Abstract
We investigated in different human cell types nuclear positioning and transcriptional regulation of the functionally unrelated genes GASZ, CFTR, and CORTBP2, mapping to adjacent loci on human chromosome 7q31. When inactive, GASZ, CFTR, and CORTBP2 preferentially associated with the nuclear periphery and with perinuclear heterochromatin, whereas in their actively transcribed states the gene loci preferentially associated with euchromatin in the nuclear interior. Adjacent genes associated simultaneously with these distinct chromatin fractions localizing at different nuclear regions, in accordance with their individual transcriptional regulation. Although the nuclear localization of CFTR changed after altering its transcription levels, the transcriptional status of CFTR was not changed by driving this gene into a different nuclear environment. This implied that the transcriptional activity affected the nuclear positioning, and not vice versa. Together, the results show that small chromosomal subregions can display highly flexible nuclear organizations that are regulated at the level of individual genes in a transcription-dependent manner.
Keywords: CFTR; nuclear architecture; gene positioning; chromatin organization; chromosome territory
Introduction
Mammalian gene loci display a nonrandom positioning within cell nuclei, which is related to their functional regulation. In particular, association with the nuclear periphery and pericentric heterochromatin seems to play a role in the regulation of transcription and recombination (Brown et al., 1997, 1999, 2001; Skok et al., 2001; Kosak et al., 2002). Also, the positioning of gene loci at specific regions of the respective chromosome territory appears to play a role in their functional regulation (Cremer et al., 1993, 1995; Volpi et al., 2000; Cremer and Cremer, 2001; Mahy et al., 2002a; Williams et al., 2002). Currently, it is unclear how the association of gene loci with specific nuclear domains relates to their organization within chromosome territories.
An important question is at which level the functional organization of chromosomal loci in the nucleus is regulated. Previous reports suggested that whole chromosomes (Croft et al., 1999; Boyle et al., 2001) or large chromosomal subregions in the size range of several hundred kb pairs up to several Mb pairs (Ferreira et al., 1997; Croft et al., 1999; Sadoni et al., 1999; Volpi et al., 2000; Williams et al., 2002) displayed a specific nuclear positioning. Also, the results of recent FISH analyses implied that region-specific gene density and transcriptional activity, rather than the activity of individual genes, influenced the organization of chromosome territories (Mahy et al., 2002a,b).
To address the question of whether nuclear positioning is indeed regulated at the level of larger chromosomal subregions rather than at the level of individual genes, we investigated the nuclear positioning of three adjacent genes in the cystic fibrosis transmembrane conductance regulator (CFTR) region on human chromosome 7q31. The genes evaluated besides CFTR were the germ cell–specific expression, presence of ANK, SAM, and basic leucine zipper domains (GASZ) gene localized ∼50 kb upstream of CFTR (Yan et al., 2002), and the cortactin-binding protein 2 (CORTBP2) gene localized ∼45 kb downstream of CFTR (Cheung et al., 2001). The CFTR gene codes for a cyclic AMP-dependent chloride channel that is expressed in a number of epithelial tissues, in particular in the respiratory tract epithelium (Engelhardt et al., 1994; Li et al., 1988). Mutations in this gene cause cystic fibrosis, which is the most common severe autosomal recessive disease amongst the Caucasian population. GASZ displays germ cell–specific expression and may represent a cytoplasmic signal transducer mediating protein–protein interactions during germ cell maturation (Yan et al., 2002). CORTBP2 codes for a cortactin-binding protein of unknown function and is expressed in a variety of human tissues (Cheung et al., 2001). Our results show that the nuclear positioning of GASZ, CFTR, and CORTBP2 and their association with distinct chromatin fractions is regulated at the level of individual genes in a transcription-dependent manner.
Results
Nuclear positioning of GASZ, CFTR, and CORTBP2
First, we investigated nuclear positioning of CFTR in human SH-EP N14 neuroblastoma cells, where this gene is not transcribed (for levels of transcription see Table I; also see Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1), and in Calu-3 adenocarcinoma cells, where CFTR is highly expressed. CFTR was detected by FISH using the prokaryotic artificial chromosome CF1 from the 5′ region of the CFTR gene (Fig. 1). This probe was used in all FISH experiments for detection of CFTR unless otherwise indicated. We investigated formaldehyde-fixed nuclei, which were also immunostained with antibodies against the lamina-associated polypeptide 2β (LAP2β) or histone H4 acetylated at lysine 8 (H4Ac8). H4Ac8 is enriched in the early replicating and transcriptionally active euchromatin in the nuclear interior, whereas later replicating and transcriptionally inactive heterochromatin, including perinuclear heterochromatin, is depleted in H4Ac8 (Sadoni et al., 1999). LAP2β is an integral protein of the inner nuclear membrane, which binds to chromatin (Furukawa et al., 1995; Nili et al., 2001). A fraction of cells was also pulse labeled with FITC-dUTP in order to label distinct fractions of heterochromatin and euchromatin displaying a distinct replication timing and localizing at specific nuclear regions (Sadoni et al., 1999; Zink et al., 2003).
Table I. Test gene expression as percentage of α-actin expression.
| Cell type | CORTBP2 | CFTR | GASZ |
|---|---|---|---|
| T-lymphocytes | 0.33 ± 0.58% | Zero | Zero |
| HEK 293 | 6.9 ± 4.8% (n = 4) | Zero (n = 4) | Zero |
| SH-EP N14 | Zero (n = 4) | Zero (n = 4) | Zero |
| Calu-3 | 0.27 ± 0.29% | 57.6 ± 23.4% (n = 5) | Zero |
| HT1080 | 0.53 ± 0.45% | Zero | Zero |
| Testis | 2.2 ± 1.7% | 2.1 ± 0.4% (n = 4) | Detectable |
| Nasal epithelium | 0.67 ± 0.12% | 1.3 ± 0.3% | Zero |
| Calu-3 control | - | 77.5 ± 13.0% | - |
| Calu-3 plus DRB | - | 34.7 ± 19.2% | - |
The values are based on average percentages of experimental gene expression while control gene (α-actin) is still in exponential phase of amplification. The values shown are means ± SD for n = 3 replications, unless otherwise shown. “Zero” indicates that the experimental peak was undetectable within log phase of α-actin amplification and within 40 cycles of normal monoplex PCR amplification. For GASZ, expression in testis was only detected after α-actin amplification had entered the plateau phase, and was confirmed by monoplex PCR.
Figure 1.
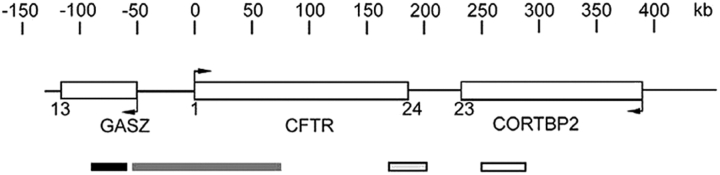
Schematic drawing of the CFTR region. CFTR, GASZ, and CORTBP2 are symbolized by open boxes. Arrows indicate the directions of transcription and the numbers of exons are given by numbers below the genes. The scale at the top shows the DNA length in kb pairs. The boxes below the gene loci indicate lengths and positions of the probes used for FISH specific for GASZ (black), the 5′ end of CFTR (CF1, dark gray), the 3′ end of CFTR (light gray), and CORTBP2 (white).
After immuno-FISH, nuclei were imaged by confocal microscopy. In SH-EP N14 cells, 87% of the CFTR-specific FISH signals (n = 100; n always refers to the numbers of FISH signals analyzed) were associated with the nuclear periphery, as visualized by the different labeling methods used (Fig. 2). At the nuclear periphery, CFTR was embedded in the perinuclear heterochromatin not enriched in H4Ac8 (Fig. 2 a; Sadoni et al., 1999) and replicating during the second half of S-phase (Fig. 2 e). Immunostaining of LAP2β revealed a very close association of CFTR with the nuclear periphery (Fig. 2 c). Also, FISH signals, appearing to localize to the nuclear interior, were associated with invaginations of the nuclear periphery forming long intrusions into the nuclear interior in this cell type (Fig. 2 c). Counterstaining of DNA with propidium iodide confirmed the perinuclear localization of CFTR and revealed that it was not associated with perinucleolar heterochromatin in SH-EP N14 cells (Fig. 2 f).
Figure 2.
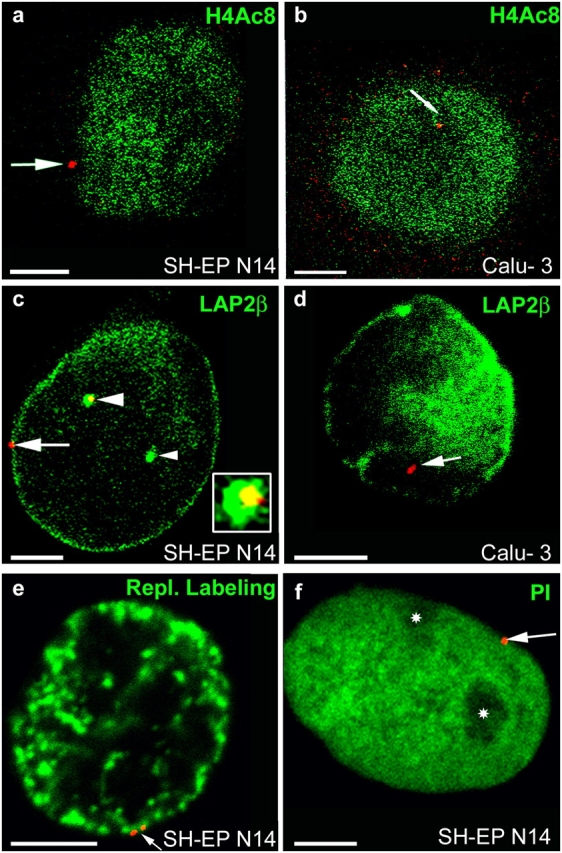
Nuclear localization of CFTR. FISH (red signals) was performed with nuclei from SH-EP N14 (a, c, e, and f) and Calu-3 cells (b and d). Nuclei were immunostained with antibodies against H4Ac8 (a and b, green) or LAP2β (green in c and d). Nuclei were also labeled by replicational pulse labeling (e, green) or were counterstained with propidium iodide (f, green false color). Single light-optical sections are shown and often only one CFTR allele is present in the displayed nuclear plane. (a and b) Arrows point to CFTR alleles associated at the nuclear periphery with chromatin not enriched in H4Ac8 (a, SH-EP N14) or embedded in the nuclear interior into chromatin enriched in H4Ac8 (b, Calu-3). (c) One CFTR allele (arrow) associates at the nuclear periphery with high concentrations of LAP2β. The other CFTR allele (large arrowhead) associates with an invagination of the nuclear periphery into the interior (cross section, green), also containing high concentrations of LAP2β. An enlargement of the FISH signal associated with the invagination is shown in the inset. Only the yellow section of the stained area corresponds to the FISH signal (colocalization of the red FISH signal with the invagination stained in green). The small arrowhead points to a second invagination not associated with a FISH signal. (d) In Calu-3 cells, CFTR (arrow) does not associate with high local concentrations of LAP2β. (e) The replication labeling pattern (green) is typical for the second half of S-phase, with strongly stained perinuclear heterochromatin. The arrow points to a CFTR allele (doublet signal) embedded within the perinuclear heterochromatin. (f) Nucleoli (asterisks) appear unstained in the propidium iodide counterstain (green). The CFTR allele (arrow) is associated with the nuclear periphery, but not with the nucleolar peripheries. Bars, 5 μm.
In Calu-3 cells, 88% of the CFTR-specific FISH signals were embedded in the nuclear interior within the transcriptionally active euchromatin, which is specifically enriched in H4Ac8 (Fig. 2 b, n = 26; Sadoni et al., 1999), and most of the FISH signals (82%) were not associated with high local concentrations of LAP2β (Fig. 2 d; n = 27). Together, these results revealed that CFTR associated in its inactive state in SH-EP N14 cells with the nuclear periphery and here with later replicating perinuclear heterochromatin depleted in H4Ac8. In contrast, in its actively transcribed state in Calu-3 cells, CFTR did not associate with the nuclear periphery, but with euchromatin enriched in H4Ac8 occupying the nuclear interior.
Next, we investigated the association of GASZ, CFTR, and CORTBP2 with perinuclear heterochromatin, highlighted by replicational pulse labeling, in SH-EP N14, Calu-3, and HEK 293 cells. Cells were fixed with formaldehyde and imaged by confocal microscopy. In SH-EP N14 cells, where none of the genes were transcribed (Table I), all three genes preferentially associated with perinuclear heterochromatin (Fig. 3, a, b, and e; ∼80–90%). GASZ and CFTR were not transcribed in 293 cells, and also here 80–90% of the corresponding FISH signals associated with perinuclear heterochromatin (Fig. 3, c and e). In contrast, CORTBP2 was transcribed at relatively high levels in this cell type and only ∼10% of the corresponding FISH signals associated with the perinuclear heterochromatin (Fig. 3 e). Closer inspection of the data revealed that although CORTBP2 was clearly not associated with the perinuclear heterochromatin in ∼90% of the cases, it still occupied relatively peripheral positions and was typically located just behind the border to the adjacent euchromatin (Fig. 3 d).
Figure 3.
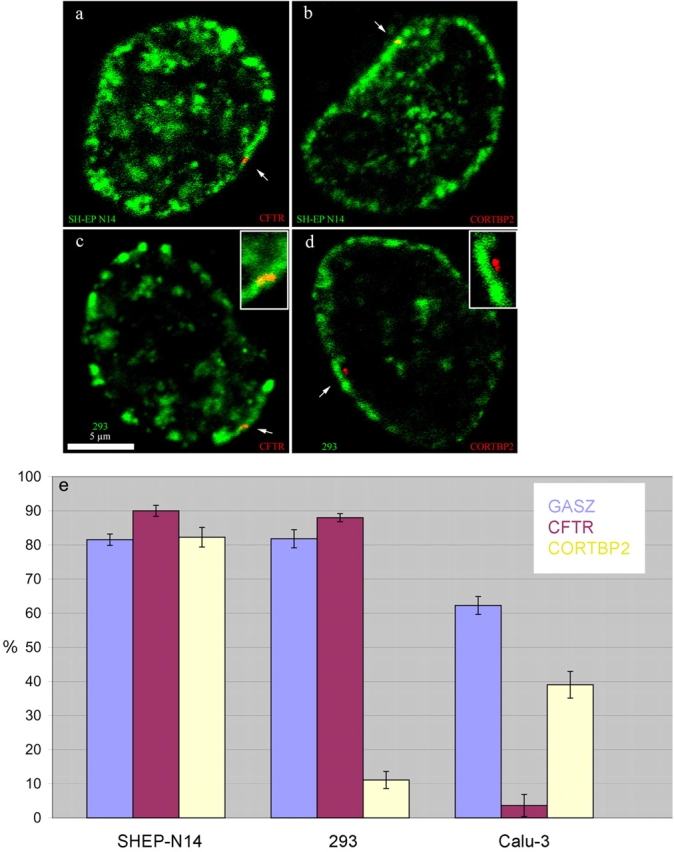
Spatial arrangements of GASZ, CFTR, and CORTBP2. (a–d) Panels show light-optical sections of formaldehyde-fixed nuclei from SH-EP N14 (a and b) and HEK 293 (c and d) cells (bar, 5 μm). CFTR (a and c) and CORTBP2 (b and d) were detected by FISH (red signals). Perinuclear and perinucleolar heterochromatin has been visualized by replication labeling (green). The arrows in a, b, and c, respectively, point to CFTR and CORTBP2 genes embedded within the perinuclear heterochromatin. The arrow in d points to the CORTBP2 locus (doublet signal) localizing within the adjacent euchromatin. The insets in c and d show the corresponding FISH signals and their nuclear environments enlarged. (e) The fractions of FISH signals (violet bars, GASZ; red bars, CFTR; yellow bars, CORTBP2; average ± SD) colocalizing with perinuclear heterochromatin were determined in the cell types indicated. Replication-labeled nuclei as shown in panels a–d were evaluated after FISH. In each case ∼50–60 FISH signals were evaluated.
In Calu-3 cells, only GASZ was not transcribed. Here, a bias toward association with perinuclear heterochromatin could be observed (Fig. 3 e; ∼60%), although in Calu-3 cells the preferential association of GASZ with perinuclear heterochromatin was not as pronounced as in the other cell types examined. CORTBP2, which was expressed in Calu-3 cells, did not preferentially associate with perinuclear heterochromatin, but also here the tendency was relatively weak (∼40% of the FISH signals still associated with perinuclear heterochromatin). In accordance with the results described above (Fig. 2), CFTR was in the vast majority of cases not associated with perinuclear heterochromatin in Calu-3 cells (Fig. 3 e; ∼95%).
In summary, the results did show that GASZ, CFTR, and CORTBP2 preferentially associated with perinuclear heterochromatin in their inactive states, but not when actively transcribed. When preferential association with perinuclear heterochromatin was observed, the genes always fully colocalized with the heterochromatin and were surrounded by it (Fig. 2 e and Fig. 3, a–c). As the layer of perinuclear heterochromatin is relatively thin (<1 μm), this suggests that the distances of the inactive gene loci to the nuclear envelope were <1 μm. A very tight association with the nuclear envelope and direct physical contact was also suggested by the results obtained after additional immunostaining of LAB2β (Fig. 2 c).
Although we did not quantitate whether the alleles present in one nucleus always showed the same behavior, it is obvious that at least in those cases, where a very pronounced preferential localization was observed (all three genes in SH-EP N14 and 293 cells and CFTR in Calu-3 cells), the alleles of a given gene in one nucleus must have shown the same behavior in the vast majority of nuclei. This can be also observed on several images shown (Fig. 2 c; see Fig. 5 a; also see Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1).
Figure 5.
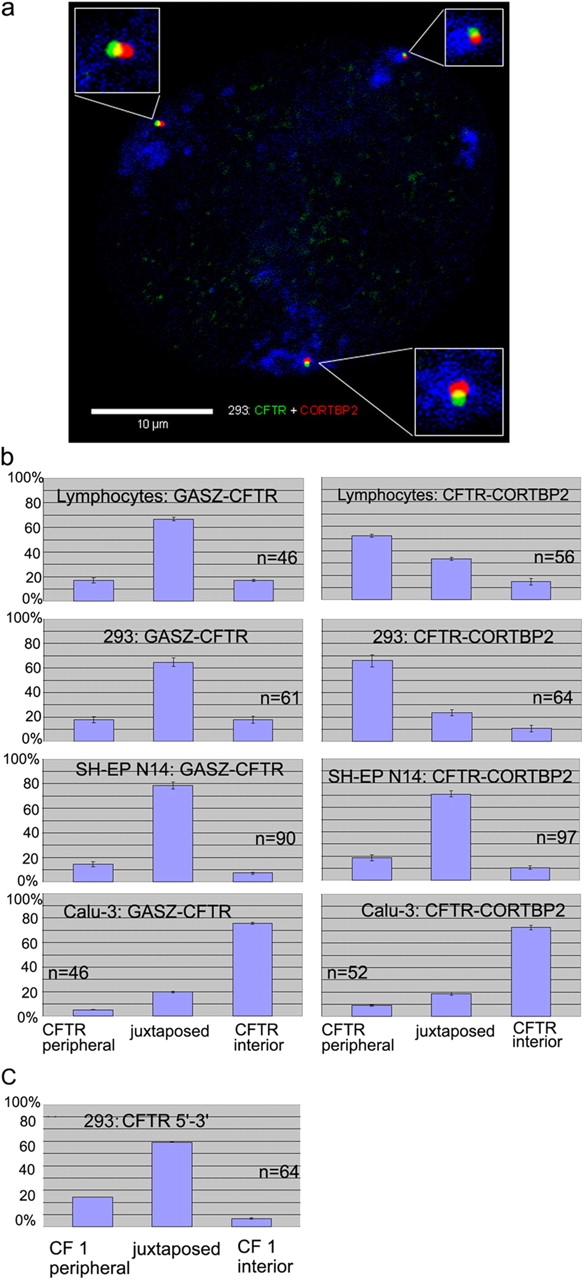
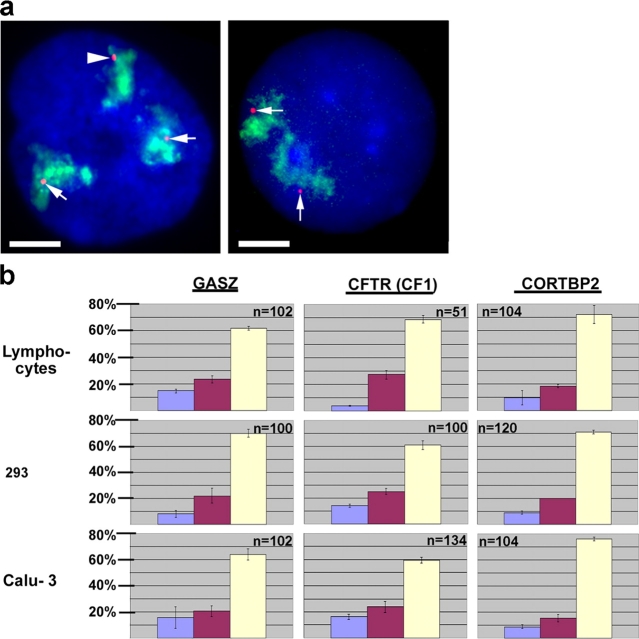
Spatial arrangements of loci as revealed by dual-color FISH. (a) CFTR (green) and CORTBP2 (red) were simultaneously detected by dual-color FISH in a 293 nucleus (enlargements shown in the insets). Chromosome 7 territories are shown in blue (293 nuclei harbor three chromosome 7 territories). (b) The nuclear positions of adjacent loci simultaneously detected in the same nucleus by dual-color FISH were determined with respect to each other. The combinations of gene loci evaluated and the cell types analyzed are indicated. It was evaluated in each case whether CFTR was located more peripheral or more interior, respectively, with regard to the other locus, or whether both loci were juxtaposed with no locus being more interior or peripheral. In all cases for CFTR detection, the CF1 probe from the 5′ region of CFTR was used. A second CFTR-specific probe from the 3′ region of CFTR (compare with Fig. 1) was used in 293 cells, and the orientation of the corresponding FISH signal relative to the CF1-specific signal is shown in c. The bars show for each combination the percentages displaying the indicated orientation (average ± SD). n indicates the number of pairs of loci examined.
Erosion analyses (Fig. 4) allow a relatively fast evaluation of nuclear gene distributions and are ideally suited for investigating large sample numbers. The results of erosion analyses can not, per se, be interpreted in terms of the nuclear distribution of FISH signals or their associations with specific nuclear regions. However, by relating results from erosion analyses to known distributions of FISH signals in the three-dimensional (3D) nuclear space, it becomes possible to interpret the data.
Figure 4.
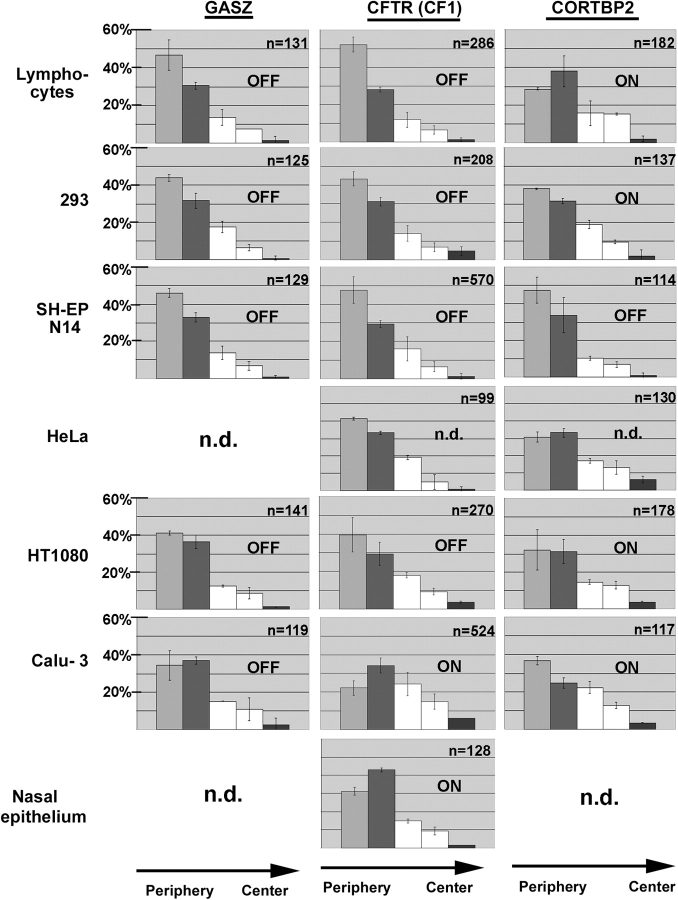
Summary of the erosion analysis results. Nuclei of the seven cell types indicated were hybridized with probes specific for GASZ (left-hand diagrams), CFTR (diagrams in the middle), and CORTBP2 (right-hand diagrams). Nuclei were imaged by epifluorescence microscopy and the 2D image of each nucleus was subdivided into five concentric areas with each area having a thickness of 20% of the nuclear radius as defined by the DAPI counterstain. The positions of FISH signals with regard to these five areas were determined. The five bars in each diagram indicate the fractions of FISH signals in the five concentric areas (average ± SD). The very left bar in each diagram corresponds to the most peripheral area (1), whereas the right bars correspond to the central area (5). For each combination (gene and cell type), 2–8 independent experiments have been performed. n indicates the number of FISH signals evaluated. In addition, on each panel it is indicated whether the gene analyzed was transcriptionally active (on) or inactive (off), as revealed by the RT-PCR analyses (Table I).
To analyze larger sample numbers we performed erosion analyses with regard to GASZ, CFTR, and CORTBP2 in the following cell types: primary nasal epithelial cells and primary T-lymphocytes obtained from healthy donors, 293 cells, HeLa cells, SH-EP N14 cells, HT1080 fibrosarcoma cells, and Calu-3 cells. The results (Fig. 4) revealed that a close association of a gene locus with the nuclear periphery and perinuclear heterochromatin, as observed for GASZ, CFTR, and CORTBP2 in SH-EP N14 cells and GASZ and CFTR in 293 cells (Fig. 2 and Fig. 3), was indicated in the erosion analysis by a peak in the number of FISH signals in the first (outermost) shell, with at least 40% of the FISH signals being located there (note that the “true” fraction of FISH signals associating with the nuclear periphery [80–90%] is largely underestimated by the erosion analysis). A second feature indicating a close association with the nuclear periphery is a constant decline in the numbers of FISH signals toward the nuclear center, giving rise to a typical “staircase-like” arrangement of the bars in the corresponding diagram. In all cases where one of the three genes was not transcribed a corresponding distribution was observed, with the exception of GASZ in Calu-3 cells (Fig. 4). In this case, our previous analyses indicated a less pronounced association with perinuclear heterochromatin (Fig. 3 e).
In all situations where a given gene was expressed, the erosion analysis indicated a more interior positioning (Fig. 4). Also, dissociation from perinuclear heterochromatin of the active CORTBP2 in 293 cells (Fig. 3) was reliably indicated by the erosion analysis by a drop in the numbers of FISH signals in the outermost shell below 40%. Nevertheless, the erosion analysis also indicated a relatively peripheral positioning of CORTBP2 in 293 cells, in accordance with our previous results (Fig. 3). Also, the more interior positioning of CFTR in Calu-3 cells (Figs. 2 and 3; also see Fig. S1) was reliably indicated by the erosion analysis. A comparable interior positioning of CFTR was observed in primary nasal epithelial cells (Fig. 4), where CFTR was also expressed. This showed that interior positioning of the active CFTR gene was not an aberrant feature of Calu-3 adenocarcinoma cells.
Additional controls showed that the relatively interior positioning of all three gene loci in Calu-3 cells was not due to disruption of the peripheral heterochromatin or disruption of the interactions of the chromosome 7 territory with the nuclear periphery (unpublished data). As a further control, we evaluated the nuclear positioning of the unrelated β-globin locus by erosion analysis in 293, SH-EP N14, and Calu-3 cells (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1). The nuclear positioning of the β-globin locus was not significantly different in Calu-3 cells in comparison to 293 or SH-EP N14 cells (unpaired t test; P < 0.05).
In summary, comparison of the results obtained with the erosion analysis to known gene distributions in the 3D nuclear space allowed to interpret the data and showed that the erosion analysis reliably indicated even subtle changes in gene distributions (e.g., CORTBP2 in 293 cells). Together, the data showed that in all cases transcriptionally inactive loci preferentially associated with the nuclear periphery, whereas the genes resided at more interior positions in their active states. It should also be noted that with regard to the active CORTBP2 gene, no correlation between its levels of transcription (Table I) and its positions in the nuclear interior could be observed. For example, CORTBP2 was transcribed at relatively high levels in 293 cells, although it occupied in this cell type more peripheral positions than in T-lymphocytes, where it was transcribed at substantially lower levels (Table I and Fig. 4). Together with the finding that CORTBP2 did not associate with perinuclear heterochromatin but resided just behind the border to the euchromatin in 293 cells (Fig. 3 d), the results suggested that association with euchromatin plays a more important role than the absolute distance to the nuclear periphery. Our finding that CORTBP2 is expressed in 293 cells at relatively high levels just behind the border to the adjacent euchromatin is in accordance with previous findings (Fakan, 1994; Cmarko et al., 1999; Verschure et al., 1999), showing that nascent RNA synthesis takes place at the surfaces of condensed chromatin domains.
Simultaneous detection of neighboring genes
To further address the spatial arrangements of CFTR and adjacent genes and the question of whether neighboring genes associated simultaneously with different regions of the same nucleus, we performed dual-color FISH analyses. Two adjacent genes (GASZ and CFTR, or CFTR and CORTBP2, respectively) were simultaneously detected together in the same nucleus with two different fluorochromes (Fig. 5). Next, it was determined with regard to lymphocytes, 293, SH-EP N14, and Calu-3 cells, how frequently one of these two genes was located more peripherally than the other. In addition, it was determined how frequently both genes were juxtaposed to each other, with neither locus locating more peripheral.
The data summarized in Fig. 5 b show that in T-lymphocytes and 293 cells, the majority of GASZ and CFTR signals were juxtaposed to each other, with no locus being more peripheral. In contrast, CFTR preferentially showed a more peripheral position than CORTBP2. These results were in accordance with the previous results (Fig. 3 and Fig. 4), showing GASZ and CFTR closely associated with the nuclear periphery of T-lymphocytes and 293 cells, in contrast to CORTBP2.
In SH-EP N14 cells, GASZ and CFTR, as well as CFTR and CORTBP2, respectively, were preferentially juxtaposed to each other (Fig. 5 b). All three loci closely associated with the nuclear periphery and perinuclear heterochromatin in this cell type (Fig. 3 and Fig. 4). In contrast, in Calu-3 cells CFTR was located more to the interior than GASZ or CORTBP2, respectively (Fig. 5 b). Also, these findings were in accordance with our previous results (Fig. 3 and Fig. 4). Although all results were in accordance with the data obtained by the previous analyses, the dual-color FISH analysis demonstrated that adjacent gene loci simultaneously associated with different nuclear regions.
Next, we addressed the question of which parts of the chromosomal region provided the flexible linker between CFTR and CORTBP2. Therefore, we performed with 293 nuclei dual-color FISH experiments with the CF1 probe specific for the 5′ region of CFTR and another probe specific for the 3′ region of CFTR (Fig. 1). The results (Fig. 5 c) revealed that the 5′ and 3′ regions of CFTR were juxtaposed next to each other in the majority of cases. Together with the finding that CORTBP2 occupied more interior positions than CF1 (Fig. 5 b), the results suggested that mainly the intergenic region between CFTR and CORTBP2 provided the flexible linker. This was also suggested by the fact that FISH signals obtained with the probe specific for the 3′ region of CFTR showed the same distribution in the erosion analysis as signals obtained with the 5′-specific probe (CF1) in cases where the neighboring CORTBP2 gene showed a markedly different distribution (T-lymphocytes and Calu-3 cells; unpublished data).
To further address the spatial organization of the CFTR region, we analyzed the distances of the gene loci (Fig. 6). Again, GASZ/CFTR and CFTR/CORTBP2 pairs were detected simultaneously with two different fluorochromes in the same nucleus. The smallest distances were measured in SH-EP N14 cells, where all three gene loci were juxtaposed next to each other at the nuclear periphery (Figs. 3–5). The slightly larger distances measured in SH-EP N14 cells between CFTR and CORTBP2, as compared with GASZ and CFTR (Fig. 6 a), likely reflected the larger distances of the probes used for FISH (Fig. 1). Although the distances between GASZ and CFTR still did not exceed the 200-nm interval in 293 cells, as in SH-EP N14 cells, larger distances were measured between CFTR and CORTBP2 in 293 cells as compared with SH-EP N14 cells. In 293 cells, CORTBP2 occupied more interior positions (Figs. 3–5), whereas GASZ and CFTR were both associated with the nuclear periphery (Fig. 3 and Fig. 4). Regarding primary T-lymphocytes, in 90% of the cases the distances between GASZ and CFTR did not exceed 200 nm. In ∼10% of the cases larger distances were measured, but did not exceed 400 nm. However, between CFTR and CORTBP2 a marked shift toward increased distances was observed (up to 1,100 nm). In primary T-lymphocytes CORTBP2 occupied substantially more interior positions than CFTR (Fig. 4 and Fig. 5). As CFTR is closely associated with the nuclear periphery in T-lymphocytes and 293 cells, the distance measurements relative to CFTR also roughly indicate the distance of the active CORTBP2 locus from the nuclear periphery.
Figure 6.

Distance measurements. Adjacent loci were detected by dual-color FISH and the distances between the simultaneously detected loci were measured. The pairs of loci evaluated and the cell types analyzed are indicated. The bars (average ± SD) show for each combination the percentages with distances within a given interval (0–100 nm, 101–200 nm, etc.). The numbers below the x axes denote the distance intervals (nm). (a) 2D distances were measured on projections of the focal planes of nuclei fixed with methanol/acetic acid. (b) 3D distances were measured on the individual focal planes of Calu-3 nuclei fixed with formaldehyde. n indicates the number of pairs of loci analyzed.
The largest distances between GASZ and CFTR, as well as between CFTR and CORTBP2, respectively, were observed in Calu-3 cells. In this cell type CFTR displayed a markedly interior positioning, in contrast to the other two gene loci (Figs. 2–5). However, we wondered whether these increased distances were due to a more decondensed chromatin structure in Calu-3 cells. Therefore, we measured in the different cell types the sizes of the nuclei and the relative nuclear areas occupied by chromosome 7 territories. It was found that Calu-3 cells did have the smallest nuclei of all cell types analyzed (unpublished data), and that the chromosome 7 territories in this cell type occupied the smallest percentage of the nuclear area (4.29% in Calu-3 cells, compared with 4.44% in SH-EP N14 cells, 5.06% in HEK 293 cells, and 4.72% in T-lymphocytes, respectively). These findings suggest that the large distances measured in Calu-3 cells do not reflect a generally more decondensed chromatin structure, or a more decondensed structure of chromosome 7.
The relatively extreme distances observed in Calu-3 cells gave rise to the question of how far the neighboring gene loci could move apart. The 2D distance measurements described above were performed in methanol/acetic acid-fixed nuclei and the absolute distances between two loci might be overestimated by using this method. Therefore, we measured the 3D distances in formaldehyde-fixed Calu-3 nuclei imaged by confocal microscopy. It should be noted that applying the FISH technique might lead to topological changes compared with the live cell situation, although generally a good preservation of the spatial organization of chromatin domains is observed at the level of light microscopy in formaldehyde-fixed nuclei (Robinett et al., 1996; Verschure et al., 1999; Solovei et al., 2002)
The results from Fig. 6 b revealed that the largest distances between GASZ and CFTR were in the 300-nm range, whereas the largest distances between CFTR and CORTBP2 were in the 500-nm range. Distances were measured between the intensity centers of the FISH signals. Considering that the distances between the middlepoints of the gene sequences covered by the respective probes used for FISH were ∼85 kb (GASZ/CFTR) and ∼255 kb (CFTR/CORTBP2) apart from each other (Fig. 1), the corresponding degrees of compaction would be ∼100-fold (GASZ/CFTR) and ∼175-fold (CFTR/CORTBP2), respectively (assuming a length of 3.4 × 105 nm per Mb DNA). This would be a higher degree of compaction as displayed by nucleosomal DNA or the 30-nm fiber. Nucleosomal DNA displays a packaging ratio of 1:7, whereas the 30-nm fiber is assumed to have an additional packaging ration of 1:6 (Belmont et al., 1989).
How are nuclear positioning and transcriptional activity of CFTR related to each other?
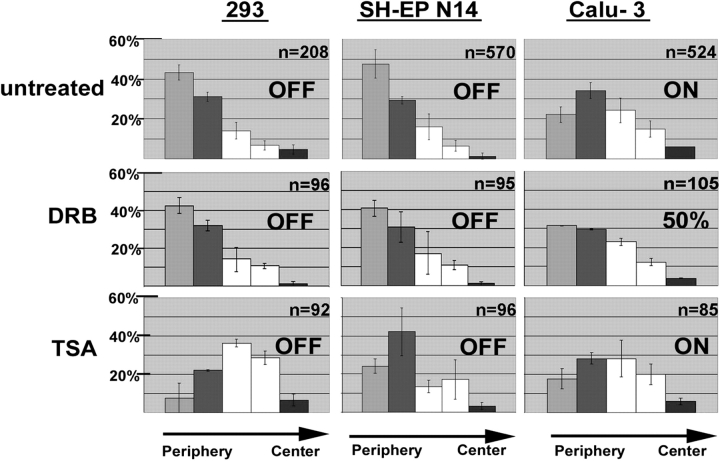
To address the question of how transcriptional regulation and nuclear positioning of CFTR were related to each other, we treated cells with the transcriptional inhibitor 5,6-dichlorobenzimidazole riboside (DRB). The nuclear localization of CFTR was determined in 293, SH-EP N14, and Calu-3 cells by erosion analysis (Fig. 7). After DRB treatment, the perinuclear localization of CFTR remained unchanged in SH-EP N14 and 293 cells (Fig. 7). As revealed by RT-PCR analyses, these cell types did not express CFTR at detectable levels, neither before nor after DRB treatment. In contrast, in Calu-3 cells a significant increase in the numbers of FISH signals was observed in the outermost shell after DRB treatment (compared with untreated Calu-3 cells; Fig. 7, unpaired t test, P < 0.05). RT-PCR analysis revealed that the level of CFTR transcription was reduced to ∼50% after DRB treatment (Table I).
Figure 7.
Positioning of CFTR after DRB and TSA treatment. FISH was performed with the CF1 probe on the cell types indicated. Cells were treated with the transcriptional inhibitor DRB or the histone deacetylase inhibitor TSA. For comparison, the results obtained for untreated cells are also included. The localization of FISH signals was determined by erosion analysis. The erosion analysis was performed and the corresponding diagrams were arranged as described in the legend of Fig. 4. In addition, on each panel it is indicated whether the gene analyzed was transcriptionally active (on) or inactive (off), as revealed by RT-PCR analysis. The level of CFTR transcription in Calu-3 cells was reduced to ∼50% after DRB treatment (Table I).
To rule out that general changes of the chromatin structure due to DRB treatment in Calu-3 cells led to the positional change of CFTR, we analyzed as a control the positioning of the β-globin locus in this cell type with and without DRB treatment. The erosion analysis revealed that the β-globin locus resided in the nuclear interior in Calu-3 cells and that its positioning was not significantly different before and after DRB treatment (Fig. S3; unpaired t test, P < 0.05). Together, the results revealed that treatment with the transcriptional inhibitor DRB only altered the positioning of CFTR in the active state in Calu-3 cells, where its transcription levels were reduced by DRB treatment, but not in the inactive state in SH-EP N14 and 293 cells. Also, the positioning of the unrelated β-globin locus was not affected. These findings strongly suggest that positioning of the active CFTR gene in the nuclear interior depends on its transcriptional activity.
The finding that CFTR associated with chromatin containing hyperacetylated histone H4 in Calu-3 cells (Fig. 2) suggested that also the histone acetylation levels might be involved in CFTR positioning. Therefore, we treated cells with the drug trichostatin A (TSA), which leads to histone hyperacetylation. In Calu-3 cells, where CFTR was embedded in the chromatin fraction containing hyperacetylated histone H4, no significant change in its positioning was observed after TSA treatment (Fig. 7; unpaired t test, P < 0.05). However, in 293 and SH-EP N14 cells, where CFTR was normally embedded within the hypoacetylated, perinuclear heterochromatin (Fig. 2 and Fig. 3), a dramatic change in CFTR positioning was observed. After TSA treatment, CFTR did not show perinuclear localization anymore, but occupied significantly more interior positions (Fig. 7; unpaired t test, P < 0.05). It should be noted that TSA treatment specifically affected only the positioning of CFTR when it was associated with hypoacetylated chromatin (293 and SH-EP N14 cells). Exactly in this situation CFTR positioning was not affected by DRB treatment. And vice versa, CFTR positioning was not affected by TSA treatment when CFTR was transcriptionally active and associated with hyperacetylated chromatin (Calu-3 cells), whereas this was the only situation when CFTR positioning was affected by DRB treatment. Also, these findings suggested that the positional changes induced by the different drugs were due to their specific effects on transcription and histone acetylation, and not due to general effects on chromatin structure.
Although the results obtained by TSA treatment suggest that the level of histone acetylation at the CFTR locus might provide positional information, additional careful analyses will be necessary to answer this question more clearly. Nevertheless, CFTR in its inactive state was driven from a repressive into an active environment by TSA treatment. Therefore, the question arose whether this might be accompanied by transcriptional activation of CFTR. RT-PCR analyses revealed that CFTR was not detectably transcribed in 293 and SH-EP N14 cells, neither before nor after TSA treatment. This result demonstrated that driving CFTR into a permissive environment was not sufficient for its transcriptional activation. Together, the results obtained by DRB and TSA treatment suggested that the nuclear positioning of CFTR was dependent on its transcriptional activity, whereas the transcriptional activity of CFTR was not primarily influenced by its nuclear localization.
Localization of CFTR with respect to α-satellite DNA
It has been described for a number of genes that they specifically associate with pericentromeric heterochromatin in their inactive states (Brown et al., 1997, 1999, 2001; Schübeler et al., 2000). This applied also to the β-globin locus, which was found to associate specifically with pericentromeric satellite DNA in T-lymphocytes (Fig. S4 a, available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1), in accordance with previous data (Brown et al., 2001). To investigate whether this also applied to CFTR, we performed dual-color FISH experiments with the CF1 probe and a probe specific for α-satellite DNA. The numbers of CF1 signals associated with α-satellite DNA were evaluated (Fig. S4 b and Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1). In all six cell types, more than 80% of the CF1 signals were not associated with α-satellite DNA. In addition, a relatively high degree of association (17%) was observed in Calu-3 cells, where CFTR is highly expressed (Table S1). These results did not reveal a correlation between transcriptional silencing of CFTR and its association with pericentromeric satellite DNA.
Positioning of GASZ, CFTR, and CORTBP2 with respect to chromosome 7 territories
Finally, we investigated the relationships between the expression patterns of GASZ, CFTR, and CORTBP2 and their positioning with respect to the chromosome 7 territory in primary T-lymphocytes, 293 cells, and Calu-3 cells (Fig. 8). Regarding the positioning, three different classes were defined: (1) localization of the gene-specific signal within; (2) at the periphery; or (3) outside of the chromosome 7 territory as defined by the FISH signal of the painting probe (Fig. 8 a). The localizations of the three genes (Fig. 8 b) did not show any correlation to their levels of transcription.
Figure 8.
Positioning of GASZ, CFTR, and CORTBP2 with respect to the chromosome 7 territory. T-lymphocytes, 293 cells, and Calu-3 cells were hybridized with the gene-specific probes (CF1 used for CFTR) and in addition with a painting probe specific for human chromosome 7. The localization of the gene-specific FISH signals was determined with regard to the chromosome 7 territory, as defined by the FISH signal of the painting probe. Gene-specific signals were assigned to three different categories: (1) localization within; (2) at the periphery; or (3) outside of the chromosome 7 territory. (a) Panels show examples (gene-specific signals, red; chromosome 7 territory, green; DAPI counterstain, blue; bars, 5 μm). The arrows in the left-hand panel point to CORTBP2-specific signals positioned within the territories, whereas the arrowhead points to a signal located at the periphery of the chromosome territory. This panel shows a Calu-3 nucleus containing three chromosome 7 territories. Both CORTBP2-specific signals (arrows) shown on the right-hand panel (T-lymphocyte nucleus) were classified as positioned outside of the corresponding chromosome territories. (b) The diagrams summarize the results obtained for the different probes and cell types indicated. Bars indicate the fractions of FISH signals (averages ± SD) located outside (left bars in the diagrams), at the periphery (bars in the middle), or within (right-hand bars) the chromosome 7 territories.
Discussion
We investigated nuclear positioning and transcriptional regulation of the three adjacent human genes GASZ, CFTR, and CORTBP2. Our results demonstrated that adjacent genes can associate simultaneously with distinct nuclear regions and chromatin fractions, in accordance with their individual transcriptional activity. These findings, showing that chromosomal subregions display highly flexible nuclear arrangements regulated at the level of individual genes, seem to be in contrast to previous data demonstrating that whole chromosomes or larger chromosomal subregions show a specific nuclear positioning (Ferreira et al., 1997; Croft et al., 1999; Sadoni et al., 1999; Volpi et al., 2000; Boyle et al., 2001; Mahy et al., 2002a,b; Williams et al., 2002). However, the position of a chromosomal subregion or of a whole chromosome reflects the sum of the positions of all individual genes and sequences comprised. Therefore, our data are in accordance with data showing that the bulk of a chromosome/chromosomal subregion containing many expressed genes localizes more to the interior than the bulk of a chromosome/chromosomal subregion comprising low numbers of genes and many nonexpressed genes (Ferreira et al., 1997; Croft et al., 1999; Sadoni et al., 1999; Boyle et al., 2001). Such data are also in accordance with the finding that larger chromosomal regions containing functionally related genes display a concerted positioning (Volpi et al., 2000; Williams et al., 2002). It should be noted that recent reports (Mahy et al., 2002a,b) suggesting rather region-specific than gene-specific positioning investigated nuclear positioning with respect to the chromosome territory. In agreement with these results, we did not find correlations between the transcriptional activity of individual genes and their positioning with respect to the chromosome territory.
Our findings with regard to endogenous loci are in agreement with the results of a previous work investigating transgenic sequences. It was demonstrated that a lac operator containing chromosome locus moved from the periphery to the nuclear interior after targeting of the transcription factor VP16 and transcriptional activation (Tumbar and Belmont, 2001). Interestingly, when a lac operator–containing construct was integrated in an extremely gene-rich region of human chromosome 1, the transgenic sequences constitutively resided in the nuclear interior and did not change their nuclear localization in conjunction with transcriptional activation (Janicki et al., 2004). These findings suggest that transcription units do not always localize in agreement with their transcriptional regulation, and that gene-rich regions might dominate the positioning of comprised inactive sequences. It will be most interesting to compare in future analyses the positioning of adjacent sequences from gene-poor and gene-rich regions in relation to their transcriptional regulation.
Our data suggested that rather than the positioning within the chromosome territory, the association with specific chromatin fractions and the nuclear periphery/lamina seems to be related to transcriptional regulation. When addressing associations with heterochromatin, our results revealed gene-specific associations with defined subfractions of heterochromatin. For example, the β-globin locus associated specifically with pericentric heterochromatin, but not with the nuclear periphery. This is in accordance with previous reports demonstrating for the human β-globin locus, as well as for other gene loci, specific associations with pericentric heterochromatin (Brown et al., 1997, 1999, 2001; Schübeler et al., 2000). In contrast, genes from the CFTR region associated specifically with perinuclear heterochromatin, but not with pericentric or perinucleolar heterochromatin. This suggests that different fractions of heterochromatin are not equivalent and play a different role in gene regulation. This view is supported by the findings that the chromatin structure of mammalian pericentric heterochromatin appears to differ from the structure of other fractions of heterochromatin (Maison et al., 2002), and that the transcription factor Ikaros, required for the successful development of lymphocytes, is specifically enriched in pericentric heterochromatin, with which lymphoid-specific genes associate in their inactive states (Brown et al., 1997). A recent report did show that targeting of HP1-proteins to a transgene in mouse cells leads to silencing and recruitment to HP-1–rich pericentric heterochromatin (Ayyanathan et al., 2003). Interestingly, repression and changes in chromatin structure remained highly localized, suggesting that also recruitment to pericentric heterochromatin did not affect closely adjacent gene loci (Ayyanathan et al., 2003), although this possibility was not experimentally tested.
It remains to be determined which role associations with different chromatin fractions and the nuclear periphery play in the transcriptional regulation of CFTR. Cystic fibrosis is one of the most important models for gene therapy. The correlations observed between spatial nuclear arrangements and transcriptional regulation indeed imply that nuclear architecture must add an important level of regulation. However, the results of the experiments involving TSA and DRB treatment do not support this view and suggest that nuclear architecture does not play a primary role in transcriptional regulation of CFTR. However, our data do not allow conclusions with regard to the cell types targeted during gene-therapy of CF, nor do they allow conclusions regarding long-term effects and the role of nuclear positioning in the stable maintenance of CFTR expression. Therefore, it will be an important task to find out more about the question of how the nuclear organization of CFTR affects its long-term regulation in the lung tissue.
Materials and methods
Cell culture and fixation
For details of cell culture see supplemental data. Cells were treated with 10 ng/ml TSA for 10 h and with 50 μg/ml DRB for 5 h. For the erosion analyses, 2D distance measurements, determining the orientation of loci in dual-color FISH experiments, and the analyses regarding the positioning with respect to chromosome 7 territories and α-satellite DNA, cells were fixed with methanol/acetic acid (3:1) after hypotonic treatment (70 mM KCl for 20 min). For 3D analyses, cells were fixed with formaldehyde (3.7% in PBS) for 10 min at RT.
DNA probes and probe labeling
As probes for FISH we used four different probes from the CFTR region (Fig. 1). In addition, probes specific for the β-globin locus and α-satellite DNA were used, as well as a painting probe for chromosome 7. For further details see supplemental data.
FISH
FISH was essentially performed as described previously (Zink et al., 1998). For further details see supplemental data.
Immuno-FISH
Immunostaining was performed with the antibody R232/8 specific for histone H4Ac8 as described previously (Sadoni et al., 1999) or with an antibody specific for LAP2β (BD Biosciences). Primary antibodies were detected with FITC- or Cy5-conjugated goat anti–rabbit or goat anti–mouse antibodies (Dianova). Immunostaining was performed with formaldehyde-fixed cells, which were fixed again with formaldehyde before in situ hybridization.
Replication labeling
Scratch replication labeling (Schermelleh et al., 2001) was performed with FITC-dUTP. Cells were fixed 30 min after scratch replication labeling with formaldehyde. After fixation, the usual FISH procedure was performed. BrdU pulse labeling was performed as described previously (Zink et al., 1998).
Imaging
Confocal imaging was performed as described previously (Eils et al., 1996). Epifluorescence imaging was performed with a microscope (Axiovert; Carl Zeiss MicroImaging, Inc.) equipped with a CCD camera. Further details are provided in the supplemental data.
Erosion analyses, calculations, distance measurements, and statistical analyses
Erosion analyses and the arrangements of images were performed with Adobe Photoshop (version 7.01). Calculations and the arrangement of diagrams were performed with Microsoft Excel 2000. For microscope calibration and shift correction, fluorescent microspheres (Molecular Probes, Inc.) were used. 2D distances were measured between the intensity centers of signals as determined by the MetaMorph software (version 4.6) and were performed on projections of the focal planes. 3D distances were measured between the intensity centers of signals in the different focal planes by taking advantage of the Image J software (version 1.3). For statistical analyses we took advantage of the StatView 5.0 software.
Quantitative RT-PCR
Human adult testis RNA has been obtained from the BioCat GmbH (Heidelberg, Germany). Other RNA was extracted from the various cell lines by the TRIzol method (Invitrogen Life Technologies), and oligo(dT)-primed reverse transcription reaction was performed with 5 μg total RNA using Superscript III reverse transcriptase (Invitrogen). Duplex PCR was performed to amplify each of the three target genes (CORTBP2, GASZ, and CFTR) simultaneously with α-actin as described previously (Amaral et al., 2004). For further details see supplemental data.
Online supplemental material
The supplemental materials describe details of the materials and methods used. The supplemental figures show a gallery of confocal sections of a replicational pulse-labeled Calu-3 nucleus after detection of CFTR and examples of relative quantitative PCR amplifications of CFTR and CORTBP2 in Calu-3 and HEK 293 cells, respectively. Furthermore, they show results of the erosion analysis with regard to the β-globin locus. Also, results of dual-color FISH experiments addressing the association of CFTR and the β-globin locus with α-satellite DNA are presented. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200404107/DC1.
Acknowledgments
We thank Bernadette Pöllinger, Andreas Laner, Susanne Christan, and Jeannette Koch, as well as Anabela Ramalho and Aleksandra Norek for contributions to the experimental work. We thank Bryan M. Turner (University of Birmingham, Birmingham, UK) for providing antibodies, and Mark Groudine and Agnes Telling (Fred Hutchinson Cancer Research Center, Seattle, WA) for providing the probe for the β-globin locus.
This work was supported by research grants from Fundaçáo para a Ciência e a Tecnologia (Portugal, POCTI/MGI/35737/2000) and from the Forschungsgemeinschaft Mukoviszidose and Vaincre la Mucoviszidose to D. Schindelhauer; by a grant from the European Commission, Fifth Framework Programme to J. Rosenecker; and by a grant from the Volkswagenstiftung to D. Zink.
S. Lang's present address is Max Planck Institute for Neurobiology, Am Klopferspitz 18, 82152 Martinsried, Germany.
K. Luther's present address is Max von Pettenkofer Institute, 80336 Munich, Germany.
Abbreviations used in this paper: 3D, three-dimensional; CFTR, cystic fibrosis transmembrane conductance regulator; CORTBP2, cortactin-binding protein 2; DRB, 5,6-dichlorobenzimidazole riboside; GASZ, germ cell–specific expression, presence of ANK, SAM, and basic leucine zipper domains; H4Ac8, H4 acetylated at lysine 8; LAP2β, lamina-associated polypeptide 2β; TSA, trichostatin A.
References
- Amaral, M.D., L.A. Clarke, A.S. Ramalho, S. Beck, F. Broackes-Carter, R. Rowntree, A. Harris, M. Tzetis, B. Steiner, J. Sanz, et al. 2004. Quantitative methods for the analysis of CFTR transcripts/splicing variants. J. Cystic Fibrosis. 3 Suppl 2:17–23. [DOI] [PubMed] [Google Scholar]
- Ayyanathan, K., M.S. Lechner, P. Bell, G.G. Maul, D.C. Schultz, Y. Yamada, K. Tanaka, K. Torigoe, and F.J. Rauscher III. 2003. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17:1855–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont, A.S., M.B. Braunfeld, J.W. Sedat, and D.A. Agard. 1989. Large-scale chromatin structural domains within mitotic and interphase chromosomes in vivo and in vitro. Chromosoma. 98:129–143. [DOI] [PubMed] [Google Scholar]
- Boyle, S., S. Gilchrist, J.M. Bridger, N.L. Mahy, J.A. Ellis, and W.A. Bickmore. 2001. The spatial organization of human chromosomes within cell nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., S.S. Guest, S.T. Smale, K. Hahm, M. Merkenschlager, and A.G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., J. Baxter, D. Graf, M. Merkenschlager, and A.G. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 3:207–217. [DOI] [PubMed] [Google Scholar]
- Brown, K.E., S. Amoils, J.M. Horn, V. Buckle, D.R. Higgs, M. Merkenschlager, and A.G. Fisher. 2001. Expression of α- and β-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 3:602–606. [DOI] [PubMed] [Google Scholar]
- Cheung, J., E. Petek, K. Nakabayashi, L.C. Tsui, J.B. Vincent, and S.W. Scherer. 2001. Identification of the human cortactin-binding protein-2 gene from the autism candidate region at 7q31. Genomics. 78:7–11. [DOI] [PubMed] [Google Scholar]
- Cmarko, D., P. Verschure, T.E. Martin, M.E. Dahmus, S. Krause, X.-D. Fu, R. van Driel, and S. Fakan. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell. 10:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292–301. [DOI] [PubMed] [Google Scholar]
- Cremer, T., A. Kurz, R. Zirbel, S. Dietzel, B. Rinke, E. Schroeck, M.R. Speicher, U. Mathieu, A. Jauch, P. Emmerich, et al. 1993. The role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb. Symp. Quant. Biol. 58:777–792. [DOI] [PubMed] [Google Scholar]
- Cremer, T., S. Dietzel, R. Eils, P. Lichter, and C. Cremer. 1995. Chromosome territories, nuclear matrix filaments and inter-chromatin channels: a topological view on nuclear architecture and function. In Kew Chromosome Conference IV. Royal Botanic Gardens, Kew. 63–81.
- Croft, J.A., J.M. Bridger, S. Boyle, P. Perry, P. Teague, and W.A. Bickmore. 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145:1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils, R., S. Dietzel, E. Bertin, E. Schroeck, M.R. Speicher, T. Ried, M. Robert-Nicoud, C. Cremer, and T. Cremer. 1996. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 135:1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt, J.F., M. Zepeda, J.A. Cohn, J.R. Yankaskas, and J.M. Wilson. 1994. Expression of the cystic fibrosis gene in adult human lung. J. Clin. Invest. 93:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan, S. 1994. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 4:86–90. [DOI] [PubMed] [Google Scholar]
- Ferreira, J., G. Paolella, C. Ramos, and A.I. Lamond. 1997. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J. Cell Biol. 139:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa, K., N. Pante, U. Aebi, and L. Gerace. 1995. Cloning of a cDNA for lamina-associated polypeptide 2 (LAP2) and identification of regions that specify targeting to the nuclear envelope. EMBO J. 14:1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki, S.M., T. Tsukamoto, S.E. Salghetti, W.P. Tansey, R. Sachidanandam, K.V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R.H. Singer, and D.L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell. 116:683–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak, S.T., J.A. Skok, K.L. Medina, R. Riblet, M.M. Le Beau, A.G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. [DOI] [PubMed] [Google Scholar]
- Li, M., J.D. McCann, C.M. Liedtke, A.C. Nairn, P. Greengard, and M.J. Welsh. 1988. Cyclic AMP-dependent protein kinase opens chloride channels in normal but not cystic fibrosis airway epithelium. Nature. 331:358–360. [DOI] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, and W.A. Bickmore. 2002. a. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy, N.L., P.E. Perry, S. Gilchrist, R.A. Baldock, and W.A. Bickmore. 2002. b. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, C., D. Bailly, A.H.F.M. Peters, J.-P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334. [DOI] [PubMed] [Google Scholar]
- Nili, E., G.S. Cojocaru, Y. Kalma, D. Ginsberg, N.G. Copeland, D.J. Gilbert, N.A. Jenkins, R. Berger, S. Shaklai, N. Amariglio, et al. 2001. Nuclear membrane protein LAP2β mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J. Cell Sci. 114:3297–3307. [DOI] [PubMed] [Google Scholar]
- Robinett, C.C., A. Straight, G. Li, C. Wilhelm, G. Sudlow, A. Murray, and A.S. Belmont. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoni, N., S. Langer, C. Fauth, G. Bernardi, T. Cremer, B.M. Turner, and D. Zink. 1999. Nuclear organization of mammalian genomes. Polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146:1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh, L., I. Solovei, D. Zink, and T. Cremer. 2001. Two-color fluorescence labeling of early and mid-to-late replicating chromatin in living cells. Chromosome Res. 9:77–80. [DOI] [PubMed] [Google Scholar]
- Schübeler, D., C. Francastel, D.M. Cimbora, A. Reik, D.I.K. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev. 14:940–950. [PMC free article] [PubMed] [Google Scholar]
- Skok, J.A., K.E. Brown, V. Azuara, M.L. Caparros, J. Baxter, K. Takacs, D. Dillon, D. Gray, R.P. Perry, M. Merkenschlager, and A.G. Fisher. 2001. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2:848–854. [DOI] [PubMed] [Google Scholar]
- Solovei, I., A. Cavallo, L. Schermelleh, F. Jaunin, C. Scasselati, D. Cmarko, C. Cremer, S. Fakan, and T. Cremer. 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization. Exp. Cell Res. 276:10–23. [DOI] [PubMed] [Google Scholar]
- Tumbar, T., and A.S. Belmont. 2001. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3:134–139. [DOI] [PubMed] [Google Scholar]
- Verschure, P.J., I. van der Kraan, E.M.M. Manders, and R. van Driel. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi, E.V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R.D. Campbell, M. Goldsworthy, S.H. Powis, J. Ragoussis, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576. [DOI] [PubMed] [Google Scholar]
- Williams, R.R.E., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163–175. [DOI] [PubMed] [Google Scholar]
- Yan, W., A. Rajkovic, M.M. Viveiros, K.H. Burns, J.J. Eppig, and M.M. Matzuk. 2002. Identification of Gasz, an evolutionary conserved gene expressed exclusively in germ cells and encoding a protein with four ankyrin repeats, a sterile-α motif, and a basic leucine zipper. Mol. Endocrinol. 16:1168–1184. [DOI] [PubMed] [Google Scholar]
- Zink, D., T. Cremer, R. Saffrich, R. Fischer, M.F. Trendelenburg, W. Ansorge, and E.H.K. Stelzer. 1998. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 102:241–251. [DOI] [PubMed] [Google Scholar]
- Zink, D., N. Sadoni, and E. Stelzer. 2003. Visualizing chromatin and chromosomes in living cells. Methods. 29:42–50. [DOI] [PubMed] [Google Scholar]