Abstract
Schwann cell factor 1 (SC1), a p75 neurotrophin receptor–interacting protein, is a member of the positive regulatory/suppressor of variegation, enhancer of zeste, trithorax (PR/SET) domain-containing zinc finger protein family, and it has been shown to be regulated by serum and neurotrophins. SC1 shows a differential cytoplasmic and nuclear distribution, and its presence in the nucleus correlates strongly with the absence of bromodeoxyuridine (BrdU) in these nuclei. Here, we investigated potential transcriptional activities of SC1 and analyzed the function of its various domains. We show that SC1 acts as a transcriptional repressor when it is tethered to Gal4 DNA-binding domain. The repressive activity requires a trichostatin A–sensitive histone deacetylase (HDAC) activity, and SC1 is found in a complex with HDACs 1, 2, and 3. Transcriptional repression exerted by SC1 requires the presence of its zinc finger domains and the PR domain. Additionally, these two domains are involved in the efficient block of BrdU incorporation by SC1. The zinc finger domains are also necessary to direct SC1's nuclear localization. Lastly, SC1 represses the promoter of a promitotic gene, cyclin E, suggesting a mechanism for how growth arrest is regulated by SC1.
Keywords: p75NTR; cell cycle; HDAC; transcriptional repressor; PR/SET domains
Introduction
Schwann cell factor 1 (SC1) is a p75 neurotrophin receptor (NTR)–interacting protein (Chittka and Chao, 1999). It belongs to a small group of proteins referred to as the retinoblastoma-interacting zinc finger (RIZ) family of transcription factors. These proteins are characterized by the presence of zinc finger and positive regulatory/suppressor of variegation, enhancer of zeste, trithorax (PR/SET) domains, and are involved in cell differentiation and tumorigenesis (for review see Huang et al., 1998). Proteins containing the PR/SET domain include the lymphocyte PRDI-BF1/BLIMP protein (Keller and Maniatis, 1991), RIZ (Buyse et al., 1995), and the EVI1 gene (Fears et al., 1996). The presence of the PR/SET domain makes these proteins distinct from a multitude of other zinc finger motif-bearing proteins. The PR/SET domain is a modified SET domain. SET domain proteins possess protein methyltransferase activity and have been shown to methylate histones, thus regulating chromatin structure and transcription (Rea et al., 2000; Strahl et al., 2002). However, close inspection of the amino acid sequences of the PR domains reveals striking differences in the highly conserved residues necessary for the methyltransferase activity of the SET domain (Kouzarides, 2002). The zinc finger domains and the PR/SET domain of SC1 imply that this protein may mediate transcriptional activities. Thus, transcriptional activities of repressive and activating nature have been described for RIZ (Xie et al., 1997; Abbondanza et al., 2000; Steele-Perkins et al., 2001), Blimp-1/PRDI-BF1 (Lin et al., 1997; Ren et al., 1999; Yu et al., 2000), and Evi-1 (Kurokawa et al., 1998; Izutsu et al., 2001; Palmer et al., 2001). SC1 is the first member of this family that is implicated in the neurotrophin signaling.
Neurotrophins, such as NGF, BDNF, and NT-3, are important survival and differentiation factors. The neurotrophins interact with two distinct classes of receptors, members of the Trk tyrosine kinase receptor subfamily and p75NTR, a member of the TNF receptor superfamily (Chao and Hempstead, 1995). There is evidence that p75NTR can function as a cell death receptor (Casaccia-Bonnefil et al., 1996; Frade et al., 1996; Bamji et al., 1998; Frade and Barde, 1998). Several proteins that interact with p75NTR have been shown to transduce apoptotic signaling. These include NRIF (Casademunt et al., 1999), NRAGE (Salehi et al., 2000), and NADE (Mukai et al., 2000). Interestingly, p75NTR is expressed very early during development (Yan and Johnson, 1987, 1988), before differentiation of many precursor cells of the nervous system and before the onset of programmed neuronal cell death. This observation makes it a candidate for a receptor functioning in the control of precursor cell proliferation and differentiation. Supporting this idea is the observation that several proteins which interact with p75NTR induce growth arrest. Expression of SC1, for example, was previously found to be correlated with a decrease in BrdU incorporation (Chittka and Chao, 1999).
To elucidate the mechanism by which SC1 transduces the neurotrophin signaling, we began to analyze its potential transcriptional activity. In a reporter gene assay, SC1 acted as a transcriptional repressor. Both the zinc finger domains as well as the PR/SET domain are necessary for SC1 to act as a repressor, and are needed to effectively block BrdU incorporation into cell nuclei. The repression exerted by SC1 required the activity of trichostatin A (TSA)–sensitive histone deacetylases (HDACs), and SC1 was found in the complex with HDAC 1, 2, and 3. Further, we show that SC1 behaves as a transcriptional repressor upon NGF application to the cells transfected with both SC1 and either p75NTR or TrkA. We found the zinc finger domains of SC1 are necessary for its nuclear localization. Finally, our analysis of genes transcriptionally regulated by SC1 revealed that SC1 down-regulates the expression of a promitotic gene, cyclin E, consistent with its ability to block DNA replication as measured by BrdU incorporation. These results implicate SC1 as a potential transcriptional mediator of NGF signaling that may be involved in modifying the chromatin structure during differentiation.
Results
Regulation of the subcellular distribution of SC1
SC1 is localized to both the nucleus and the cytoplasm of COS1 and Schwann cells (Chittka and Chao, 1999). We sought to determine which domains of SC1 were responsible for this differential distribution. To address this question, we generated a series of deletion mutants of SC1 fused to GFP. These include full-length SC1 or deletions lacking either the acidic COOH terminus, the zinc finger domains, or the PR/SET domain (see Fig. 1 for a schematic representation of the constructs used in these experiments). These constructs were then transfected into COS1 cells, and the distribution of each protein was investigated. The results of these experiments are shown in Fig. 1 (a–e). GFP alone distributed to the cytoplasm and nucleus (Fig. 1 a). Full-length SC1 as well as SC1ΔPR and SC1ΔC were found predominantly in the nucleus (Fig. 1, b, c, and e, respectively), as can be visualized by their overlapping distribution with the Hoechst stain in the nuclei. Strikingly, the deletion of zinc finger domains resulted in a cytoplasmic distribution of SC1 (Fig. 1 d). Therefore, we concluded that zinc finger domains are necessary for nuclear localization of SC1.
Figure 1.
The zinc finger domains of SC1 are necessary for nuclear localization. Localization of GFP, GFP-SC1, GFP-SC1ΔPR, GFP-SC1ΔZF, and GFP-SC1ΔC was assayed 48 h after transient transfection in COS1 cells. Cells were assessed for Hoechst staining (blue) to visualize nuclei, and for GFP fluorescence to visualize overexpressed proteins. COS1 cells were transfected with (a) GFP, (b) GFP-SC1, (c) GFP- SC1ΔPR, (d) GFP-SC1ΔZF, and (e) GFP-SC1ΔC. Bar, 20 μm.
SC1 is an HDAC-dependent transcriptional repressor
The localization of SC1 is highly regulated, and translocation of the protein to the nucleus was closely correlated with a loss of BrdU incorporation (Chittka and Chao, 1999). These results suggest that SC1 may be involved in transcriptional events associated with growth arrest. To assess whether SC1 possessed the ability to modulate gene transcription, we fused the coding sequence of SC1 to the Gal4 DNA-binding domain (DBD; see Fig. 2 B for the schematic representation of the effector and reporter constructs). Fusion of these truncated SC1 proteins with Gal4-DBD, which contains an endogenous NLS, ensured nuclear localization of the proteins (see Fig. 5). A luciferase reporter under transcriptional control of Gal4 upstream activating sequences was used for measuring transcription relative to a control effector without SC1 (i.e., Gal4 alone). Gal4-SC1 or Gal4 and the reporter DNAs were cotransfected into HEK293 cells, and luciferase activity was measured 48 h later.
Figure 2.
SC1 is a repressor that requires its zinc finger domains and the PR/SET domain for repression. (A) Histogram of representative relative luciferase units after a cotransfection of Gal4 or Gal4SC1 with the Gal4-luc reporter (lanes 1 and 2, respectively) or TATA-luc reporter (lanes 3 and 4, respectively) in HEK293 cells. Whole-cell extracts were used in the luciferase assays and β-galactosidase measurements. The basal promoter activity was measured in the presence of Gal4 alone (black bar), and SC1's regulation of the basal promoter was compared with it (open bar). A schematic map of the reporter construct is represented on the top. The graph shows the result of one experiment performed in triplicate that was reproduced in several independent experiments. (B) Left: histogram of relative luciferase units measured after cotransfection of various truncated mutants of SC1 fused to the Gal4-DBD represented schematically on the right. The numbers on the schematic drawings specify the amino acids of SC1. PR, PR/SET domain; ZF, zinc finger domain; Ac, an acidic COOH-terminal portion of SC1 protein. Black bar represents relative luciferase units in a control cotransfection experiment using Gal4 alone as an effector; white bars represent the results of cotransfection with various Gal4SC1 mutants. (C) Western blot showing the expression of Gal4-SC1 fusion proteins in HEK293 cells, as detected by anti-Flag antibody. Fusion proteins were immunoprecipitated from whole-cell lysates with anti-Flag antibody and separated on a 7.5% SDS-PAGE before blotting and detection with anti-Flag antibodies. The graph below demonstrates the results of the control transfections where increased amount of cDNA coding for the deleted SC1 proteins with lower expression levels were tested for their ability to influence luciferase measurements. The amounts of DNA used are shown below the appropriate construct (compared with the 0.3 μg normally used in these experiments).
Figure 5.

Correlation of transcriptional activity with SC1's ability to block BrdU incorporation. NIH3T3 (A and B) and HEK293 (C and D) cells transiently transfected with Gal4SC1 fusion protein constructs. (A) SC1-expressing cells (green) are visualized by FITC-coupled anti-Flag antibodies. Bottom: the same field showing BrdU-labeled cells. (B) Histogram represents the percentage of cells transfected with Gal4, Gal4SC1FL, Gal4SC1Δ754–798, and Gal4SC1Δ583–798, which incorporated BrdU (mean and SD of at least three different experiments are shown). C and D are the same as A and B for HEK293 cells. Arrows indicate the cells transfected with SC1 constructs and the corresponding BrdU incorporation into their nuclei. Bars, 20 μm.
A reduction in luciferase activity was consistently observed when SC1 was fused to the Gal4-DBD and cotransfected with the reporter as compared with the luciferase activity measured in the presence of Gal4-DBD only (Fig. 2 A, lanes 1 and 2). We used a reporter construct where the luciferase gene was under the control of the TATA element only to verify the specificity of the results observed when luciferase is driven by the Gal4 upstream activating sequence. No repression was measured when SC1 was compared with Gal4 alone in these reporter assays (Fig. 2 A, lanes 3 and 4), supporting the conclusion that SC1 acts as a transcriptional repressor with a responsive promoter.
To define the domains of SC1 responsible for the repressive activity, we created truncated fusion proteins of NH2-terminally Flag-tagged SC1 with the Gal4-DBD and used them for transfection of HEK293 cells in our reporter assays. Three truncated proteins were made: (1) Δ754−798 lacks the extreme COOH terminus, which contains a highly acidic domain; (2) Δ583−798 lacks the six zinc finger domains in addition to the COOH-terminal domain; and (3) Δ404−798 lacks the PR domain and the entire COOH terminus. (see Fig. 2 B for the schematic representation of the fusion proteins). Expression of these proteins was verified by transfection of HEK293 cells and subsequent immunoprecipitation of the proteins using anti-Flag antibodies (Fig. 2 C).
We observed that deletion of the zinc finger domains from SC1 leads to a loss of its repressive activity (Fig. 2 B, compare lane Δ583–798 lacking the zinc fingers with Gal4 alone), whereas deletion of the acidic-rich COOH terminus had no influence on the repression by SC1 (Fig. 2 B, compare lane Δ754–798 lacking the COOH-terminus with Gal4 alone). Interestingly, deletion of the PR domain rendered SC1 a transcriptional activator, as can be seen from an increase in luciferase activity over the control levels (Fig. 2 B, compare lane Δ404–798 with Gal4). Such behavior has been observed for Blimp/PRDI-BF1 (Ren et al., 1999; Yu et al., 2000). We noticed that the truncations of the zinc fingers and the PR/SET domain led to a reduction of the total protein levels detected. To exclude the possibility that the increase in the luciferase activity observed upon the truncations of SC1 is due to the decrease of the protein level, we performed a series of experiments where an increasing amount of DNA encoding these proteins was transfected and luciferase measurement was performed. We obtained similar results in these experiments (Fig. 2 C, bottom graph).
As many transcriptional repressors recruit HDACs and need this activity to exert their repression (Grunstein, 1997; Pazin and Kadonaga, 1997; Wolffe, 1997), we tested whether TSA, a potent inhibitor of HDACs, influences the repressive activity of SC1 in the transcriptional reporter assays. Fig. 3 A shows that the repressive activity of SC1 was completely abolished by the addition of 50 ng/ml TSA, indicating that SC1's repression relies on the activity of TSA-sensitive HDACs. To directly test the interaction between SC1 and HDACs, we cotransfected Flag-tagged SC1 with HA-tagged HDAC 1, 2, or 3 into HEK293 cells and performed a coimmunoprecipitation assay. Anti-Flag Sepharose was used to bring down the Flag-tagged SC1. Expression of the fusion proteins was monitored by Western blot analysis. Cotransfection of Flag-vector with HDACs was used as a negative control. The results of these experiments are presented in Fig. 3 B, which demonstrated that SC1 can be found in a complex containing all the tested HDACs. Thus, SC1 associates with the class I HDACs and is likely to exert its repression by the recruitment of these proteins to the appropriate promoter sites.
Figure 3.

SC1 forms a complex with HDACs 1, 2, and 3, and its repression is TSA sensitive. (A) Repression of the Gal4-luc promoter is abolished by the addition of 50 ng/ml TSA to the transfected HEK293 cells. Relative luciferase activity was measured as described in Fig. 2 A. Black bars represent the control measurement with Gal4 only as an effector; open bars represent the measurement with Gal4SC1 as an effector. (B) SC1 coprecipitates with HDACs 1, 2, and 3 when cotransfected in HEK293 cells. Proteins from transfected cell lysates were immunoprecipitated using anti-Flag M2 Sepharose, separated by SDS-PAGE, and immunoblotted with either anti-Flag M2 antibody for SC1 or anti-HA antibody for HDACs. The bottom panel shows input of HDACs.
The zinc finger domains and the PR/SET domain of SC1 are necessary to block BrdU incorporation
The zinc finger domains of SC1 are required for both nuclear localization and transcriptional repression, whereas the PR/SET domain is involved in the modulation of the repressive activity by SC1. In this work, we investigated whether the deletion of zinc finger domains or the PR/SET domain would influence BrdU incorporation. Full-length and truncated forms of SC1 fused to either GFP or Gal4-DBD were used for this investigation. Fig. 4 summarizes the results from transfections of COS1 cells and Schwann cells with the GFP-SC1 fusion proteins. The data were normalized with respect to the BrdU incorporation of cells expressing GFP alone (Fig. 4, A and B). The quantification of nuclei that have incorporated BrdU upon transfection with the various SC1 truncated proteins revealed that the deletion of zinc finger or the PR/SET domains resulted in an inability to block BrdU incorporation. Fig. 4 A contains the results for ΔPR and ΔZF in COS1 cells. Fig. 4 B contains the results for ΔPR and ΔZF in Schwann cells and the respective quantifications. Overexpression of both the full-length and COOH-terminally truncated SC1 protein led to a block of DNA synthesis to the same extent (Fig. 4, A and B). These experiments verified that the zinc fingers are necessary for nuclear entry of SC1. On the other hand, deletion of the PR/SET domain led to a loss of BrdU incorporation, even though the SC1ΔPR protein remained in the nucleus. Hence, the PR/SET domain is involved in the regulation of BrdU incorporation by SC1.
Figure 4.
Zinc finger and the PR/SET domains of SC1 are required for its ability to block BrdU incorporation. COS1 (A) and Schwann (B) cells transiently transfected with GFP and GFP-SC1 fusion protein constructs. (a) GFP transfected cells, (b) GFP-SC1 transfected cells, (c) GFP-SC1ΔPR transfected cells, (d) GFP-SC1ΔZF transfected cells, (e) GFPΔC transfected cells. Cells were stained with antibodies against GFP and BrdU. GFP is labeled with FITC-coupled secondary antibodies (green), and BrdU with rhodamine-coupled secondary antibodies (red). Arrowheads indicate cells positive for BrdU incorporation after expression of SC1. Histograms represent the percentage of cells that express GFP or GFP-SC1 and incorporate BrdU. Data were normalized with respect to the BrdU incorporation of cells expressing GFP (mean and SD of at least three different experiments are shown). Bars, 20 μm.
To characterize the transcriptional activity of SC1 further, we used the Gal4-SC1 constructs used previously for the reporter gene assays. In contrast to the GFP fusion proteins used for experiments with COS1 and Schwann cells, SC1 deletion proteins were tethered to the Gal4-DBD with an endogenous NLS (see Fig. 5, A and C, for immunocytochemical detection of overexpressed proteins). This allowed us to establish a correlation between SC1's repressive activity and its ability to influence BrdU incorporation. As described in the previous paragraph, we normalized the BrdU incorporation data with respect to Gal4-DBD–transfected cells. Again, we observed a loss of SC1's capacity to block BrdU incorporation in the absence of its zinc finger domains (Fig. 5, B and D, lane SC1Δ583–798 for the respective quantification of BrdU incorporation in NIH3T3 and HEK293 cells, respectively). Overexpression of both full-length SC1 (Fig. 5, B and D, lane SC1FL for NIH3T3 and HEK293 cells, respectively) and a COOH-terminally truncated SC1 (Fig. 5, B and D, lane SC1Δ754–798 for NIH3T3 and HEK293 cells, respectively) resulted in much lower BrdU incorporation in both cell lines. Thus, SC1's repressive transcriptional activity could be involved in the implementation of the growth arrest in these cells.
The influence of NGF on the transcriptional activity of SC1
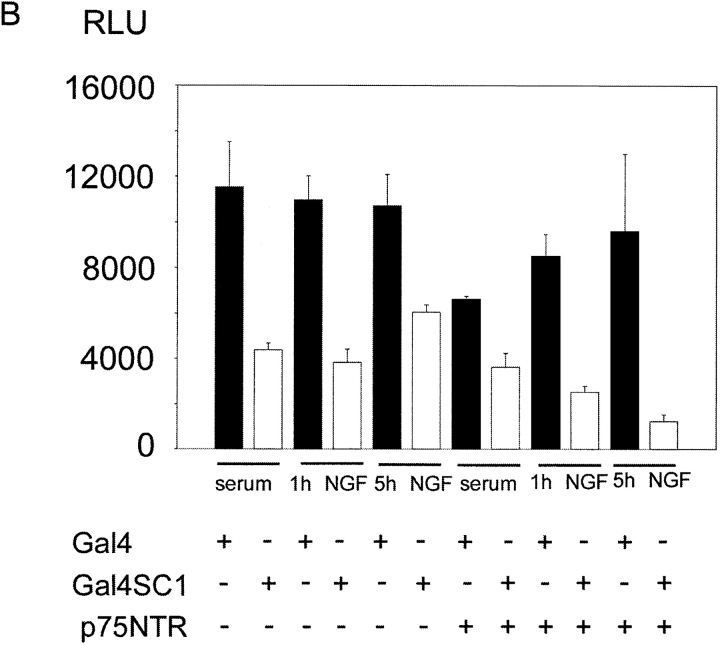
SC1 was originally isolated as a protein that interacts with the cytoplasmic domain of p75NTR, and its subcellular distribution was shown to be regulated by NGF. Therefore, we sought to investigate whether its transcriptional activity is regulated by the addition of NGF. To investigate this, we transfected HEK293 cells with Gal4-SC1 fusion protein as well as either TrkA or p75NTR and luciferase cDNAs under the control of Gal4 promoter as a reporter. Expression of p75NTR or TrkA was monitored by Western blotting after transfection (unpublished data). NGF was applied for either 1 or 5 h, and the luciferase activity was measured afterwards. We observed that coexpression of TrkA with SC1 led to a potentiation of the repressive activity of SC1 (see Fig. 6 A for the graphic representation of the relative luciferase units measured in these experiments). To verify the specificity of TrkA's influence on the repressive activity of SC1, we used K252a, a potent inhibitor of Trk signaling, and SC1's transcriptional output was then evaluated. Addition of K252a led to a loss of the potentiation of SC1's repressive activity as measured by the reporter assay (refer to Fig. 6 A for the graphic representation of these results). This observation implicates TrkA in the regulation of SC1's transcriptional activity. In the cotransfection experiments with p75NTR, SC1's repressive activity also correlated with the addition of NGF to the transfected cells. Thus, the addition of NGF for 1 h activated the repressive activity of SC1, and this activity increased over time as can be seen from the 5-h measurement (see Fig. 6 B for the graphic representation of SC1's transcriptional activity in the presence of p75NTR and NGF). Hence, SC1 is a transducer of both TrkA and p75NTR receptor signaling, and its activity is regulated by these two receptors.
Figure 6.
Regulation of SC1's repressive activity by TrkA, p75NTR, and NGF. (A) HEK293 cells were transiently transfected with TrkA and Gal4DBD-SC1, and Gal4-luc was used as a reporter. Cultures were either left in low serum-containing medium (1% FCS) or treated with NGF for the indicated amount of time. Relative luciferase activity (RLU) was measured in these lysates and normalized for transfection efficiency by cotransfection with β-galactosidase and by measuring the latter's activity. (B) Same as in A, except that p75NTR was used in these cotransfection experiments instead of TrkA. The results represent a mean of an experiment performed in triplicate and its SD; these were reproduced in several independent experiments.
SC1 represses transcription of genes that drive cell cycle progression
The fact that SC1's transcriptional activity influenced BrdU incorporation led us to test the possibility that SC1 may repress the expression of promitotic genes. To this end, we used a reporter gene system where the expression of cell cycle–related genes was assayed using a luciferase reporter assay. We used cyclin E, cyclin A, and cyclin B promoters for this analysis. The cyclin E promoter contained 1.4 kb of the sequence upstream of the transcription start site, which has been shown to regulate its transcription during G1 phase (Geng et al., 1996); the cyclin A promoter contained 3.2 kb of the sequence upstream of the transcription start site known to regulate its transcription during S phase (Yoshizumi et al., 1995); and the cyclin B1 promoter contained 3.8 bp upstream of the transcription start site. This sequence has been shown to enhance its transcription during G2 phase (Cogswell et al., 1995). Flag-tagged full-length SC1 was used as an effector, and the cyclin promoter-specific driven luciferase constructs were used as reporters. NIH3T3 cells were synchronized by serum withdrawal (see the Materials and methods section) for 48 h and released from growth arrest by the addition of serum-containing medium.
First, we determined the time course of expression of cyclins E, A, and B in these cultures by performing Northern blots at 0, 4, 8, 12, 16, 20, and 24 h after serum addition. Cyclin E could be detected at 12 h first and persisted until 24 h, cyclin A could first be detected at 20 h, and cyclin B was first detected at 24 h after serum addition of the growth-arrested cells (unpublished data). The cells were transfected with either SC1-bearing plasmid or a control plasmid, and lysates were collected at the following time points after serum addition: at 12 h for cyclin E measurement; at 20 h for cyclin A measurement; and at 24 h for cyclin B measurement. Subsequently, luciferase activity was measured in these lysates. We observed that cyclin E was down-regulated as measured by the luciferase activity, and the repression of the cyclin E promoter was comparable in magnitude to the repression exerted by SC1 when it is tethered to the Gal4 moiety (compare Fig. 7 A with Fig. 2 A). There was also a slight reduction of cyclin B expression as measured by the luciferase activity (Fig. 7 C). Expression of cyclin A, the major cyclin acting during S phase of the cell cycle, was not down-regulated by SC1 in these experiments (Fig. 7 B). To verify that the cyclin genes are expressed in these cells, we performed RT-PCR to monitor the endogenous expression of the cyclins (Fig. 7). Then, we investigated whether the levels of endogenous cyclin E protein were affected by the overexpression of SC1. To this end, we analyzed the protein lysates from NIH3T3 cells that had been transfected with either SC1 or a control Flag-bearing plasmid and treated as described above. Equal amounts of total protein were loaded on the gels, which were then transferred for Western blot analysis. We used an anti-cyclin E antibody to detect the levels of total cyclin E protein in these lysates, and normalized these by probing the blots with an anti-actin antibody (Fig. 7 D). The images were analyzed using a densitometer to determine the relative levels of cyclin E and actin present in transfected and mock-transfected cells, and were corrected for the transfection efficiency of these cells (which was routinely between 30 and 40%). We calculated a reduction of the total cyclin E protein level by a factor of 2.5 in cells overexpressing SC1, supporting our previous observations on the repression of the cyclin E promoter in the reporter assays (see Fig. 7 A).
Figure 7.
SC1 represses transcription of cyclin E in NIH3T3 cells. Flag-SC1FL constructs were used as effectors in this work, and were cotransfected with the indicated cyclin-specific driven luciferase gene. The two right bars on the graphs are shown to indicate that pGL reporter (luciferase under the basic promoter) is not influenced by the expression of SC1 as compared with control Flag plasmid. (A) Histogram of representative relative luciferase units measured after a cotransfection of SC1 with cyclin E-luc reporter. Luciferase activity was measured upon cotransfection with Flag-only vector as a control (black bars) and with Flag-SC1 fusion protein (white bars). The schematics of reporter promoters are shown below the histograms for each pair of effectors. An inset shows the results of a cyclin E–specific RT-PCR; +, reactions where RT was performed; −, the reaction where no RT was performed before PCR. (B) Same as in A, but with the cyclin A-luc reporter. (C) Same as in A, but with the cyclin B-luc reporter. (D) A Western blot showing the levels of endogenous cyclin E upon transfection with either SC1 or a control vector (top). Lane 1 shows the cyclin E levels before release from cell cycle arrest, lane 2 shows cyclin E in mock-transfected cells, and lane 3 shows cyclin E levels in SC1-transfected cells 12 h after release from the cell cycle block. Bottom: levels of actin in the same lysates; the numbering of lanes is the same as for the top panels.
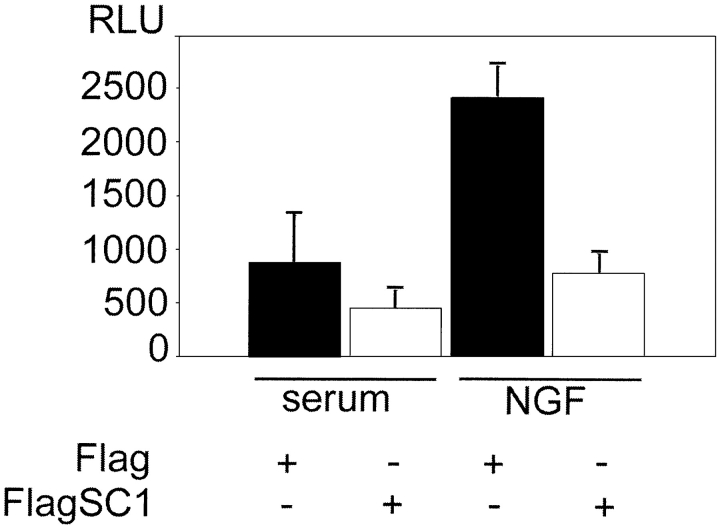
Our observations that SC1 represses the expression of cyclin E and that its activity is regulated by both TrkA and p75NTR led us to test the validity of these observations in a well-characterized cell line used in studying neurotrophin signaling, namely PC12 cells. Thus, the cells were synchronized using the method of Rudkin et al. (1989) by serum withdrawal, released from the cell cycle block by the addition of different factors, and samples to be analyzed were taken at ∼50 h after transfection for luciferase measurement (expression of the endogenous cyclin E at this time was verified by performing RT-PCR; unpublished data). We tested the effects of NGF on the repression of the cyclin E promoter by SC1. The results of these experiments are summarized in Fig. 8. Application of NGF to these cells enhanced the repression of cyclin E promoter by SC1. These observations implicate SC1 in the control of cell cycle progression and make it an important component of NGF signaling.
Figure 8.
SC1 represses transcription of cyclin E in PC12 cells. Flag-SC1FL or Flag-only expression plasmids were used in these experiments. Cells were treated as indicated, and the luciferase activity was measured ∼50 h after transfection and release from the cell cycle block. The results represent the mean of an experiment performed in triplicate and its SD; these were reproduced in several independent experiments.
Next, we investigated the role of the endogenous SC1 in regulating cyclin E expression in PC12 cells. To this end, we used the small interfering RNA (siRNA) method to block the expression of endogenous SC1 in PC12 cells, and measured the levels of cyclin E proteins in the lysates of transfected or mock-transfected cells. In parallel experiments with GFP expression plasmids, transfection rates in PC 12 cells were in the range of 30–50%, and thus resembled those in a recent paper (Rossoll et al., 2003). This corresponded to a reduction of SC1 by 40–60% in our experiments (Fig. 9). Down-regulation of SC1 leads to a significant increase in cyclin E expression, whereas the levels of cyclin A are not modified (Fig. 9), consistent with our observations in NIH3T3 cells as measured by luciferase reporter gene assays (Fig. 7, A and B). The quantification of the expression levels of SC1, cyclin E, and cyclin A proteins in PC12 cell lysates transfected with either SC1 siRNA or a scrambled siRNA construct is presented in Fig. 9 B. These experiments clearly show the involvement of endogenous SC1 in the regulation of transcription of cyclin E, but not cyclin A in PC12 cells.
Figure 9.
Endogenous SC1 down-regulation by siRNA in PC12 cells increases the expression of cyclin E, but not cyclin A. (A) PC12 cells were transfected with scrambled (mock) or SC1 siRNA (SC1), serum starved for 24 h, and collected 24–48 h after serum addition. Western blotting was performed to detect SC1, cyclin E, cyclin A, and actin. Representative blots are shown. (B) Quantification of SC1, cyclin E, and cyclin A expression in SC1 siRNA-transfected cells. Relative amounts were normalized with respect to the amounts of the corresponding cyclin or SC1 in mock siRNA-transfected cells (100%). The results are the mean ± SD of three independent experiments. Note the reduction in the amount of SC1, which corresponds to the increase in the amount of cyclin E in extracts from SC1 siRNA-treated PC12 cells. Significance of these results was tested using a t test with P < 0.05 for cyclin E and SC1. n.s., not significant.
Discussion
Transcriptional regulation through chromatin modification is emerging as one of the crucial ways to control differentiation during development. In this paper, we describe a function of SC1, a novel p75NTR-interacting zinc finger protein with a PR/SET domain. We present evidence that SC1 can act as a transcriptional repressor, it is found in a complex with HDACs 1, 2, and 3, and its activity is sensitive to TSA. Additionally, we demonstrate here that zinc finger domains are required for multiple functions of SC1 (i.e., nuclear localization, transcriptional repression, and SC1's ability to negatively influence DNA replication). The PR/SET domain is involved in transcriptional repression and decreases BrdU incorporation. Further, we present evidence that SC1 can down-regulate the expression of cyclin E, which is instrumental for the S-phase entry during cell cycle (Ewen, 2000). This down-regulation is observed in both NIH3T3 cells and in PC12 cells where the effect is enhanced by NGF.
Analysis of the transcriptional activity by SC1 revealed that it acts as a transcriptional repressor both in the context of being tethered to a DBD of another protein, Gal4, and by itself in transfection experiments. Our data indicate that the six zinc finger domains and the PR/SET domain are required for transcriptional repression by SC1. This implicates the zinc finger's and PR/SET domain's direct involvement in transcriptional activity either by allowing DNA binding of SC1 or by recruitment of other proteins that may modulate transcriptional repression. Consistent with the latter hypothesis, we observed that SC1 exerts its repressive activity by recruiting TSA-sensitive HDACs. Complexes involving HDACs have been implicated in silencing neuronal-specific genes (Naruse et al., 1999; Ballas et al., 2001). SC1 is an HDAC-dependent repressor, which may be involved in actively predisposing cells for differentiation by blocking their proliferative potential through down-regulation of promitotic genes.
Interestingly, truncation of the region of SC1 that contains the PR/SET domain transforms SC1 into a transcriptional activator, thus pointing to an involvement of this domain in repressive activity of SC1. Similar observations were made when Blimp-1's PR domain was removed (Yu et al., 2000). In this respect, it is noteworthy that two related proteins (RIZ and MDS1-EVI1) encode two different transcripts, which differ only in the presence of the PR/SET domain in the respective proteins (Bartholomew and Ihle, 1991; Liu et al., 1997). The absence of the PR/SET domain correlates with a weaker repressive activity in RIZ protein (Xie et al., 1997), suggesting a modulatory role of PR/SET domains during repression. Additionally, Blimp-1/PRDF-BF1 can exert repression through its PR/SET domain (Ghosh et al., 2001).
SET domains are found in chromosomal proteins that modulate gene expression and chromatin structure (for review see Jenuwein, 2001). Several proteins containing SET domains possess lysine histone methyltransferase activity (O'Carroll et al., 2000; Rea et al., 2000; Strahl et al., 2002). These histone modifications can lead to transcriptional repression (Firestein et al., 2000), but other enzymes are likely to be required, e.g., HDACs, to act together with methyltransferases. We do not know whether the PR/SET domain of SC1 possesses a protein methyltransferase activity. However, given SC1's ability to repress transcription in an HDAC-dependent manner, it would make SC1 a prime candidate for developmental regulation and subsequent maintenance of differentiation. Such a role is further substantiated by the observation that SC1's action is correlated with a block of BrdU incorporation, suggesting a role in cell differentiation and possible maintenance of the differentiated state.
One of the genes whose transcription is negatively regulated by SC1 is cyclin E, the major cyclin at G1–S phase transition. The down-regulation of cyclin E is consistent with the hypothesis that SC1 may play a major role at the G1–S decision making stage, and possibly during terminal mitosis as the cells prepare to enter a differentiative program. A down-regulation of cyclin E mRNA was observed by Tramtrack protein during glial development in Drosophila melanogaster, blocking entry into S phase and thus regulating glial cell proliferation (Badenhorst, 2001). Additionally, the activity of cyclin E together with CDK2, its binding partner, is instrumental for the regulation of cell cycle progression in neural and glial progenitor cells, and its block leads to an efficient cell cycle arrest (Casaccia-Bonnefil et al., 1999; Ferguson et al., 2000). Thus, SC1 may be one of the key regulators determining the decision of proliferating cells to exit cell cycle in the nervous system.
Several interesting points arise concerning the possible role of SC1 in neurotrophin signaling. It has been shown that in PC12 cells, the responsiveness to NGF is cell cycle stage-dependent (Rudkin et al., 1989), i.e., they will respond by differentiating when in G1 phase, but progress through the cell cycle when exposed to NGF during the other cell cycle phases (van Grunsven et al., 1996a,b). This responsive difference to NGF can be partially explained by the different temporal cell surface expression of TrkA and p75NTR (Urdiales et al., 1998). Thus, TrkA is observed on the cell surface during late M/early G1 phase, whereas p75NTR is highly expressed at the surface during the remaining time of cell cycle. Consistent with this temporal expression of the two NGF receptors, SC1 may act as a transducer of anti-proliferative signaling by NGF at an appropriate cell cycle stage, i.e., at G1–S transition as a blocker of further S phase entry. We observed that both TrkA and p75NTR enhanced the repressive activity of SC1 in our cotransfection experiments, implying the role of SC1 as the transducer of NGF signaling by these two receptors. This observation is particularly interesting in view of our data, showing that NGF enhances the repressive activity of SC1 at the cyclin E promoter in PC12 cells. As such, SC1 could play a decisive role during the differentiation of PC12 cells upon NGF addition. Specifically, as the decision to exit the cell cycle is made during G1 phase in these cells and both TrkA and p75NTR are expressed at this stage, they would be involved in mediating the anti-mitogenic response through SC1 as one of the key players in this process. The observations presented here offer novel venues for probing the molecular mechanism of NGF action as it is transduced by both TrkA and p75NTR through the activation of SC1.
Materials and methods
Cells lines, transfections, and reporter gene assays
HEK293 cells and NIH3T3 cells were kept in DME supplemented with 10% FCS unless otherwise indicated. We used a subline of PC12 cells, PC6-3 (a gift of Jonathan Ham, Institute of Child Health, London, UK; Pittman et al., 1993). These were grown in DME with 10% horse serum and 5% FCS, except for the synchronization experiments and treatment with growth factors where low serum was used (1% horse serum and 0.5% FCS). Synchronization was done according to a published protocol (Rudkin et al., 1989) where cells were kept without serum for 3 d and then released from the cell cycle block by the addition of either 100 ng/ml NGF or serum. For synchronization of NIH3T3, we used a previously published method (Kerkhoff and Rapp, 1997). In brief, the cells were arrested by keeping them in low serum (0.5% FCS) for 48 h, and were released from growth arrest by the addition of full serum-containing medium. In the experiments with K252a, the latter was added to the cells at a final concentration of 0.2 μM for 30 min before NGF addition. Transfections were performed using the LipofectAMINE™ 2000 kit according to the manufacturer's instructions (Invitrogen). 24-well plates were used for all the experiments, and the amount of DNA was kept constant, as suggested by the manufacturer. For TSA (Upstate Biotechnology) treatment of transfected cells, TSA was dissolved in DMSO and added directly to the transfected cells at a final concentration of 50 ng/ml for 16 h. DMSO was added to mock-treated cells in control reactions. Cells were lysed in the luciferase assay buffer as suggested by the manufacturer (Promega), and luciferase activity was measured using a luminometer (Berthold). As an internal control for transfection efficiency, β-galactosidase–expressing plasmid (pCMV-βgal) was cotransfected with the reporter and effector plasmids. The data were normalized to the β-galactosidase activity. Luciferase activity measurement was corrected for the transfection efficiency by calculating the ratio of luciferase units to β-galactosidase units, represented as the relative luciferase units. Transfections of COS1 and primary Schwann cells were performed using the Effectene Kit (QIAGEN) following the instructions of the vendor. Primary Schwann cells were isolated from P1 sciatic nerve according to a previously published protocol (Tikoo et al., 2000).
Plasmids
We used 0.75 μg 5xGal4 UAS-luciferase DNA (a gift of Al Fisher, Cornell University Medical College, New York, NY; Catron et al., 1995); 0.25 μg Gal4SC1 or other deletion mutants of SC1, and 0.05 μg pCMV-βgal DNA for transfections of HEK293 cells. The same relative ratios and amounts of plasmid DNA were used for transfections with cyclin-specific luciferase constructs. Reporters were used as follows: pGL2-cyclin E-luc containing 1.4 kb of cyclin E promoter; pGL2-cyclin B-luc containing 3.8 kb of the cyclin B promoter; and pGL2-cyclin A-luc containing ∼3.5 kb of cyclin A promoter (all these were gifts of E. Kerkhoff, University of Würzburg). As an effector, pSC1 construct was used, which is an NH2-terminal fusion of full-length SC1 with a Flag tag (Chittka and Chao, 1999). GFP fusion proteins were generated as follows: full-length SC1, SC1ΔZF (deletion from H608 to H744) and SCΔC (deletion from K749 to the end of the protein) were subcloned into pEGFP-C3 (CLONTECH Laboratories, Inc.) using HindIII and BamHI restriction sites. SC1ΔZF was created by generating an NdeI site in the full-length cDNA. Subsequent digestion with NdeI resulted in a construct without the zinc fingers. SC-1ΔPR was created by generating a PstI restriction site in the full-length cDNA. Digestion with PstI resulted in a construct without the PR domain. HA-tagged HDAC 1-, 2-, and 3-bearing plasmids were gifts of R. Evans (The Salk Institute for Biological Studies, La Jolla, CA). The TATA-luc plasmid was a gift of Heike Boemmel (University of Würzburg).
Immunocytochemistry
For the experiments using NIH3T3 and HEK293 cells, BrdU was purchased from Sigma-Aldrich and used according to a previously published protocol (Krek and DeCaprio, 1995). Cells were plated on sterile glass coverslips coated with poly-ornithine and were pulsed for 2–3 h with 10 μM BrdU before fixation with cold 50:50 methanol/acetone for 2–3 min. Immunostaining was performed as suggested by the manufacturer of the anti-BrdU antibody kit (Zymogen). Immunostaining of labeled cells with a monoclonal anti-FLAG FITC-conjugated antibody (Upstate Biotechnology) to visualize SC1-expressing cells was performed according to the manufacturer's instructions. Coverslips were embedded in Mowiol after washing. COS1 and Schwann cells were plated as above, except that poly-d-lysine was used for coating the coverslips. They were pulsed for 24 h with 10 μM BrdU before processing for immunocytochemistry. The cells were fixed in 4% PFA in PBS. Primary anti-BrdU antibody from DakoCytomation was used, with subsequent visualization by application of anti–mouse rhodamine-conjugated secondary antibody from Jackson ImmunoResearch Laboratories. Scans of stained cells were made using a confocal microscope (model TCS; Leica) for NIH3T3, COS1, and Schwann cells with identical settings for pinhole and voltage for any panel of analysis, and a fluorescent microscope (model IX70; Olympus) was used for HEK293 cells.
RT-PCR for cyclin-specific mRNA
RNA was extracted from the cells using TRIzol® (Invitrogen), according to the manufacturer's instructions. RT-PCR was performed using the SuperScript™ II kit (Invitrogen). Primers were used as follows: cyclin E, forward: 5′-GTGAAAAGCGAGGATAGCAG-3′, reverse: 5′-TGTTGTGATGCCATGTAACG-3′; cyclin B, forward: 5′-GGCACTGTTAAAGCCCTACC-3′, reverse: 5′-GTTCTGCATGAACCATC-3′; and cyclin A, forward: 5′-AAGGACCTTCCTATAAAC-3′, reverse: 5′-TTCTCCCACCTCAACCAG-3′. Cycling was performed as follows: for cyclin E and cyclin B: 5 min at 94°C for 1 cycle; 30 s at 94°C, 30 s at 53°C, 30 s at 72°C for 30 cycles; 10 min at 72°C for 1 cycle. For cyclin A, the annealing was performed at 50°C.
Co-immunoprecipitation experiments and Western blotting
Co-immunoprecipitations of Flag-SC1 and HDACs were performed as follows: HEK293 cells were transfected with either Flag-SC1 or Flag alone and HDAC 1, 2, or 3, and were harvested 48 h after transfection. Cells were lysed using RIPA buffer on ice and centrifuged to get rid of cell debris. Lysates were then precleared using protein A/G beads (Amersham Biosciences) and Flag-tagged proteins were precipitated using anti-Flag M2 Sepharose (Sigma-Aldrich). The beads were then washed with RIPA buffer 4–5 times and the immune complexes were separated by SDS-PAGE. Half of the reaction was run out on a 10% gel, blotted and probed for HDACs using anti-HA antibody (Santa Cruz Biotechnology, Inc.); another half was separated on a 7.5% gel, blotted and probed with anti-Flag M2 antibody (Sigma-Aldrich) to detect SC1. For Western blotting with anti-cyclin E antibody, we used a polyclonal antibody (Santa Cruz Biotechnology, Inc.); the anti-actin mouse monoclonal antibody was from DakoCytomation.
siRNA experiments
SC1 siRNA was obtained using the Silencer™ siRNA Cocktail Kit (RNase III; Ambion). As a template for the in vitro transcription, a fragment of 1.2 kb corresponding to nucleotides 1–1200 of the rat SC1 sequence was subcloned in pcDNA3 in both orientations. PC12 cells were transfected with SC1 siRNA or scrambled siRNA (0.5 μg/well) using LipofectAMINE™ 2000. 24 h later, cells were serum starved for 24 h. Serum-containing medium was added, and 24–48 h later, cells were collected directly in protein loading buffer and boiled for 5 min. Western blotting was performed to determine levels of SC1, cyclin E, cyclin A, and actin.
SC1 antibodies
Antibodies against SC1 were obtained from rabbits injected with the peptide GSMTTEGCRMSSAVYSADESLSAHKC coupled to KLH. Antibodies were purified using an affinity column with the peptide coupled to CNBr Sepharose (Roche). Antibodies were used at 2 μg/ml for Western blotting.
Acknowledgments
We thank Wilfried Rossoll for help with the luciferase measurements and discussion, and Ulrich Schweizer for plentiful discussions during the course of this work. We are greatly indebted to Martin Koltzenburg for letting us carry out the work in his laboratory at the Institute of Child Health in London. Additionally, we would like to thank Jonathan Ham for providing us with the cells and reagents necessary to carry out the experiments with PC12 cells.
This work was supported by the Deutsche Forschungsgemeinschaft grants SFB487, TPC4, and SFB465, TPA3; J.C. Arevalo was supported by a postdoctoral fellowship from the Spanish Ministry of Education; M.V. Chao was supported by National Institutes of Health grant CA56490; and Pilar Pérez was supported by Junta de Castilla y León (CSI5/02).
Abbreviations used in this paper: DBD, DNA-binding domain; HDAC, histone deacetylase; NTR, neurotrophin receptor; PR/SET, positive regulatory/suppressor of variegation, enhancer of zeste, trithorax; RIZ, retinoblastoma-interacting zinc finger; SC1, Schwann cell factor 1; siRNA, small interfering RNA; TSA, trichostatin A.
References
- Abbondanza, C., N. Medici, V. Nigro, V. Rossi, L. Gallo, G. Piluso, A. Belsito, A. Roscigno, P. Bontempo, A.A. Puca, et al. 2000. The retinoblastoma-interacting zinc-finger protein RIZ is a downstream effector of estrogen action. Proc. Natl. Acad. Sci. USA. 97:3130–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst, P. 2001. Tramtrack controls glial number and identity in the Drosophila embryonic CNS. Development. 128:4093–4101. [DOI] [PubMed] [Google Scholar]
- Ballas, N., E. Battaglioli, F. Atouf, M.E. Andres, J. Chenoweth, M.E. Anderson, C. Burger, M. Moniwa, J.R. Davie, W.J. Bowers, et al. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 31:353–365. [DOI] [PubMed] [Google Scholar]
- Bamji, S.X., M. Majdan, C.D. Pozniak, D.J. Belliveau, R. Aloyz, J. Kohn, C.G. Causing, and F.D. Miller. 1998. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J. Cell Biol. 140:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew, C., and J.N. Ihle. 1991. Retroviral insertions 90 kilobases proximal to the Evi-1 myeloid transforming gene activate transcription from the normal promoter. Mol. Cell. Biol. 11:1820–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyse, I.M., G. Shao, and S. Huang. 1995. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc. Natl. Acad. Sci. USA. 92:4467–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil, P., B.D. Carter, R.T. Dobrowsky, and M.V. Chao. 1996. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature. 383:716–719. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil, P., R.J. Hardy, K.K. Teng, J.M. Levine, A. Koff, and M.V. Chao. 1999. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 126:4027–4037. [DOI] [PubMed] [Google Scholar]
- Casademunt, E., B.D. Carter, I. Benzel, J.M. Frade, G. Dechant, and Y.A. Barde. 1999. The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. EMBO J. 18:6050–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catron, K.M., H. Zhang, S.C. Marshall, J.A. Inostroza, J.M. Wilson, and C. Abate. 1995. Transcriptional repression by Msx-1 does not require homeodomain DNA-binding sites. Mol. Cell. Biol. 15:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, M.V., and B.L. Hempstead. 1995. p75 and Trk: a two-receptor system. Trends Neurosci. 18:321–326. [PubMed] [Google Scholar]
- Chittka, A., and M.V. Chao. 1999. Identification of a zinc finger protein whose subcellular distribution is regulated by serum and nerve growth factor. Proc. Natl. Acad. Sci. USA. 96:10705–10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell, J.P., M.M. Godlevski, M. Bonham, J. Bisi, and L. Babiss. 1995. Upstream stimulatory factor regulates expression of the cell cycle-dependent cyclin B1 gene promoter. Mol. Cell. Biol. 15:2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen, M.E. 2000. Where the cell cycle and histones meet. Genes Dev. 14:2265–2270. [DOI] [PubMed] [Google Scholar]
- Fears, S., C. Mathieu, N. Zelenik-Le, S. Huand, J.D. Rowley, and G. Nucifora. 1996. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. USA. 93:1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, K.L., S.M. Callaghan, M.J. O'Hare, D.S. Park, and R.S. Slack. 2000. The Rb-CDK4/6 signalling pathway is critical in neural precursor cell cycle regulation. J. Biol. Chem. 275:33593–33600. [DOI] [PubMed] [Google Scholar]
- Firestein, R., X. Cui, P. Huie, and M.L. Cleary. 2000. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol. 20:4900–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade, J.M., and Y.A. Barde. 1998. Microglia-derived Nerve Growth Factor causes cell death in the developing retina. Neuron. 20:35–42. [DOI] [PubMed] [Google Scholar]
- Frade, J.M., A. Rodriguez Tebar, and Y.A. Barde. 1996. Induction of cell death by endogenous nerve growth factor through its p75 receptor. Nature. 383:166–168. [DOI] [PubMed] [Google Scholar]
- Geng, Y., E.N. Eaton, M. Picon, J.M. Roberts, A.S. Lundberg, A. Gifford, C. Sardet, and R.A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 12:1173–1180. [PubMed] [Google Scholar]
- Ghosh, N., I. Gyory, G. Wright, J. Wood, and K.L. Wright. 2001. Positive regulatory domain I binding factor 1 silences class II transactivator expression in multiple myeloma cells. J. Biol. Chem. 276:15264–15268. [DOI] [PubMed] [Google Scholar]
- Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature. 389:349–352. [DOI] [PubMed] [Google Scholar]
- Huang, S., G. Shao, and L. Liu. 1998. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 273:15933–15939. [DOI] [PubMed] [Google Scholar]
- Izutsu, K., M. Kurokawa, Y. Imai, K. Maki, K. Mitani, and H. Hirai. 2001. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signalling. Blood. 97:2815–2822. [DOI] [PubMed] [Google Scholar]
- Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266–273. [DOI] [PubMed] [Google Scholar]
- Keller, A.D., and T. Maniatis. 1991. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5:868–879. [DOI] [PubMed] [Google Scholar]
- Kerkhoff, E., and U.R. Rapp. 1997. Induction of cell proliferation in quiescent NIH 3T3 cells by oncogenic c-Raf-1. Mol. Cell. Biol. 17:2576–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198–209. [DOI] [PubMed] [Google Scholar]
- Krek, W., and J.A. DeCaprio. 1995. Cell synchronization. Methods Enzymol. 254:114–124. [DOI] [PubMed] [Google Scholar]
- Kurokawa, M., K. Mitani, K. Irie, T. Matsuyama, T. Takahashi, S. Chiba, Y. Yazaki, K. Matsumoto, and H. Hirai. 1998. The oncoprotein Evi-1 represses TGF-beta signalling by inhibiting Smad3. Nature. 394:92–96. [DOI] [PubMed] [Google Scholar]
- Lin, Y., K. Wong, and K. Calame. 1997. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 276:596–599. [DOI] [PubMed] [Google Scholar]
- Liu, L., G. Shao, G. Steele-Perkins, and S. Huang. 1997. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J. Biol. Chem. 272:2984–2991. [DOI] [PubMed] [Google Scholar]
- Mukai, J., T. Hachiya, S. Shoji-Hoshino, M.T. Kimura, D. Nadano, P. Suvanto, T. Hanaoka, Y. Li, S. Irie, L.A. Greene, and T.A. Sato. 2000. NADE, a p75NTR-associated cell death executor, is involved in signal transduction mediated by the common neurotrophin receptor p75NTR. J. Biol. Chem. 275:17566–17570. [DOI] [PubMed] [Google Scholar]
- Naruse, Y., T. Aoki, T. Kojima, and N. Mori. 1999. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA. 96:13691–13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Carroll, D., H. Scherthan, A.H. Peters, S. Opravil, A.R. Haynes, G. Laible, S. Rea, M. Schmid, A. Lebersorger, M. Jerratsch, et al. 2000. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 20:9423–9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, S., J.P. Brouillet, A. Kilbey, R. Fulton, M. Walker, M. Crossley, and C. Bartholomew. 2001. Evi-1 transforming and repressor activities are mediated by CtBP co-repressor proteins. J. Biol. Chem. 276:25834–25840. [DOI] [PubMed] [Google Scholar]
- Pazin, M.J., and J.T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell. 89:325–328. [DOI] [PubMed] [Google Scholar]
- Pittman, R.N., W. Songli, A.J. DiBenedetto, and J.C. Mills. 1993. A system for characterizing cellular and molecular events in programmed neuronal cell death. J. Neurosci. 13:3669–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S., F. Eisenhaber, D. O'Carroll, B.D. Strahl, Z.W. Sun, M. Schmid, S. Opravil, K. Mechtler, C.P. Ponting, C.D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 406:593–599. [DOI] [PubMed] [Google Scholar]
- Ren, B., K.J. Chee, T.H. Kim, and T. Maniatis. 1999. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13:125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll, W., S. Jablonka, C. Andreassi, A.-K. Kröning, K. Karle, U.R. Monani, and M. Sendtner. 2003. Smn, the spinal muscular atrophy–determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163:801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudkin, B.B., P. Lazarovici, B.-Z. Levi, Y. Abe, K. Fujita, and G. Guroff. 1989. Cell cycle-specific action of nerve growth factor in PC12 cells: Differentiation without proliferation. EMBO J. 8:3319–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi, A.H., P.P. Roux, C.J. Kubu, C. Zeindler, A. Bhakar, L.L. Tannis, J.M. Verdi, and P.A. Barker. 2000. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 27:279–288. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins, G., W. Fang, X.H. Yang, M. Van Gele, T. Carling, J. Gu, I.M. Buyse, J.A. Fletcher, J. Liu, R. Bronson, et al. 2001. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev. 15:2250–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl, B.D., P.A. Grant, S.D. Briggs, Z.W. Sun, J.R. Bone, J.A. Caldwell, S. Mollah, R.G. Cook, J. Shabanowitz, D.F. Hunt, and C.D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikoo, R., G. Zanazzi, D. Shiffman, J. Salzer, and M.V. Chao. 2000. Cell cycle control of Schwann cell proliferation: role of cyclin-dependent kinase-2. J. Neurosci. 20:4627–4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdiales, J.L., E. Becker, M. Andrieu, A. Thomas, J. Jullien, L.A. van Grunsven, S. Menut, G.I. Evan, D. Martin-Zanca, and B.B. Rudkin. 1998. Cell cycle phase-specific surface expression of nerve growth factor receptors TrkA and p75(NTR). J. Neurosci. 18:6767–6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Grunsven, L.A., N. Billon, P. Savatier, A. Thomas, J.L. Urdiales, and B.B. Rudkin. 1996. a. Effect of nerve growth factor on the expression of cell cycle regulatory proteins in PC12 cells: dissection of the neurotrophic response from the anti-mitogenic response. Oncogene. 12:1347–1356. [PubMed] [Google Scholar]
- van Grunsven, L.A., A. Thomas, J.L. Urdiales, S. Machenaud, P. Choler, I. Durand, and B.B. Rudkin. 1996. b. Nerve growth factor-induced accumulation of PC12 cells expressing cyclin D1: evidence for a G1 phase block. Oncogene. 12:855–862. [PubMed] [Google Scholar]
- Wolffe, A.P. 1997. Transcriptional control. Sinful repression. Nature. 387:16–17. [DOI] [PubMed] [Google Scholar]
- Xie, M., G. Shao, I.M. Buyse, and S. Huang. 1997. Transcriptional repression mediated by the PR domain zinc finger gene RIZ. J. Biol. Chem. 272:26360–26366. [DOI] [PubMed] [Google Scholar]
- Yan, Q., and E.M. Johnson, Jr. 1987. A quantitative study of the developmental expression of nerve growth factor (NGF) receptor in rats. Dev. Biol. 121:139–148. [DOI] [PubMed] [Google Scholar]
- Yan, Q., and E.M. Johnson, Jr. 1988. An immunohistochemical study of the nerve growth factor receptor in developing rats. J. Neurosci. 8:3481–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi, M., C.M. Hsieh, F. Zhou, J.C. Tsai, C. Patterson, M.A. Perrella, and M.E. Lee. 1995. The ATF site mediates downregulation of the cyclin A gene during contact inhibition in vascular endothelial cells. Mol. Cell. Biol. 15:3266–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., C. Angelin-Duclos, J. Greenwood, J. Liao, and K. Calame. 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20:2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]