W.C. Wigley, P.J. Thomas, and G.N. DeMartino. 2001. Interferon γ regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J. Cell Sci. 114:29–36. [DOI] [PubMed] [Google Scholar]
- Frey, M.R., and A.G. Matera. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsova, B., P. Novak, J. Kafkova, and P. Hozak. 2002. Nuclear DNA helicase II is recruited to IFN-α-activated transcription sites at PML nuclear bodies. J. Cell Biol. 158:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, A.D., J. Borrow, P.S. Freemont, and E. Solomon. 1991. Characterization of a zinc finger gene disrupted by the t(15;17) in acute promyelocytic leukemia. Science. 254:1371–1374. [DOI] [PubMed] [Google Scholar]
- Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. Paolo Pandolfi. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730–736. [DOI] [PubMed] [Google Scholar]
- Isogai, Y., and R. Tjian. 2003. Targeting genes and transcription factors to segregated nuclear compartments. Curr. Opin. Cell Biol. 15:296–303. [DOI] [PubMed] [Google Scholar]
- Jensen, K., C. Shiels, and P.S. Freemont. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 20:7223–7233. [DOI] [PubMed] [Google Scholar]
- Jolly, C., L. Konecny, D.L. Grady, Y.A. Kutskova, J.J. Cotto, R.I. Morimoto, and C. Vourc'h. 2002. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 156:775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka, A., W.H. Miller, Jr., K. Umesono, R.P. Warrell, Jr., S.R. Frankel, V.V. Murty, E. Dmitrovsky, and R.M. Evans. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 66:663–674. [DOI] [PubMed] [Google Scholar]
- Kiesslich, A., A. von Mikecz, and P. Hemmerich. 2002. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 140:167–179. [DOI] [PubMed] [Google Scholar]
- Koken, M.H., G. Linares-Cruz, F. Quignon, A. Viron, M.K. Chelbi-Alix, J. Sobczak-Thepot, L. Juhlin, L. Degos, F. Calvo, and H. de The. 1995. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 10:1315–1324. [PubMed] [Google Scholar]
- LaMorte, V.J., J.A. Dyck, R.L. Ochs, and R.M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA. 95:4991–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.H., M.Y. Chang, J.I. Ahn, D.H. Yu, S.S. Jung, J.H. Choi, Y.H. Noh, Y.S. Lee, and M.J. Ahn. 2002. Differential gene expression in retinoic acid-induced differentiation of acute promyelocytic leukemia cells, NB4 and HL-60 cells. Biochem. Biophys. Res. Commun. 296:1125–1133. [DOI] [PubMed] [Google Scholar]
- Lehembre, F., S. Muller, P.P. Pandolfi, and A. Dejean. 2001. Regulation of Pax3 transcriptional activity by SUMO-1-modified PML. Oncogene. 20:1–9. [DOI] [PubMed] [Google Scholar]
- Luciani, M.F., F. Denizot, S. Savary, M.G. Mattei, and G. Chimini. 1994. Cloning of two novel ABC transporters mapping on human chromosome 9. Genomics. 21:150–159. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, S. Gilchrist, R.A. Baldock, and W.A. Bickmore. 2002. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson, J.C. 2000. Acute promyelocytic leukemia. From morphology to molecular lesions. Clin. Lab. Med. 20:83–103. [PubMed] [Google Scholar]
- Muto, A., M. Kizaki, C. Kawamura, H. Matsushita, Y. Fukuchi, A. Umezawa, T. Yamada, J. Hata, N. Hozumi, K. Yamato, et al. 2001. A novel differentiation-inducing therapy for acute promyelocytic leukemia with a combination of arsenic trioxide and GM-CSF. Leukemia. 15:1176–1184. [DOI] [PubMed] [Google Scholar]
- Negorev, D., and G.G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 20:7234–7242. [DOI] [PubMed] [Google Scholar]
- Regad, T., and M.K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene. 20:7274–7286. [DOI] [PubMed] [Google Scholar]
- Roix, J.J., P.G. McQueen, P.J. Munson, L.A. Parada, and T. Misteli. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat. Genet. 34:287–291. [DOI] [PubMed] [Google Scholar]
- Ruggero, D., Z.G. Wang, and P.P. Pandolfi. 2000. The puzzling multiple lives of PML and its role in the genesis of cancer. Bioessays. 22:827–835. [DOI] [PubMed] [Google Scholar]
- Seeler, J.S., and A. Dejean. 1999. The PML nuclear bodies: actors or extras? Curr. Opin. Genet. Dev. 9:362–367. [DOI] [PubMed] [Google Scholar]
- Shiels, C., S.A. Islam, R. Vatcheva, P. Sasieni, M.J. Sternberg, P.S. Freemont, and D. Sheer. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114:3705–3716. [DOI] [PubMed] [Google Scholar]
- Shopland, L.S., M. Byron, J.L. Stein, J.B. Lian, G.S. Stein, and J.B. Lawrence. 2001. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol. Biol. Cell. 12:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.P., P.T. Moen, K.L. Wydner, J.R. Coleman, and J.B. Lawrence. 1999. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J. Cell Biol. 144:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto, T., N. Hashiguchi, S.M. Janicki, T. Tumbar, A.S. Belmont, and D.L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871–878. [DOI] [PubMed] [Google Scholar]
- Vallian, S., K.V. Chin, and K.S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147–7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden, C.M., D.S. Lapointe, M.X. Luong, D. Peric-Hupkes, B. Cho, J.L. Stein, A.J. van Wijnen, and G.S. Stein. 2002. Gene profiling of cell cycle progression through S-phase reveals sequential expression of genes required for DNA replication and nucleosome assembly. Cancer Res. 62:3233–3243. [PubMed] [Google Scholar]
- van Endert, P.M. 1999. Genes regulating MHC class I processing of antigen. Curr. Opin. Immunol. 11:82–88. [DOI] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- von Mikecz, A., S. Zhang, M. Montminy, E.M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis, K., S. Rambaud, C. Lavau, J. Jansen, T. Carvalho, M. Carmo-Fonseca, A. Lamond, and A. Dejean. 1994. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 76:345–356. [DOI] [PubMed] [Google Scholar]
- Williams, R.R., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175. [DOI] [PubMed] [Google Scholar]
- Wu, W.S., Z.X. Xu, and K.S. Chang. 2002. The promyelocytic leukemia protein represses A20-mediated transcription. J Biol Chem. 277: 31734–31739. [DOI] [PubMed] [Google Scholar]
- Zheng, P., Y. Guo, Q. Niu, D.E. Levy, J.A. Dyck, S. Lu, L.A. Sheiman, and Y. Liu. 1998. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 396:373–376. [DOI] [PubMed] [Google Scholar]