Abstract
Misfolded proteins retained in the endoplasmic reticulum (ER) are degraded by the ER-associated degradation pathway. The mechanisms used to sort them from correctly folded proteins remain unclear. Analysis of substrates with defined folded and misfolded domains has revealed a system of sequential checkpoints that recognize topologically distinct domains of polypeptides. The first checkpoint examines the cytoplasmic domains of membrane proteins. If a lesion is detected, it is retained statically in the ER and rapidly degraded without regard to the state of its other domains. Proteins passing this test face a second checkpoint that monitors domains localized in the ER lumen. Proteins detected by this pathway are sorted from folded proteins and degraded by a quality control mechanism that requires ER-to-Golgi transport. Although the first checkpoint is obligatorily directed at membrane proteins, the second monitors both soluble and membrane proteins. Our data support a model whereby “properly folded” proteins are defined biologically as survivors that endure a series of distinct checkpoints.
Keywords: ER-associated degradation; misfolded proteins; protein folding; protein trafficking; endoplasmic reticulum
Introduction
Most proteins secreted from the cell or resident along the secretory pathway must first traverse the membranes of the ER. Nascent polypeptides are translocated in the unfolded state so folding and oligomerization occur in the lumen of the ER. For this reason, the ER maintains a full complement of chaperones and enzymes required for folding. In addition, maturation of many secretory proteins requires posttranslational modifications that are performed by enzymes resident in the ER. To prevent the transport of polypeptides to their sites of function until they are properly folded, cells evolved mechanisms termed collectively “ER quality control” (Brodsky and McCracken, 1999; Ellgaard and Helenius, 2003). Proteins irreversibly misfolded are degraded by the ER-associated degradation (ERAD) pathway, an aspect of ER quality control (Hampton, 2002; Kostova and Wolf, 2003).
Although the mechanisms underlying ER quality control are incompletely understood, the emerging view describes a series of requisite events that culminate with degradation. The first step requires sorting of folded proteins from folding intermediates and misfolded proteins. How the cell distinguishes the vast array of possible conformations and places them into these broad categories is unclear. Some folded proteins bind specific cargo receptor molecules for sorting into COPII-coated vesicles destined for the Golgi apparatus (Belden and Barlowe, 2001). By extension, it is conceivable that unfolded proteins remain in the ER by default. This type of mechanism would require that receptors recognize folded conformations, which is not yet established. In mammalian cells, the ER lectins calnexin and calreticulin retain unfolded glycoproteins reglucosylated by UDP-glucose–glycoprotein glucosyltransferase (GT; Hebert et al., 1995; Trombetta and Helenius, 1999). Thus, GT appears to be a factor capable of distinguishing folded and unfolded glycoproteins. In yeast, there is no GT equivalent but mounting evidence suggests that the ER chaperone BiP could play a role in conformational sorting. BiP mutants are defective in ERAD and exhibit aggregation of misfolded proteins (Plemper et al., 1997; Brodsky et al., 1999; Nishikawa et al., 2001).
Once sorted, misfolded proteins must be targeted for degradation. The best available information suggests that the key target is the Sec61 translocon, an ER membrane pore complex known for its role in ER protein import (Johnson and van Waes, 1999). Thus, the translocon appears to mediate both forward and reverse translocation of proteins across ER membranes. How the substrate is delivered to the translocon is unclear but recent works suggest that the folding catalyst protein disulfide isomerase could play a key role (Gillece et al., 1999; Tsai et al., 2001). For glycoproteins, the putative lectin Htm1p/Mnl1p (EDEM in mammals) could play a role in presenting substrates to the translocon (Jakob et al., 2001; Nakatsukasa et al., 2001; Molinari et al., 2003; Oda et al., 2003). On the cytosolic side of the membrane, substrate retrotranslocation and extraction is assisted by the Cdc48–Ufd1–Npl4 complex (Ye et al., 2001; Jarosch et al., 2002; Rabinovich et al., 2002). The substrate is next ubiquitylated by specific E2 ubiquitin conjugating enzymes and E3 ubiquitin ligases located at the ER membrane (Hampton et al., 1996; Hiller et al., 1996; Bordallo et al., 1998; Bays et al., 2001). Misfolded glycoproteins are deglycosylated by the cytosolically localized enzyme protein N-glycanase (Suzuki et al., 2000; Hirsch et al., 2003). Finally, the processed substrate is deubiquitinated and degraded by the 26S proteasome (Ward et al., 1995; Hampton et al., 1996; Hiller et al., 1996).
The wide array of ERAD substrates confounds the idea that a single pathway is sufficient for all the needs of ER quality control. Proteins can be soluble in the lumen or integrated in the ER membrane. Furthermore, membrane proteins can be composed of three distinct domains: luminal, membrane, and cytosolic. Therefore, it is difficult to envision how a single surveillance mechanism can inspect all classes of aberrant secretory proteins. From studies in yeast and mammals, several pathways operate simultaneously for quality control. Some proteins are transported to the Golgi apparatus and retrieved to the ER for degradation, whereas others are statically retained (Hammond and Helenius, 1994; Caldwell et al., 2001; Vashist et al., 2001; Yamamoto et al., 2001). In yeast, certain misfolded proteins bypass ER quality control entirely and are directly transported to the vacuole (the yeast lysosome) for degradation (Hong et al., 1996; Holkeri and Makarow, 1998; Luo et al., 2002). This pathway, termed “Golgi quality control” (Arvan et al., 2002), also serves to degrade excess substrates of the ER quality control pathway in cells under stress (Spear and Ng, 2003).
Precisely why some misfolded proteins are selected for one pathway over others is unknown. Studies examining multiple classes of ERAD substrates have provided some clues. In yeast, substrates requiring ER-to-Golgi transport for degradation are typically soluble, whereas those statically retained are integral membrane proteins (Caldwell et al., 2001; Vashist et al., 2001). From these data, we hypothesized that misfolded soluble proteins are sorted to the retrieval pathway, whereas membrane proteins are statically retained before degradation. Although the notion was appealing because of its simplicity, the relatively small number of substrates examined left open the possibility of other models.
Here, we tested our hypothesis directly by designing a series of ERAD substrates with defined misfolded domains. Our data show that proteins are not sorted on the basis of their association with the membrane. Instead, the site of the lesion is the most important determinant. Membrane proteins with lesions in their cytosolic domains are retained, whereas proteins with solely luminal lesions are sorted for retrieval. This suggests the existence of distinct luminal and cytosolic surveillance mechanisms for ER quality control. Proteins with both cytosolic and luminal lesions exclusively use the retention pathway showing that it precedes the retrieval pathway. Together, our data support a model by which the quality control of secretory protein folding is comprised of a series of checkpoints. Proteins halted at any checkpoint are degraded. Proteins that pass all checkpoints are deemed as “folded” and transported to their sites of function.
Results
Two distinct ERAD pathways are defined by specific kinetic and genetic parameters
Two soluble proteins, mutant carboxypeptidase Y (CPY*) and yeast Kar2p signal sequence fused to the simian virus 5 HA-Neuraminidase ectodomain (KHN), and two membrane proteins, Sec61-2p (mutant ER translocon component) and Ste6-166p (mutant a-factor transporter), have been studied as model substrates for ER quality control in the budding yeast Saccharomyces cerevisiae (Finger et al., 1993; Sommer and Jentsch, 1993; Loayza et al., 1998; Vashist et al., 2001). Diagnostic of their identity as ERAD substrates is the requirement of ERAD-specific ubiquitylation genes for degradation and dispensability of genes required for vacuolar proteolytic function (Table I , CUE1, HRD1/DER3, and DOA10 ubiquitylation genes and the PEP4 gene required for vacuolar protease activation).
Table I. Membrane and soluble proteins differ in their requirements for degradation.
| Membrane proteins
|
Soluble proteins
|
||||
|---|---|---|---|---|---|
| Sec61-2p | Ste6-166p | KHN | CPY* | ||
| t 1/2 (min) | 12 | 8 | 35 | 27 | |
| Requires | CUE1 | yes | yes | yes | yes |
| PEP4 | no | no | no | no | |
| SEC12 | no | no | yes | yes | |
| SEC18 | no | no | yes | yes | |
| DER1 | no | no | yes | yes | |
| HRD1/DER3 | yes | no | yes | yes | |
| DOA10 | ND | yes | no | no | |
Data complied from Vashist et al., 2001, and this work.
Before degradation, the soluble substrates are sorted into a pathway that requires ER-to-Golgi transport, whereas the membrane substrates are retained statically in the ER. This difference is reflected by whether the ER-to-Golgi transport genes SEC12 and SEC18 are required for degradation (Table I) (Caldwell et al., 2001; Vashist et al., 2001). They also differ in their requirement for DER1, a gene required for the degradation of the soluble substrates but dispensable for the membrane substrates (Table I). In addition to differences in genetic requirements, the two classes of substrates can be distinguished by their turnover rates. The membrane substrates (t 1/2, 8–12 min) are degraded more rapidly than the soluble substrates (t 1/2, 27–35 min). This difference is probably a reflection of their distinctive sorting mechanisms. Thus, the pathway used by a substrate can be characterized by the genetic and kinetic parameters as outlined in Table I.
From the accumulated data, the simplest view suggests that soluble substrates require the pathway involving transport between the ER and Golgi apparatus, whereas membrane substrates are degraded independently of transport (Table I). However, the locations of genetic lesions raised a second possibility. Both Sec61-2p and Ste6-166p contain mutations mapped in or near their cytosolic domains (Sec61-2p: Gly213 to Asp; Ste6-166p: Gln1249 to stop that truncates 42 aa), whereas misfolded portions of the soluble substrates are entirely luminal (Finger et al., 1993; Loayza et al., 1998; Nishikawa et al., 2001). This suggests that the site of lesion with respect to the ER membrane could be a major determinant. To distinguish these possibilities, we constructed and analyzed a series of chimeric molecules with defined folded and misfolded domains.
The site of lesion determines the pathway used
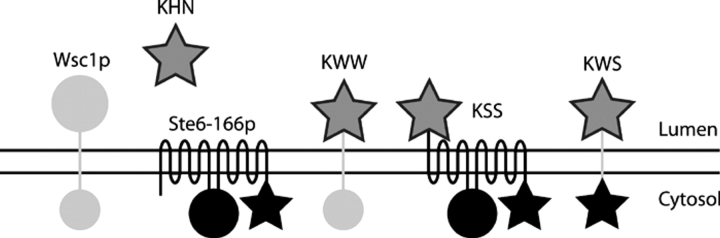
First, we analyzed an integral membrane substrate designed to be misfolded exclusively in the luminal domain. If all types of misfolded membrane proteins are statically retained as a rule, we expect it to be degraded independently of vesicular transport. Dependence on SEC12 and SEC18 would support the alternative model. This substrate was created by replacing the luminal/extracellular domain of Wsc1p with KHN. Wsc1p is a nonessential signaling protein anchored by a single transmembrane domain in the type I orientation (NoutCin; Lodder et al., 1999). As a plasma membrane protein, it does not contain intrinsic ER retention signals that might indirectly influence its targeting for ERAD. KHN was selected for the luminal domain because it contains O-linked sugars that are modified after transport to the Golgi apparatus. The modification provides a convenient localization marker that can be monitored by a characteristic shift in gel mobility (Vashist et al., 2001). The chimeric molecule, designated KHN luminal domain/Wsc1p transmembrane domain/Wsc1p cytosolic domain (KWW), is schematically depicted in Fig. 1 .
Figure 1.
Schematic representation of substrates. The engineered ERAD substrates bear three letter designations that describe their composition. The first, second, and third letters represent the luminal, transmembrane, and cytosolic domains, respectively. For each protein, KHN is symbolized by dark gray stars, Wsc1p portions by light gray bars (transmembrane) and circles (folded cytosolic and luminal domains), and Ste6-166p by black bars/loops (transmembrane), black circles (folded cytosolic domain), and black stars (misfolded cytosolic domain).
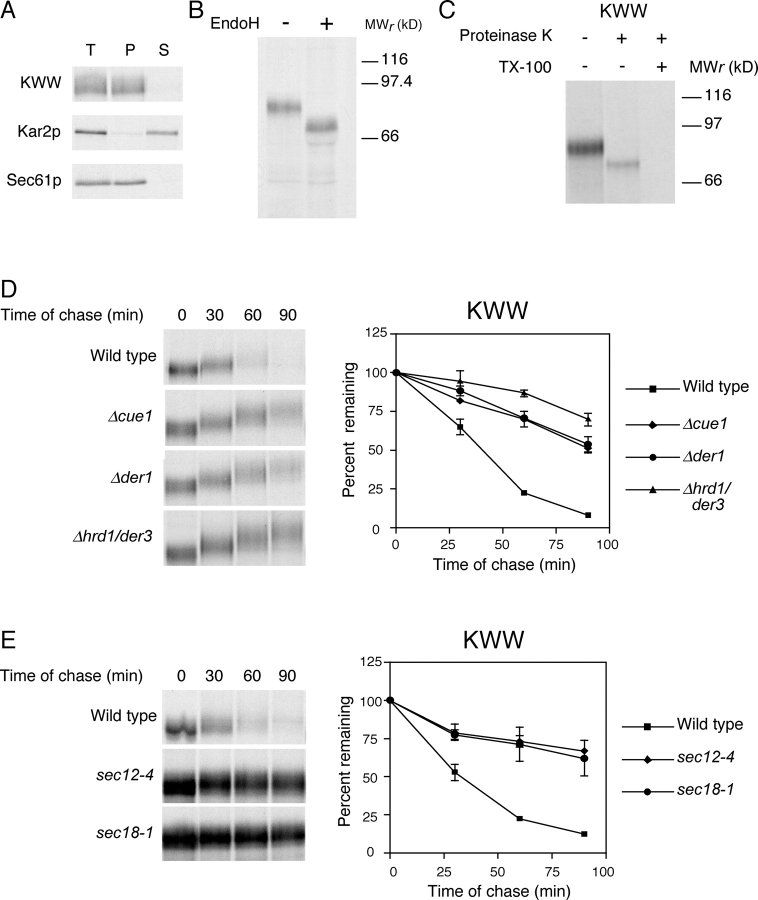
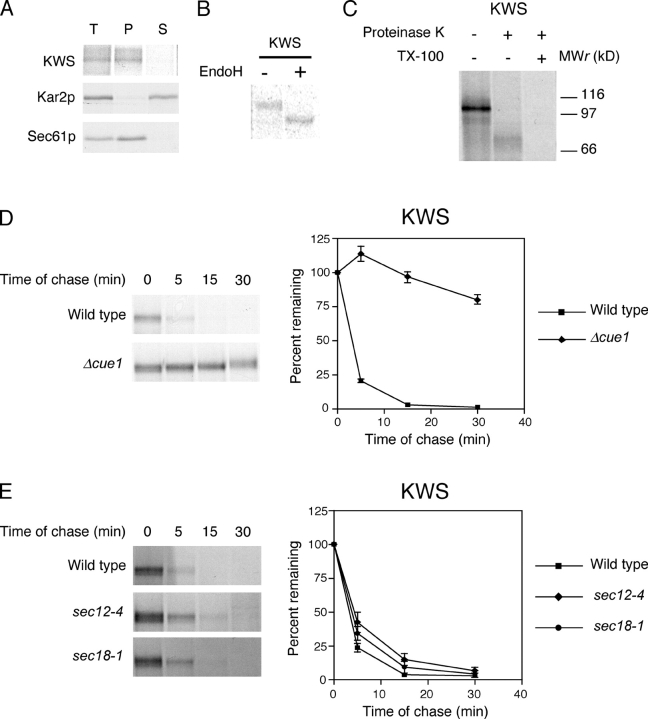
To determine if KWW is indeed an integral membrane protein, we prepared a zirconium bead–disrupted extract from wild-type cells expressing the protein and subjected it to alkali. Under these conditions, soluble and peripherally associated membrane proteins are extracted from membranes. These can be subsequently separated from integral membrane proteins by centrifugation. As shown in Fig. 2 A, KWW was found exclusively in the pellet fraction along with the Sec61p membrane protein control. The soluble luminal protein Kar2p partitioned into the supernatant fraction, as expected. These data confirm that KWW is a bona fide integral membrane protein.
Figure 2.
KWW is degraded by the ERAD-L pathway. (A) Cells expressing KWW were metabolically labeled with [35S]methionine/cysteine for 20 min. Cell lysates were prepared and incubated in 0.1 M of sodium carbonate, pH 11.0, for 30 min at 4°C and the membrane fraction separated by centrifugation. Proteins were immunoprecipitated from total (T), pellet (P), and supernatant (S) fractions. Sec61p and Kar2p serve as controls for integral membrane and soluble proteins, respectively. KWW was immunoprecipitated using anti-HA mAb (HA.11; Covance) and resolved by electrophoresis using 10% SDS polyacrylamide gels. (B) Cells expressing KWW were labeled with [35S]methionine/cysteine for 10 min. KWW immunoprecipitates were mock treated (−) or digested (+) with Endo H, resolved by SDS-PAGE, and visualized by autoradiography. (C) Protease protection assay. Wild-type cells expressing KWW were pulse labeled for 10 min and a fraction containing cytosol and membranes was prepared. The extracts were split and treated with proteinase K in the presence or absence of Triton X-100 or mock treated. KWW was immunoprecipitated and analyzed by SDS-PAGE (8%) and autoradiography. (D) Wild-type and mutant strains expressing KWW were pulse labeled for 10 min with [35S]methionine/cysteine followed by cold chase for times indicated. Immunoprecipitated proteins were analyzed by electrophoresis on 8% gels and visualized by autoradiography. (E) Wild-type, sec12-4, and sec18-1 strains were grown to log phase at 22°C and shifted to the restrictive temperature (37°C) for 30 min before pulse-chase analysis as described for D. The data were quantified by PhosphorImager analysis and plotted as mean values with SDs of two independent experiments (D and E). Representative autoradiograms are shown.
KWW was designed to maintain a type I membrane orientation by including Wsc1p membrane flanking sequences that are important determinants (Hartmann et al., 1989). We confirmed the orientation using two methods. First, we analyzed the N-linked glycosylation pattern of KWW. Each N-linked carbohydrate decreases the gel mobility of yeast glycoproteins by ∼2 kD. KHN contains four N-linked glycosylation sites, whereas the cytosolic domain of Wsc1p contains only a single cryptic site. Thus, correctly oriented KWW exhibits a substantial shift when deglycosylated, whereas a minor shift would be observed if misoriented. Wild-type cells expressing KWW were pulse labeled, KWW immunoprecipitated, and digested with endoglycosidase H (Endo H) to remove N-linked sugars. As shown in Fig. 2 B, deglycosylated KWW migrated at a position consistent with an 8-kD shift. Notably, glycosylated KWW migrated as a single species indicating only one membrane orientation. As a second approach, we applied a microsome protease-protection assay. Because the portion carboxyl-proximal to the transmembrane domain contributes 10 kD to KWW, whereas the amino-terminal portion contributes 67 kD (not including N- and O-linked sugars if properly oriented), the size of the protected fragment provides an unambiguous determination of membrane orientation. A membrane fraction was prepared from cells expressing KWW and digested with proteinase K or mock treated. As shown in Fig. 2 C, protease treatment generated a protected KWW fragment that migrates in SDS-PAGE consistent with a 10-kD reduction in size. Recognition of this fragment by the HA mAb further confirms the luminal orientation of the KHN domain as the epitope is located there. These data show that KWW is a type I integral membrane protein.
To determine which pathway degrades KWW, we monitored its turnover in wild-type and mutant strains using metabolic pulse-chase experiments. With a t 1/2 of 35 min, KWW was degraded at a rate similar to KHN and CPY* and much slower than the membrane substrates Ste6-166p and Sec61-2p (Fig. 2 D). Strains deleted of the ERAD-related genes CUE1 and HRD1/DER3, required for substrate ubiquitylation, stabilized KWW confirming its degradation by the ERAD pathway. Interestingly, KWW also required DER1, an ERAD gene required by many misfolded soluble proteins (Fig. 2 D and Table I; Knop et al., 1996a; Vashist et al., 2001). These data provide the first evidence that DER1 plays a role in degrading misfolded integral membrane proteins. We also noticed that KWW acquired a mobility shift during the time course suggesting that some molecules were transported to the Golgi apparatus. To support this assertion and demonstrate that KWW degradation requires vesicular transport, we measured its turnover in sec12-4 and sec18-1 mutant strains. SEC12 is required for vesicle budding from the ER and SEC18 is required for vesicle fusion to the Golgi apparatus (Eakle et al., 1988; Nakano et al., 1988; Barlowe and Schekman, 1993). In both strains, KWW was stabilized confirming that it uses the same pathway as the soluble substrates (Fig. 2 E). Furthermore, the lack of a mobility shift in these strains confirmed that it was not subject to static ER retention. These data rule out the hypothesis that the retrieval pathway is specific for soluble substrates, whereas membrane substrates exclusively use the static retention pathway. As a membrane protein misfolded only in its luminal domain, KWW shows that the site of lesion is the major determinant used for sorting substrates.
Sequential checkpoints are used to monitor protein folding
The data suggest the existence of at least two surveillance mechanisms for ER protein folding: one that inspects luminal domains (soluble or membrane proteins) and another that inspects cytosolic domains (membrane proteins). We propose to use the designations ERAD-L (ERAD-Luminal) and ERAD-C (ERAD-Cytosolic) to describe these pathways. Because the two pathways ultimately converge at the proteasomal degradation step, we wondered about their functional relationship. Do they compete for substrates or do they maintain an ordered relationship whereby one pathway precedes the other (the checkpoint model)?
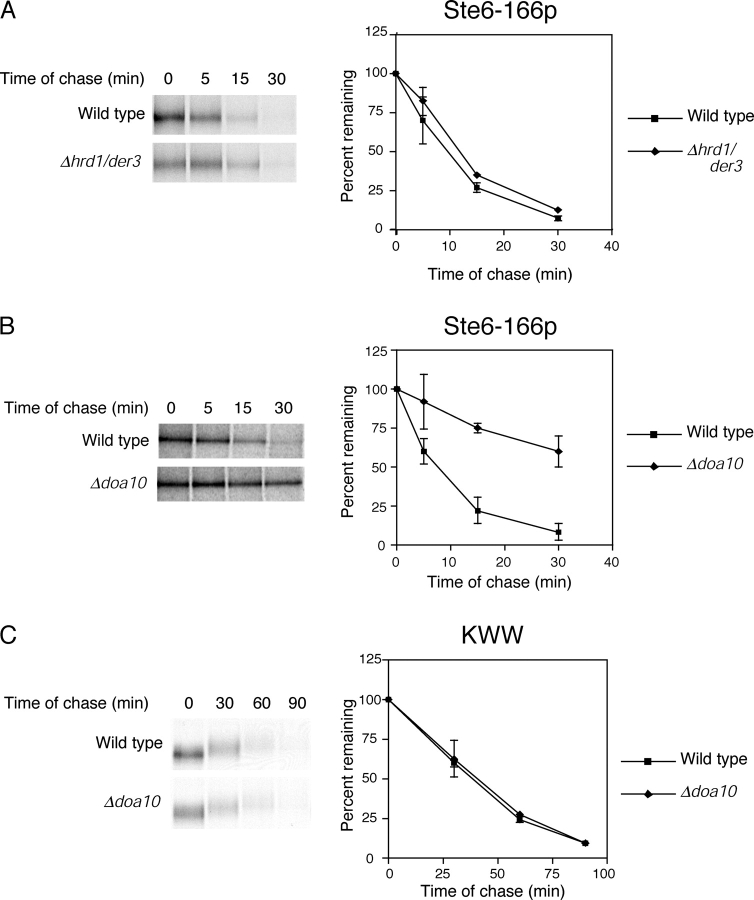
To test this question, we created a substrate containing determinants for both pathways. For this, we fused KHN to Ste6-166p. We selected Ste6-166p because it is retained by ER quality control rather than by an intrinsic signal, which is possible for Sec61-2p. Unlike the other substrates, the E3 ubiquitin ligase for Ste6-166p was not known as it is degraded efficiently in a strain lacking Hrd1p/Der3p (Fig. 3 A). We searched for the E3 enzyme required by Ste6-166p as it might lead to the identification of a factor devoted to the surveillance of cytosolic domains. It was reported previously that the ER-anchored E3 ligase Doa10p is required for the degradation of Ubc6p, an ER membrane protein with its globular domain cytosolically oriented (Swanson et al., 2001). To determine whether it might also play a role in the degradation of misfolded membrane proteins, we analyzed the turnover of Ste6-166p in a Δdoa10 strain. As shown in Fig. 3 B, Ste6-166p was stabilized in Δdoa10 cells. Moreover, a double Δhrd1/der3Δdoa10 mutant exhibited the strongest stabilization suggesting that Hrd1p/Der3p can partly compensate when Doa10p is absent (unpublished data). By contrast, DOA10 is not required for the degradation of ERAD-L substrates CPY* (Swanson et al., 2001), KWW (Fig. 3 C), or KHN (unpublished data).
Figure 3.
Ste6-166p requires the Doa10p E3 ubiquitin ligase for degradation. (A and B) Ste6-166p turnover was measured in wild-type, Δhrd1/der3, and Δdoa10 strains by pulse-chase assay as described in Fig. 2, except that cells were pulse labeled for 5 min followed by shorter times of chase. (C) The rate of KWW degradation was determined in wild-type and Δdoa10 cells as described in Fig. 2.
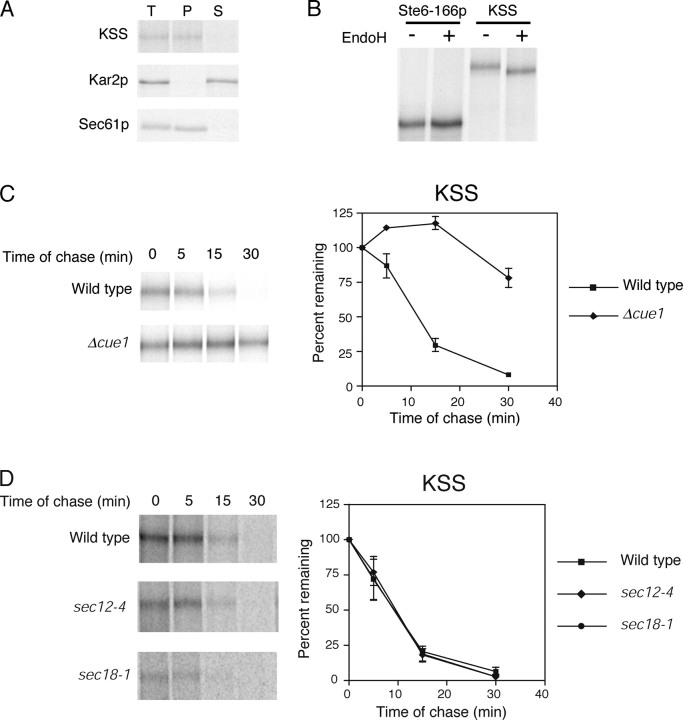
To construct KSS (KHN luminal/Ste6p transmembrane domains/Ste6-166p cytosolic mutant domain), we fused KHN to the second transmembrane domain of Ste6-166p to direct KHN to the luminal side (Fig. 1). Translocation of KSS is directed by the cleaved Kar2p signal sequence located at the amino terminus of the KHN portion (Vashist et al., 2001). Alkali carbonate extraction experiments confirmed that KSS is a bona fide integral membrane protein (Fig. 4 A). To confirm that the KHN portion of KSS is luminal, Endo H analysis was performed as described in Fig. 2 B. Fig. 4 B shows that KSS is glycosylated, whereas the control, Ste6-166p, is nonglycosylated. This experiment shows that the KHN portion is localized in the lumen.
Figure 4.
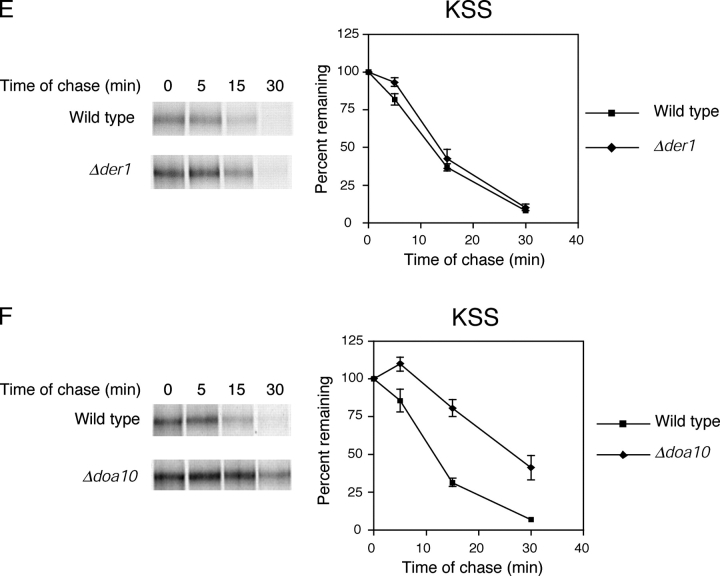
KSS is degraded by the ERAD-C pathway. (A) Carbonate alkali extraction assay was performed on wild-type cells expressing KSS as described in Fig. 2. (B) Cells expressing Ste6-166p or KSS were labeled with [35S]methionine/cysteine for 5 min and substrates immunoprecipitated from detergent lysates. N-linked carbohydrates were removed by Endo H digestion, proteins resolved by SDS-PAGE (7% gel), and visualized by autoradiography. (C) Wild-type and Δcue1 cells expressing KSS were labeled for 5 min with [35S]methionine/cysteine and chased for times indicated. KSS was resolved by electrophoresis using a 7% SDS polyacrylamide gel. (D) ER-to-Golgi transport is not required for degradation of KSS. Wild-type, sec12-4, and sec18-1 strains were grown to log phase at 22°C and then shifted to the restrictive temperature (37°C) for 30 min. (E and F) KSS turnover in Δder1 and Δdoa10 cells was performed as described for C.
Next, we determined the nature of KSS degradation. In wild-type cells, KSS was degraded very rapidly with a t 1/2 of ∼10 min (Fig. 4 C). This rate is similar to the ERAD-C substrates Ste6-166p and Sec61-2p, and much faster than KWW despite KSS being more than twice as large. The degradation is ERAD specific because it was strongly stabilized in the Δcue1 strain (Fig. 4 C) and degraded with wild-type kinetics in a Δpep4 strain (unpublished data). In sec12-4 and sec18-1 mutant strains, the degradation rates were indistinguishable to wild type indicating that KSS uses the same pathway as Ste6-166p (Fig. 4 D). Notably, KSS degradation was independent of DER1 despite containing the KHN domain that, by itself or anchored to a normal transmembrane protein, is DER1 dependent (Fig. 4 E). Like Ste6-166p, KSS degradation was dependent on DOA10 (Fig. 4 F). These data show that KSS is degraded exclusively by the ERAD-C pathway despite displaying determinants for both pathways. We conclude that the ERAD-C pathway precedes the ERAD-L pathway as two sequential checkpoints of ER quality control.
Because the ERAD-L pathway inspects the folding of luminal domains, we wished to determine whether the ERAD-C pathway inspects cytosolic domains of membrane proteins. The sites of the sec61-2 and ste6-166 mutations and our experiments with KSS support this notion. However, it remained possible that the lesions disrupted interactions between the membrane segments, which, in turn, were recognized as aberrant. To determine if the misfolded cytosolic domain of Ste6-166p is sufficient for recognition by the ERAD-C pathway, we replaced the native cytosolic domain of KWW with the cytosolic domain of Ste6-166p containing the mutation. The resulting molecule, designated KHN luminal domain/Wsc1p transmembrane domain/Ste6-166p mutant cytosolic domain (KWS), is schematically depicted in Fig. 1.
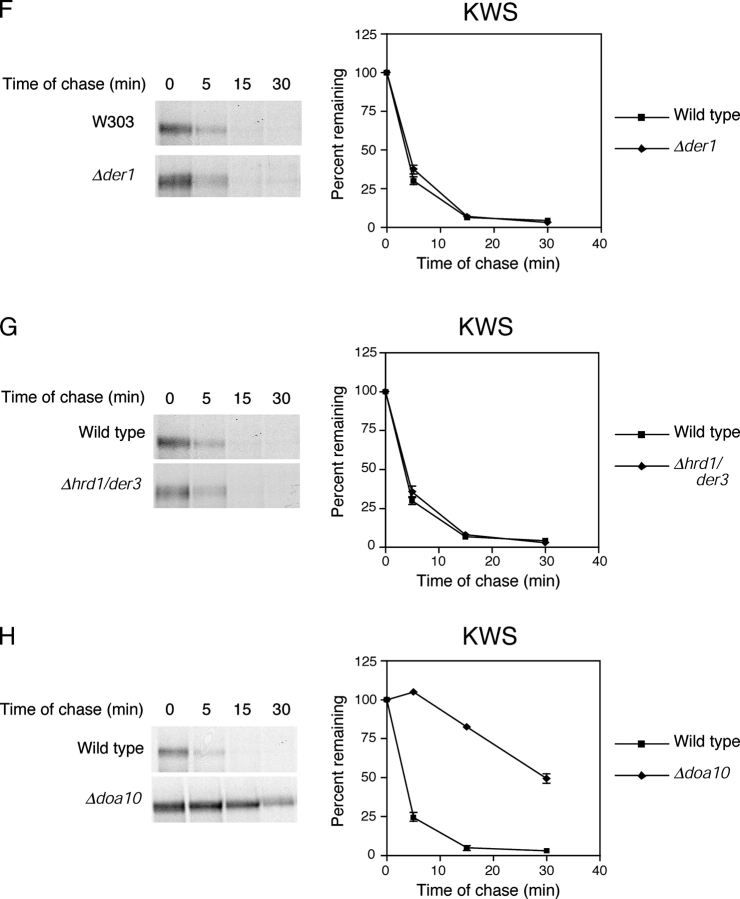
Using the assays used for KWW, we confirmed that KWS is a type I integral membrane protein (Fig. 5 , A–C). When expressed in wild-type cells, KWS was degraded rapidly with a t 1/2 of <5 min and stabilized in the Δcue1 ERAD mutant (Fig. 5 D). Interestingly, KWS is the most rapidly degraded ERAD substrate we have examined so far. This might reflect its smaller size compared with Ste6-166p and KSS, or alternatively, as a single pass membrane protein it might be easier to extract from the membrane. Its rapid kinetics is consistent with degradation by the ERAD-C pathway. This notion was confirmed by the observation that ER-to-Golgi blocks have no effect on their turnover (Fig. 5 E). Furthermore, tests with Δder1, Δhrd1/der3, and Δdoa10 mutant strains show that KWS share the same requirements as Ste6-166p and KSS (Fig. 5, F–H; Table II) . Because KWS has determinants for both pathways, these data show that a misfolded cytosolic domain is sufficient to direct a substrate to the ERAD-C pathway of ER quality control.
Figure 5.
A misfolded cytosolic domain directs entry to the ERAD-C pathway. (A) Carbonate alkali extraction assay of KWS expressed in wild-type cells was performed as described in Fig. 2. T, total; P, pellet; S, supernatant. (B) KWS expressed in wild-type cells was pulse labeled for 5 min, immunoprecipitated, mock treated (−) and digested (+) with Endo H, and resolved by SDS-PAGE. (C) Protease protection assay was performed to determine the orientation of KWS in wild-type cells as described in Fig. 2, except that the proteins were resolved using a 10% polyacrylamide gel. (D–H) KWS stability was measured in wild-type and indicated mutant strains as described in Fig. 4.
Table II. Degradation profiles of substrates used in this work.
| ERAD-C substrates
|
ERAD-L substrates
|
|||||
|---|---|---|---|---|---|---|
| Ste6-166p | KSS | KWS | KHN | KWW | ||
| t 1/2 (min) | 8 | 10 | 4 | 35 | 35 | |
| Requires | CUE1 | yes | yes | yes | yes | yes |
| PEP4 | no | no | no | no | no | |
| SEC12 | no | no | no | yes | yes | |
| SEC18 | no | no | no | yes | yes | |
| DER1 | no | no | no | yes | yes | |
| HTM1/MNL1 | no | no | no | yes | yes | |
| HRD1/DER3 | no | no | no | yes | yes | |
| DOA10 | yes | yes | yes | no | no | |
Htm1p/Mnl1p is required for glycoprotein degradation in the ERAD-L pathway but dispensable for the ERAD-C pathway
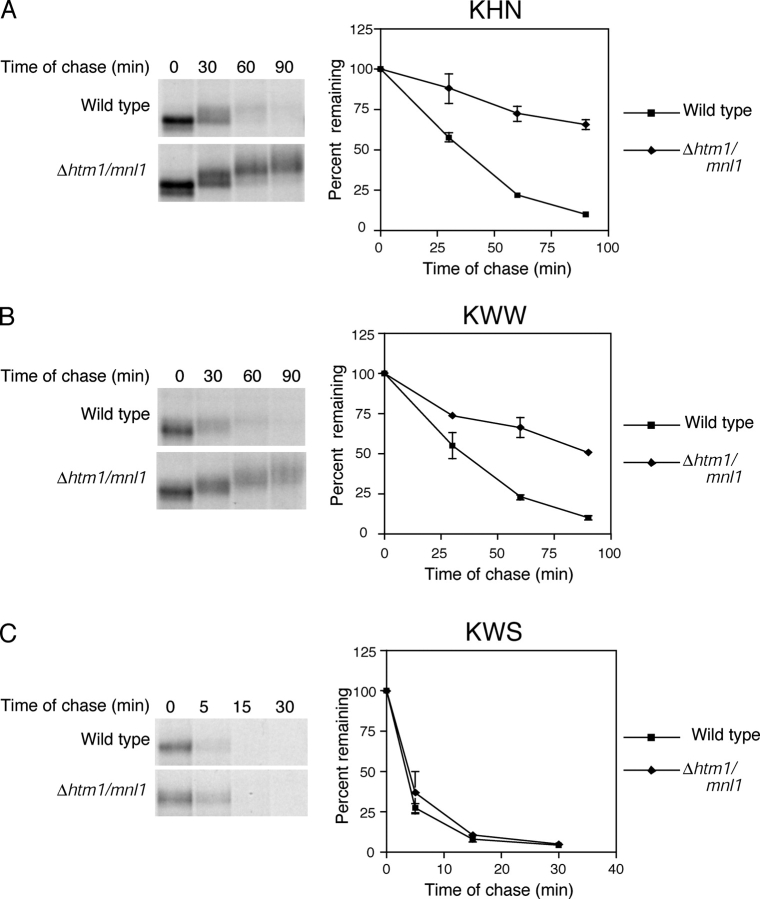
Recently, an ER-localized lectin called Htm1p/Mnl1p (EDEM in mammals) was found to be required for the degradation of misfolded glycoproteins (Jakob et al., 2001; Nakatsukasa et al., 2001; Molinari et al., 2003; Oda et al., 2003). Because the lectin domain of Htm1p/Mnl1p orients to the ER lumen, we wondered whether it functions specifically in the ERAD-L pathway or if it is required for the degradation of all misfolded glycoproteins. For this question, we analyzed a strain deleted of the HTM1/MNL1 gene. As expected, KHN and KWW degradation was defective in Δhtm1/mnl1 cells (Fig. 6, A and B) . This result is in agreement with its proposed role in the processing of both soluble and membrane-integrated glycoproteins for ERAD (Jakob et al., 2001). However, KWS was degraded as efficiently in Δhtm1/mnl1 cells as wild type (Fig. 6 C). This occurred despite KWS containing the misfolded glycoprotein domain that renders KHN and KWW dependent on HTM1/MNL1. These data provide evidence that Htm1p/Mnl1p functions in the ERAD-L pathway. However, it is not required for substrates subject to the ERAD-C pathway even if the substrate contains a large glycosylated luminal domain.
Figure 6.
The Htm1p/Mnl1p ER lectin functions in the ERAD-L pathway. (A–C) Wild-type and Δhtm1/mnl1 mutant strains expressing KHN, KWW, or KWS were analyzed for substrate turnover as described for Fig. 4.
Discussion
It is now well established that cells use multiple mechanisms to monitor the folded states of secretory proteins (Arvan et al., 2002). Remarkably, these mechanisms extend beyond the ER. Nonnative versions of λ repressor, NGF receptor, and the plasma membrane ATPase (Pma1p, specific variants) are not subject to ERAD but transported directly to the vacuole for degradation (Hong et al., 1996; Holkeri and Makarow, 1998; Luo et al., 2002). In most cases, the vacuolar sorting machinery at the Golgi apparatus is required for trafficking these molecules. Therefore, the Golgi apparatus serves as a distal checkpoint capable of sorting misfolded proteins that escaped detection by ER quality control or became damaged after transport (Arvan et al., 2002). Beyond the Golgi apparatus exists yet another checkpoint. Some nonnative proteins that evade both ER and Golgi quality control mechanisms are able to reach the plasma membrane. However, once there, residency is transient as they are rapidly endocytosed and degraded by vacuolar/lysosomal proteases (Ferreira et al., 2002; Fayadat and Kopito, 2003). The mechanism for their recognition is not known. Together, these studies show that quality control is composed of multiple checkpoints throughout the secretory pathway, with each layer contributing added stringency.
Under certain circumstances, ERAD substrates will use the distal Golgi checkpoint. During severe ER stress, when the unfolded protein response is activated, excess ERAD substrates are transported to the vacuole for degradation (Spear and Ng, 2003). Genes constituting the ER-to-vacuole transport pathway including vacuolar proteases are activated by the unfolded protein response suggesting that this alternative route is preprogrammed into the ER stress response (Travers et al., 2000). Indeed, mutants of the BST1 and ERV29 genes that are needed for the vesicular trafficking of misfolded proteins are hypersensitive to ER stress (Spear and Ng, 2003). Together, the studies show that although the Golgi checkpoint is needed for substrates not recognized by ER quality control, it also serves as an alternative degradation pathway when ERAD is saturated.
In this paper, we demonstrate that two previously recognized pathways of ER quality control constitute two sequential checkpoints. From these and other studies, we propose the following model. At the ER, ERAD-C pathway monitors the folding state of cytosolic domains of membrane proteins and rapidly clears detected proteins from the ER. This occurs without regard to the state of the luminal domain. Should the conformation of cytosolic domains pass the ERAD-C checkpoint, the ERAD-L pathway will monitor the state of luminal domains. If a lesion is detected, the protein is processed for ERAD using a distinct set of factors not required for the ERAD-C pathway. These include the vesicular trafficking machinery, Bst1p, BiP and cofactors, Der1p, the P-type ATPases Cod1p and Pmr1p, Mns1p, and Htm1p/Mnl1p (Knop et al., 1996a,b; Durr et al., 1998; Brodsky et al., 1999; Cronin et al., 2000; Jakob et al., 2001; Vashist et al., 2001). Naturally, all soluble proteins bypass the ERAD-C pathway, as they are entirely luminal. Once passing these, and possibly other, ER checkpoints, non-ER proteins are transported to the Golgi apparatus where they face another checkpoints. Once all checkpoints are passed, the protein is finally transported to its site of function.
Asymmetric recognition of substrates requires a complement of factors on both sides of the membrane. However, the degradation machinery itself is located exclusively on the cytosolic face of the ER membrane. Thus, cytosolic determinants have the potential to form direct contacts with extraction and degradation factors, whereas proteins with exclusively luminal determinants must first be targeted to membrane channels for dislocation before they can be degraded. This might account for the difference in degradation kinetics of the two pathways. Despite this mechanistic difference, analogous luminal and cytosolic factors required for ERAD have been identified. The ER chaperone BiP (luminal Hsp70 homologue) is required for CPY* degradation and not cystic fibrosis transmembrane conductance regulator (CFTR). Conversely, cytosolic Hsp70 is required for CFTR and not CPY* (Zhang et al., 2001). Because CPY* is a substrate of the ERAD-L pathway and CFTR is a substrate of the ERAD-C pathway in yeast (Fu and Sztul, 2003), it is tempting to speculate that these chaperones are responsible for recognition and sorting of misfolded proteins for ER quality control. They certainly are qualified to play the role as both are known to preferentially bind unfolded proteins.
Although the proposed model accounts for a range of substrates, there is evidence of additional checkpoints. Our studies explored the role of misfolded luminal and cytosolic domains but did not examine the effects of misfolded or unassembled transmembrane domains, a key determinant for some substrates. For example, unassembled α-subunits of the T cell receptor are retained in the ER and rapidly degraded by ERAD. The determinant (termed “degron”) has been mapped to specific residues in the transmembrane domain that are responsible for recognition by ER quality control when the segment is not partnered with other subunits (Bonifacino et al., 1991). The mechanism for the surveillance of membrane segments is unknown. Thus, the existence of a distinct ERAD-Membrane pathway remains a possibility that awaits further study. For soluble substrates, the nonglycosylated form of pro–α factor, does not require ER-to-Golgi transport for degradation in vitro (McCracken and Brodsky, 1996) or in vivo (unpublished data). It also does not require ubiquitylation but is degraded by the proteasome. It requires the ER lectin calnexin, whereas substrates of the ERAD-L pathway like CPY* do not (McCracken and Brodsky, 1996). Thus, it likely represents a distinct ER checkpoint for soluble proteins. Its faster rate of degradation suggests that it might precede the ERAD-L checkpoint described in our work.
In a recently published work, membrane-bound versions of CPY* were expressed and assessed for their genetic requirements in ERAD (Taxis et al., 2003). The analysis allowed investigators to propose a general ERAD machinery composed of the ubiquitin conjugating enzymes Ubc1p and Ubc7p, ubiquitin ligase complex Hrd1p–Der3p–Hrd3p, the Cdc48p–Ufd1p–Npl4p complex, and the proteasome. Our studies show that additional ubiquitylation machinery is required for other classes of substrates, notably of the ERAD-C pathway. Doa10p is required for the degradation of Ste6-166p, KSS, and KWS, whereas Hrd1p/Der3p is dispensable. Interestingly, the molecules membrane-bound CPY* lacking a cytosolic domain (CT*) and CT* with GFP as its cytosolic domain (CTG*) were degraded without DER1 or KAR2 functions, which are requirements of soluble CPY*. Because these substrates would be predicted to bypass ERAD-C (no cytosolic domain in CT* and folded GFP in CTG*), these data do not conform to our model. However, differences in the experimental designs could account for the apparent discrepancy. The simplest explanation is that the CT* and CTG* proteins were overexpressed from the constitutive TDH3 promoter (Taxis et al., 2003). Previous studies have shown that cell stress caused by increasing the load of CPY* activates degradation pathways that can function independently of HRD and DER genes (Haynes et al., 2002; Spear and Ng, 2003). All the substrates examined in the current work were expressed at levels that do not saturate the basal ERAD machinery. A second difference lies in the design of the chimeric substrates. The single transmembrane segment of CT* and CTG* was acquired from the multi-spanning membrane protein Pdr5p. Thus, it seems plausible that the transmembrane segment of CT* and CTG*, out of its native environment (of likely interactions with other Pdr5p segments), contributes to the degradation that is independent of Der1p and Kar2p. In support of this possibility, it is well established that transmembrane segments with significant hydrophilic content are determinants of ER retention and/or degradation when isolated from their partner sequences (Bonifacino et al., 1991; Letourneur and Cosson, 1998; Sato et al., 2003). Indeed, inspection of the selected Pdr5p transmembrane core sequence revealed a high number of polar residues (4 tyrosine and 2 asparagine). By contrast, the hydrophobic core of the sole Wsc1p transmembrane domain is devoid of hydrophilic residues.
With the ever-increasing number of misfolded protein substrates examined, it is clear that the variety of determinants or degrons is extensive. Correspondingly, it is now appreciated that protein quality control is comprised of multiple mechanisms housed in several organelles and the cytosol. Although the mechanisms remain poorly defined, the list of players is growing rapidly. Placement of key factors into defined pathways with their client substrates is an important step toward the goal of fully understanding how “bad” proteins are sorted from “good” proteins for destruction.
Materials and methods
Plasmids used in this work
Plasmids were constructed using standard cloning protocols. HA epitope-tagged Ste6-166p was expressed from pSM1083 and was a gift from S. Michaelis (Johns Hopkins University, Baltimore, MD; Loayza et al., 1998). All ERAD substrates used in this work contain an engineered HA epitope tag. KHN (pSM70) is the same as KHNt described previously (Vashist et al., 2001). The three-letter designation is used for simplicity and to avoid confusion with the designations of other substrates.
pSM101 (KWW): The gene encoding KWW was constructed by ligating the coding sequences of KHN (pSM70; Vashist et al., 2001) to sequences encoding transmembrane and cytosolic domains of WSC1. A 1.6-kb fragment containing KHN was amplified using Vent DNA polymerase and primers N193 (5′-GGGATCGATAGTTTAATAACCCAAAAGCA-3′) and N253 (5′-GCACTGAGCAGCGTAATCTGG-3′). A 500-bp fragment was released from the 3′ end after digestion with BstEII and purified. A second fragment was prepared by amplifying a 697-bp fragment of WSC1 using the primers N254 (5′-AAGAAAGCCAATGTAGGGGCA-3′) and N256 (5′-GCGTCTAGATATTAGGTGGTATTCATTTCA-3′) and digested with XbaI. Both fragments were ligated into pSM70 digested with BstEII and XbaI to generate pSM101. A triple HA epitope tag is located at the junction of the gene fusion from fragment 1.
pSM114 (KSS): KSS is a fusion between KHN and Ste6-166p. This chimera was generated in three steps. First, a fusion was created between KHN and the sequence encoding amino acids 69–403 of Ste6-166p using the PCR-based approach described above. A 1.6-kb fragment was amplified using N193 and N253 primers and digested with ClaI. A second 1.0-kb fragment was amplified from STE6 using N294 (5′-TCCCAACTAGTACAGAGG-3′) and N295 (5′-GTTGGATAATGTAGATTTACC-3′) and digested with XbaI. Both fragments were inserted into pDN121 (a variant of pRS314) digested with ClaI and XbaI to form the KHN/Ste6p junction. The remaining 3′ coding sequences of Ste6-166p was inserted into the intermediate construct to generate pSM108. A GAS1 promoter was inserted upstream of the coding sequences to generate pSM114.
pSM118 (KWS): A 0.9-kb fragment was amplified from pSM101 using primers N360 (5′-CATACTATAGCAGCTGGTTTGGC-3′) and N390 (5′-TCTGACAATCAACAAGATACAAAGAGC-3′), digested with BstEII and the 0.6-kb fragment purified. A fragment encoding the mutant cytosolic domain of Ste6-166p was amplified from pSM1083 using primers N392 (5′-ATACCCGATATAAGTAGAGGCCAACGT-3′) and N410 (CGCTCTAGATTATTCACTATGCGTTATAACC) and digested with XbaI. These fragments were simultaneously inserted into pSM101 digested with BstEII and XbaI.
Strains and antibodies
Strains used in this work are listed in Table III . Anti-HA mAb was purchased from BabCo. Polyclonal antisera specific for Kar2p and Sec61p were provided by P. Walter (University of California, San Francisco, San Francisco, CA).
Table III. Strains used in this work.
| Strain | Genotype | Source |
|---|---|---|
| W303 | Mat a , leu2-3,112, his3-11, trp1-1, can1-100, ade2-1 | P. Waltera |
| SMY489 | Mat a, W303, pSM101 | This paper |
| SMY490 | Mat a, cue1::TRP1, pSM101, W303 background | This paper |
| SMY491 | Mat a, der1::KANMX, pSM101, W303 background | This paper |
| SMY492 | Mat a, hrd1/der3::KANMX, pSM101, W303 background | This paper |
| SMY628 | Mat a, doa10::KANMX, pSM101, W303 background | This paper |
| SMY496 | Mat a, sec12-4, pSM101, W303 background | This paper |
| SMY497 | Mat a, sec18-1, pSM101, W303 background | This paper |
| SMY371 | Mat a, W303, pSM1083 | This paper |
| SMY613 | Mat a, hrd1/der3::KANMX, pSM1083, W303 background | This paper |
| SMY601 | Mat a, doa10::KANMX, pSM1083, W303 background | This paper |
| SMY610 | Mat a, W303, pSM114 | This paper |
| SMY614 | Mat a, cue1::TRP1, pSM114, W303 background | This paper |
| SMY611 | Mat a, der1::KANMX, pSM114, W303 background | This paper |
| SMY615 | Mat a, hrd1/der3::KANMX, pSM114, W303 background | This paper |
| SMY612 | Mat a, doa10::KANMX, pSM114, W303 background | This paper |
| SMY626 | Mat a, sec12-4, pSM114, W303 background | This paper |
| SMY627 | Mat a, sec18-1, pSM114, W303 background | This paper |
| SMY639 | Mat a, W303, pSM118 | This paper |
| SMY641 | Mat a, cue1::TRP1, pSM118, W303 background | This paper |
| SMY656 | Mat a, der1::KANMX, pSM118, W303 background | This paper |
| SMY661 | Mat a, hrd1/der3::KANMX, pSM118, W303 background | This paper |
| SMY652 | Mat a, doa10::KANMX, pSM118, W303 background | This paper |
| SMY645 | Mat a, sec12-4, pSM118, W303 background | This paper |
| SMY647 | Mat a, sec18-1, pSM118, W303 background | This paper |
| SMY500 | Mat a, pep4::HIS3, pSM101, W303 background | This paper |
| WKY753 | Mat a, htm1/mnl1::KANMX4, pSM70, W303 background | This paper |
| WKY754 | Mat a, htm1/mnl1::KANMX4, pSM101, W303 background | This paper |
| WKY755 | Mat a, htm1/mnl1::KANMX4, pSM118, W303 background | This paper |
University of California, San Francisco, San Francisco, CA.
Cell labeling and immunoprecipitation
Typically, 3 OD600 U of log phase cells were pelleted and resuspended in synthetic media lacking uracil, methionine, and cysteine. After 30 min of incubation at the appropriate temperature, 480 μCi of Trans 35S-label was added to initiate a pulse. Cells were labeled for 5 or 10 min as described in the figure legends. Cold methionine and cysteine were added (to a final concentration of 2 mM each) 30 s before end of the pulse to initiate the chase. Termination of the chase was performed by the addition of TCA (final concentration of 10%). TCA precipitates were resuspended and boiled in 100 mM Tris, pH 11.0, and 3% SDS for 5 min. Immunoprecipitations were performed by incubating in immunoprecipitation solution (50 mM Tris, pH 7.5, 1% Triton X-100, 150 mM NaCl) and appropriate antibody for 1 h at 4°C. The samples were centrifuged at 20,000 g for 20 min and transferred to fresh tubes containing protein A–Sepharose beads. The samples were incubated with beads on a rocking platform for 2 h at 4°C. After incubation, the samples were washed twice with high salt immunoprecipitation solution (50 mM Tris, pH 7.5, 1% Triton X-100, 300 mM NaCl), twice with low salt solution (50 mM Tris, pH 7.5, 1% Triton X-100, 150 mM NaCl) and twice with PBS. Proteins were separated by electrophoresis using SDS-polyacrylamide gels of varying composition as indicated in the figure legends and visualized by autoradiography. Quantification was performed from PhosphorImager scans of polyacrylamide gels using Imagequant software (Molecular Dynamics).
Protein deglycosylation
Immunoprecipitated proteins on washed protein A–Sepharose beads were denatured in SDS and digested with Endo H (New England Biolabs, Inc.) according to the manufacturer's protocol. Control samples were processed identically except that the enzyme was omitted during incubation.
Alkaline carbonate extraction
10 OD600 U of cells expressing KWW, KSS, or KWS were labeled using Trans 35S-label for times indicated in the legend to Fig. 2. The pulse was terminated by addition of 20 mM sodium azide. Cells were pelleted by 20,000 g centrifugation, washed once with ice-cold water, and resuspended in buffer (10 mM sodium phosphate, pH 7.0, 1 mM PMSF, and 1× yeast protease inhibitor cocktail; Sigma-Aldrich). Cell disruption was performed by vortexing at maximum setting with zirconium beads at 4°C. Unbroken cells and debris were separated from membranes by two successive low speed centrifugation steps at 500 g for 5 min each. The supernatant fraction was treated with sodium carbonate, pH 11.0, for 30 min on ice. One third was removed, precipitated in 10% TCA on ice for 30 min, and saved. The remaining was fractionated by centrifugation at 100,000 g for 30 min at 4°C. The pellet fraction was washed once in water, resuspended in 3% SDS, 100 mM Tris, pH 7.5, 3 mM DTT, and heated for 5 min at 95°C. The supernatant fraction was incubated in 10% TCA and precipitates were collected by centrifugation. TCA precipitated proteins were resuspended in 100 mM Tris, pH 11.0, 3% SDS, 3 mM DTT, and heated to 95° C for 5 min. Detergent lysates were adjusted to reflect equivalent cell numbers, immunoprecipitated, and analyzed by SDS-PAGE.
Protease protection assay
To determine membrane orientation of substrate proteins, 12 OD600 U of cells were grown to log phase and metabolically labeled with Trans 35S-label for 10 min. Labeling was terminated with the addition of sodium azide to a final concentration of 20 mM. Cells were harvested by low speed centrifugation and washed once in ice-cold water. Cells were then resuspended in 25 mM sodium phosphate, pH 7.0, and disrupted by vortexing at full speed in the presence of zirconium beads. Cell debris was separated using two successive centrifugation steps, each at 500 g for 5 min, and discarded. The remaining fraction containing membranes and cytosol was split into three aliquots. To two aliquots, proteinase K (5 μg/ml final concentration; IBI) was added, with or without Triton X-100 (1% final concentration). The third was mock treated. All samples were incubated on ice for 10 min. To terminate the reaction, PMSF was added to final concentration of 10 mM. Proteins were precipitated by the addition of TCA (10% final concentration). Detergent lysates were prepared and immunoprecipitations were performed as described above.
Acknowledgments
We thank the members of the Ng Lab for discussion and comments on this work. We thank Susan Michaelis and Peter Walter for gifts of plasmids and antibodies. We thank Woong Kim for constructing the Δhtm1/mnl1 strains. We thank Chris Graham for his contribution to this work during his laboratory rotation.
This work was supported by a grant from the National Institutes of Health to D.T.W. Ng (GM059171).
Abbreviations used in this paper: CFTR, cystic fibrosis transmembrane conductance regulator; CPY*, mutant carboxypeptidase Y; CT*, membrane-bound CPY* lacking a cytosolic domain; CTG*, CT* with GFP as its cytosolic domain; Endo H, endoglycosidase H; ERAD, ER-associated degradation; ERAD-C, ERAD-Cytosolic; ERAD-L, ERAD-Luminal; GT, glucosyltransferase; KHN, yeast Kar2p signal sequence fused to the simian virus 5 HA-Neuraminidase ectodomain; KWS, KHN luminal domain/Wsc1p transmembrane domain/Ste6-166p mutant cytosolic domain; KWW, KHN luminal domain/Wsc1p transmembrane domain/Wsc1p cytosolic domain.
References
- Arvan, P., X. Zhao, J. Ramos-Castaneda, and A. Chang. 2002. Secretory pathway quality control operating in Golgi, plasmalemmal, and endosomal systems. Traffic. 3:771–780. [DOI] [PubMed] [Google Scholar]
- Barlowe, C., and R. Schekman. 1993. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature. 365:347–349. [DOI] [PubMed] [Google Scholar]
- Bays, N.W., R.G. Gardner, L.P. Seelig, C.A. Joazeiro, and R.Y. Hampton. 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER- associated degradation. Nat. Cell Biol. 3:24–29. [DOI] [PubMed] [Google Scholar]
- Belden, W.J., and C. Barlowe. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 294:1528–1531. [DOI] [PubMed] [Google Scholar]
- Bonifacino, J.S., P. Cosson, N. Shah, and R.D. Klausner. 1991. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 10:2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo, J., R.K. Plemper, A. Finger, and D.H. Wolf. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell. 9:209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and A.A. McCracken. 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10:507–513. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., E.D. Werner, M.E. Dubas, J.L. Goeckeler, K.B. Kruse, and A.A. McCracken. 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274:3453–3460. [DOI] [PubMed] [Google Scholar]
- Caldwell, S.R., K.J. Hill, and A.A. Cooper. 2001. Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 276:23296–23303. [DOI] [PubMed] [Google Scholar]
- Cronin, S.R., A. Khoury, D.K. Ferry, and R.Y. Hampton. 2000. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J. Cell Biol. 148:915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr, G., J. Strayle, R. Plemper, S. Elbs, S.K. Klee, P. Catty, D.H. Wolf, and H.K. Rudolph. 1998. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol. Biol. Cell. 9:1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakle, K.A., M. Bernstein, and S.D. Emr. 1988. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol. Cell. Biol. 8:4098–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191. [DOI] [PubMed] [Google Scholar]
- Fayadat, L., and R.R. Kopito. 2003. Recognition of a single transmembrane degron by sequential quality control checkpoints. Mol. Biol. Cell. 14:1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, T., A.B. Mason, M. Pypaert, K.E. Allen, and C.W. Slayman. 2002. Quality control in the yeast secretory pathway: a misfolded PMA1 H+-ATPase reveals two checkpoints. J. Biol. Chem. 277:21027–21040. [DOI] [PubMed] [Google Scholar]
- Finger, A., M. Knop, and D.H. Wolf. 1993. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 218:565–574. [DOI] [PubMed] [Google Scholar]
- Fu, L., and E. Sztul. 2003. Traffic-independent function of the Sar1p/COPII machinery in proteasomal sorting of the cystic fibrosis transmembrane conductance regulator. J. Cell Biol. 160:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillece, P., J.M. Luz, W.J. Lennarz, F.J. de La Cruz, and K. Romisch. 1999. Export of a cysteine-free misfolded secretory protein from the endoplasmic reticulum for degradation requires interaction with protein disulfide isomerase. J. Cell Biol. 147:1443–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, C., and A. Helenius. 1994. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J. Cell Biol. 126:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton, R. 2002. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14:476–482. [DOI] [PubMed] [Google Scholar]
- Hampton, R.Y., R.G. Gardner, and J. Rine. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 7:2029–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, E., T.A. Rapoport, and H.F. Lodish. 1989. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA. 86:5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, C.M., S. Caldwell, and A.A. Cooper. 2002. An HRD/DER-independent ER quality control mechanism involves Rsp5p- dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert, D.N., B. Foellmer, and A. Helenius. 1995. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 81:425–433. [DOI] [PubMed] [Google Scholar]
- Hiller, M.M., A. Finger, M. Schweiger, and D.H. Wolf. 1996. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 273:1725–1728. [DOI] [PubMed] [Google Scholar]
- Hirsch, C., D. Blom, and H.L. Ploegh. 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22:1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holkeri, H., and M. Makarow. 1998. Different degradation pathways for heterologous glycoproteins in yeast. FEBS Lett. 429:162–166. [DOI] [PubMed] [Google Scholar]
- Hong, E., A.R. Davidson, and C.A. Kaiser. 1996. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 135:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob, C.A., D. Bodmer, U. Spirig, P. Battig, A. Marcil, D. Dignard, J.J. Bergeron, D.Y. Thomas, and M. Aebi. 2001. Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosch, E., C. Taxis, C. Volkwein, J. Bordallo, D. Finley, D.H. Wolf, and T. Sommer. 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4:134–139. [DOI] [PubMed] [Google Scholar]
- Johnson, A.E., and M.A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799–842. [DOI] [PubMed] [Google Scholar]
- Knop, M., A. Finger, T. Braun, K. Hellmuth, and D.H. Wolf. 1996. a. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15:753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop, M., N. Hauser, and D.H. Wolf. 1996. b. N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast. 12:1229–1238. [DOI] [PubMed] [Google Scholar]
- Kostova, Z., and D.H. Wolf. 2003. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 22:2309–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letourneur, F., and P. Cosson. 1998. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J. Biol. Chem. 273:33273–33278. [DOI] [PubMed] [Google Scholar]
- Loayza, D., A. Tam, W.K. Schmidt, and S. Michaelis. 1998. Ste6p mutants defective in exit from the endoplasmic reticulum (ER) reveal aspects of an ER quality control pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 9:2767–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder, A.L., T.K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics. 152:1487–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W.J., X.H. Gong, and A. Chang. 2002. An ER membrane protein, Sop4, facilitates ER export of the yeast plasma membrane [H+]ATPase, Pma1. Traffic. 3:730–739. [DOI] [PubMed] [Google Scholar]
- McCracken, A.A., and J.L. Brodsky. 1996. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari, M., V. Calanca, C. Galli, P. Lucca, and P. Paganetti. 2003. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 299:1397–1400. [DOI] [PubMed] [Google Scholar]
- Nakano, A., D. Brada, and R. Schekman. 1988. A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa, K., S. Nishikawa, N. Hosokawa, K. Nagata, and T. Endo. 2001. Mnl1p, an alpha -mannosidase-like protein in yeast Saccharomyces cerevisiae, is required for endoplasmic reticulum-associated degradation of glycoproteins. J. Biol. Chem. 276:8635–8638. [DOI] [PubMed] [Google Scholar]
- Nishikawa, S.I., S.W. Fewell, Y. Kato, J.L. Brodsky, and T. Endo. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153:1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, Y., N. Hosokawa, I. Wada, and K. Nagata. 2003. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 299:1394–1397. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., S. Bohmler, J. Bordallo, T. Sommer, and D.H. Wolf. 1997. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 388:891–895. [DOI] [PubMed] [Google Scholar]
- Rabinovich, E., A. Kerem, K.U. Frohlich, N. Diamant, and S. Bar-Nun. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, K., M. Sato, and A. Nakano. 2003. Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell. 14:3605–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, T., and S. Jentsch. 1993. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 365:176–179. [DOI] [PubMed] [Google Scholar]
- Spear, E.D., and D.T. Ng. 2003. Stress tolerance of misfolded carboxypeptidase y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell. 14:2756–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., H. Park, N.M. Hollingsworth, R. Sternglanz, and W.J. Lennarz. 2000. PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J. Cell Biol. 149:1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis, C., R. Hitt, S.H. Park, P.M. Deak, Z. Kostova, and D.H. Wolf. 2003. Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J. Biol. Chem. 278:35903–35913. [DOI] [PubMed] [Google Scholar]
- Travers, K.J., C.K. Patil, L. Wodicka, D.J. Lockhart, J.S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. [DOI] [PubMed] [Google Scholar]
- Trombetta, E.S., and A. Helenius. 1999. Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum. EMBO J. 18:3282–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, B., C. Rodighiero, W.I. Lencer, and T.A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell. 104:937–948. [DOI] [PubMed] [Google Scholar]
- Vashist, S., W. Kim, W.J. Belden, E.D. Spear, C. Barlowe, and D.T. Ng. 2001. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 155:355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, C.L., S. Omura, and R.R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 83:121–127. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., R. Fujii, Y. Toyofuku, T. Saito, H. Koseki, V.W. Hsu, and T. Aoe. 2001. The KDEL receptor mediates a retrieval mechanism that contributes to quality control at the endoplasmic reticulum. EMBO J. 20:3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Y., H.H. Meyer, and T.A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., G. Nijbroek, M.L. Sullivan, A.A. McCracken, S.C. Watkins, S. Michaelis, and J.L. Brodsky. 2001. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 12:1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]