Abstract
Hypocalcemia and hyperphosphatemia caused by parathyroid hormone (PTH)-resistance are the only discernible abnormalities in pseudohypoparathyroidism type Ib (PHP-Ib). Because mutations in the PTH/PTH-related peptide receptor, a plausible candidate gene, had been excluded previously, we conducted a genome-wide search with four PHP-Ib kindreds and established linkage to a small telomeric region on chromosome 20q, which contains the stimulatory G protein gene. We, furthermore, showed that the genetic defect is imprinted paternally and thus is inherited in the same mode as the PTH-resistant hypocalcemia in kindreds with PHP-Ia and/or pseudo-pseudohypoparathyroidism, two related disorders caused by different stimulatory G protein mutations.
The term pseudohypoparathyroidism (PHP) first was used in the description of patients with parathyroid hormone (PTH)-resistant hypocalcemia and hyperphosphatemia (1) in whom exogenous PTH fails to increase urinary phosphate and cAMP excretion (2–4). Besides an abnormal regulation of mineral ion homeostasis, affected patients show other endocrine deficiencies and characteristic physical stigmata, now collectively referred to as Albright’s hereditary osteodystrophy (AHO). This syndrome, now termed PHP type Ia (PHP-Ia), is caused by heterozygous inactivating mutations in the GNAS 1 gene encoding the α subunit of the stimulatory G protein (Gsα). These mutations lead to an ≈50% reduction in Gsα activity/protein and thus explain, at least partially, the resistance toward PTH and other hormones that mediate their actions through G protein-coupled receptors (2–4). A similar reduction in Gsα activity/protein also is found in patients with pseudo-pseudohypoparathyroidism, who show the same physical appearance as in PHP-Ia but have no endocrine abnormalities. This suggested that mutations in the Gsα gene are necessary but not sufficient to explain fully either PHP-Ia or pseudo-pseudohypoparathyroidism (2–7). Both disorders typically are found within the same kindred, and recent studies have shown that resistance toward PTH and other hormones is imprinted paternally; that is, PHP-Ia occurs only if the defective gene is inherited from a female affected by either form of the two disorders (8, 9).
Another form of pseudohypoparathyroidism, PHP type Ib (PHP-Ib), also is characterized by PTH-resistant hypocalcemia and hyperphosphatemia. However, in contrast to the findings in PHP-Ia or pseudo-pseudohypoparathyroidism, patients affected by PHP-Ib have normal Gsα activity, lack developmental defects, and typically show, besides resistance toward PTH, no additional endocrine abnormalities (2–4). These differences in clinical and laboratory presentation suggested that PHP-Ia and PHP-Ib are unrelated disorders and therefore were thought to be caused by distinct molecular defects. Because of the selective resistance toward a single hormone, inactivating mutations in the receptor for PTH, i.e., the PTH/PTH-related peptide (PTHrP) receptor (10–12), initially were thought to be responsible for PHP-Ib (13, 14). However, in a considerable number of PHP-Ib patients, such mutations were excluded for all coding and noncoding exons of the PTH/PTHrP receptor gene (15, 16), and analysis of the receptor’s mRNA provided no evidence for splice variants that could have offered an explanation for the disorder (17, 18).
Inactivating PTH/PTHrP receptor mutations were, however, found in patients with Blomstrand lethal chondrodysplasia, a rare autosomal recessive disorder characterized by advanced skeletal maturation and accelerated chondrocyte differentiation (19–21), leading to developmental abnormalities that are similar to those in PTH/PTHrP receptor-ablated mice (22). However, in humans and mice, the lack of only one functional PTH/PTHrP receptor allele does not result in obvious abnormalities, indicating that heterozygous inactivating receptor mutations are unlikely to cause an autosomal dominant disorder such as PHP-Ib. Furthermore, individuals with PHP-Ib show normal osseous response to PTH or even evidence for increased bone turnover and osteoclastic resorption, indicating that not all PTH-dependent actions on osteoblasts are impaired (2, 3, 23). Moreover, PHP-Ib patients lack obvious abnormalities in the metaphyseal growth plates and thus show normal longitudinal growth, indicating that the PTHrP-dependent regulation of chondrocyte growth and differentiation is normal (22, 24).
Taken together, all available data imply that PHP-Ib is caused by a tissue- or cell-specific defect in PTH/PTHrP receptor expression or by a defect in a protein that mediates the PTH-dependent signaling events downstream. To identify the genetic locus of PHP-Ib and to gain, through the identification of the underlying molecular defect, novel insights into the regulation of calcium homeostasis, we performed a genome-wide search using genomic DNA from one large kindred with the disorder.
METHODS
PHP-Ib Kindreds.
One or several members of the investigated kindreds had been diagnosed with PHP-Ib several years or decades ago, and several of us (J.D.C., D.E.C.C., M.L.L., and T.K. and H.K., respectively) were involved in their long-term medical care; none of the affected members in either kindred show(ed) clinical evidence for AHO. The North American kindreds (F, P, and D) were Caucasian of Western European origin; one family (T) was from Japan. Genomic DNA was extracted from peripheral blood leukocytes as described (15); the study was approved by the Subcommittee on Human Studies of the Massachusetts General Hospital (Accession No. 92–7338).
Genotype Analysis.
The random genome scan was performed with ≈350 microsatellite markers of the Weber 6.0 primer set (Research Genetics, Huntsville, AL); additional primer pairs (www.CHLC.org and www.genome.wi.mit.edu; ref. 25) either were purchased from Research Genetics or were synthesized at the Polymer Core Facility, Massachusetts General Hospital, Boston. For the telomeric region of chromosome 20q, we used previously described markers (26, 27) and markers developed by The Sanger Centre, Cambridge, U.K. (www.sanger.ac.uk/HGP/Chr20). PCR reactions using 20 ng of genomic DNA were performed as described in 96-well microtiter plates (28); the number of different alleles was assessed, and results for each marker were scored by two independent observers; discrepancies were resolved by re-examination of the gels or by repeating the analysis for a particular marker.
Linkage Analysis.
Two point logarithm of the odds of linkage (lod) scores were calculated by using the mlink option of the linkage package software, version 5.1 (29, 30), assuming a disease frequency of 0.1%, equal allele frequencies, and autosomal dominant inheritance with full penetrance.
Southern Blot Analysis of Genomic DNA.
Genomic DNA from an affected and an unaffected individual of each PHP-Ib kindred was digested with one of several different restriction endonucleases (PvuII, XbaI, EcoRI, and BamHI). After separation on an 0.8% agarose gel and transfer onto nitrocellulose, the blot was probed with a cDNA fragment encoding most of the human Gsα protein (IMAGE Consortium; clone ID: 359224), which was 32P-labeled as described by random priming (15).
Southern Blot Analysis of Phage Artificial Chromosome (PAC) Clones.
PAC clones dJ588K17, dJ588H16, dJ552A20, dJ746H22, dJ614C15, dJ746G23, and dJ884F15, kindly provided by Panos Deloukas (The Sanger Centre, Cambridge, U.K.), were grown overnight in Luria–Bertani medium (35 ml) containing 25 μg/ml kanamycin, and DNA was extracted by alkaline lysis as described (31). PAC-derived genomic DNA was digested with BamHI and was separated on an 0.8% agarose gel, and the resulting blots were probed with 32P-end-labeled primers as described (15).
RESULTS
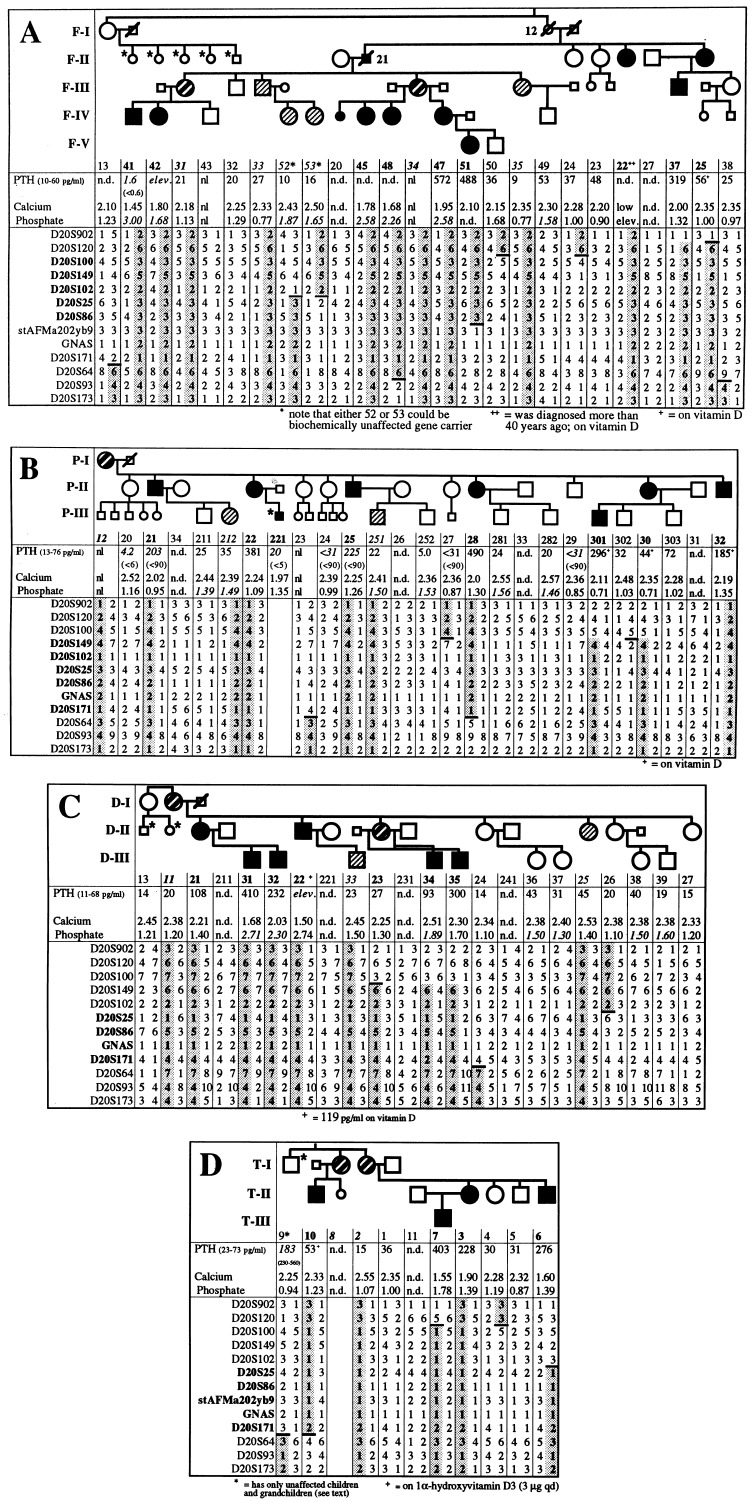
PHP-Ib Kindreds. Four kindreds were investigated in which PHP-Ib followed an autosomal dominant mode of inheritance, albeit with variable penetrance (Fig. 1A–D). Furthermore, the severity of the disease appeared to vary from one family to another, and, even within a single kindred, some individuals showed significant hypocalcemia and consequently severe secondary hyperparathyroidism whereas others showed elevated PTH concentrations as their only evidence for abnormal regulation of mineral ion homeostasis. For example, F-IV/41 and F-V/51 had hypocalcemic seizures at age 8 and 3 years, respectively, whereas F-III/37 was not diagnosed until age 25, when he was evaluated for the present study (calcium, 2.00 mmol/liter; phosphate, 1.32 mmol/liter; and PTH, 319 pg/ml). However, recurrent muscle cramps, particularly during mild gastrointestinal infections, had been noted since childhood. Similarly, P-III/301 had recurrent hypocalcemic seizures before treatment with calcium and vitamin D, initiated at age 10, whereas P-II/22 was not diagnosed until age 52 (calcium 2.24 mmol/liter/phosphate 1.09 mmol/liter/PTH 381 pg/ml).
Figure 1.
(A–D) Laboratory findings and haplotype analysis for markers on the telomeric end of chromosome 20q13. Most laboratory results of individuals affected by PHP-Ib were obtained at the time of diagnosis, and all other results were obtained for the present study; adult normal range for calcium is 2.15–2.60 mmol/liter, and for phosphate is 0.80–1.50 mmol/liter; phosphate measurements in children are shown in italics (pediatric normal range is 1.30–1.80 mmol/liter); normal range for PTH is indicated for each kindred; results obtained with earlier radioimmunoassay systems are shown in italics (respective normal range in parenthesis underneath). n.d., not determined; nl, within normal limits. The haplotype associated with the disorder is shown in bold and is highlighted by shading; recombinations in the allele inherited from obligate gene carriers are indicated by –; markers within the linked region are in bold; • or ■ and bold identification numbers, affected individuals; ○ or □ and regular numbers, healthy individuals; or ▧ and bold italic numbers, unaffected obligate gene carriers; and or ▧ and italic numbers, individuals identified in this study as carriers of the disease haplotype; individuals that were not available for testing and are affected or unaffected by history only are depicted by small symbols; some individuals were tested only for laboratory abnormalities (an asterisk next to a small symbol); \, deceased individual.
In family D, the propositus, D-II/22, was diagnosed with PHP-Ib in 1976 at age 13 after a hypocalcemic seizure (calcium, 1.50 mmol/liter; and phosphate, 2.74 mmol/liter). One of his sister’s sons, D-III/31, presented in 1995 at age 11 with muscle weakness and pain but without other clinical symptoms despite severe hypocalcemia (calcium, 1.68 mmol/liter; phosphate, 2.71 mmol/liter; and PTH, 410 pg/ml). D-III/32, D-III/34, and D-III/35 were completely asymptomatic when evaluated for the present study but showed either hypocalcemia associated with hyperphosphatemia and secondary hyperparathyroidism (D-III/32) or normal blood calcium levels but elevated phosphate and PTH (D-III/34 and D-III/35). Furthermore, D-III/35 (age 17), but not his younger brother D-III/34 (age 6), exhibited significant basal ganglia calcifications. D-II/21 had an elevated PTH whereas her sister D-II/23 (the mother of D-III/34 and D-III/35) showed no laboratory abnormalities. Thus, the females in kindred D were particularly mildly affected, and the presence or absence of PHP-Ib was determined primarily on the basis of laboratory findings in their children.
In kindred T, individuals T-II/3, T-II/6, and T-III/7 complained of paresthesias and recurrent muscle cramps since 4–5 years of age. T-II/10 had recurrent seizures before the diagnosis of PHP-Ib was established in the early 1970s and treatment with calcium and vitamin D analogs was initiated. T-I/2 and T-I/8 were asymptomatic; similarly, T-I/9 showed no clinical or laboratory evidence for PHP-Ib, and all of his five children and 12 grandchildren are healthy.
Based on the above findings, individuals were classified as obligate carriers of the PHP-Ib gene if (i) biochemical abnormalities were documented, i.e., low calcium, elevated phosphate, and/or elevated PTH, or (ii) an individual without obvious laboratory abnormalities had affected children and other affected relatives.
Genome-Wide Search for the PHP-Ib Gene Locus.
The PTH/PTHrP receptor (12, 32, 33), the PTH2-receptor (34), and the GNAS 1 gene (3, 27, 35), initially considered as potential candidate genes, had been excluded in preliminary studies by others (36, 37). We, therefore, decided to perform a genome-wide scan using genomic DNA from 15 members of kindred F and ≈350 microsatellite markers that were spaced at <10-centimorgan intervals.
Nine markers gave a positive lod score of ≈1 or more; however, reanalyses with additional markers and additional family members excluded eight of these chromosomal regions (data not shown). Only the telomeric region of chromosome 20q, initially screened with markers D20S100 and D20S102, could not be excluded and gave a maximal lod score of 2.20 (marker D20S102; θ = 0); no recombinants were identified for this marker, but several affected and unaffected family members were uninformative. We therefore generated a more detailed linkage map of the region between markers D20S902 and D20S173 for all four kindreds and obtained with fully informative markers maximal lod scores of 2.51 (D20S149; θ = 0.1) for kindred F, 2.90 (marker D20S149; θ = 0.1) for kindred P, 1.51 (marker D20S86; θ = 0.1) for kindred D, and 2.07 (marker D20S171; θ = 0) for kindred T; note that all offspring of affected males showed normal blood chemistries and therefore were classified as unaffected. When using only the affected members (and available unaffected spouses) for calculations, maximal lod scores (all with θ = 0.0) for fully informative markers in all four kindreds were identical to the maximal theoretical lod scores (Table 1). Although none of the markers was equally informative for all kindreds, the combined maximal lod scores was 5.09 (marker D20S25) (combined maximal theoretical lod score: 6.89), strongly indicating that the PHP-Ib gene is localized within the chromosomal region 20q13.3.
Table 1.
lod score calculations using only the data obtained with affected members of each PHP-Ib kindred
| Maximal theoretical lod scores | Kindred
F
|
Kindred P
|
Kindred
D
|
Kindred T
|
Combined 6.89 | θ combined | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2.73 | θ | 1.49 | θ | 1.50 | θ | 1.17 | θ | |||
| D20S902 | 1.30 | 0.2 | 0.24 | 0.1 | 0.13 | 0.2 | 0.49 | 0 | 2.17 | 0.5 |
| D20S120 | 1.80 | 0 | 0.24 | 0.1 | 0.90 | 0 | −0.31 | 0.2 | 2.63 | 0.3 |
| D20S100 | 2.16 | 0 | 0.24 | 0.1 | 0.32 | 0.2 | 0.09 | 0.2 | 2.82 | 0.5 |
| D20S149 | 2.73 | 0 | 1.49 | 0 | 0.40 | 0 | 0.00 | 0.2 | 4.62 | 0.2 |
| D20S102 | 2.39 | 0 | 0.10 | 0 | 1.49 | 0 | −0.11 | 0.2 | 3.88 | 0.2 |
| D20S25 | 2.37 | 0 | 0.08 | 0 | 1.50 | 0 | 1.14 | 0 | 5.09 | 0 |
| D20S86 | 0.90 | 0 | 1.49 | 0 | 1.50 | 0 | 0.30 | 0 | 4.19 | 0 |
| stAFMa202yb9 | 0.07 | 0 | 0.20 | 0 | 0.53 | 0 | 0.80 | 0 | ||
| GNAS | 0.34 | 0.1 | 1.44 | 0 | 0.40 | 0 | 0.26 | 0 | 2.44 | 0.1 |
| D20S171 | 1.42 | 0.1 | 1.49 | 0 | 0.40 | 0 | 1.17 | 0 | 4.47 | 0.1 |
| D20S64 | 1.50 | 0.1 | 0.52 | 0.1 | 1.50 | 0 | 0.08 | 0.2 | 3.60 | 0.4 |
| D20S93 | 0.47 | 0.2 | 0.52 | 0.1 | 1.50 | 0 | 0.19 | 0.2 | 2.68 | 0.5 |
| D20S173 | 1.25 | 0.1 | 0.24 | 0.2 | 1.20 | 0 | 1.10 | 0 | 3.79 | 0.3 |
Maximal lod scores (and the recombination fraction) for the different markers are shown for each kindred, as are the combined maximal lod scores; results identical to the maximal theoretical lod scores are in bold; the highest combined lod score is in bold italic.
The PHP-Ib Gene Is Paternally Imprinted.
In each kindred, one or several individuals carried the allele associated with PHP-Ib but showed no biochemical evidence for the disorder; these unaffected gene carriers were always offspring of obligate male gene carriers. For example, haplotype analysis of the offspring of the affected male F-II/21 showed that four of his five children (F-III/31, F-III/33, F-III/34, and F-III/35; see Fig. 1A) had inherited the same paternal allele but showed no clinical or laboratory abnormalities and consequently never required treatment with calcium and/or vitamin D analogs. However, F-III/31 and F-III/34 have affected children and grandchildren, respectively, indicating that they are obligate gene carriers. Similarly, in kindreds D and P, three unaffected individuals (P-III/212, P-III/251, and D-III/33) had inherited the disease allele from their affected fathers (P-II/21, P-II/25, and D-II/22, respectively). In kindred T, the biochemically unaffected sisters T-I/2 and T-I/8 have affected children, suggesting that both are carriers of the PHP-Ib gene; their unaffected brother T-I/9 has five unaffected children and twelve unaffected grandchildren (note that nine of his grandchildren are from daughters), suggesting that he is not a carrier of the genetic defect.
Because of these findings and because several genes on chromosome 20q, including GNAS 1, are known to be expressed from only one parental allele (8, 38), we assessed whether paternal imprinting could account for the incomplete penetrance observed in PHP-Ib. In the four PHP-Ib kindreds, 17 females and 16 males were considered to be obligate gene carriers (Table 2). Although none of the obligate male carriers had affected children, obligate female carriers had 18 affected children (P < 0.001). These findings suggested that the PHP-Ib gene is imprinted paternally and, furthermore, indicated that the females F-I/12, P-I/12, and D-I/11 and the sisters T-I/2 and T-I/8 were/are unaffected gene carriers who inherited the disease allele from their respective fathers; this conclusion is strongly supported by the finding that these individuals have, in the linked region, the same haplotype as their affected descendants (see Fig. 1 A–D).
Table 2.
Paternal imprinting of the PHP-Ib gene
| Kindred | Parent | Offspring
of obligate male
carriers
|
Total | Parent | Offspring of obligate
female carriers
|
Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Male
offspring
|
Female offspring
|
Male
offspring
|
Female offspring
|
|||||||||
| Affected | Unaffected | Affected | Unaffected | Affected | Unaffected | Affected | Unaffected | |||||
| F | II-21 | 2 | 3 | 5 | II-25 | 1 | 1 | 2 | ||||
| III-31 | 1 | 1 | 1 | 3 | ||||||||
| III-34 | 4 | 4 | ||||||||||
| IV-47 | 1 | 1 | 2 | |||||||||
| P | II-21 | 1 | 1 | 2 | II-22 | 1 | 1 | |||||
| II-25 | 2 | 2 | II-28 | 2 | 2 | |||||||
| II-30 | 1 | 2 | 3 | |||||||||
| D | II-22 | 1 | 1 | II-21 | 2 | 2 | ||||||
| II-23 | 2 | 2 | ||||||||||
| J | I-2 | 1 | 1 | 1 | 1 | 4 | ||||||
| I-8 | 1 | 1 | 2 | |||||||||
| II-3 | 1 | 1 | ||||||||||
| Total analyzed | 10 | 28 | ||||||||||
| Unaffected | 6 | 4 | 10 | 7 | 3 | 10 | ||||||
| Affected | 0 | 0 | 0 | 11 | 7 | 18 | ||||||
Offspring of obligate male or female carriers (see text for details) were classified as affected (bold letters) or unaffected (regular letters) according to their laboratory findings.
lod Score Calculations Assuming Paternal Imprinting of the PHP-Ib Gene.
Because of the incomplete penetrance of the PHP-Ib phenotype caused by paternal imprinting, lod scores were recalculated but without considering offspring of affected males that have no affected children or grandchildren (i.e., F-III/32, F-III/33, F-III/35, D-III/33, P-III/211, P-III/212, P-III/251, and P-III/252). Furthermore, we excluded two females of kindred D (D-II/26 and D-II/27) who are unaffected biochemically and have no children. With these exclusions, maximal lod scores, all at θ = 0, were for each kindred identical to the maximal theoretical lod scores (Table 3). The combined maximal lod score was 8.88 (marker D20S149); note that this marker was almost completely uninformative for kindred D and that one affected individual in kindred T was recombinant at this locus.
Table 3.
Recalculations of lod scores assuming paternal imprinting of the PHP-Ib gene
| Marker | Kindred | lod scores at different
recombination fractions
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.01 | 0.02 | 0.03 | 0.04 | 0.05 | 0.06 | 0.07 | 0.10 | 0.20 | ||
| D20S149 | F | 4.63 | 4.55 | 4.46 | 4.37 | 4.28 | 4.19 | 4.10 | 4.01 | 3.73 | 2.76 |
| P | 4.52 | 4.45 | 4.38 | 4.30 | 4.23 | 4.16 | 4.09 | 4.01 | 3.78 | 2.96 | |
| D | 0.04 | 0.04 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 | 0.01 | |
| T | −∞ | −0.26 | 0.01 | 0.16 | 0.25 | 0.32 | 0.36 | 0.40 | 0.46 | 0.43 | |
| Combined | 8.77 | 8.88 | 8.87 | 8.80 | 8.70 | 8.58 | 8.45 | 8.00 | 6.17 | ||
| D20S86 | F | 0.96 | 0.94 | 0.93 | 0.91 | 0.89 | 0.87 | 0.85 | 0.83 | 0.77 | 0.59 |
| P | 3.31 | 3.26 | 3.21 | 3.15 | 3.10 | 3.04 | 2.99 | 2.93 | 2.76 | 2.15 | |
| D | 2.71 | 2.66 | 2.61 | 2.56 | 2.52 | 2.47 | 2.42 | 2.37 | 2.21 | 1.67 | |
| T | 0.30 | 0.29 | 0.28 | 0.27 | 0.26 | 0.25 | 0.24 | 0.23 | 0.20 | 0.11 | |
| Combined | 7.29 | 7.16 | 7.02 | 6.89 | 6.76 | 6.63 | 6.49 | 6.36 | 5.94 | 4.52 | |
| GNAS | F | −∞ | 0.76 | 0.99 | 1.08 | 1.13 | 1.15 | 1.14 | 1.13 | 1.04 | 0.48 |
| P | 4.49 | 4.42 | 4.35 | 4.28 | 4.21 | 4.13 | 4.06 | 3.99 | 3.76 | 2.94 | |
| D | −0.74 | −0.67 | −0.61 | −0.56 | −0.51 | −0.47 | −0.44 | −0.40 | −0.32 | −0.15 | |
| T | 0.30 | 0.29 | 0.28 | 0.27 | 0.26 | 0.25 | 0.24 | 0.23 | 0.20 | 0.11 | |
| Combined | 4.80 | 5.01 | 5.07 | 5.08 | 5.06 | 5.01 | 4.94 | 4.67 | 3.38 | ||
| D20S171 | F | −∞ | 1.64 | 1.87 | 1.97 | 2.03 | 2.05 | 2.06 | 2.05 | 1.98 | 1.48 |
| P | 3.01 | 2.96 | 2.91 | 2.86 | 2.82 | 2.77 | 2.71 | 2.66 | 2.51 | 1.94 | |
| D | 0.04 | 0.04 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.02 | 0.01 | |
| T | 2.07 | 2.03 | 1.99 | 1.95 | 1.91 | 1.87 | 1.83 | 1.79 | 1.66 | 1.21 | |
| Combined | 6.67 | 6.81 | 6.83 | 6.79 | 6.72 | 6.63 | 6.53 | 6.18 | 4.66 | ||
Results that were obtained with some fully informative markers, and with GNAS were reanalyzed by using most available affected and unaffected members of the four PHP-Ib kindreds; with the exception of F-III/31 and F-III/34, all offspring of obligate male carriers, and D-II/25 and D-II/27 were excluded for these calculations (see text for details). lod scores are shown at different recombination fractions; results identical to the maximal theoretical lod scores are in bold; combined maximal lod score is in bold italic (combined maximal theoretical lod score is 13.9).
Fine Mapping of the Linked Region and Haplotype Analyses.
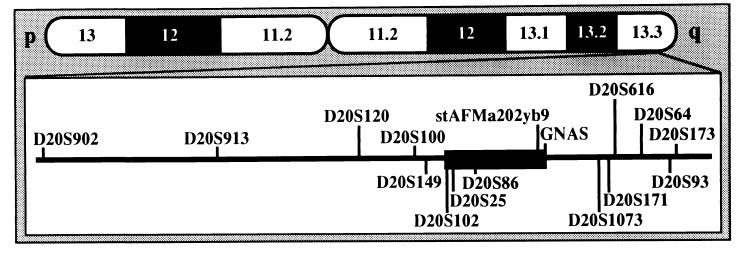
To minimize the number of recombination events for both alleles of all investigated individuals, the previously described location of some markers (www.CHLC.org and www.sanger.ac.uk/HGP/Chr20; refs. 25–27) was altered (Fig. 2; see also Fig. 1 A–D). For example, marker D20S93 was placed telomeric of D20S171 and not centromeric, as suggested by radiation hybrid mapping (wwwsanger.ac.uk/HGP/Chr20), because three individuals in kindred F (F-I/13, F-IV/48, and F-III/38) would otherwise be double recombinants.
Figure 2.
Schematic presentation of chromosome 20 and the region linked to PHP-Ib. The linked region is indicated (■), and the locations of microsatellite markers are based on mapping data produced by the Chromosome 20 Mapping Group at the Sanger Centre, Cambridge, U.K. (www.sanger.ac.uk/HGP/Chr20) and the Whitehead Institute, Cambridge, MA (www.genome.wi.mit.edu), by haplotype analysis of the four PHP-Ib kindreds (see Fig. 1 A–D), and by Southern blot analysis of selected PAC clones provided by the Sanger Centre (see Results for details).
Haplotype analysis of all four kindreds showed that the centromeric boundary of the linked region is located between markers D20S102 and D20S25 (Fig. 2; see also Fig. 1 A–D). For kindreds P, D, and T, the telomeric boundary of the linked region was placed between markers D20S64 and D20S171; in kindred F, one affected individual (F-V/51) was recombinant at D20S171 and the intragenic GNAS marker [a dinucleotide repeat polymorphism in the third intron of the Gsα gene (39)]; the PHP-Ib gene thus resides between markers D20S102 and GNAS (≈7 centirays).
To confirm and refine the reported order for markers stAFMa202yb9, GNAS, and D20S171 (www.genome.wi.mit.edu and www.sanger.ac.uk/HGP/Chr20), seven PAC clones were probed by Southern blot analysis with different markers (see Fig. 2). Six of these PAC clones were positive for GNAS and the more telomeric marker D20S1073 (www.genome.wi.mit.edu and www.sanger.ac.uk/HGP/Chr20). Two of these clones, dJ588K17 and dJ588H16, were also positive for D20S171 and D20S616, which are even further telomeric than D20S1073; this indicates that both clones extend further toward the telomere. Two other clones, dJ746H22 and dJ746G23, were not only positive for D20S1073 and GNAS but also for stAFMa202yb9, indicating that these extend further toward the centromere. The most telomeric PAC clone, dJ884F15, was only positive for D20S616. These findings indicate that stAFMa202yb9 is located centromeric of GNAS and that D20S171 is telomeric of GNAS; the latter observation was suggested by radiation hybrid mapping (www.sanger.ac.uk/HGP/Chr20). Because of this order of the markers, and because F-V/51 was not informative for stAFMa202yb9 (see Fig. 1A), the GNAS 1 gene was not excluded completely as a candidate gene for PHP-Ib. We, therefore, performed Southern blot analysis of genomic DNA from affected and unaffected members of each kindred that had been digested with several different, infrequently cutting restriction endonucleases. However, no evidence was obtained for larger deletions and/or rearrangements in the coding exons of the Gsα gene and in the adjacent 5′ and 3′ regulatory regions (data not shown).
DISCUSSION
In the present study, we established linkage of PHP-Ib to a region on the telomeric end of chromosome 20q (20q13.3). Consistent with earlier observations for other genes in this region (38), the disease was inherited in the four investigated kindreds in a mode that strongly suggested paternal imprinting and thus resulted in biochemical abnormalities only if the defect was inherited from an obligate female gene carrier. The previously noted uncertainty regarding the mode of inheritance of PHP-Ib (see ref. 3 for review) thus can be explained through paternal imprinting of the genetic defect. However, other factors, which may include other genes or changes in the daily intake of calcium and vitamin D, are likely to contribute to the considerable variability of the disease severity. Because of these yet unknown factors, and because of the unusual mode of inheritance, it appears likely that at least some “sporadic” cases of PHP-Ib in fact may be familial and that detailed laboratory investigations of the entire family of affected individuals may lead to the identification of additional, possibly less severely affected, family members.
The telomeric end of chromosome 20q, which contains the PHP-Ib locus, comprises the gene encoding Gsα, and different heterozygous mutations in this latter gene had been identified in a large variety of patients with either PHP-Ia and/or pseudo-pseudohypoparathyroidism (3, 4, 6, 7). Although these mutations offered a plausible explanation for the equivalent reductions in Gsα activity/protein, the mechanisms leading either to AHO alone, or to AHO combined with resistance toward PTH and other hormones, remains obscure (3, 4, 6, 7). More recently, it was shown that endocrine abnormalities are only present if the Gsα-defect is inherited from a female whereas the AHO phenotype can be transmitted from either parent (8, 9).
Similar observations, i.e., paternal imprinting of PTH-resistant hypocalcemia, were now made in patients with PHP-Ib. Furthermore, linkage was obtained to the chromosomal region that comprises the Gsα gene locus, indicating that a common mechanism could be responsible for the abnormal regulation of mineral ion homeostasis in PHP-Ib and PHP-Ia. However, because one affected individual (F-V/51; see Fig. 1A) was recombinant within the GNAS 1, at least portions of this gene appear to be excluded. Furthermore, no gross deletions or rearrangements of the Gsα gene were detected by Southern blot analysis. It is, therefore, possible that two distinct genes, the Gsα gene and the “PHP-Ib gene,” are involved independently in the regulation of mineral ion homeostasis. However, these two genes would have to reside in the same or in adjacent, paternally imprinted regions and could be in close enough physical proximity to allow the sharing of regulatory elements and/or coding nucleotide sequences, as observed with other imprinted genes (40–44).
Because the recombination event (F-V/51; see Fig. 1A) in the third intron did not exclude the entire Gsα locus, this gene remains a plausible candidate, and it appears possible that the PTH-resistant hypocalcemia observed in patients with either PHP-Ia or PHP-Ib is caused by a tissue- or cell-specific defect in Gsα gene expression. In fact, recent data from Gsα gene-ablated mice indicate that offspring who inherit the mutant allele from a female show substantially decreased Gsα concentrations in the renal cortex and in adipocytes but not in the renal medulla or in other investigated tissues. Furthermore, only offspring of heterozygous, Gsα-ablated females develop hypocalcemia and secondary hyperparathyroidism (4, 45), indicating that ablation of one Gsα allele can lead in mice to changes in mineral ion homeostasis that are similar to those observed in patients with either PHP-Ia or PHP-Ib.
Of interest, recent preliminary findings suggest that a splice variant of the Gsα gene, XLαs (46), is transcribed only from the paternal allele (47). This finding provides further confirmation that the chromosomal region that comprises the GNAS locus undergoes imprinting (38). If Gsα transcripts are derived, at least in some tissues or cells, from only one parental allele, as suggested by this and other studies in humans and mice (4, 8, 9, 45), mutations in a promoter or enhancer of the Gsα gene could explain the kidney-specific resistance toward PTH and the resulting hypocalcemia in patients with PHP-Ib.
Acknowledgments
We thank Drs. Takashi Akamizu, Brian Beate, Stuart Carr, Marie Demay, Michael Econs, Yasutomo Fukunaga, Daisuke Inoue, Kyle Landt, Kiyoshi Nishio, Kimberly Strauch, Andrew Stewart, and Mika Yamauchi and Mrs. Suzanne Payant for their help in obtaining blood samples and laboratory results. Furthermore, we are grateful for expert technical assistance by Geoffrey Jensen, Jeffry Pincus, and Michael Roehrl, secretarial work by Judith Graham, and for the continuous support and interest of all participating members of PHP-Ib families. This work was supported by grants from National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Disease (Grant RO1 46718-06 to H.J.) and National Institute of Arthritis and Musculoskeletal and Skin Diseases (Grant RO1 36819 to B.R.O.), the Canadian Medical Research Council (Grant MT-9315 to G.N.H.), and by grants from the Finnish Cultural Foundation and the Academy of Finland (to M.V.).
ABBREVIATIONS
- PHP
pseudohypoparathyroidism
- PHP-Ia
PHP type Ia
- PTH
parathyroid hormone
- PTHrP
PTH-related peptide
- Gsα
α subunit of the stimulatory G protein
- AHO
Albright’s hereditary osteodystrophy
- lod
logarithm of the odds of linkage
- PAC
phage artificial chromosome
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1. Albright F, Burnett C H, Smith P H, Parson W. Endocrinology. 1942;30:922–932. [Google Scholar]
- 2.van Dop C. Semin Nephrol. 1989;9:168–178. [PubMed] [Google Scholar]
- 3.Levine M A. In: Principles of Bone Biology. Bilezikian J P, Raisz L G, Rodan G A, editors. New York: Academic; 1996. pp. 853–876. [Google Scholar]
- 4.Weinstein L S. In: G Proteins, Receptors, and Disease. Spiegel A M, editor. Totowa, NJ: Humana; 1998. pp. 23–56. [Google Scholar]
- 5.Schuster V, Eschenhagen T, Kruse K, Gierschik P, Kreth H W. Eur J Pediatr. 1993;152:185–189. doi: 10.1007/BF01956140. [DOI] [PubMed] [Google Scholar]
- 6.Miric A, Vechio J D, Levine M A. J Clin Endocrinol Metab. 1993;76:1560–1568. doi: 10.1210/jcem.76.6.8388883. [DOI] [PubMed] [Google Scholar]
- 7.Weinstein L S, Gejman P V, Friedman E, Kadowaki T, Collins R M, Gershon E S, Spiegel A M. Proc Natl Acad Sci USA. 1990;87:8287–8290. doi: 10.1073/pnas.87.21.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies A J, Hughes H E. J Med Genet. 1993;30:101–103. doi: 10.1136/jmg.30.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson, L. C., Oude-Luttikhuis, M. E. M., Clayton, P. T., Fraser, W. D. & Trembath, R. C. (1994) J. Med. Genet. 31. [DOI] [PMC free article] [PubMed]
- 10.Jüppner H, Abou-Samra A B, Freeman M W, Kong X F, Schipani E, Richards J, Kolakowski L F, Jr, Hock J, Potts J T, Jr, Kronenberg H M, et al. Science. 1991;254:1024–1026. doi: 10.1126/science.1658941. [DOI] [PubMed] [Google Scholar]
- 11.Abou-Samra A B, Jüppner H, Force T, Freeman M W, Kong X F, Schipani E, Urena P, Richards J, Bonventre J V, Potts J T, Jr, et al. Proc Natl Acad Sci USA. 1992;89:2732–2736. doi: 10.1073/pnas.89.7.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schipani E, Karga H, Karaplis A C, Potts J T, Jr, Kronenberg H M, Segre G V, Abou-Samra A B, Jüppner H. Endocrinology. 1993;132:2157–2165. doi: 10.1210/endo.132.5.8386612. [DOI] [PubMed] [Google Scholar]
- 13.Silve C, Santora A, Breslau N, Moses A, Spiegel A. J Clin Endocrinol Metab. 1986;62:640–644. doi: 10.1210/jcem-62-4-640. [DOI] [PubMed] [Google Scholar]
- 14.Silve C, Suarez F, El Hessni A, Loiseau A, Graulet A M, Gueris J. J Clin Endocrinol Metab. 1990;71:631–638. doi: 10.1210/jcem-71-3-631. [DOI] [PubMed] [Google Scholar]
- 15.Schipani E, Weinstein L S, Bergwitz C, Iida-Klein A, Kong X F, Stuhrmann M, Kruse K, Whyte M P, Murray T, Schmidtke J, et al. J Clin Endocrinol Metab. 1998;83:3373–3376. doi: 10.1210/jcem.80.5.7745008. [DOI] [PubMed] [Google Scholar]
- 16.Bettoun J D, Minagawa M, Kwan M Y, Lee H S, Yasuda T, Hendy G N, Goltzman D, White J H. J Clin Endocrinol Metab. 1997;82:1031–1040. doi: 10.1210/jcem.82.4.3906. [DOI] [PubMed] [Google Scholar]
- 17.Suarez F, Lebrun J J, Lecossier D, Escoubet B, Coureau C, Silve C. J Clin Endocrinol Metab. 1995;80:965–970. doi: 10.1210/jcem.80.3.7883858. [DOI] [PubMed] [Google Scholar]
- 18.Fukumoto S, Suzawa M, Takeuchi Y, Nakayama K, Kodama Y, Ogata E, Matsumoto T. J Clin Endocrinol Metab. 1996;81:2554–2558. doi: 10.1210/jcem.81.7.8675577. [DOI] [PubMed] [Google Scholar]
- 19.Blomstrand S, Claësson I, Säve-Söderbergh J. Pediatr Radiol. 1985;15:141–143. doi: 10.1007/BF02388725. [DOI] [PubMed] [Google Scholar]
- 20.Jobert A S, Zhang P, Couvineau A, Bonaventure J, Roume J, LeMerrer M, Silve C. J Clin Invest. 1998;102:34–40. doi: 10.1172/JCI2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, P., Jobert, A. S., Couvineau, A. & Silve, C. (1998) J. Clin. Endocrinol. Metab., in press. [DOI] [PubMed]
- 22.Lanske B, Karaplis A C, Luz A, Vortkamp A, Pirro A, Karperien M, Defize L H K, Ho C, Mulligan R C, Abou-Samra A B, et al. Science. 1996;273:663–666. doi: 10.1126/science.273.5275.663. [DOI] [PubMed] [Google Scholar]
- 23.Murray T, Gomez Rao E, Wong M M, Waddell J P, McBroom R, Tam C S, Rosen F, Levine M A. J Bone Miner Res. 1993;8:83–91. doi: 10.1002/jbmr.5650080111. [DOI] [PubMed] [Google Scholar]
- 24.Vortkamp A, Lee K, Lanske B, Segre G V, Kronenberg H M, Tabin C J. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 25.Gyapay G, Morissette J, Vignal A, Dib C, Fizames C, Millasseau P, Marc S, Bernardi G, Lathrop M, Weissenbach J. Nat Genet. 1994;7:246–334. doi: 10.1038/ng0694supp-246. [DOI] [PubMed] [Google Scholar]
- 26.Melis R, Bradley P, Elsner T, Robertson M, Lawrence E, Gerken S, Albertsen H, White R. Genomics. 1993;16:56–62. doi: 10.1006/geno.1993.1140. [DOI] [PubMed] [Google Scholar]
- 27.Gejman P V, Weinstein L S, Martinez M, Spiegel A M, Cao Q, Hsieh W T, Hoehe M R, Gershon E S. Genomics. 1991;9:782–783. doi: 10.1016/0888-7543(91)90377-q. [DOI] [PubMed] [Google Scholar]
- 28.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J H M, et al. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 29.Ott J. Analysis of Human Linkage. Baltimore: Johns Hopkins Univ. Press; 1983. [Google Scholar]
- 30.Lathrop G M, Lalouel J M, Julier C, Ott J. Proc Natl Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 32.Pausova Z, Bourdon J, Clayton D, Mattei M-G, Seldin T M F, Janicic N, Riviere M, Szpirer J, Levan G, Szpirer C, et al. Genomics. 1994;20:20–26. doi: 10.1006/geno.1994.1122. [DOI] [PubMed] [Google Scholar]
- 33.Gelbert L, Schipani E, Jüppner H, Abou-Samra A B, Segre G V, Naylor S, Drabkin H, White R, Heath H. J Clin Endocrinol Metab. 1994;79:1046–1048. doi: 10.1210/jcem.79.4.7962272. [DOI] [PubMed] [Google Scholar]
- 34.Usdin T B, Modi W, Bonner T I. Genomics. 1996;37:140–141. doi: 10.1006/geno.1996.0532. [DOI] [PubMed] [Google Scholar]
- 35.Kozasa T, Itoh H, Tsukamoto T, Kaziro Y. Proc Natl Acad Sci USA. 1988;85:2081–2085. doi: 10.1073/pnas.85.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding, C. L., Usdin, T. B., Labuda, M. & Levine, M. A. (1996) J. Bone Miner. Res. 11, Suppl. 1, M483.
- 37.Jan de Beur S M, Labuda M C, Timberlake R J, Levine M A. 79th Annual Meeting of the Endocrine Society. Minneapolis: The Endocrine Society; 1997. , P1-432 (abstr.). [Google Scholar]
- 38.Hall J G. Am J Hum Genet. 1990;46:857–873. [PMC free article] [PubMed] [Google Scholar]
- 39.Granqvist M, Xiang K, Seino M, Bell G I. Nucleic Acids Res. 1988;19:4569. doi: 10.1093/nar/19.16.4569-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohlsson R, Nyström A, Pfeifer-Ohlsson S, Töhönen V, Hedborg F, Schofield P, Flam F, Ekström T J. Nat Genet. 1993;4:94–97. doi: 10.1038/ng0593-94. [DOI] [PubMed] [Google Scholar]
- 41.Li E, Beard C, Jaenisch R. Nature (London) 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 42.Vu T H, Hoffman A R. Nature (London) 1994;371:714–717. doi: 10.1038/371714a0. [DOI] [PubMed] [Google Scholar]
- 43.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Nature (London) 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 44.Jaenisch R. Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 45.Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein L S. Proc Natl Acad Sci USA. 1998;95:8715–8720. doi: 10.1073/pnas.95.15.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kehlenbach R H, Matthey J, Huttner W B. Nature (London) 1994;372:804–809. doi: 10.1038/372804a0. [DOI] [PubMed] [Google Scholar]
- 47.Hayward, B., Kamiya, M., Takada, S., Moran, V., Strain, L., Hayashizaki, Y. & Bronthon, D. T. (1998) Eur. J. Hum. Genet.6, Suppl. 1, 36.