Abstract
We recently have identified an antigen receptor in sharks called NAR (new or nurse shark antigen receptor) that is secreted by splenocytes but does not associate with Ig light (L) chains. The NAR variable (V) region undergoes high levels of somatic mutation and is equally divergent from both Ig and T cell receptors (TCR). Here we show by electron microscopy that NAR V regions, unlike those of conventional Ig and TCR, do not form dimers but rather are independent, flexible domains. This unusual feature is analogous to bona fide camelid IgG in which modifications of Ig heavy chain V (VH) sequences prevent dimer formation with L chains. NAR also displays a uniquely flexible constant (C) region. Sequence analysis and modeling show that there are only two types of expressed NAR genes, each having different combinations of noncanonical cysteine (Cys) residues in the V domains that likely form disulfide bonds to stabilize the single antigen-recognition unit. In one NAR class, rearrangement events result in mature genes encoding an even number of Cys (two or four) in complementarity-determining region 3 (CDR3), which is analogous to Cys codon expression in an unusual human diversity (D) segment family. The NAR CDR3 Cys generally are encoded by preferred reading frames of rearranging D segments, providing a clear design for use of preferred reading frame in antigen receptor D regions. These unusual characteristics shared by NAR and unconventional mammalian Ig are most likely the result of convergent evolution at the molecular level.
At the heart of the adaptive immune system are the antigen receptors, Ig and T cell receptor (TCR), that are generated in anticipation of recognition of pathogens (1). The typical antigen receptor is composed of two polypeptide chains [heavy (H) and light (L) for Igs and α and β or γ and δ for TCRs]. Each chain, in turn, is composed of a single, variable (V) domain at the N-terminal end followed by one to seven constant (C) domains. C domains define the effector functions characteristic of a given class of Ig whereas V domains each display a unique sequence and structure defining antigen specificity. Igs can be subdivided further into Fab and Fc fragments, responsible for antigen binding and for effector function, respectively. Ig and TCR V regions are encoded by a mosaic of genes ligated together somatically during lymphocyte ontogeny (2). Specifically, single V and J elements are joined together at the DNA level for Ig L chain or TCR α and γ V regions. In Ig H chains and TCR β and δ chains, one or, occasionally, two D elements are joined between the V and J segments. Together, the V, (D), and J elements encode framework (FR, responsible for protein folding and structure) and complementarity-determining regions (CDR, responsible for antigen interactions) within the V domains.
The evolutionary origin of antigen receptors is unknown, but the first indication of their emergence phylogenetically is in cartilaginous fish (sharks, skates, and rays), where at least three types of Ig (3–9) and four TCR isotypes (10, 11) are found. Recently, we identified an antigen receptor in sharks, called the new or nurse shark antigen receptor (NAR) that, while having both transmembrane and secreted forms like Ig, is no more related in its V region sequence to Ig than to TCR and thus may be an evolutionary intermediate (3, 4).
The NAR protein has been shown to be a dimer with each chain composed of one V and five C domains (ref. 3; see Fig. 1G). No L chains or any other proteins can be demonstrated to associate with this dimer (3). The NAR V region conforms to the model of prototypic Ig superfamily domains with the predicted canonical disulfide bond connecting two β sheets and several other invariant or conserved residues involved in structural packing (3, 12, 13); nevertheless, NAR V is unique in that it has an exceptionally small CDR2 and poor conservation of those residues responsible for VH/VL and V α/β dimerization in typical Igs and TCRs, respectively (ref. 3; see Figs. 2 and 4). In addition, comparison of cDNA sequences reveals that noncanonical cysteine (Cys) residues are always found in NAR V regions. We hypothesized, therefore, that NAR V regions would be expressed as discrete structures not forming dimers in the standard Ig/TCR fashion (3). In camelids (camels and llamas) this is indeed the case as two of their three IgG subclasses contain no L chains and the unassociated VH domains interact with antigen as monomers (14, 15). We examined NAR structure by performing an electron microscopic (EM) analysis of NAR proteins and by modeling of the NAR V domain onto previously reported IgV x-ray diffraction structures. The results are discussed in an evolutionary context through comparison with Ig and TCR structure and function.
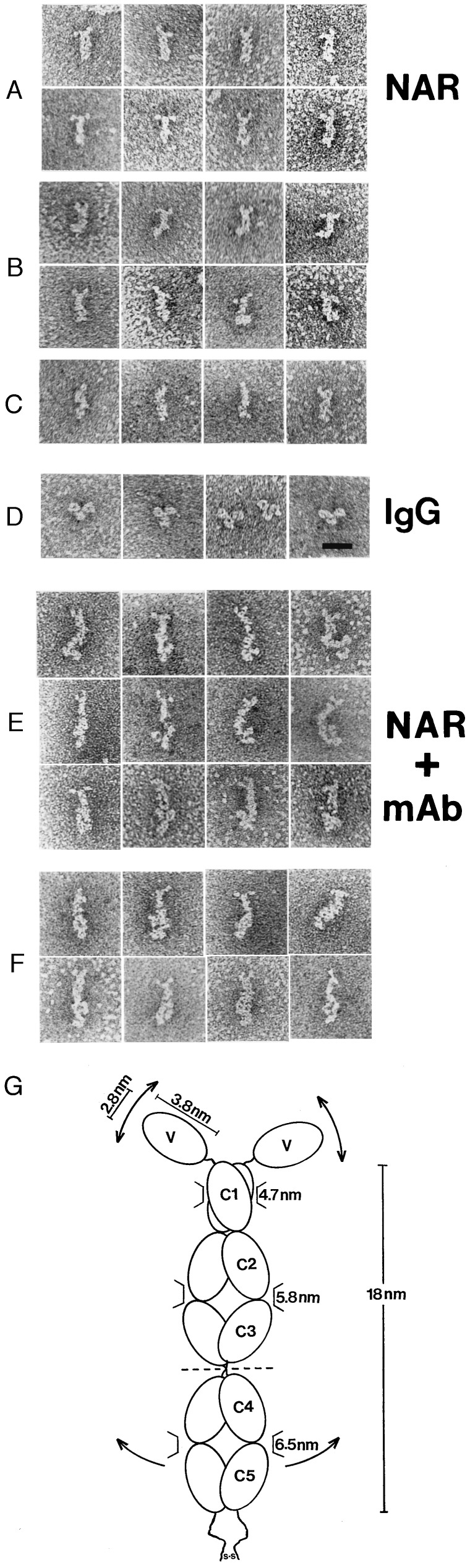
Figure 1.
EM views of NAR show it to be a dimer with single, bivalent V domains. Small, flexible knobs are present at one end of NAR (A, B, E, and F), and a torsion of the last quarter of the molecule is seen at the other end (B). The small knobs are approximately one-quarter the size of the control human IgG Fab regions (D), i.e., the size of a single Ig domain. An mAb specific for the NAR C terminus orients NAR and indicates that the small knobs are the V regions (E and F). mAb were bound either by one Fab arm (E) or both Fab arms (F). Complexes are oriented so that the mAbs are at the bottom of the images. (Bar = 20 nm.) (G) Model of NAR showing dimensions and hypothetical domain orientations and flexibility patterns.
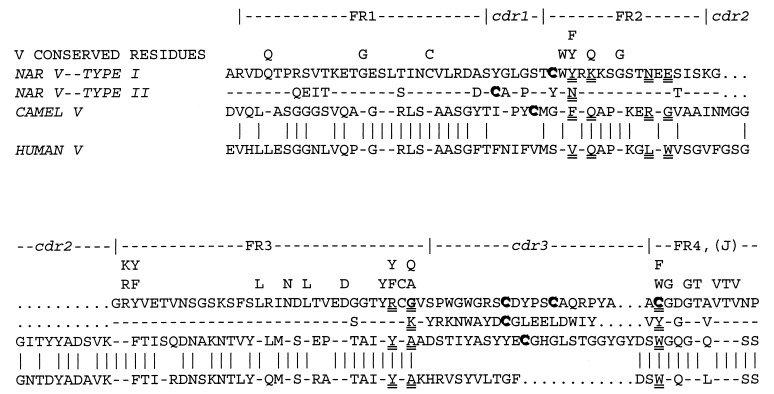
Figure 2.
NAR Type I and Type II protein V region sequences compared with each other and with human and camel V. The NAR Type I sequence is of a nonmutated (germ-line-encoded) cDNA (3). The Type II sequence is from a somatically mutated cDNA selected as a typical clone from ref. 3 (no nonmutated Type II cDNA have been observed, but the features described are common to almost every clone). Residues generally considered to be the most evolutionarily conserved among Ig and TCR V are indicated, as are the FR/CDR (13). Dash symbols indicate identity of the bottom three sequences to the NAR V Type I. Vertical lines denote identity between the camel and human sequences (same alignment as in ref. 15). Note the following: (i) presence of evolutionarily conserved or invariant residues in NAR FR regions; (ii) poor conservation of residues that interact between Ig H/L chains (double-underlined in NAR, camel, and human; see ref. 13); (iii) the very short CDR2 of NAR (shown on model in Fig. 4c); (iv) high similarity of the two NAR classes to one another on one hand and of the human to the camel on the other; (v) low identity between NAR and the mammalian sequences; (vi) the unusual Cys residues (bold and shadowed) found in FR2 (C-35), FR4 (C-107), and CDR3 of Type I NAR, and in CDR1 and CDR3 of Type II NAR and the camel sequence; and (vii) the unusual glycine residue found at the end of NAR Type I FR3 (G-84, bold and double-underlined; see Figs. 3c and 4e).
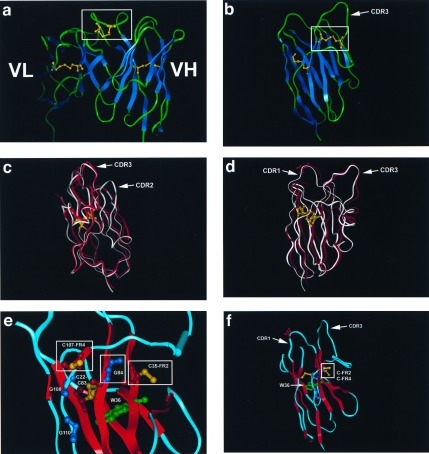
Figure 4.
Modeling of NAR V and comparison with camel and human VH structures. (a) The VH/VL of the KOL human crystal (18, 23) showing the disulfide bridge in the CDR3 VH (boxed). Note that the VL (left) binding site is obscured by the VH CDR3. Canonical intradomain disulfide bonds between the two β sheets of the H and L chains are displayed in yellow (not boxed). The boxed disulfide bridge (also yellow) is postulated to be similar to bonds that would form in NAR Type I CDR3. (b) Crystal structure of the unusual camel VH (15). The disulfide bridge between CDR1 and 3 is boxed, and the canonical intradomain disulfide bridge is displayed. The disulfide bond displayed here is postulated to be similar to those formed by CDR1 and 3 Cys residues in NAR Type II proteins. (c and d) Model of the Type I NAR V domain (red) superimposed on the camel VH (white). Note the high similarity of the two structures except in the CDR2 region, which in NAR is very small and may connect the two β sheets in a fashion similar to Ig C1 domains (33). The canonical disulfide bridge (yellow) and serine (red) in NAR CDR1 (S-28, Fig. 2) are displayed for bearings. (e and f) Potential disulfide bond formed in NAR between unconventional Cys (boxed) in FR2 (C-35) and FR 4 (C-107); see Figs. 2 and 3 a and c. The side chains of these two Cys project into the region in which conventional VH and VL interact. We propose that the small size of the highly conserved glycine at position 84 (boxed G-84, Figs. 2 and 3c) permits C-35 and C-107 to disulfide-bond; it is important to emphasize that this glycine is conserved only in Type I NAR sequences and is rarely found in Ig/TCR of other vertebrates or even in Type II NAR (3). We speculate that NAR is more compact, and, because of irregular H bonding in the G strand (FR4) of the model, it is possible that a disulfide bond can be formed between C-35 and C-107. The canonical disulfide bond (C-22, C-83) and the invariant tryptophan (W-36) forming the core of the NAR domain, as well as the invariant Gly-Xaa-Gly (ref. 18; G-108, G-110), are displayed.
MATERIALS AND METHODS
Immunoelectron Microscopy.
Immunoelectron microscopic analyses of NAR and NAR-mAb complexes were performed by negative staining using previously described procedures (16). Briefly, NAR at 1 μg/ml or NAR-mAb complexes preincubated for 20 min at room temperature at 1 μg/ml in borate-buffered saline were affixed to thin carbon membranes, stained with uranyl formate, and mounted on copper grids for analysis. Electron micrographs were recorded at ×100,000 magnification on a JEOL CX 1200 electron microscope and printed at ×258,000 magnification for analysis. Fields in which >90% of the molecules were scorable were chosen for analysis. Measurements were taken with the aid of an optical loupe fitted with a measuring graticule (Electron Microscopy Sciences, Fort Washington, PA).
NAR was purified by affinity chromatography using a mouse mAb specific for NAR (3) covalently coupled to protein-G Sepharose beads, and brought to homogeneity by HPLC over SEC 300SW (Beckman). A peptide encompassing the NAR C-terminal tail GKPSSVNVSVVLSDTVKSST (3) was prepared as a “multiantigenic peptide” (an antigen in which eight peptides are linked together on a branching lysine matrix; ref. 17), and mice were immunized as described (9) for mAb production. Positive mAb clones were tested by ELISA against the peptide and then were screened by immunoprecipitation of radiolabeled NAR protein. Protein G-purified mAbs were used in the experiment shown in Fig. 1 E and F.
Modeling.
The camel VH sequence was aligned with the type I NAR sequence (3) in look 2 (Molecular Application Group, Palo Alto, CA) and modified by hand based on conserved or invariant residues found in the framework of all antigen receptor V domains (13). An NAR three-dimensional structure was generated with look’s SegMod using a homology-based approach. Minimization and refinement of the model was a fully automated feature of this program. A second model was created by using insight ii and homology 95 (Biosym/Molecular Simulation, San Diego). Both NAR models generated by look were visualized using insight ii and for the creation of the structure figures. The protein database code for the camel VH domain (15) is 1 mel, and the human Ig from myeloma patient KOL (18) serum is 2FB4. Only the KOL VL and VH and camel VH were used.
RESULTS AND DISCUSSION
NAR Structure Revealed by EM.
EM examination of NAR (Fig. 1) reveals molecules that are rod-shaped, approximately 18 nm in length, and composed of several bead-like segments. Protruding from one end of most (73%) molecules are two ovoid, knob-like structures (3.8 × 2.8 nm), each of which is attached to the main body of the molecule by a short filamentous segment at sites slightly lateral to the long axis of the main body (Fig. 1 A and B). Some molecules (19%) display one or, in a few cases (8%, Fig. 1C), no knob-like structures. The orientation of the protruding structures varies, indicating a flexible connection to the rest of the molecule. The missing ovoid structures presumably are folded back onto the main body of the molecule or perhaps superimposed on the visible arm. Another distinctive feature is a pronounced kink (30–90o) in the main body of many (44%) of the molecules, located approximately 2/3 of the distance from the end with the knob-like structures (Fig. 1B). The length of the off-axis hook-portion of the molecule is 7.7 nm. Measurements of the diameters of the upper (toward knobs), middle, and lower regions of the main body are 4.7, 5.8, and 6.5 nm, respectively (Fig. 1G). For comparative purposes, the Fab domains of IgG (Fig. 1D), prepared under identical conditions, were found to be 7.3 × 5.5 nm.
An mAb specific for the NAR C terminus reacts with the end of the molecule opposite the knob-like structures (Fig. 1 E and F). Complexes showing only one mAb Fab arm (50%, Fig. 1E) or two arms binding to NAR (50%, Fig. 1F) are evident. The latter case demonstrates that the epitopes on the NAR tail are spaced far enough apart to minimize steric interference between Fab arms. The knob-like structures described above are surely the NAR V domains since they protrude from the opposite end of the molecule, i.e., from the N terminus. The V region dimensions are typical of single Ig domains based on x-ray crystallography data (4.0 × 2.5 nm, ref. 19) and on previous EM analysis of a mutant form of IgG displaying a protruding unpaired VL domain (20). In summary, NAR “Fab” arms are short, single domains attached to flexible hinge-like regions (Fig. 1G). A candidate hinge peptide segment of 11 aa (PGIPPSPPIVS) is present in the primary sequence immediately after the V domain (3).
The NAR Fc possesses a unique region that permits intra-Fc hinge-like folding. Though many of the observed molecules are linear throughout the Fc, proteins displaying a wide variety of angles, up to and including right angles, also are seen (Fig. 1B). Of the five mammalian Ig classes, only IgE has a bent Fc region, and it is believed to be relatively inflexible (21, 22). The distance from the C terminus to this bend in NAR (7.7 nm compared with 8.0 nm for the four-domain Ig Fab fragment) would place the joint at or near the C3–C4 junction (Fig. 1G). One can only speculate on the function of this flexible Fc, but it is a property likely to be shared by another isotype in sharks called IgW (8) or IgNARC (9), which is homologous to NAR in the four C-terminal domains.
Sequence Comparisons: Noncanonical Residues, Additional Cys Residues, Multiple D Segments and Preferred Reading Frames, and Convergence on Atypical IgV Domains.
Various peculiarities of NAR protein sequences deduced from cDNAs can be explained by our EM observation that NAR V regions do not form dimers and are free of quaternary associations. In particular, the presence of noncanonical Cys residues and changes in evolutionarily conserved amino acids that interact between VH and VL (Fig. 2; ref. 13–15) are likely to be hallmarks of single V domains.
There are only two closely related classes of expressed NAR genes in nurse sharks (Fig. 2), both types having one V, three D, and one J gene segment (3, 4). In the majority of NAR cDNAs analyzed to date, all three D regions are included in the rearrangement event (ref. 3 and Fig. 3c). Type I NAR proteins bear noncanonical Cys residues in FR 2 (Cys-35) and FR 4 (Cys-107) and in the somatically generated CDR3 (bold, shadowed residues in Fig. 2). Although varying greatly in size and sequence, Type I NAR CDR3 must be under considerable selective pressure as they almost always bear an even number of Cys residues. Most of these Cys residues are encoded by a preferred (most frequently used) RF of the rearranged D segments (Figs. 2 and 3 a and c), especially apparent in D2 and D3. However, in those cases when D2 or D3 is “read” in other RF or is not utilized in the rearrangement event, alternative Cys are encoded either by the D1 segment or by nucleotides inserted in the joins presumably through N-region addition.
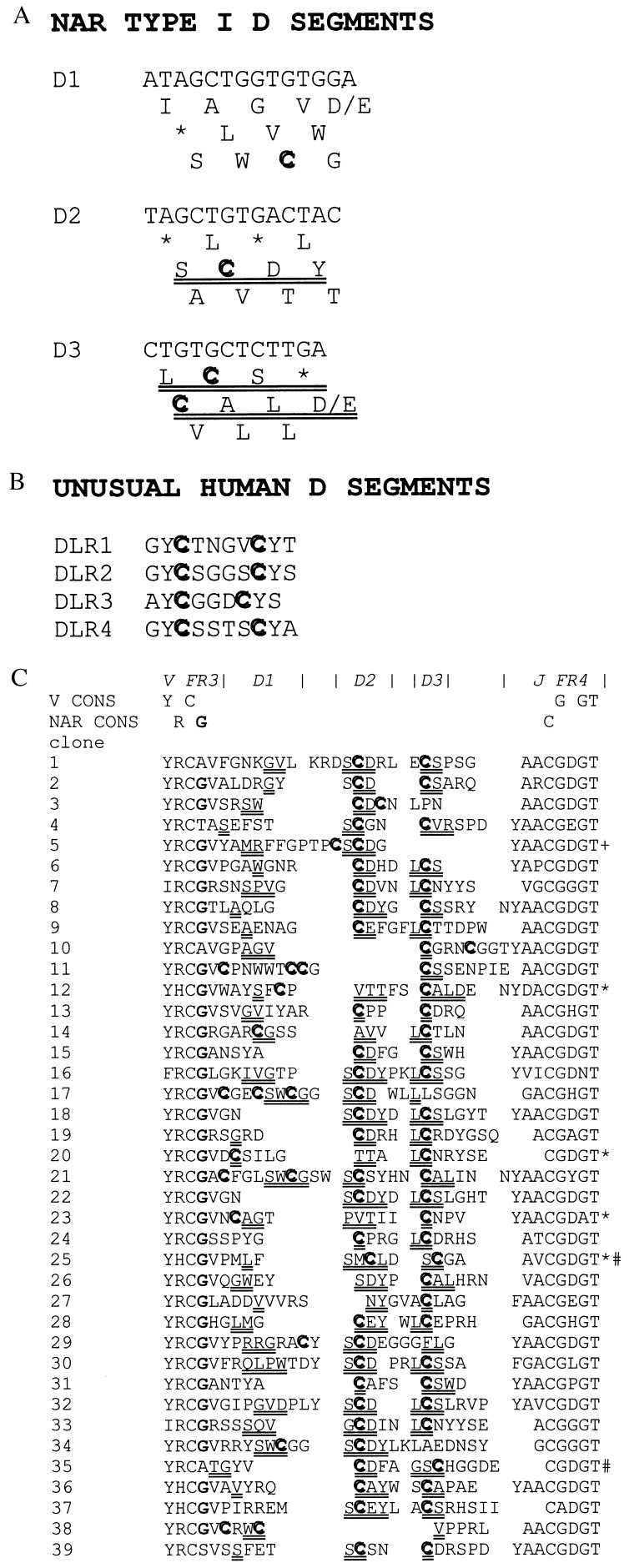
Figure 3.
Nucleotide sequence of NAR Type I D segments. (a) The three reading frames (RF) of D1, D2, and D3. Preferred RF are double underlined, Cys are in bold, and asterisks indicate stop codons. (b) Amino acid sequences of unusual human D segments that form disulfide bridges in some human IgH chains (23). (c) Amino acid sequences deduced from cDNAs of 39 Type I CDR3 junctions in NAR. Amino acids encoded by the D segments are double-underlined (at least two nucleotides of the D region had to be included in the codon encoding the amino acid). Note that sometimes the amino acid encoded by the D cannot be identified in any RF in a presumably because of somatic mutations in the particular cDNA clone or because the first nucleotide of the codon is specified by an N-region addition. In all cases but four (noted by ∗) the D2 segment is “read” in RF “SCDY” (a). D3 is “read” in two RF (“LCS” and “CAL,” a) that encode Cys (# indicates two cases in which D3 encodes Cys, but this was through somatic mutation of nucleotides in D3, not a preferred RF). +, May denote an “oligonucleotide capture” (32) of a D2 segment, i.e., not rearranged in the conventional manner, which is nevertheless “read” in the same RF as most of the other D2 segments. Conserved or invariant residues found in nearly all V domains or are specific only to NAR are indicated above. The NAR-specific glycine (bold, fourth position from left) and Cys in FR4 are displayed on the NAR model in Fig. 4e.
In those NAR CDR3 that are somewhat longer than average, four Cys sometimes are observed (clones 11, 17, and 21 in Fig. 3c). The CDR3 Cys (two or four) almost certainly form disulfide bridges within the CDR3 loop in a manner documented previously for an unusual human D segment bearing two Cys (DLR1–4, Fig. 3b and refs. 23 and 24; structure of entire Fab, Fig. 4a and see ref. 4). In these human molecules, the more rigid CDR3 blocks the remainder of the binding site; it therefore is not surprising that the RF encoding these Cys seem to be counterselected by mature human B cells (23, 24). By contrast, NAR with its single V seems to have much of its repertoire defined by diversity generated in its long CDR3. We speculate that the size and critical role in antigen recognition of NAR CDR3 likely requires the stabilizing effects of the additional disulfide bond(s). Note that in the cow, analysis of VH cDNA clones also has revealed extremely long CDR3 that almost always encode an even number of Cys residues (25).
An unusual FR2–FR4 disulfide bridge (Fig. 4 e and f) is unique to Type I NAR: modeling of this bond onto an Ig crystal structure shows that the sulfur atoms in the two exposed Cys are in position to make the disulfide bond over a small, well conserved NAR-specific glycine residue (Figs. 2, 3c, and 4 e and f). Substitution of other residues for glycine at this position probably would result in steric inhibition of disulfide bond formation.
NAR Type II genes, overall, are very similar in sequence to the Type I (Fig. 2), but instead have a Cys residue located in the center of CDR1 and another in CDR3 (Fig. 2, Cys, bold and shadowed). These NAR Type II Cys are also likely to form a disulfide bridge since residues at similar positions in the camel single V crystal structure have been shown to form such a bond (refs. 14 and 26; Figs. 2 and 4b).
Significance of Noncanonical Cys Residues.
There seems to be strong, selective pressures to preserve the various disulfide bridges in expressed NAR proteins. First, D regions are read in preferred Cys-containing RF despite the fact that other non-Cys-encoding RF are also “open” (Fig. 3 a and c). This provides evidence for a structural rationale to maintain a D segment-preferred RF (23, 27). Second, NAR is exceptional in ectothermic vertebrates in that its rearranged V, D, and J genes undergo a high frequency of somatic diversification (3): the codon encoding the FR2 Cys-35 in Type I NAR, although in a region of hypermutability, is under strong selection not to mutate to other residues (1 replacement vs. 8 silent changes in this codon out of 31 sequences analyzed; ref. 28). A simple model to interpret NAR function is to propose that diversity in the primary repertoire is concentrated principally in the long and heterogeneous CDR3. The fusion of three separate D genes with themselves and with V and J genes implies that four rearrangement events occur, generating vast diversity in the CDR3-encoding region through N- and P-region addition (3, 4). Because of the large CDR3 loop and the nonassociation of NAR V with any other domains, it is not unreasonable to assume that NAR CDR3 must be stabilized via disulfide bonds to ensure sufficient affinity and specificity. The primary CDR3-based repertoire is likely, then, to be fine-tuned by hypermutation leading to changes in CDR1 and other regions in NAR after exposure to antigen (3, 28).
Conclusions.
Evolutionary convergence at the molecular level is presumed to be widespread, but is poorly documented (29, 30). Are the structural features that we have shown and modeled for NAR—a single unassociated V domain and disulfide bridges within CDR3 in Type I genes and between CDR1 and CDR3 in Type II genes—truly convergent on the known structures of the unusual human D regions and the L chain-less camelid Ig or are they derived from common ancestors? There is every reason to believe that the camelid V regions represent bona fide mammalian VH that recently have been modified to form monomers as they have up to 75% amino acid identity with other mammalian V regions. Can the same be said for the NAR V domain? Of the various unique characteristics of NAR, two stand out. First, the overall NAR V sequence is not at all similar to conventional IgVH (25% identity) and is only somewhat more similar to VL and TCR V (3), suggesting that NAR must have diverged from Ig/TCR long ago. The origin of the second characteristic, the unpaired V domain, is less obvious. It may have been an early trait that coevolved with or perhaps influenced the overall uniqueness of the NAR V domain sequence. By this view, the unpaired V domain may represent a primordial relic that has been superseded largely by the more efficient two-domain, antigen-binding motifs. Alternatively, the dissimilarity of NAR V sequence to other Ig/TCR V sequences and its single-domain characteristic need not be directly linked: the former characteristic indicates early divergence but the latter could have been derived at any point in NAR’s evolutionary history. In any case, the singularity of NAR and camelid V domains (and perhaps a subset of V regions in another cartilaginous fish, the ratfish; ref. 31) would be independently arising and convergent characteristics. By extension, it is likely that the disulfide bridges between CDR1 and CDR3 in NAR and camel Ig (and also within CDR3 in NAR, human, and perhaps cow) also have been derived independently. The convergence of these structural features most likely has been driven by the independent development of V domains that do not form dimers with other domains.
Acknowledgments
We thank Lloyd Epstein, Abdu Azad, David Nemazee, Marilyn Diaz, and Lynn Rumfelt for critical reading of the manuscript, Ellen Hsu for calling our attention to the selection that must be imposed on the CDR3 regions, and Ms. Kimberly Riddle for assistance with EM. This work was supported by National Science Foundation Grant MCB-9304790 (K.H.R.) and National Institutes of Health Grant RR06603 (M.F.F.).
ABBREVIATIONS
- NAR
new or nurse shark antigen receptor
- TCR
T cell receptor
- CDR
complementarity-determining region
- EM
electron microscopy
- FR
framework
- RF
reading frame
Footnotes
References
- 1. Paul W E. In: Fundamental Immunology. 3rd Ed. Paul W E, editor. New York: Raven; 1993. pp. 1–7. [Google Scholar]
- 2.Tonegawa S. Nature (London) 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg A S, Avila D, Hughes M, Hughes A, McKinney E C, Flajnik M F. Nature (London) 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg A S. Ph.D. dissertation. Miami: University of Miami; 1994. [Google Scholar]
- 5.Marchalonis J J, Edelman G M. Science. 1966;154:1567–1568. doi: 10.1126/science.154.3756.1567. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi K, Tomonaga S, Kajii T A. Mol Immunol. 1984;21:397–404. doi: 10.1016/0161-5890(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 7.Harding F A, Amemiya C T, Litman R T, Cohen N, Litman G W. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernstein R M, Schluter S, Shen S, Marchalonis J J. Proc Natl Acad Sci USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenberg A S, Hughes A L, Guo J, Avila D, McKinney E C, Flajnik M F. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 10.Rast J P, Litman G W. Proc Natl Acad Sci USA. 1994;91:9248–9252. doi: 10.1073/pnas.91.20.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rast J P, Anderson M K, Strong S J, Luer C, Litman R T, Litman G W. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- 12.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 13.Chothia C, Boswell D R, Lesk A M. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spinelli S, Frenken L, Bourgeois D, de Ron L, Bos W, Verrips T, Anguille C, Cambillau C, Tegoni M. Nat Struct Biol. 1996;3:752–757. doi: 10.1038/nsb0996-752. [DOI] [PubMed] [Google Scholar]
- 15.Desmyter A, Transue T R, Ghahroudi M A, Dao Thi M-H, Poortmans F, Hamers R, Muyldermans S, Wyns L. Nat Struct Biol. 1996;3:803–810. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 16.Roux K H. Methods. 1996;10:247–256. doi: 10.1006/meth.1996.0099. [DOI] [PubMed] [Google Scholar]
- 17.Posnett D N, McGrath H, Tam J P. J Biol Chem. 1994;263:1719–1725. [PubMed] [Google Scholar]
- 18.Marquart M, Deisenhofer J, Huber R, Palm W. J Mol Biol. 1980;141:369–391. doi: 10.1016/0022-2836(80)90252-1. [DOI] [PubMed] [Google Scholar]
- 19.Roux K H, Shuford W W, Finley J W, Esselstyn J, Pankey S, Raff H V, Harris L J. Mol Immunol. 1994;31:933–942. doi: 10.1016/0161-5890(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 20.Poljak R J, Amzel L M, Avey H P, Chen B L, Phizackerley R P, Saul F. Proc Natl Acad Sci USA. 1973;70:3305–3310. doi: 10.1073/pnas.70.12.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Shopes B, Holowka D, Baird B. Biochemistry. 1992;31:7446–7456. doi: 10.1021/bi00148a004. [DOI] [PubMed] [Google Scholar]
- 22.Beavil A J, Young R J, Sutton B J, Perkins S J. Biochemistry. 1975;34:14449–14461. doi: 10.1021/bi00044a023. [DOI] [PubMed] [Google Scholar]
- 23.Raaphorst F M, Raman C S, Nall B T, Teale J M. Immunol Today. 1997;18:37–43. doi: 10.1016/s0167-5699(97)80013-8. [DOI] [PubMed] [Google Scholar]
- 24.Milili M, Schiff C, Fougereau M, Tonnelle C. Eur J Immunol. 1996;26:63–69. doi: 10.1002/eji.1830260110. [DOI] [PubMed] [Google Scholar]
- 25.Lopez O, Perez C, Wylie D. Immunol Rev. 1998;162:55–66. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]
- 26.Muyldermans S, Atarhouch T, Saldanha J, Barbosa J A R G, Hamers R. Protein Eng. 1994;7:1129–1135. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- 27.Cohn M. Annu Rev Immunol. 1994;12:1–62. doi: 10.1146/annurev.iy.12.040194.000245. [DOI] [PubMed] [Google Scholar]
- 28.Du Pasquier L, Wilson M, Greenberg A S, Flajnik M F. Current Top Microbiol Immunol. 1997;229:199–216. doi: 10.1007/978-3-642-71984-4_14. [DOI] [PubMed] [Google Scholar]
- 29.Stewart C-B, Shilling J W, Wilson A C. Nature (London) 1987;330:401–404. doi: 10.1038/330401a0. [DOI] [PubMed] [Google Scholar]
- 30.Sharp P M. Nature (London) 1997;385:111–112. doi: 10.1038/385111a0. [DOI] [PubMed] [Google Scholar]
- 31.Rast J P, Amemiya C T, Litman R T, Strong S J, Litman G W. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- 32.Schuler W, Ruetsch N R, Amsler M, Bosma M J. Eur J Immunol. 1991;21:589–596. doi: 10.1002/eji.1830210309. [DOI] [PubMed] [Google Scholar]
- 33.Harpaz Y, Chothia C. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]