Abstract
Assembly at the mother–bud neck of a filamentous collar containing five septins (Cdc3, Cdc10, Cdc11, Cdc12, and Shs1) is necessary for proper morphogenesis and cytokinesis. We show that Cdc10 and Cdc12 possess GTPase activity and appropriate mutations in conserved nucleotide-binding residues abrogate GTP binding and/or hydrolysis in vitro. In vivo, mutants unable to bind GTP prevent septin collar formation, whereas mutants that block GTP hydrolysis do not. GTP binding-defective Cdc10 and Cdc12 form soluble heteromeric complexes with other septins both in yeast and in bacteria; yet, unlike wild-type, mutant complexes do not bind GTP and do not assemble into filaments in vitro. Absence of a p21-activated protein kinase (Cla4) perturbs septin collar formation. This defect is greatly exacerbated when combined with GTP binding-defective septins; conversely, the septin collar assembly defect of such mutants is suppressed efficiently by CLA4 overexpression. Cla4 interacts directly with and phosphorylates certain septins in vitro and in vivo. Thus, septin collar formation may correspond to septin filament assembly, and requires both GTP binding and Cla4-mediated phosphorylation of septins.
Keywords: Saccharomyces cerevisiae; guanine nucleotide; mutants; p21-activated protein kinase; filaments
Introduction
Septins are a protein family conserved from fungi to humans. Its first members were identified as cdc (cell division cycle) mutants in the yeast Saccharomyces cerevisiae that were defective in cytokinesis and displayed elongated buds (Hartwell, 1971). Septins play a role in cytokinesis in other organisms (Neufeld and Rubin, 1994; Kinoshita et al., 1997). Mounting evidence indicates septin functions in actin recruitment, secretion, and membrane deposition (Adam et al., 2000; Dent et al., 2002; Kinoshita et al., 2002).
During mitosis, S. cerevisiae expresses five septins: Cdc3, Cdc10, Cdc11, Cdc12, and Shs1/Sep7 (Gladfelter et al., 2001). Early in the cell cycle, septins assemble into a patch at the incipient bud site. As the cell cycle progresses, septins reorganize at the mother–bud neck into an hourglass-shaped collar of cytoplasmic filaments that subtend the plasma membrane (Byers and Goetsch, 1976; Frazier et al., 1998). The collar remains at the neck throughout the rest of the cycle, and splits into two rings at cytokinesis, which then disassemble. Various proteins colocalize with septins at specific stages of the cell cycle and, at the bud neck, associate with the daughter or mother (or both) sides of the collar, suggesting that septin filaments serve as a scaffold and spatial determinant. Among the proteins recruited to the daughter side is a protein-arginine methyltransferase, Hsl7, and two protein kinases, Hsl1 and Swe1 (Barral et al., 1999; Shulewitz et al., 1999; Longtine et al., 2000). Swe1 blocks mitotic entry by inhibiting B cyclin (Clb)–bound Cdc28 protein kinase. Swe1, in turn, is inactivated via concerted action of Hsl1 and Hsl7. Hsl1 and Hsl7 activity depends on their localization to properly assembled septins at the bud neck. If the collar is perturbed, Swe1 remains active, inhibits Cdc28–Clb complexes, and prevents passage through G2/M, providing a mechanism to monitor the status of septin assembly and to retard cell cycle progression if there is a defect. The molecular feature of septin collar organization deciphered by the Hsl1–Hsl7–Swe1-dependent checkpoint is not known.
The collar also forms a physical barrier between a mother cell and its bud, preventing diffusion of bud-specific proteins to the mother cell and maintaining actin-based cell polarity during bud growth (Barral et al., 2000; Takizawa et al., 2000). Collar splitting into two separate rings precedes cell division and may remove a constraint that prevents actomyosin ring contraction (Lippincott et al., 2001).
A small GTPase, Cdc42, is required for septin collar formation (Gladfelter et al., 2002). A known Cdc42 target, Cla4 (a p21-activated protein kinase [PAK]), has been implicated in collar assembly because cla4Δ mutants are elongated and septins localize predominantly to the bud tip, not to the bud neck (Cvrckova et al., 1995; Weiss et al., 2000). Two other protein kinases (Elm1 and Gin4) and Bni5 (a septin-binding protein) are also potential regulators because deletion of these genes results in aberrant septin organization (Longtine et al., 1998; Bouquin et al., 2000; Lee et al., 2002). How all these proteins contribute to septin dynamics remains unresolved.
Another unsettled question is the role of GTP binding to the guanine nucleotide-binding domain found in all septins. Isolated Drosophila septin complexes (340 kD) contained 2.6 times more GDP than GTP and displayed measurable, but very low, rates of guanine-nucleotide exchange and GTP hydrolysis (Field et al., 1996), as do similar complexes from yeast (Vrabioiu et al., 2004). Microinjection of nonhydrolyzable guanosine 5′-O-3′-thiotriphosphate (GTPγS) into cultured mouse cells prevented assembly of fibrils containing the septin Nedd5/Sept2 (Kinoshita et al., 1997), suggesting GTP hydrolysis might be important for septin assembly. One group reported that a single recombinant septin (Xenopus Sept2) polymerizes into filaments in vitro in a GTP-dependent manner (Mendoza et al., 2002), whereas two other groups showed that exogenously added guanine nucleotides had no effect on in vitro assembly of filaments from multi-septin complexes (Kinoshita et al., 2002; Sheffield et al., 2003).
As a means to resolve these somewhat contradictory findings, we used a genetic approach in yeast to assess the roles of guanine nucleotide binding and hydrolysis in septin assembly in vivo. To generate mutations that we could be assured actually perturbed these functions, we first examined which septins bind and hydrolyze GTP. Based on the phenotypes observed, we also explored the role of Cla4, and discovered that septins are direct and important in vivo targets of this protein kinase.
Results
Cdc10 and Cdc12 are active GTPases
Cdc3, Cdc10, and Cdc11 were purified to near homogeneity from bacterial cells using an NH2-terminal tandem-affinity tag (His6-GST). After detaching the tag by tobacco etch virus (TEV) protease digestion, and removal of protease and uncleaved fusion, soluble untagged septins were obtained (Fig. 1 A). Nearly homogenous preparations of soluble Cdc12-His6 were also produced. GTPase activity of these proteins was measured by release of free phosphate from γ[32P]GTP (Fig. 1 B). Only Cdc10 and Cdc12 displayed detectable GTPase activity above the background control (purified His6-GST).
Figure 1.
Purified Cdc10 and Cdc12 bind and hydrolyze GTP. (A) The indicated septin preparations (1–5 μg) were analyzed by SDS-PAGE and stained with Coomassie dye. (B) GTPase activity was measured by phosphate release from γ[32P]GTP for Cdc3 (⋄), Cdc10 (♦), Cdc11 (X) and Cdc12-His6 (•), and His6-GST (*). (C) GTPase activity of Cdc12-His6 (•), Cdc12ΔC-His6 (○), Cdc12(S43V)-His6 (▴), and Cdc12(T48N)-His6 (▪), as in B. (D) GTPase activity of Cdc12ΔC-His6 (○), Cdc12ΔC(S43V)-His6 (Δ), and Cdc12ΔC(T48N)-His6 (□), as in B. (E) [32P]GTPγS binding by His6-tagged Cdc12 (•), Cdc12(S43V) (▴), and Cdc12(T48N) (▪). (F) [32P]GTPγS binding by His6-tagged Cdc12ΔC (○), Cdc12ΔC(S43V) (Δ), and Cdc12ΔC(T48N) (□). For B–F, representative data are shown for experiments that were each repeated two to three (or more) times with different septin preparations and varying substrate or ligand concentrations.
Cdc10, the only septin lacking a COOH-terminal coiled-coil, reproducibly displayed more robust GTPase activity than Cdc12, suggesting that catalytic function of Cdc12 might be restrained by its COOH-terminal coiled-coil. Hence, we purified a truncated version, Cdc12(Δ339–407)-His6 (hereafter Cdc12ΔC; Fig. 1 A). Cdc12ΔC displayed a rate of GTP hydrolysis four- to fivefold higher than that of full-length Cdc12 and comparable to Cdc10 (Fig. 1 C), consistent with the idea that its COOH-terminal coiled-coil is a negative regulatory domain. However, Cdc11 lacking its coiled-coil, Cdc11(Δ357–416), did not display any GTPase activity above the nonspecific background (unpublished data). As expected for an authentic GTPase, chelation of Mg2+ with excess EDTA reduced Cdc12ΔC activity to the background level (unpublished data). The apparent Km of Cdc12ΔC for GTP was ∼0.6 μM (unpublished data).
To further confirm the intrinsic GTPase activity of Cdc12, we introduced mutations into this septin at positions analogous to those that prevent GTP binding and/or GTP hydrolysis by a canonical GTPase, Ras. Cdc12(S43V) alters a residue equivalent to that in H-Ras(G12V). H-Ras(G12V) binds GTP, but is deficient in GTP hydrolysis (McGrath et al., 1984). Cdc12(T48N) alters a residue equivalent to that in H-Ras(S17N), which is unable to bind any nucleotide (Feig, 1999). As expected, both mutations largely abrogated the GTPase activity of both full-length Cdc12 (Fig. 1 C) and Cdc12ΔC (Fig. 1 D). To verify that the phosphate release assay reflects conversion of GTP to GDP, we used α[32P]GTP as substrate and a TLC-based assay to examine product formation. Indeed, GDP was formed with time as the amount of GTP diminished; moreover, the S43V mutation drastically reduced the rate of product formation, to the same extent as was observed using the phosphate release assay (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1).
Mutation T48N prevents GTP binding to Cdc12
Binding of [35S]GTPγS to Cdc12, Cdc12ΔC, and the derived site-directed mutants was assessed as described in the Materials and methods. Both full-length Cdc12 and Cdc12ΔC formed stable complexes with GTPγS over time (Fig. 1, E and F). The S43V mutation did not prevent GTP binding or alter the kinetics of GTP binding to either Cdc12 or Cdc12ΔC (Fig. 1, E and F), even though it did prevent GTP hydrolysis (Fig. 1, C–E). In contrast, the T48N mutation abolished the ability of Cdc12 and Cdc12ΔC to bind GTP (Fig. 1, E and F). Thus, both mutations caused effects in vitro consonant with those of analogous mutations in other GTPases, and thus provided tools for discriminating the roles of GTP binding and GTP hydrolysis by Cdc12 in vivo.
GTP binding to Cdc12 is required for normal bud morphology and cytokinesis
Cdc12 is indispensable for viability (Cvrckova et al., 1995). To examine the effect of the cdc12(S43V) and cdc12(T48N) mutations, each was integrated at the CDC12 locus. At 30°C, cells expressing Cdc12(S43V) or cells expressing Cdc12(T48N) showed no obvious defect. At elevated temperature (37°C), the GTP binding-defective mutant, cdc12(T48N), displayed abnormalities in both morphology (Fig. 2, A and B) and cytokinesis (Fig. 2 A and Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1). The GTP hydrolysis-defective mutant, cdc12(S43V), showed no detectable aberration, even at 37°C (Fig. 2, A and B), and even when overexpressed from a multi-copy plasmid (unpublished data). At 37°C, expression of Cdc12(T48N) was lower than that of normal Cdc12 (Fig. S3), which might have accounted for its phenotype. However, even when overexpressed from a multi-copy (2 μm DNA) plasmid (Fig. S3), the same morphological and cytokinesis defects were observed at 37°C (Fig. 2 A) Presence of wild-type CDC12 on a CEN plasmid restored a normal phenotype to cdc12(T48N) cells (Fig. 2 A); thus, the defect is recessive.
Figure 2.

GTP binding-dependent functions of Cdc12 and Cdc10 overlap. (A) Wild-type (BY4741) transformed with empty vector (YCplac33), cdc12(S43V) (YMVB2), cdc12(T48N) (YMVB1), cdc12(T48N) (YMVB1) carrying an episomal vector overexpressing cdc12(T48N) (pMVB49), and cdc12(T48N) (YMVB3) carrying a CEN plasmid with CDC12 (pMVB39), were grown at 30 or 37°C for 16 h under selective conditions. Cells were stained with DAPI and examined by Nomarski optics (differential interference contrast) or fluorescence microscopy (DAPI). Bar, 5 μm. (B) Wild-type (YMVB25), cdc10(S46N) (YMVB6), cdc12(T48N) (YMVB24), cdc10(S46N) cdc12(T48N) (YMVB8), and cdc10(S41V) cdc12(S43V) (YMVB60) cells were grown in YPD to mid-exponential phase at 26, 30, or 37°C, and morphology of the cells was examined by bright-field microscopy. Bar, 5 μm.
GTP binding-dependent functions of Cdc10 and Cdc12 overlap
The fact that Cdc12(T48N) lacked a phenotype at 30°C raised the possibility that GTP binding to other septins might partially mask its effects. The septin with the greatest homology to Cdc12 is Cdc10 (40% identity). Moreover, Cdc10 was the only other active GTPase in vitro (Fig. 1 B). To test for functional overlap, the equivalent mutation to prevent GTP binding, cdc10(S46N), was constructed and integrated at the CDC10 locus in a cdc10Δ mutant. As observed for cdc12(T48N), cdc10(S46N) cells had no phenotype at 30°C, but displayed aberrant bud morphology (Fig. 2 B) and cytokinesis defects (Fig. S2) at 37°C. A cdc10(S46N) cdc12(T48N) double mutant showed dramatic morphology defects even at 30°C, and was inviable at 37°C (see also Fig. 6 A), whereas both single mutants to grow at 37°C. This additive phenotype indicates redundancy in the function of GTP binding to Cdc10 and Cdc12, and indicates that lack of GTP binding causes a defect in septin function even at 30°C, but that defect is exacerbated at higher temperature. In marked contrast, a double GTP hydrolysis-defective mutant, cdc10(S41V) cdc12(S43V), did not display any aberration at any temperature tested (Fig. 2 B).
Figure 6.
Synergistic roles for Cla4 phosphorylation and GTP binding in septin collar assembly. (A) A cla4Δ mutant (YMVB12), cdc12(T48N) (YMVB3), cla4Δ cdc12(T48N) (YMVB14), cdc12(T48N) cdc10(S46N) (YMVB50), and cla4Δ cdc12(T48N) cdc10(S46N) (YMVB51) were grown to mid-exponential phase on YPD at 26, 30, or 37°C, and examined by bright-field microscopy. (B) The cdc12(T48N) cdc10(S46N) (YMVB50) double mutant carrying either empty vector (YCplac33) or CEN vectors expressing CLA4 (pMVB113), SKM1 (YCpUG-Myc-SKM1), STE20 (pCJ160), or kinase-dead CLA4(K495A) (pMVB112) under control of the GAL1-promoter were grown on SC-Ura+Raf at 26°C overnight, transferred to SC-Ura+Gal for 3 h, shifted to 37°C, and examined as in A at the indicated times. Bars, 5 μm.
Formation of heteromeric septin complexes does not require GTP binding
An inherent property of septins is the ability to form heteromeric complexes with other septins (Frazier et al., 1998; Kinoshita et al., 2002; Sheffield et al., 2003). We used three independent approaches, all of which showed that GTP binding to Cdc12 is not necessary for its association with other septins. First, when immunoprecipitated with anti-Cdc12ΔC antibody, Cdc12 and Cdc12(T48N) coimmunoprecipitated equivalent amounts of Cdc3-GFP, Cdc10-GFP, and Cdc11-GFP (Fig. 3 A), even from extracts of yeast cultures that had been shifted to a temperature (37°C) at which the cdc12(T48N) mutant cells display a pronounced phenotype. Second, purified Cdc12 and Cdc12(T48N) interacted equally well with the three other septins (Cdc3, Cdc10, and Cdc11), with Cdc12 itself, and with the septin-binding fragment of Hsl1 (Barral et al., 1999; Shulewitz et al., 1999), which were all purified from bacteria as GST fusions and immobilized on glutathione-agarose beads, but did not bind to immobilized GST (Fig. 3 B). Incubation at different temperatures (4, 25, or 37°C), or preincubation with 0.1 mM GDP, GTP, or GTPγS, did not affect the affinity of Cdc12 or Cdc12(T48N) for any of the other septins, itself, or Hsl1(ΔN) (unpublished data). Thus, GTP binding to Cdc12 is not required for its ability to associate with other septin partners and does not require any other yeast proteins to do so. In contrast, Cdc12ΔC showed greatly reduced binding to GST-Cdc3 (Fig. 3 B), indicating that its coiled-coil contributes to this interaction.
Figure 3.
Septin–septin interactions do not require GTP binding. (A) Wild-type (BY4741) or cdc12(T48N) cells (YMVB3) transformed with CEN plasmids containing Cdc3-GFP, Cdc11-GFP (pSB5), or Cdc10-GFP (pLA-10) were grown to mid-exponential phase at 30°C and shifted to 37°C for 8 h. Extracts were subjected to immunoprecipitation with anti-Cdc12ΔC antibody. The inputs and resulting immune complexes were solubilized in SDS-PAGE sample buffer and analyzed by immunoblotting. (B) Cdc12-His6 and Cdc12(T48N)-His6 (0.1 μM) were incubated with 0.1 mM GTP for 1 h, and their ability to bind the indicated proteins (immobilized as GST fusions) was assessed. Input represents 20% of the total amount of Cdc12-His6 and Cdc12(T48N)-His6 initially added in each binding reaction. (C) Septin complexes were purified on Ni2+-saturated NTA-agarose from E. coli cells coexpressing His6-Cdc12 and the other septins indicated. Eluates were resolved on SDS-PAGE, stained with Coomassie blue. (D) GTP binding to the His6Cdc12–Cdc3–Cdc10–Cdc11 complex (•), the His6Cdc12(T48N)–Cdc3–Cdc10(S46N)–Cdc11 complex (▪), and His6-GST (negative control; ▴), immobilized on Ni2+-NTA beads. Values represent the average (error bars indicate the range) for the results of a typical experiment, where each measurement was performed in duplicate.
Third, to further confirm that GTP binding is not required for formation of multi-septin complexes, we coexpressed His6-Cdc12 with one, two, or three other septins (Cdc3, Cdc3 + Cdc10, and Cdc3 + Cdc10 + Cdc11) in Escherichia coli at 37°C using two compatible, bicistronic vectors. His6-tagged Cdc12 and any associated proteins then were purified on Ni2+-nitrilotriacetate (NTA) beads. As judged by Coomassie staining, stoichiometric complexes of His6-Cdc12 with the other coexpressed septins were observed in every case (Fig. 3 C). Inclusion of 0.1 mM GDP, GTP, or GTPγS in the buffers during purification did not affect the stoichiometry or yield of these complexes (unpublished data). When His6-Cdc12(T48N) and Cdc10(S46N) were used instead of their wild-type counterparts, equivalent complexes were observed in every case (Fig. 3 C). Examination of nucleotide binding in vitro showed that eliminating GTP binding to Cdc10 and Cdc12 was sufficient to abrogate GTP binding to the heterotetrameric complex (Fig. 3 D). Thus, even in the context of a heterotetrameric complex, Cdc10 and Cdc12 seem to be the only septins capable of GTP binding.
GTP binding is required for septin filament assembly in vitro
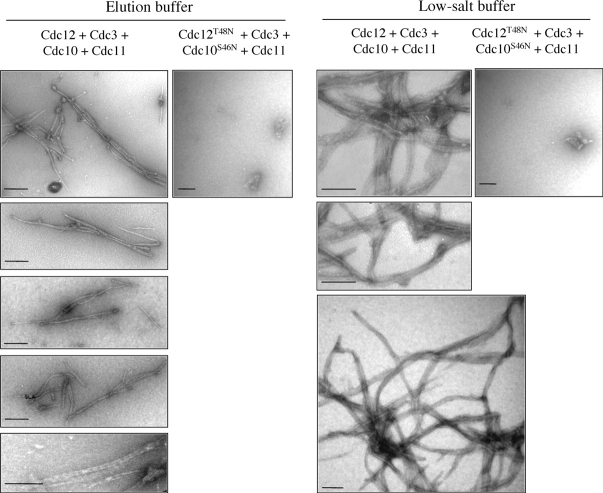
Dialysis into low salt buffer (50 mM KCl) promotes assembly of multi-septin complexes into filaments (Frazier et al., 1998; Kinoshita et al., 2002). We used two different assays to examine filament formation by our preparations of recombinant heterotetrameric complexes before and after dialysis into low salt. First, purified heterotetrameric complexes were applied in high salt elution buffer or after dialysis into low salt to polylysine-coated glass slides, stained with anti-Cdc12ΔC antibody and visualized using TRITC-conjugated secondary antibody. Wild-type complexes displayed prominent filament-like structures even in elution buffer, and the quantity, size, and brightness of these structures dramatically increased upon dialysis into low salt buffer (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1). No such structures formed when Cdc10 or Cdc11 was absent, such as in His6Cdc12–Cdc3, His6Cdc12–Cdc3–Cdc10, and His6Cdc12–Cdc3–Cdc11 complexes (unpublished data), consistent with the reported inability of septin complexes lacking either Cdc10 or Cdc11 isolated from yeast cells to form filaments in vitro (Frazier et al., 1998). By this method, His6-Cdc12(T48N)–Cdc3–Cdc10(S46N)–Cdc11 heterotetramers failed to form filament-like structures, either before or after dialysis (Fig. S4). Heterotetrameric complexes containing just Cdc12(T48N), but wild-type Cdc3, Cdc10, and Cdc11, did form filament-like forms, but much shorter in length than those produced by wholly wild-type heterotetrameric complexes (unpublished data).
These conclusions were confirmed at the ultrastructural level by EM. In elution buffer, wild-type heterotetramers contained filaments; individual filaments were ∼10 nm wide and 200 nm–2 μm in length. The filaments were often paired or formed hairpins, and sometimes had lateral striations (Fig. 4). The dimensions and appearance of the filaments reconstituted from recombinant proteins are quite similar to those reported for filaments prepared from septin complexes isolated from yeast (Frazier et al., 1998). Our results show that the above-mentioned characteristics are intrinsic to the septins and do not require any other yeast protein. After dialysis into low salt buffer, wild-type heterotetramers assembled into very long paired filaments, many with numerous apparent branches (Fig. 4). His6Cdc12 (T48N)–Cdc3–Cdc10(S46N)–Cdc11 heterotetramers never formed any filaments observable by EM under any condition examined (Fig. 4). These findings suggest that GTP binding to Cdc10 and Cdc12 is necessary to induce (or stabilize) a conformation of the heterotetrameric complex that supports its ability to self-associate into filaments.
Figure 4.
Septin filament assembly in vitro requires GTP binding. Filament formation of the indicated complexes (see Fig. 3 C) in elution buffer or after dialysis against low salt buffer was visualized by negative-stain transmission EM. Bars, 200 nm.
GTP binding to Cdc10 and Cdc12 is required for septin collar assembly
To determine what aspect of septin organization fails in vivo in the absence of GTP binding, Cdc12-GFP and Cdc12(T48N)-GFP were expressed in either wild-type or cdc12(T48N) cdc10(S46N) cells. At 26°C, both the normal and mutant fusions localized properly to the bud neck in either normal or mutant cells. After a shift to 37°C, localization of both fusions was not perturbed in the normal cells. However, by 4 h after shift to 37°C, Cdc12(T48N)-GFP in the cdc12(T48N) cdc10(S46N) strain (or GFP-tagged versions of any of the other three normal septins) was disorganized or absent altogether from the bud neck in ≥95% of the cells, even though the mutant protein was stably expressed, as judged by immunoblotting (unpublished data). In contrast, localization of septins in a GTP hydrolysis-defective double mutant, cdc10(S41V) cdc12(S43V), at 37°C was indistinguishable from the wild type (unpublished data). One explanation for absence of proper septin organization at the neck at 37°C in the GTP binding-defective septin mutants is that lack of GTP binding to Cdc10 and Cdc12 makes the septin collar unstable at that temperature. Alternatively, lack of GTP binding could prevent efficient assembly of the septin collar at elevated temperature.
To distinguish these possibilities, we examined septin collar assembly in synchronized cells. Cells were arrested in G1 by treatment with α-factor at 26°C and released from the block at 37°C (Fig. 5 A). We compared Cdc12-GFP in wild-type cells to Cdc12(T48N)-GFP in cdc12(T48N) cdc10(S46N) cells. By 20 min after release, both Cdc12 and Cdc12(T48N) assembled into a patch at the incipient bud site. In wild-type cells, by 40 min after release, Cdc12 formed into a circular structure around the neck of the emerging bud and, by 60 min had formed a prominent collar at the neck. By 80 min after release, the collar split into two rings and the cells underwent cytokinesis. In the mutant cells, Cdc12(T48N) remained as a patch at the bud tip throughout the time course of the experiment, even though the bud elongated and the cell continued to grow during this period (Fig. 5 A). In the absence of the collar, the mutant cells failed to undergo cytokinesis. An otherwise identical experiment was performed in cdc12(T48N) cdc10(S46N) cells expressing Cdc3-GFP with the same result (unpublished data). Thus, reorganization of the patch into the collar is dependent on GTP binding, whereas recruitment of septins to the presumptive bud site is not. Consistent with this view, we observed in mutant cells that, whereas the old patch of Cdc12(T48N) persisted at the previous bud site, new Cdc12(T48N) fluorescence appeared at the new presumptive bud site by 80 min after release (Fig. 5 A).
Figure 5.
GTP binding is necessary for septin collar assembly. (A) Wild-type cells (BY4741) carrying CDC12-GFP (pLP29), and cdc10(S46N) cdc12(T48N) (YMVB8) expressing cdc12(T48N)-GFP (pMVB91) were grown to mid-exponential phase on SCD(-His) at 26°C, synchronized with α-factor, released and shifted to 37°C, and samples were taken at the indicated times. (B) The same strains as in A were grown to mid-exponential phase on SCD(-His) at 26°C, synchronized with hydroxyurea, released and shifted to 37°C, and samples were taken at the indicated times. BF, bright-field microscopy; GFP, fluorescence microscopy. Bar, 5 μm.
Shifting cdc12(T48N) cdc10(S46N) cells to 37°C upon release from G1 arrest could bias the block observed to the first point in the cell cycle where GTP binding is required for septin function, but might not reveal other stages where GTP binding is needed. For this reason, the same strains were arrested in S phase at 26°C by treatment with hydroxyurea and released from the block at 37°C (Fig. 5 B). At 26°C, normal septin collars assembled in wild-type cells expressing Cdc12-GFP and in mutant cells expressing Cdc12(T48N) during the S phase block (0-min time point). Upon release at 37°C, both strains proceeded normally through the cell cycle (20-min time point) and underwent cytokinesis (40-min time point), indicating that GTP binding is not required for the stability or function of the collar once formed. In the next cycle, the wild-type cells assembled new patches at the presumptive bud site (60 min) and reorganized them into new collars in budded cells (80 min). In the ensuing cycle, the mutant cells also assembled new septin patches at the bud site (60 min). However, these patches persisted at the bud tip throughout the time course of the experiment (80 min), and were never able to form a collar at the neck of the emerging bud (Fig. 5 B). Thus, GTP binding to septins is critical for their reorganization from the patch into the collar (rather than in maintaining stability of the collar once assembled).
Cla4 protein kinase promotes GTP-dependent septin collar assembly
We were struck by the fact that cla4 mutants accumulate septins at the bud tip (Cvrckova et al., 1995; Weiss et al., 2000; Schmidt et al., 2003), similar to the phenotype of septins defective in GTP binding. If Cla4 action, like GTP binding, contributes to septin reorganization necessary for collar assembly, loss of Cla4 function should aggravate the phenotype of septin mutants defective in GTP binding. Indeed, unlike cla4Δ or cdc12(T48N) single mutants, which grow at all temperatures tested (26, 30, and 37°C) and only have a mild phenotype at 37°C, a cla4Δ cdc12(T48N) double mutant has dramatically aberrant morphology at 30°C, even more striking than cdc12(T48N) cdc10(S46N) cells, and is inviable at 37°C (Fig. 6 A). Moreover, a cla4Δ cdc12(T48N) cdc10-(S46N) triple mutant has pronounced morphological defects even at 26°C, grows in a remarkably hyphal-like fashion at 30°C, and is inviable at 37°C (Fig. 6 A).
If Cla4 and septins function in the same process, then overproduction of Cla4 might rescue the defects caused by GTP binding-deficient septins. Indeed, overexpression of CLA4 driven by the GAL promoter on a CEN plasmid largely suppressed the temperature-induced morphological aberrations and cytokinesis defects of cdc12(T48N) cdc10- (S46N) cells (Fig. 6 B). This effect required the catalytic activity of Cla4 because overexpression of a kinase-dead allele, cla4(K594A) (Tjandra et al., 1998), did not rescue the phenotype of cdc12(T48N) cdc10(S46N) cells. This effect was highly specific to Cla4 because overexpression to the same extent of each of the two other S. cerevisiae PAKs, Ste20 and Skm1 (as judged by immunoblotting [unpublished data] of NH2-terminally Myc epitope-tagged proteins), was unable to ameliorate the defects of cdc12(T48N) cdc10(S46N) cells (Fig. 6 B).
We noted that prolonged overexpression of CLA4 is detrimental to cell growth (but only well after 8 h of Gal induction). To verify that CLA4 overexpression suppresses the effects of the GTP binding-defective septin mutations by restoring septin collar assembly at bud emergence (rather than appearing to do so by simply inhibiting growth), three strains were arrested with α-factor on Gal-containing medium for 3 h at 26°C and released into Gal-containing medium at 37°C (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1). These were cdc12(T48N) cdc10(S46N) cells expressing Cdc12(T48N)-GFP, as before; cla4Δ cells expressing Cdc12-GFP; and cdc12(T48N) cdc10(S46N) cells expressing Cdc12(T48N)-GFP, but also overexpressing CLA4. As expected, the GTP binding-defective septin assembled into a patch at the bud tip, but failed to form a collar. Likewise, in cells lacking Cla4, the bulk of Cdc12-GFP remained at the bud tips, as observed previously (Weiss et al., 2000). By contrast, in the cdc12(T48N) cdc10(S46N) mutant overexpressing CLA4, the patch formed, the cells proceeded through the cell cycle, and then the collar assembled efficiently (Fig. S5). Additional evidence corroborated that rescue of collar formation is specific to Cla4; for example, overexpression of two other putative septin regulators, Gin4 protein kinase (Longtine et al., 1998) or the bud neck-localized protein, Bni5 (Lee et al., 2002), did not suppress the septin collar assembly defect of the GTP binding-defective septin mutants (unpublished data).
Cla4 physically associates with and directly phosphorylates septins
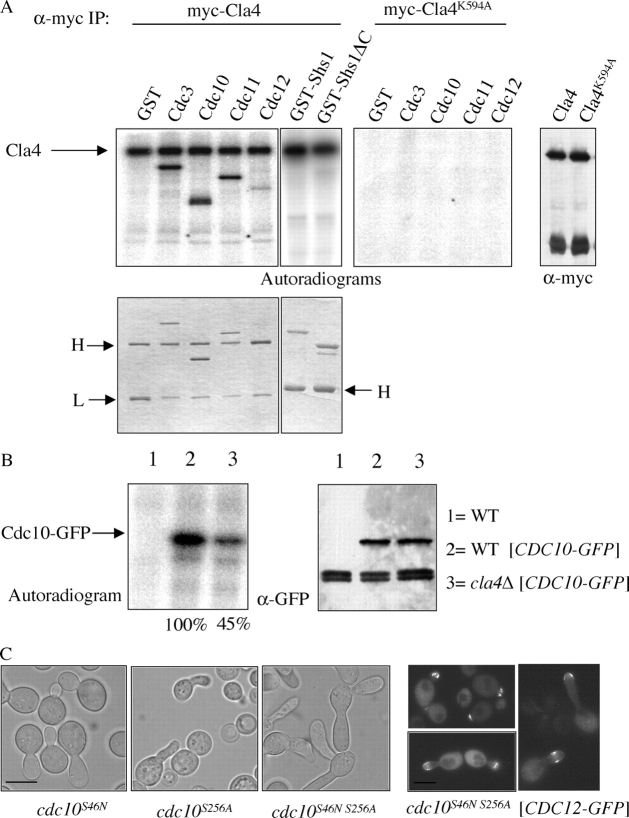
Our finding that catalytically inactive Cla4 did not suppress the phenotype of the GTP binding-defective septins (Fig. 6 B) suggested that septins might be substrates of Cla4, a possibility not heretofore explored. To test this idea, NH2-terminally Myc-tagged versions of Cla4 and Cla4(K594A) were immunopurified from yeast extracts and incubated with γ[32P]ATP and His6-tagged recombinant septins. Cla4 recovered from yeast showed potent autophosphorylation that was abrogated by the K594A mutation (Fig. 7 A). Likewise, Cla4, but not Cla4(K594A), efficiently phosphorylated Cdc3, Cdc10, Cdc11, and to a much lesser extent, Cdc12. Cla4 did not phosphorylate GST (Fig. 7 A), GFP (unpublished data), or even another yeast septin (either full-length [GST-Shs1] or COOH-terminally-truncated [GST-Shs1ΔC)]; Fig. 7 A). Cdc3, Cdc10, Cdc11, or Cdc12 were not phosphorylated by immunoprecipitated HA-tagged Elm1 or Gin4, although both kinases were active, as judged by their autophosphorylation (unpublished data).
Figure 7.
Cla4 phosphorylates septins. (A) NH2-terminally myc-tagged Cla4 (pMVB113) or Cla4(K495A) (pMVB112) were expressed in cla4Δ cells (YMVB12) and recovered by immunoprecipitation, and equal amounts, as verified by immunoblotting (outer right panel), were used in immune complex kinase assays, with the indicated substrates. Cdc12-His6 was present, but not resolved from the heavy (H) and light (L) chains of the anti-Myc mAb. The gel containing GST-Shs1 and GST-Shs1ΔC was run longer to ensure separation of GST-Shs1 from Cla4. (B) Wild-type cells carrying empty vector (YCplac33) or YCp-CDC10-GFP, and an isogenic cla4Δ mutant (YMVB12) carrying YCp-CDC10-GFP, were labeled with [32P]H3PO4, lysed, and the resulting extracts were subjected to immunoprecipitation with anti-GFP antibody. The precipitates were resolved by SDS-PAGE and incorporation was quantified using a PhosphorImager. Equal recovery of Cdc10-GFP was confirmed by immunoblotting (right). (C) The following strains, cdc10(S46N) (YMVB6), cdc10(S256A) (YMVB53), cdc10(S46N S256A) (YMVB54), and cdc10(S46N S256A) expressing CDC12-GFP, were grown at 26°C to mid-exponential phase, shifted to 37°C for 4 h, and viewed by differential interference contrast (left) or fluorescence microscopy (right). Bar, 5 μm.
To verify that Cla4 directly phosphorylates septins in vitro, GST-Cla4 and GST-Cla4(K594A) were expressed and purified from E. coli and incubated with His6-tagged septins. Again, both Cdc3 and Cdc10 served as phosphoacceptors for Cla4 and were not detectably phosphorylated by Cla4(K594A) (Fig. S6 A, available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1). However, Cdc11 (Fig. S6 A) and Cdc12 (unpublished data) did not serve as substrates for recombinant Cla4. Moreover, the isolated Cla4 kinase domain, GST-Cla4(524–842) (hereafter called Cla4ΔN), did not phosphorylate septins at all, although it was very active, as judged by its robust autophosphorylation (Fig. S6 A) and by its ability to phosphorylate a nonspecific substrate, myelin basic protein (unpublished data).
These observations suggested that a region in Cla4 upstream of its COOH-terminal kinase domain is required for its ability to recognize and efficiently phosphorylate Cdc3 and Cdc10. In agreement with the presence of a docking site(s) for Cdc3 and Cdc10 in the NH2-terminal regulatory region of Cla4, we found that full-length Cla4 interacts very tightly with Cdc10 and Cdc3 in an in vitro pull-down assay (and only weakly, if at all with Cdc11 or Cdc12), whereas Cla4ΔN did not associate with either Cdc3 or Cdc10 above the nonspecific background level (Fig. S6 B). The fact that recombinant Cla4 interacts poorly, if at all, with Cdc11 and Cdc12 provides a straightforward explanation for why these septins were not in vitro substrates for the bacterially expressed enzyme. Recruitment of Cdc11 and Cdc12 via their binding to Cla4-bound Cdc3 and Cdc10 present in Cla4 recovered from yeast likely explains phosphorylation of these septins in the immune complex kinase assay (Fig. 7 A).
To confirm that septins are phosphorylated in vivo in a Cla4-dependent manner, we examined Cdc10, the best substrate for Cla4 in vitro. We expressed Cdc10-GFP in wild-type cells or an isogenic cla4Δ mutant, labeled the cells with [32P]H3PO4, and recovered Cdc10-GFP by immunoprecipitation with anti-GFP mAb. In several independent experiments, absence of Cla4 reduced incorporation of radioactivity into Cdc10-GFP to 50% or lower than that seen in control cells (Fig. 7 B). Thus, Cdc10 is a phosphoprotein in vivo and Cla4 is responsible for at least half of its endogenous phosphorylation. Residual phosphorylation on Cdc10 in a cla4Δ mutant might be due to one or both of the other yeast PAKs, Ste20 and Skm1. Ste20 can contribute to septin localization (Cvrckova et al., 1995) and overproduction of Skm1 causes morphological aberrations (Martin et al., 1997). However, Ste20 and Skm1 phosphorylate Cdc10 in immune complex kinase assays, but much less efficiently than Cla4 (unpublished data). We mapped two primary sites for Cla4 in Cdc10 (Ser256 and Ser312) both in vitro and in vivo (see online supplemental Materials and methods). Preventing phosphorylation of the major site by a cdc10(S256A) mutation results in an elongated bud phenotype at 37°C (Fig. 7 C), which is not significantly worse in a cdc10(S256A S312A) double mutant (unpublished data). Strikingly, preventing both GTP binding to Cdc10 and phosphorylation at Ser256 in the double mutant cdc10-(S46N S256A) synergistically caused a severe morphological defect at 37°C (Fig. 7 C). Moreover, in these cells, Cdc12-GFP localized exclusively to the tips and lateral aspects of buds and did not form a collar at the neck (Fig. 7 C). First, these data confirm the physiological importance of Cla4 phosphorylation of Cdc10 in vivo, and second, demonstrate that Cla4-mediated phosphorylation of septins acts in concert with GTP binding to regulate the septin reorganization necessary for transition from the patch to the collar.
Discussion
Cdc10 and Cdc12 bind and hydrolyze GTP
To assess GTPase activity and to avoid contamination by other septins or septin-associated proteins, we purified each of four yeast septins from E. coli. Cdc10 and Cdc12 had GTPase activity in vitro, whereas Cdc3 and Cdc11 did not. Lack of activity in Cdc3 and Cdc11 could reflect presence of a bound but nonexchangeable GTP, or the need for a yeast-specific cofactor or modification. We found that Cdc10 and Cdc12 are solely responsible for GTP binding even by heterotetrameric complexes containing Cdc3 and Cdc11. Hence, we favor the view that only a subset of septins (Cdc10 and Cdc12) are intrinsically active GTPases. In hindsight, this principle may be more general. In a multi-septin complex (Pnut, Sep1, and Sep2) isolated from Drosophila embryos, Sep2 incorporated much less GTP than either Pnut or Sep1, although GTP exchange into the complex was very slow (∼0.0001 min−1; Field et al., 1996). Similarly, two groups found that mammalian Sept7 and Sept2 showed measurable GTP binding, whereas Sept6 did not (Kinoshita et al., 1997; Sheffield et al., 2003). Likewise, and in striking confirmation of our observations, in a septin complex purified from yeast, GTP exchange was slow, but occurred primarily on Cdc10 and Cdc12 (Vrabioiu et al., 2004). Our observation that mutating the GTP-binding sites in Cdc10 and Cdc12 abrogates the ability of septin heterotetramers to form filaments in vitro, and prevents formation of the filamentous septin collar in vivo, suggests that nucleotide binding to these two septins alone has a critical role in septin filament assembly.
Turnover numbers for Cdc10 (0.001 min−1) and Cdc12 (full length, 0.0005 min−1; Cdc12ΔC, 0.002 min−1) are an order of magnitude lower than H-Ras measured under similar conditions (0.028 min−1) in the absence of Ras-GTPase-activating protein (Krengel et al., 1990). In H-Ras, Glu61 orients an H2O molecule for nucleophilic attack on the γ phosphate of GTP (Scheffzek et al., 1998). Four of the mitotic septins have Phe (and Shs1 has Ile) at the equivalent position. Thus, low intrinsic GTPase activity is not unexpected. Indeed, an E61L mutation in H-Ras reduces its turnover number to 0.0013 min−1 (Krengel et al., 1990).
GTP binding is necessary for septin collar assembly during bud emergence
In vitro, both S43V and T48N reduced GTPase activity of Cdc12 to background levels. However, unlike S43V, the T48N mutation abrogated GTP binding. In vivo the cdc12(T48N) allele caused temperature-sensitive morphology and cytokinesis defects, whereas the cdc12(S43V) allele did not have a phenotype under any condition or in any genetic background tested. Cdc11(G29A G32A G34A) with a triple mutation in its P-loop, a motif necessary for GTP binding in other G-proteins, has a mild temperature-dependent bud morphology and cytokinesis phenotype (Casamayor and Snyder, 2003); however, to examine GTP binding, epitope-tagged Cdc11 was immunoprecipitated from yeast cells and hence, the contribution of associated septins to the GTP binding observed could not be ruled out. In our hands, bacterially expressed, highly purified, soluble Cdc11 does not bind GTP (unpublished data). In the same work, another Cdc11 mutation (N40E) situated just downstream of the P-loop caused a defect in self-association, as assessed by the yeast two-hybrid method (Cdc11(G29A G32A G34A) was not examined), which was interpreted to mean that GTP binding may regulate septin–septin interaction (yet GTP binding to Cdc11(N40E) was not examined). Our findings also directly contradict this idea because GTP binding-defective Cdc10 and Cdc12 are incorporated stoichiometrically into heterotetrameric septin complexes.
Using synchronized cells, we found that GTP binding to Cdc10 and Cdc12 is critical for assembly of the septin collar at the bud neck. In contrast, GTP binding was dispensable for recruitment of septins to the bud site (patch formation), for stability of the collar once formed, and for disassembly of the collar at cytokinesis. Our findings are the first demonstration that GTP binding has such a defined role in septin organization in any in vivo system. Our observations (Fig. 8) suggest that the filaments that comprise the septin collar are formed by GTP-dependent reorganization of preexisting septin complexes that can form independently of GTP binding. Indeed, we found that septin–septin interactions and heteromeric septin complex formation are unaffected by nucleotide binding, whereas assembly of those complexes into filaments in vitro is prevented when the GTP-binding sites of Cdc10 and Cdc12 are mutated. It was reported that Xenopus Sept2 binds and hydrolyzes GTP in vitro, and that binding of either GTP or GTPγS is sufficient to induce formation of homopolymeric filaments in vitro, indicating that GTP binding (rather than GTP turnover) promotes septin polymerization (Mendoza et al., 2002), consistent with our results. However, presence of excess GDP or GTP did not affect the efficiency of filament formation by our recombinant heterotetrameric septin complexes (unpublished data). In our preparations, we assume that complexes containing wild-type Cdc10 and Cdc12, but not those containing Cdc10(S46N) and Cdc12(T48N), bind sufficient nucleotide during their expression and purification to enable filament assembly. To deplete GTP from reconstituted septin heterotetramers and show that readdition of GTP can induce filament assembly will require identification of conditions or factors that enhance nucleotide exchange and/or hydrolysis in those complexes.
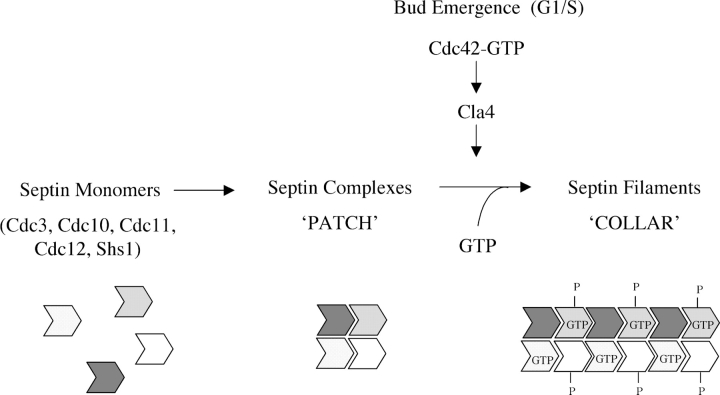
Figure 8.
GTP binding and Cla4-mediated phosphorylation in filament assembly and septin collar formation. The diagram summarizes our results (see Discussion), but is not meant to imply when GTP binding occurs (during nascent synthesis and folding of each individual septin, or at a later time) or a regulatory role for GTP-for-GDP exchange in triggering filament assembly.
The overlapping function of Cdc10 and Cdc12 revealed in our work suggests that GTP binding to either septin is sufficient for reasonably efficient septin collar formation, consistent with a model (Fig. 8) wherein filament formation requires that GTP bind to septins preassembled in heteromeric complexes (rather than to free septin monomers). The transition from a patch to a collar implies a structural rearrangement. Indeed, septins localize to the incipient bud site early in the cell cycle (as detected by GFP fusions or immunofluorescence), but pronounced septin filaments (detected by thin-section EM) are only observed in cells with medium to large buds (Byers and Goetsch, 1976; Frazier et al., 1998). FRAP indicates that septin mobility is high in the patch in unbudded cells, whereas septin movement is extremely slow in the collars at the neck of budded cells (Caviston et al., 2003; Dobbelaere et al., 2003), also consistent with immobilization of septin complexes via their assembly into septin filaments.
Our observations suggest that the reorganization of septins necessary for collar formation corresponds to polymerization of preformed heteromeric septin complexes into filaments and that this assembly requires that GTP be bound to Cdc10 and Cdc12 (Fig. 8). Whether subsequent GTP hydrolysis has a role in filament depolymerization (or in septin ring disassembly after cytokinesis) remains an open question. The absence of a phenotype, even for a cdc10(S41V) cdc12(S43V) double mutant that is unable to hydrolyze GTP, argues that GTP turnover does not play a switch-like regulatory role in septin function. Thus, GTP binding may serve a more structural role, akin to nucleotide binding to α-tubulin in microtubule formation (Nogales, 2001). Indeed, using mass spectrometry, Vrabioiu et al. (2004) found that nucleotide turnover on septins in vivo is too slow for GTP hydrolysis to have a role in septin dynamics during the cell cycle. Other mechanisms, such as phosphorylation (Dobbelaere et al., 2003; see next section) or SUMOylation (Johnson and Blobel, 1999), may impose temporal control.
A PAK phosphorylates septins and regulates collar assembly
Prior work indicated that Cla4 might be an important regulator of septin assembly or localization (Cvrckova et al., 1995; Weiss et al., 2000; Dobbelaere et al., 2003; Schmidt et al., 2003). First, we found that absence of Cla4 dramatically exacerbated all the phenotypes of the GTP binding-deficient septin mutants, and conversely, that overexpression of CLA4 suppressed even the septin collar assembly defect of the GTP-binding defect mutants. These strong genetic interactions suggest that GTP binding and Cla4 phosphorylation regulate the same septin-dependent process, namely the changes necessary to promote septin filament formation and septin collar assembly (Fig. 8). Second, we found that Cdc3 and Cdc10 serve as direct substrates of Cla4 in vitro and showed that, at least for Cdc10, phosphorylation in vivo is largely Cla4-dependent. Thus, septins constitute an important new target for Cla4.
How do GTP binding and Cla4 phosphorylation act in concert to promote septin collar assembly? It is unlikely that GTP-bound septins stimulate Cla4 because when recovered from cdc10(S46N) cdc12(T48N) cells shifted to 37°C, the specific activity of Cla4 is not reduced compared with Cla4 recovered from wild-type cells (unpublished data). Conversely, it is unlikely that GTP binding makes a septin a better substrate for Cla4 because both Cdc10 and Cdc10(S46N) are phosphorylated equivalently by Cla4, and heterotetrameric septin complexes composed of wild-type or GTP binding-defective Cdc10 and Cdc12 interact with Cla4 to the same extent (unpublished data). Revealingly though, mutations in a single septin (Cdc10) that reduce both GTP binding (S46N) and Cla4 phosphorylation (S256A) caused a potent synergistic defect in septin collar assembly, suggesting that GTP binding and Cla4-mediated phosphorylation act in parallel, but via independent mechanisms, to promote filament assembly (Fig. 8).
Based on genetic findings, it was proposed that Cdc42 acts as a direct assembly factor for septin polymerization (Gladfelter et al., 2002). However, no interaction between septins and Cdc42 has been reported, and we failed to detect any interaction between Cdc3, Cdc10, Cdc11, or Cdc12 (as GST fusions) and His6-tagged Cdc42 (either GTP or GDP bound; unpublished data). It has also been suggested that Cdc42 GTPase-activating proteins (Bem3, Rga1, and Rga2) have a role in septin collar formation as effectors rather than regulators of Cdc42 (Caviston et al., 2003). Our demonstration that Cla4 directly phosphorylates septins makes it more likely that Cdc42 regulates septin organization via its activation of Cla4. Cdc42-mediated membrane recruitment of Cla4 and activation of Cla4-dependent phosphorylation of septins could provide a means to couple assembly of the septin collar with bud emergence both spatially and temporally (Fig. 8). Our demonstration that Cla4 interacts directly with Cdc3 and Cdc10 provides an additional mechanism for stable recruitment of Cla4 to sites of septin assembly. By analogy, a PAK may also phosphorylate septins and regulate their assembly into filaments in animal cells.
Materials and methods
Strains and growth conditions
Yeast strains are listed in Table I. Rich (YP) and defined minimal (SC) media (Sherman et al., 1986) contained either 2% dextrose (D), 2% galactose (Gal), or 2% raffinose plus 0.2% sucrose (Raf). Low phosphate medium was prepared by adding 10 ml of 1 M MgSO4 and 10 ml of NH4OH (28% wt/vol) to 1 l YP, removing the precipitated phosphate salts by filtration through 3 MM paper (Whatman), and adjusting final pH to 5.8 with HCl. For G1 synchronization and release, cells were pregrown in appropriate selective medium, arrested with 2 μg/ml α-factor at A600nm = 0.05 in YPD or YPGal (adjusted to pH 4.0) for 3.5 h at 26°C, then washed three times with water, and resuspended in prewarmed (37°C) YPD or YPGal. For S-phase arrest and release, cells were incubated in YPD or YPGal with 0.2 M hydroxyurea for 3 h at 26°C, then resuspended in prewarmed (37°C) YPD or YPGal.
Table I. Yeast strains used in this work.
| Strain | Relevant genotype | Source/reference |
|---|---|---|
| BY4741 | MAT a his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics, Inc. |
| YMVB1a | BY4741 cdc12(T48N)::URA3 | This paper |
| YMVB2a | BY4741 cdc12(S43V)::URA3 | This paper |
| YMVB3b | BY4741 cdc12(T48N)::ura3 | This paper |
| YMVB5c | BY4741 cdc10Δ(::KANMX) | This paper |
| YMVB15c | BY4741 cdc10Δ(::KANMX) cdc12(T48N)::URA3 | This paper |
| YMVB25d | BY4741 cdc10Δ(::KANMX) CDC10::LEU2 | This paper |
| YMVB24d | BY4741 cdc10Δ(::KANMX) CDC10::LEU2 cdc12(T48N)::URA3 | This paper |
| YMVB6d | BY4741 cdc10Δ(::KANMX) cdc10(S46N)::LEU2 | This paper |
| YMVB8d | BY4741 cdc10Δ(::KANMX) cdc10(S46N)::LEU2 cdc12(T48N)::URA3 | This paper |
| YMVB60d | BY4741 cdc10Δ(::KANMX) cdc10(S41V)::LEU2 cdc12(S43V)::URA3 | This paper |
| YMVB12e | BY4741 cla4Δ(::LEU2) | This paper |
| YMVB14e | BY4741 cla4Δ(::LEU2) cdc12(T48N)::ura3 | This paper |
| YMVB48f | BY4741 cdc10Δ(::KANMX) cdc10(S46N)::LEU2 cdc12(T48N)::URA3::HIS3 | This paper |
| YMVB49f | BY4741 cdc12(T48N)::URA3::HIS3 | This paper |
| YMVB50g | BY4741 cdc12(T48N)::URA3::HIS3 cdc10(S46N) | This paper |
| YMVB51g | BY4741 cdc12(T48N)::URA3::HIS3 cdc10(S46N) cla4Δ(::LEU2) | This paper |
| YMVB53h | BY4741cdc10Δ(::KANMX) cdc10(S256A) | This paper |
| YMVB54h | BY4741 cdc10Δ(::KANMX) cdc10(S46N S256A) | This paper |
To replace the wild-type CDC12 locus with the GTPase mutants, pMVB21 or pMVB22 was digested with EcoRI and SpHI and transformed to BY4741. A PCR was performed to confirm correct replacement of CDC12 with the URA3 containing CDC12 cassette, and this PCR product was subsequently sequenced to verify introduction of the correct mutation.
Ura mutants were selected on agar medium containing 0.5 mg/ml 5-fluoro-orotic acid (Boeke et al., 1987).
The exact CDC10 open reading frame was deleted using the PCR-based “Short Flanking Homology” method, using vector pFA6, containing the kanMX4 cassette (Wach et al., 1994). The resulting PCR product was transformed into BY4741, YMVB1, or YMVB2, G418-resistant colonies were selected and replacement of the CDC10 ORF was verified by PCR.
The XhoI-digested pMVB41 (containing wild-type CDC10) was integrated at the promoter locus of CDC10 in YMVB5 and YMVB15. To replace the wild type CDC10 locus with the GTPase mutant, cdc10(S46N), the XhoI-digested pMVB40 was integrated at the promoter locus of CDC10 in YMVB5 or YMVB15. To replace the wild type CDC10 with the mutant, cdc10(S41V), the XhoI-digested pMVB155 was integrated in YMVB16 (BY4741 cdc10Δ cdc12(S43V)::URA3).
To delete CLA4, FD26, a construct described by Cvrckova et al. (1995) containing LEU2 as the selection marker, was transformed into BY4741 or YMVB3.
Marker swap plasmid pUH7 digested with SmaI (Cross, 1997) was used to replace the URA3 marker with HIS3 in YMVB1 and YMVB8.
To introduce the cdc10(S46N) mutant, YIplac211-cdc10(S46N) (YMVB69) was integrated at the CDC10 locus using the XhoI enzyme; Ura-transformants were selected on plates containing 5-FOA, and mutants were the cdc10(S46N) allele was retained, were selected: first by testing the temperature-sensitive phenotype of the mutants, then by sequencing.
To replace the wild type CDC10 locus with cdc10(S256A) or cdc10(S46N S256A) the XhoI-digested pMVB143 or pMVB144 was integrated at the promoter locus of CDC10 in YMVB5.
Plasmids and recombinant DNA methods
Plasmids (Table II) were constructed by standard methods in E. coli strain DH5α. Lack of PCR errors, correct in-frame junctions, and incorporation of site-directed mutations were verified by nucleotide sequencing. Unless stated otherwise, template for all PCR amplifications was genomic DNA of strain BY4741.
Table II. Plasmids used in this work.
| Plasmid | Description | Source/reference |
|---|---|---|
| pMVB12a | CDC12-His6 | This paper |
| pMVB13a | CDC12ΔC-His6 | This paper |
| pMVB14b | CDC12(S43V)-His6 | This paper |
| pMVB15b | CDC12(T48N)-His6 | This paper |
| pMVB16b | CDC12ΔC(S43V)-His6 | This paper |
| pMVB17b | CDC12ΔC(T48N)-His6 | This paper |
| pMVB120c | CDC11-His6 | This paper |
| pDT72d | pET24d-His6 vector | D. Truckses (this laboratory) |
| pBEG2e | His6-CDC3 | B. Gullbrand (this laboratory) |
| pBEG3e | His6-CDC10 | B. Gullbrand (this laboratory) |
| pMVB150 | His6-CDC10(S256A) | This paper |
| pMVB151 | His6-CDC10(S256A S312A) | This paper |
| pKM263 | His6-GST vector | Melcher, 2000 |
| pMVB7f | His6-GST-CDC3 | This paper |
| pMVB8f | His6-GST-CDC10 | This paper |
| pMVB10f | His6-GST-CDC11 | This paper |
| pGST-CDC11 | GST-CDC11 | D. Kellogg (University of California, Santa Cruz, Santa Cruz, CA) |
| pMVB24g | GST-CDC10 | This paper |
| pMVB25g | GST-CDC12 | This paper |
| pMVB27g | GST-CDC3 | This paper |
| pGEX-HSL1ΔN | GST-HSL1(833-1518) | Shulewitz, 2000 |
| pMVB80g | GST-CLA4 | This paper |
| pMVB82g | GST-CLA4(524–842) | This paper |
| pMVB81b | GST-CLA4(K594A) | This paper |
| pMVB83b | GST-CLA4(524–842)(K594A) | This paper |
| pLP17 | CEN, LEU2, CDC12-GFP | Lippincott and Li, 1998 |
| pLP29 | CEN, HIS3, CDC12-GFP | Lippincott and Li, 1998 |
| pMVB32b | CEN, LEU2, CDC12(T48N)-GFP | This paper |
| pMVB91b | CEN, HIS3, CDC12(T48N)-GFP | This paper |
| pMVB19h | pUC18-CDC12 | This paper |
| pMVB64h | pUC18-CDC12(S43V) | This paper |
| pMVB65h | pUC18-CDC12(T48N) | This paper |
| pMVB21i | pUC18-CDC12(S43V)-URA3 | This paper |
| pMVB22i | pUC18-CDC12(T48N)-URA3 | This paper |
| YCplac33 | CEN, URA3 vector | Gietz and Sugino, 1988 |
| pMVB39j | CEN, URA3, CDC12 | This paper |
| YEplac181 | 2 μm, LEU2 vector | Gietz and Sugino, 1988 |
| pMVB49k | 2 μm, LEU2, CDC12(T48N) | This paper |
| YIplac128 | LEU2 vector | Gietz and Sugino, 1988 |
| pMVB41l | LEU2, CDC10 | This paper |
| pMVB40b | LEU2, CDC10(S46N) | This paper |
| pMVB155b | LEU2, CDC10(S41V) | This paper |
| YIplac211 | URA3 vector | Gietz and Sugino, 1988 |
| pMVB69m | URA3, CDC10(S46N) | This paper |
| pMVB143m | LEU2, CDC10(S256A) | This paper |
| pMVB144m | LEU2, CDC10(S46N S256A) | This paper |
| pLA10 | CEN, URA3, CDC10-GFP | Cid et al., 1998 |
| pSB5 | CEN, URA3, CDC11-GFP | S. Bahmanyar (this laboratory) |
| CDC3-GFP | CEN, URA3, CDC3-GFP | B. Haarer |
| pCJ160 | CEN, URA3, GAL1-myc-STE20 | Shulewitz, 2000 |
| YCpUG-SKM1 | CEN, URA3, GAL1-myc-SKM1 | Shulewitz, 2000 |
| YEpUG-CLA4 | 2 μm, URA3, GAL1-myc-CLA4 | Shulewitz, 2000 |
| pMVB113n | CEN, URA3, GAL1-myc-CLA4 | This paper |
| pMVB112o | CEN, URA3, GAL1-myc-CLA4(K594A) | This paper |
| pMVB121p | pETDuet-His6 CDC12 | This paper |
| pMVB122p | pETDuet-His6 CDC12-CDC3 | This paper |
| pMVB123p | pACYDuet-CDC10 | This paper |
| pMVB124p | pACYDuet-CDC10-CDC11 | This paper |
| pMVB125p | pETDuet-His6 CDC12T48N-CDC3 | This paper |
| pMVB126p | pACYDuet-CDC10S46N-CDC11 | This paper |
CDC12 was amplified using a forward primer, 5′-GGAATTCCAT ATGAGTGCTGCCACTGC-3′ (NdeI site in italics, start codon in bold) and as reverse primer, 5′-TATCCGCTCGAGAGATCCACGTGGAACCAGTTTTAAATGGGATTTTTTTACTT-3′ (XhoI site in italics). CDC12(Δ339–407), abbreviated Cdc12ΔC, was amplified using the same forward primer and a different reverse primer, 5′-TATCCGCTCGAGAGAGCCACGCGGAACTAAGTGTGACAATTTCCTTGCT-3′. Both reverse primers encode a thrombin cleavage site. The resulting CDC12 and CDC12ΔC derivatives were inserted as NdeI–XhoI fragments into the corresponding sites of pET-24b (Novagen) upstream and in-frame with a His6 tag.
These constructs were created via site-directed mutagenesis of the indicated codons using mismatch primers.
Derived from pET-23d (Novagen).
The NcoI–BamHI fragment of pET-24d was replaced with an NcoI–BamHI fragment of pBH4 (a gift of K. Prehoda and W. Lim, UCSF) that contains six tandem His codons and a sequence encoding the cleavage site for TEV.
Derived from pDT72.
Derived from pKM263.
Derived from pGEX3 (Amersham Biosciences).
CDC12, including 500 bp of its promoter region, was amplified and inserted in pUC18. Mutations S43V and T48N were introduced separately into Cdc12, as described above. To permit subsequent integration at the CDC12 locus on chromosome VIII, 446 bp of the 3′-untranslated region of CDC12 was inserted in the corresponding constructs.
To provide a selectable marker for integration, the URA3 gene, excised from pJJ244 (Jones and Prakash, 1990) was inserted into the naturally occurring SnaBI site present in the 3′-flanking region of CDC12.
A fragment of pMVB19 carrying CDC12 and its promoter was inserted into the corresponding sites in YCplac33.
A fragment of pMVB22 carrying CDC12(T48N) and its promoter was inserted into the corresponding sites in YEplac181.
CDC10, including 500 bps of its promotor region and 250 bp 3′ to its stop codon, was amplified and cloned into YIplac128.
A fragment containing cdc10S46N and its promotor was subcloned from pMVB40 into YIplac211. The cdc10S256A mutation was introduced via site-directed mutagenesis.
The GAL1-Myc-CLA4 fragment from YEpUG-Myc-CLA4 was subcloned into YCplac33.
GAL1-Myc-CLA4 was subcloned from pMVB113 to pUC18, which was used as a template in a site directed mutagenesis reaction, changing Lys 594 to Ala; GAL1-Myc-CLA4(K594A) was then inserted into YCplac33.
These vectors are based on pETDuet-1 and pACYDuet-1 (Novagen). The His6 tag in pACYDuet was entirely deleted in pACYDuet1 using PCR mutagenesis. The GTP binding mutants were introduced by site-directed mutagenesis.
Protein purification and binding to immobilized GST fusions
His6-tagged septins were expressed in E. coli, purified using Ni2+-NTA beads, eluted with imidazole, and immediately desalted over Sephadex® G-25. For eluates containing purified His6-tagged septin complexes, imidazole was removed by dialysis overnight in low salt buffer. Recombinant proteins with a tandem affinity tag (His6-GST-TEV) were purified on glutathione-agarose and released from the matrix by cleavage with TEV protease. GST fusions were immobilized on glutathione-agarose beads, incubated with purified potential binding partners, and after extensive washing, bead-bound proteins were solubilized in SDS-PAGE sample buffer.
GTP binding and hydrolysis
After incubation of [35S]GTPγS with purified septins, septin-bound nucleotide was separated from free nucleotide by filtration; after binding of α[32P]GTP to purified septin complexes immobilized on Ni2+-NTA beads, excess nucleotide was removed by washing. GTPase activity was measuring as free radiolabeled phosphate released from γ[32P]GTP after charcoal adsorption, or as formation of α[32P]GDP from α[32P]GTP, resolved by TLC.
Antibodies
Polyclonal antisera were raised in adult female New Zealand white rabbits against purified Cdc12(Δ339–407)-His6 (primary inoculation, 0.5 mg; secondary boost, 0.25 mg). The IgG fraction from serum was enriched by ammonium sulfate fractionation (Harlow and Lane, 1988). His6-tagged proteins were detected with mouse anti-His(C-term) mAb (Invitrogen) or anti-His6 (BMG-His-1; Roche). Other antisera were mouse anti-c-Myc mAb 9E10 and anti-GFP (mixture of clones 7.1 and 13.1; Roche).
Protein kinase activity and metabolic labeling
Immune complex kinase assays (using Myc-tagged Cla4), kinase assays (using recombinant GST-Cla4), and metabolic labeling of yeast cultures with [32P]H3PO4 were performed as described in detail in the online supplemental material.
EM
0.25-μg protein samples were applied to polylysine-treated, glow-discharged, carbon-coated copper grids, negatively stained with 1% uranyl acetate in 50% methanol, and viewed in a transmission electron microscope (Tecnai 12; Philips).
Fluorescence microscopy
To visualize GFP fusions, live cells were washed once with PBS and viewed under an epifluorescence microscope (model BH-2; Olympus) using a 100× objective equipped with a GFP band-pass filter (Chroma Technology Corp.). Images were collected using a charged-coupled device camera (Olympus), processed with Magnafire SP imaging software (Optronics) and Adobe Photoshop®. To stain nuclei, cells were fixed briefly in 70% ethanol, washed several times with PBS, and incubated with DAPI (0.2 μg/ml final). Cytokinesis was quantified according to Hartwell (1971) after fixation with formaldehyde and subsequent zymolyase treatment (Lippincott and Li, 1998).
Online supplemental material
Detailed methods and additional results, described in the text, are provided in the online supplemental material (available at http://www.jcb.org/cgi/content/full/jcb.200312070/DC1).
Acknowledgments
We thank T. Alber, P. Barth, V. Cid, D. Kellogg, R. Li, and E. Weiss for communication of unpublished results, reagents, and advice, D. Truckses, B. Gullbrand, F. Roelants, D. Ballon, and other Thorner Lab members for plasmids and helpful discussion, and Reena Zalpuri for expert assistance with EM.
This work was supported by a long-term fellowship from the Human Frontier Science Program Organization and an Advanced NATO Fellowship (to M. Versele), by facilities provided by the Berkeley campus Cancer Research Laboratory, and by National Institutes of Health research grant GM21841 (to J. Thorner).
The online version of this article includes supplemental material.
Abbreviations used in this paper: GTPγS, guanosine 5′-O-3′-thiotriphosphate; NTA, nitrilotriacetate; PAK, p21-activated protein kinase; TEV, tobacco etch virus.
References
- Adam, J.C., J.R. Pringle, and M. Peifer. 2000. Evidence for functional differentiation among Drosophila septins in cytokinesis and cellularization. Mol. Biol. Cell. 11:3123–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., M. Parra, S. Bidlingmaier, and M. Snyder. 1999. Nim1-related kinases coordinate cell-cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, Y., V. Mermali, M.S. Mooseker, and M. Snyder. 2000. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol. Cell. 5:841–851. [DOI] [PubMed] [Google Scholar]
- Boeke, J.D., J. Trueheart, G. Natsoulis, and G.R. Fink. 1987. 5-Flouroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164–175. [DOI] [PubMed] [Google Scholar]
- Bouquin, N., Y. Barral, R. Courbeyrette, M. Blondel, M. Snyder, and C. Mann. 2000. Regulation of cytokinesis by the Elm1 protein kinase in Saccharomyces cerevisiae. J. Cell Sci. 113:1435–1445. [DOI] [PubMed] [Google Scholar]
- Byers, B., and L. Goetsch. 1976. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69:717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor, A., and M. Snyder. 2003. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization and function. Mol. Cell. Biol. 23:2762–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J.P., M. Longtine, J.R. Pringle, and E. Bi. 2003. The role of Cdc42 GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid, V.J., L. Adamikova, R. Cenamor, M. Molina, M. Sanchez, and C. Nombela. 1998. Cell integrity and morphogenesis in a budding yeast septin mutant. Microbiology. 144:3463–3474. [DOI] [PubMed] [Google Scholar]
- Cross, F.R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 13:647–653. [DOI] [PubMed] [Google Scholar]
- Cvrckova, F., C. De Virgilio, E. Manser, J.R. Pringle, and K. Nasmyth. 1995. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 9:1817–1830. [DOI] [PubMed] [Google Scholar]
- Dent, J., K. Kato, X.R. Peng, C. Martinez, M. Cattaneo, C. Poujol, P. Nurden, A. Nurden, W.S. Trimble, and J. Ware. 2002. A prototypic platelet septin and its participation in secretion. Proc. Natl. Acad. Sci. USA. 99:3064–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere, J., M.S. Gentry, R.L. Hallberg, and Y. Barral. 2003. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell. 4:345–357. [DOI] [PubMed] [Google Scholar]
- Feig, L.A. 1999. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases. Nat. Cell Biol. 1:E25–E27. [DOI] [PubMed] [Google Scholar]
- Field, C.M., O. al-Awar, J. Rosenblatt, M.L. Wong, B. Alberts, and T.J. Mitchinson. 1996. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133:605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier, J.A., M.L. Wong, M.S. Longtine, J.R. Pringle, M. Mann, T.J. Mitchison, and C. Field. 1998. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., J.R. Pringle, and D.J. Lew. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681–689. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., I. Bose, T.R. Zyla, E.S. Bardes, and D.J. Lew. 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: a Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 298–299.
- Hartwell, L. 1971. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69:265–276. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.S., and L. Prakash. 1990. Yeast Saccharomyces cerevisiae selectable markers in pUC18 polylinkers. Yeast. 6:363–366. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M., S. Kumar, A. Mizoguchi, C. Ide, A. Kinoshita, T. Haraguchi, Y. Hiraoka, and M. Noda. 1997. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11:1535–1547. [DOI] [PubMed] [Google Scholar]
- Kinoshita, M., C.M. Field, M.L. Coughlin, A.F. Straight, and T.J. Mitchison. 2002. Self- and actin-templated assembly of mammalian septins. Dev. Cell. 3:791–802. [DOI] [PubMed] [Google Scholar]
- Krengel, U., L. Schlichting, A. Scherer, R. Schumann, M. Frech, J. John, W. Kabsch, E.F. Pai, and A. Wittinghofer. 1990. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 62:539–548. [DOI] [PubMed] [Google Scholar]
- Lee, P.R., S. Song, H.S. Ro, C.J. Park, J. Lippincott, R. Li, J.R. Pringle, C. De Virgilio, M.S. Longtine, and K.S. Lee. 2002. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:6906–6920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 1998. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143:1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., K.B. Shannon, W. Shou, R.J. Deshaies, and R. Li. 2001. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114:1379–1386. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., H. Fares, and J.R. Pringle. 1998. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J. Cell Biol. 143:719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., C.L. Theesfeld, J.N. McMillan, E. Weaver, J.R. Pringle, and D.J. Lew. 2000. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, H., A. Mendoza, J.M. Rodriguez-Pachon, M. Molina, and C. Nombela. 1997. Characterization of SKM1, a Saccharomyces cerevisiae gene encoding a novel Ste20/PAK- like protein kinase. Mol. Microbiol. 23:431–444. [DOI] [PubMed] [Google Scholar]
- McGrath, J.P., D.J. Capon, D.V. Goeddel, and A.D. Levinson. 1984. Comparative biochemical properties of normal and activated human ras p21 protein. Nature. 310:644–649. [DOI] [PubMed] [Google Scholar]
- Melcher, K. 2000. A modular set of prokaryotic and eukaryotic expression vectors. Anal. Biochem. 277:109–120. [DOI] [PubMed] [Google Scholar]
- Mendoza, M., A.A. Hyman, and M. Glotzer. 2002. GTP binding induces filament assembly of a recombinant septin. Curr. Biol. 12:1858–1863. [DOI] [PubMed] [Google Scholar]
- Neufeld, T.P., and G.M. Rubin. 1994. The Drosphila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 77:371–379. [DOI] [PubMed] [Google Scholar]
- Nogales, E. 2001. Structural insights into microtubule function. Annu. Rev. Biophys. Biomol. Struct. 30:397–420. [DOI] [PubMed] [Google Scholar]
- Scheffzek, K., M.R. Ahmadian, and A. Wittinghofer. 1998. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257–262. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., A. Varma, T. Drgon, B. Bowers, and E. Cabib. 2003. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell. 14:2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield, P.J., C.J. Oliver, B.E. Kremer, S. Sheng, Z. Shao, and I.G. Macara. 2003. Borg/Septin interactions and the assembly of mammalian septin heterodimers, trimers and filaments. J. Biol. Chem. 278:3483–3488. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 163–165.
- Shulewitz, M.J. 2000. Septin assembly regulates cell cycle progression through activation of a protein kinase signaling pathway. Ph.D. thesis. University of California, Berkeley, CA. 172 pp.
- Shulewitz, M.J., C.J. Inouye, and J. Thorner. 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, P.A., J.L. DeRisi, J.E. Wilhelm, and R.D. Vale. 2000. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 290:341–344. [DOI] [PubMed] [Google Scholar]
- Tjandra, H., J. Compton, and D. Kellogg. 1998. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK family. Curr. Biol. 8:991–1000. [DOI] [PubMed] [Google Scholar]
- Vrabioiu, A.M., S.A. Gerber, S.P. Gygi, C.M. Field, and T.J. Mitchison. 2004. The majority of the Saccharomyces cerevisiae septin complexes do not exchange guanine nucleotides. J. Biol. Chem. 279:3111–3118. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10:1793–1808. [DOI] [PubMed] [Google Scholar]
- Weiss, E.L., A.C. Bishop, K.M. Shokat, and D.G. Drubin. 2000. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat. Cell Biol. 2:677–685. [DOI] [PubMed] [Google Scholar]