Abstract
Although the microtubule-depolymerizing KinI motor Kif2a is abundantly expressed in neuronal cells, we now show it localizes to centrosomes and spindle poles during mitosis in cultured cells. RNAi-induced knockdown of Kif2a expression inhibited cell cycle progression because cells assembled monopolar spindles. Bipolar spindle assembly was restored in cells lacking Kif2a by treatments that altered microtubule assembly (nocodazole), eliminated kinetochore–microtubule attachment (loss of Nuf2), or stabilized microtubule plus ends at kinetochores (loss of MCAK). Thus, two KinI motors, MCAK and Kif2a, play distinct roles in mitosis, and MCAK activity at kinetochores must be balanced by Kif2a activity at poles for spindle bipolarity. These treatments failed to restore bipolarity to cells lacking the activity of the kinesin Eg5. Thus, two independent pathways contribute to spindle bipolarity, with the Eg5-dependent pathway using motor force to drive spindle bipolarity and the Kif2a-dependent pathway relying on microtubule polymer dynamics to generate force for spindle bipolarity.
Keywords: mitotic spindle; kinetochore; microtubule; centrosome, poleward flux
Introduction
The spindle is a microtubule-based structure responsible for accurate chromosome segregation during mitosis and meiosis (McIntosh and Koonce, 1989; Hyman and Karsenti, 1996; Compton, 2000). Organization of microtubules into a fusiform, bipolar spindle requires numerous nontubulin accessory proteins including several members of the kinesin gene family (Vale and Fletterick, 1997; Sharp et al., 2000). Kinesin-related proteins with the conserved motor domain positioned at the NH2 terminus (KinN kinesins) display plus-end–directed motility and perform diverse functions during spindle morphogenesis including kinetochore–microtubule interactions (CENP-E; Yao et al., 1997), generation of polar ejection force (Kid; Levesque and Compton, 2001), and spindle bipolarity (Eg5; Sawin et al., 1992). Kinesin-related proteins with the conserved motor domain positioned at the COOH terminus (KinC kinesins) display minus-end–directed motility and participate in focusing microtubule minus ends at spindle poles (Endow et al., 1994; Mountain et al., 1999). Finally, kinesin-related proteins with the conserved motor domain positioned in the middle of the protein (KinI kinesins) lack motility and use their catalytic motor domains to induce conformation changes in protofilament structure at microtubule ends, which stimulates microtubule depolymerization by promoting catastrophe (Desai et al., 1999; Moores et al., 2002; Ogawa et al., 2004). MCAK/XKCM1/Kif2c (hereafter referred to as MCAK) is the best characterized member of the KinI subgroup. MCAK localizes to kinetochores and spindle microtubules (Wordeman and Mitchison, 1995; Walczak et al., 1996) where it regulates microtubule dynamics essential for proper chromosome attachment to spindle microtubules, chromosome movement, and correction of attachment errors (Maney et al., 1998, 2001; Kline-Smith and Walczak, 2002; Walczak et al., 2002; Hunter et al., 2003; Ohi et al., 2003; Kline-Smith et al., 2004). However, MCAK is not the only member of KinI kinesin-related proteins. Kif2a is a closely related KinI kinesin-related protein that plays a role in neurogenesis (Noda et al., 1995; Debernardi et al., 1997; Morfini et al., 1997; Homma et al., 2003). In this paper we demonstrate that Kif2a is essential for bipolar spindle assembly during mitosis in cultured vertebrate cells.
Results and discussion
To build Kif2a-specific antibodies we immunized rabbits with the unique 119 amino acids of the NH2-terminal end of human Kif2a. A single protein species of 110 kD was detected by immunoblot analysis with these antibodies in total cell extracts prepared from human HeLa, hamster CHO, and frog A6 cells (Fig. 1 A). The molecular mass of this protein is consistent with the predicted size of Kif2a, indicating that the antibodies are specific for Kif2a, they cross react to Kif2a from several vertebrate species, and Kif2a is expressed in cultured vertebrate cells. Kif2a localized to centrosomes in interphase and mitotic human CFPAC-1 cells (Fig. 1, B–F), but also to spindle microtubules being concentrated at poles (Fig. 1, D–F). In anaphase and telophase, Kif2a localized to the spindle midzone and midbody in addition to spindle poles (Fig. 1 F). We detected Kif2a at kinetochores, but the signal was just slightly above background levels and was only observed after depolymerization of microtubules with nocodazole (unpublished data). This localization pattern for Kif2a is different from that reported for the related KinI motor MCAK (Wordeman and Mitchison, 1995; Walczak et al., 1996).
Figure 1.
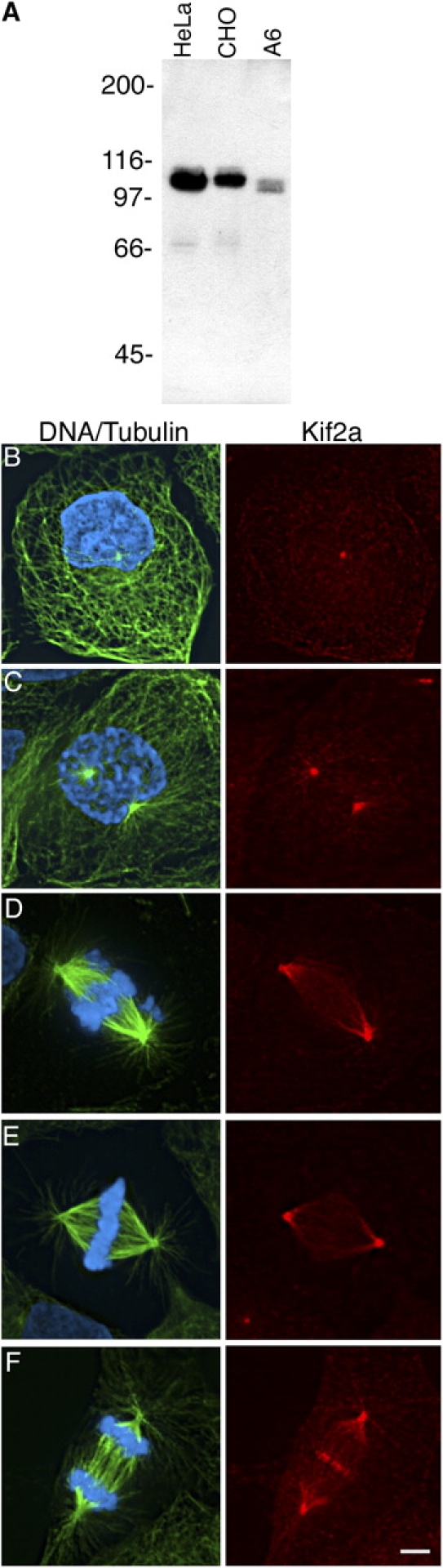
Characterization of Kif2a in cultured vertebrate cells. (A) Total cell protein from ∼100,000 human HeLa, frog A6, or hamster CHO cells was separated by SDS-PAGE and blotted with Kif2a polyclonal antibodies. Positions of myosin (200), β-galactosidase (116), phosphorylase B (97), albumin (66), and ovalbumin (45) are shown in kilodaltons. (B–F) Human CFPAC-1 cells were processed for indirect immunofluorescence microscopy using Kif2a- (red) and tubulin-specific antibodies (green), as well as the DNA-specific dye DAPI (blue). Cells are in interphase (B), prophase (C), prometaphase (D), metaphase (E), and anaphase (F). Bar, 10 μm.
To determine if Kif2a is required for mitotic spindle assembly or function, we used siRNA to knock down Kif2a protein levels in human U2OS cells (Fig. 2). 48 h after transfection, immunoblot analysis demonstrated that Kif2a levels were reduced >90% in cells transfected with Kif2a-specific RNA relative to cells transfected with nonspecific RNA (Fig. 2 A). At this time point, cells transfected with Kif2a-specific RNA had an average mitotic index of ∼36% in contrast to the mitotic index of ∼3% observed after transfection with nonspecific RNA. Most (>99%) mitotic cells transfected with nonspecific RNA had bipolar spindles with Kif2a localized at spindle poles (Fig. 2, B, D, and F). In contrast, >90% of mitotic cells transfected with Kif2a-specific RNA had monopolar spindles (Fig. 2, C and E; and see Fig. 5 H), and microtubule attachment to kinetochores was detected by electron microscopy (not depicted). Kif2a was undetectable on these monopolar spindles by indirect immunofluorescence, which is consistent with the results of immunoblot analysis (Fig. 2 G). Centrosome staining demonstrated that initial separation of centrosomes at prophase in cells lacking Kif2a was not maintained when cells entered prometa/metaphase (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200404012/DC1).
Figure 2.
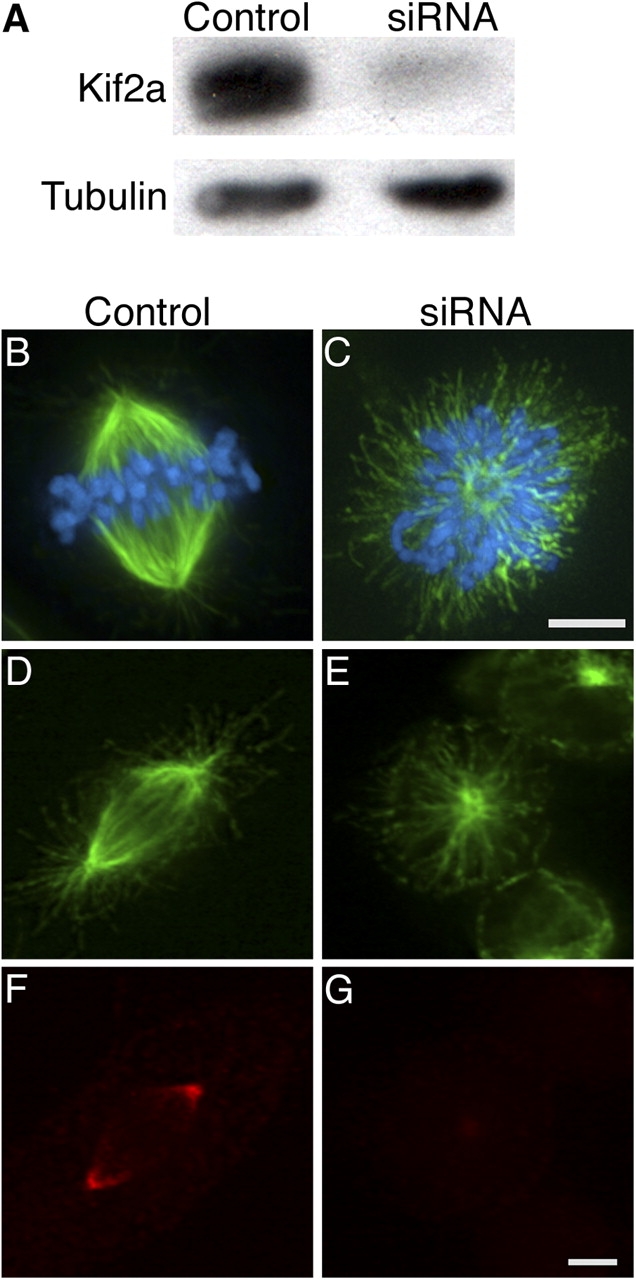
Kif2a is essential for spindle bipolarity. (A) Human U2OS cells were transfected with either nonspecific (control) or Kif2a-specific (siRNA) RNAs. Total cell protein was separated by size using SDS-PAGE and blotted with Kif2a-specific antibodies and tubulin antibodies. (B–G) Human U2OS cells were transfected with either nonspecific (control) or Kif2a-specific (siRNA) RNAs, fixed with either glutaraldehyde (B and C) or methanol (D–G), and stained for tubulin (green), DNA (blue), and/or Kif2a (red). Bars, 10 μm.
Figure 5.
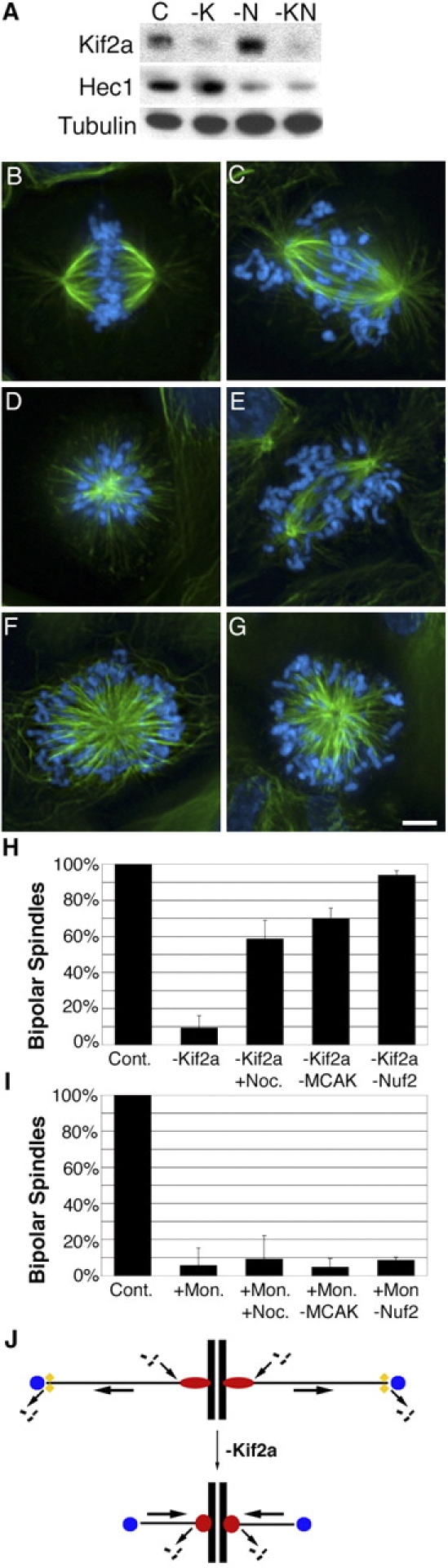
Nuf2 knockdown restores spindle bipolarity to cells lacking Kif2a. (A) U2OS cells were transfected with nonspecific (C), Kif2a-specific (-K), Nuf2-specific (-N), or both Kif2a- and Nuf2-specific (-KN) RNAs. Total cell protein was separated by size using SDS-PAGE and blotted with Kif2a-, Hec1-, and tubulin-specific antibodies. (B–E) Human U2OS cells were stained for tubulin (green) or DNA (blue) after transfection with control RNA (B), Kif2a-specific RNA (D and E), Nuf2-specific RNA (C, E, and G), or treatment with 100 μM monastrol (F and G). Bar, 10 μM. (H and I) Quantification of the percentage of mitotic U2OS cells with bipolar spindles under various conditions after Kif2a knockdown (H) or monastrol treatment (I). Error bars represent SD. (J) Kif2a depolymerizes microtubule minus ends associated with poleward microtubule flux. Flux generates tension that induces kinetochores to add tubulin subunits. When flux is perturbed in the absence of Kif2a, tension at kinetochores is lost and kinetochores depolymerize microtubules drawing centrosomes together into monopolar arrays.
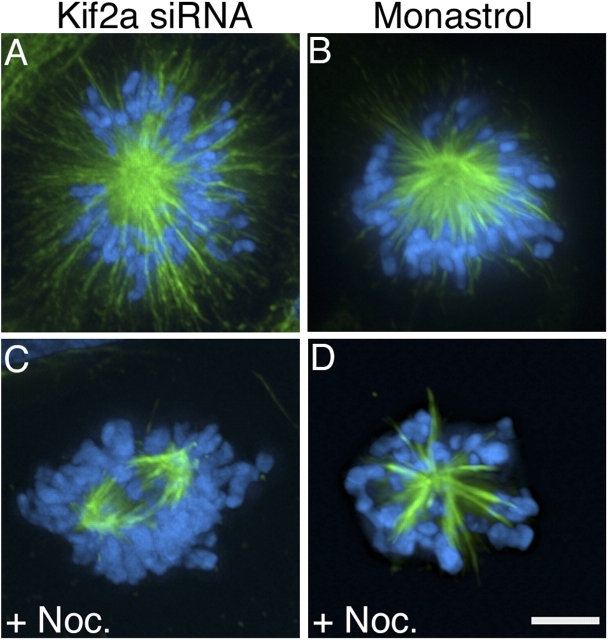
Kif2a has been shown to catalyze microtubule depolymerization in the absence of motile function (Desai et al., 1999). To determine if altered microtubule dynamics might contribute to the failure to maintain centrosome separation in mitotic cells lacking Kif2a, we suppressed microtubule dynamics by addition of 100 nM nocodazole (Fig. 3). Whereas only ∼8% of mitotic cells transfected with Kif2a-specific RNA alone formed bipolar spindles, ∼58.6% of mitotic cells transfected with Kif2a-specific RNA and treated with nocodazole for 5 h before fixation assembled bipolar spindles (Fig. 3 C and see Fig. 5 H, n = 232, five independent experiments). Chromosomes did not align efficiently on these bipolar spindles, which is consistent with disruption of microtubule dynamics by nocodazole treatment. Restoration of spindle bipolarity by nocodazole treatment was specific to cells lacking Kif2a activity because nocodazole treatment did not alter the percentage of cells with monopolar spindles induced with 100 μM monastrol (Fig. 3 D and see Fig. 5 I, n = 282, six independent experiments), a cell-permeable inhibitor of Eg5 function (Mayer et al., 1999). Microtubules in monopolar spindles induced by Eg5 inhibition end abruptly near chromosomes in contrast to microtubules in monopolar spindles induced by the lack of Kif2a, which extend beyond chromosomes to the cell cortex (Fig. 3, A and B). A similar difference between cells lacking Kif2a or Eg5 activity was observed with low doses of taxol, as 53.6% (n = 41) of mitotic cells lacking Kif2a and treated with 15 nM taxol had either multipolar or bipolar spindles, whereas 94.3% (n = 35) of mitotic cells treated with both 100 μM monastrol and 15 nM taxol had monopolar spindles.
Figure 3.
Nocodazole treatment restores spindle bipolarity to cells lacking Kif2a. U20S cells stained for tubulin (green) and DNA (blue) after transfection with Kif2a-specific RNA (A and C) or treatment with 100 μM monastrol (B and D) in the presence (C and D) or absence (A and B) of 100 nM nocodazole. Bar, 10 μm.
To alter microtubule dynamics in mitotic cells through a mechanism independent of microtubule drugs, we used siRNA to knock down expression of the microtubule destabilizing protein MCAK (Fig. 4). 48 h after transfection, immunoblot analysis indicated that both MCAK and Kif2a were reduced to levels equivalent to when each protein was knocked down alone (Fig. 4 A). In contrast to the monopolar spindles induced by knockdown of Kif2a alone (Fig. 4 B), most cells (∼91%) lacking MCAK alone had bipolar spindles (Cassimeris and Morabito, 2004; Holmfeldt et al., 2004). Simultaneous knockdown of MCAK and Kif2a resulted in bipolar spindles with aligned chromosomes in a majority of cells (70%, n = 200, four independent experiments; Fig. 4 D and see Fig. 5 H). Astral microtubules were extremely long in these spindles and often extended from one spindle pole to the cell cortex beyond the opposite spindle pole. Anaphase cells were observed and the mitotic index was ∼6%, a significant decrease from that observed after diminution of Kif2a alone. Reduction of MCAK levels failed to prevent monopolarity in monastrol-treated cells, as only ∼5% had bipolar spindles (Fig. 4 E and see Fig. 5 I; n = 84, two independent experiments).
Figure 4.
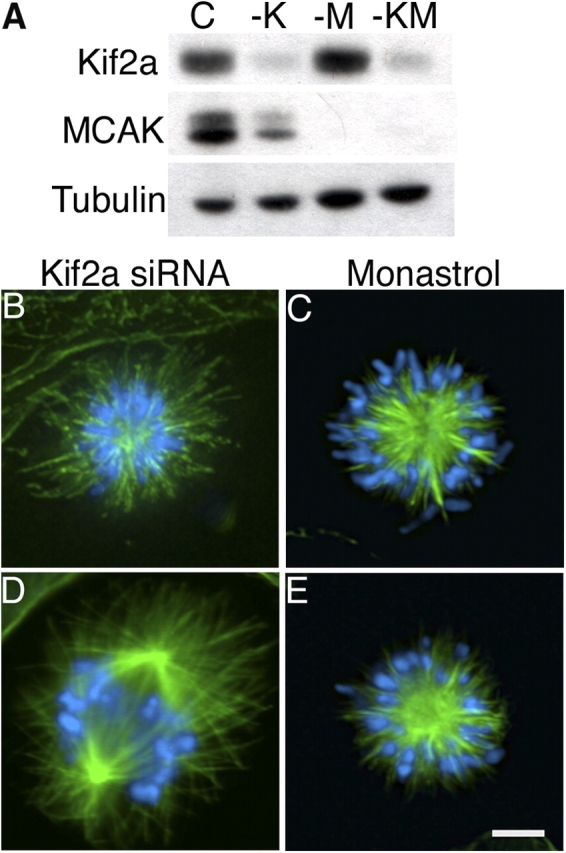
MCAK knockdown restores spindle bipolarity to cells lacking Kif2a. (A) U2OS cells were transfected with nonspecific (C), Kif2a-specific (-K), MCAK-specific (-M), or both Kif2a- and MCAK-specific (-KM) RNAs. Total cell protein was separated by size using SDS-PAGE and blotted with Kif2a-, MCAK-, and tubulin-specific antibodies. (B–E) Human U2OS cells were stained for tubulin (green) or DNA (blue) after transfection with Kif2a-specific RNA alone (B), transfection with both Kif2a- and MCAK-specific RNA (D), treatment with 100 μM monastrol alone (C), or transfection with MCAK-specific RNA and treatment with 100 μM monastrol (E). Bar, 10 μM.
These results demonstrate that MCAK and Kif2a play functionally different roles during mitosis in cultured cells, a conclusion paralleled by analyses of Klp59C and Klp10A in fruit flies (Goshima and Vale, 2003; Rogers et al., 2003). MCAK is necessary for correction of chromosome attachment errors on spindles (Maney et al., 1998; Walczak et al., 2002; Kline-Smith et al., 2004), but not bipolar spindle assembly (Cassimeris and Morabito, 2004; Holmfeldt et al., 2004). In contrast, Kif2a is essential for bipolar spindle assembly. Also, the relative contributions made by MCAK and Kif2a to spindle assembly differ between mitotic and meiotic systems. MCAK plays a dominant role in regulating microtubule plus end dynamic instability in meiotic systems through antagonism of TOGp/XMAP215 activity (Tournebize et al., 2000; Kinoshita et al., 2001) such that massive monopolar arrays form if it is inhibited (Walczak et al., 1996). Perturbation of Kif2a also leads to monopolar arrays in meiotic extracts (Gaetz and Kapoor, 2004). Meiotic systems have far greater pools of tubulin subunits relative to mitotic cells, and perturbation of either MCAK or Kif2a most likely result in monopolar arrays in meiotic systems because excess microtubule polymer overwhelms the machinery that organizes microtubules into bipolar arrays. Finally, MCAK activity must be appropriately balanced by Kif2a activity for bipolar spindle assembly in cultured cells. This unexpected functional relationship between two KinI kinesin-related proteins indicates that MCAK and Kif2a activities are spatially restricted in spindles with MCAK regulating microtubule plus ends at kinetochores and Kif2a regulating microtubule minus ends at poles.
To test if kinetochore-induced microtubule depolymerization is necessary for monopolar spindle assembly in the absence of Kif2a, we eliminated kinetochore–microtubule attachment using siRNA to knock down expression of Nuf2, a component of a conserved, essential kinetochore complex containing Nuf2 and Hec1 (Fig. 5; DeLuca et al., 2003). We used Hec1-specific antibodies to monitor the efficiency of Nuf2 knockdown because the stability of Hec1 relies on the presence of Nuf2 (DeLuca et al., 2003), and Nuf2-specific antibodies were not available. 48 h after transfection, immunoblot analysis indicated that Kif2a and Hec1 were reduced to levels equivalent to when each protein was knocked down alone (Fig. 5 A). Most (∼98%; n = 77, three independent experiments) cells lacking Nuf2 alone had bipolar spindles with unattached chromosomes (Fig. 5 C), which is consistent with previous findings (DeLuca et al., 2003). In contrast to the monopolar spindles formed in cells lacking Kif2a alone (Fig. 5 D), ∼94% (n = 213, three independent experiments) of cells lacking both Kif2a and Nuf2 assembled bipolar spindles with unattached chromosomes (Fig. 5, E and H). Knockdown of Nuf2 failed to restore bipolarity to cells lacking Eg5 activity (Fig. 5, F, G, and I; n = 83, three independent experiments), and knockdown of both MCAK and Nuf2 showed no detectable difference from knockdown of Nuf2 alone (not depicted).
These results demonstrate that Kif2a and Eg5 act through independent pathways to promote spindle bipolarity, and that each pathway is necessary but not sufficient for bipolar spindle assembly in cultured cells. Eg5 uses motor force to slide antiparallel microtubules relative to one another to promote spindle bipolarity (Sharp et al., 2000) in a mechanism that is insensitive to treatments that alter microtubule dynamics and independent of kinetochore–microtubule interactions. In contrast, Kif2a is a microtubule-depolymerizing enzyme that promotes spindle bipolarity in a mechanism that is sensitive to treatments that alter microtubule dynamics. Similar differences have been noted for the Cin8 and Kip3 (and Kar3) kinesin-related proteins in yeast (Cottingham et al., 1999). It is unlikely that loss of Kif2a leads to monopolar spindles due to microtubules being excessively long because cells lacking both Kif2a and MCAK have unusually long microtubules but still assemble bipolar spindles (Fig. 4 D). Instead, kinetochore-derived forces draw centrosomes together in the absence of Kif2a. This finding explains why centrosome separation in the absence of Kif2a is normal until nuclear envelope break down (Fig. S1), and why treatments that stabilize microtubule plus ends, like low doses of nocodazole or loss of MCAK (Jordan et al., 1992; Vasquez et al., 1997; Kinoshita et al., 2001; Kline-Smith and Walczak, 2002; Ohi et al., 2003), or eliminate kinetochore–microtubule attachment promote spindle bipolarity in the absence of Kif2a. We cannot discount the possibility that Kif2a acts directly at kinetochores, but that is inconsistent with Kif2a localization to poles. Alternatively, recent data shows that disassembly of microtubule minus ends associated with poleward microtubule flux requires the targeting of Klp10A (Rogers et al., 2003) and Kif2a (Gaetz and Kapoor, 2004) to spindle poles. Force from poleward flux generates tension that induces kinetochores to add tubulin subunits to microtubule plus ends (Maddox et al., 2003). In that context, disruption of flux by the loss of Kif2a would relax tension at kinetochores. Consequently, kinetochores would switch to the depolymerization state and remove tubulin subunits from microtubule plus ends, drawing centrosomes together into monopolar spindles (Fig. 5 J). Although stabilization of microtubule plus ends at kinetochores through loss of MCAK is sufficient to restore kinetochore tension and prevent monopolarity, other kinetochore proteins execute microtubule depolymerization during chromosome movement because chromosome velocity is unaffected by depletion of MCAK from centromeres (Kline-Smith et al., 2004). Together, these data lead us to speculate that force from microtubule polymer dynamics (for review see Inoue and Salmon, 1995) contributes to bipolar spindle assembly.
Materials and methods
Cell culture
Human HeLa, CFPAC-1, and U2OS cells were maintained in Dulbecco's modified medium, Iscove's modified Dulbecco's medium, and McCoy's medium, respectively, containing 10% FBS, 50 IU/ml penicillin, and 50 μg/ml streptomycin.
Antibodies
The region of the cDNA encoding the NH2-terminal 119 aa of human Kif2a (GenBank/EMBL/DDBJ accession no. BG721281) was amplified using forward (5′-ATCGGATCCTGCTGCTCCAGATGAGGT-3′) and reverse (5′-CTTCTCTGGGGCACGGGAAATTCTTAAG-3′) primers and inserted into the BamHI–EcoRI sites of pRSET-B plasmid (Invitrogen). The His-tagged recombinant protein was purified by affinity chromatography and used to immunize two rabbits (Covance Research Products).
Other antibodies used in this work included MCAK (Mack and Compton, 2001), DM1α (Sigma-Aldrich), Hec1 (Novus Biologicals), and human anticentrosome antibody (provided by J.B. Rattner, University of Calgary, Alberta, Canada).
Indirect immunofluorescence microscopy
Cells were extracted in microtubule-stabilizing buffer (4 M glycerol, 100 mM Pipes, pH 6.9, 1 mM EGTA, 5 mM MgCl2, and 0.5% Triton X-100) followed by fixation in methanol (Kif2a staining) or 1% glutaraldehyde (microtubule staining). Subsequent antibody incubations and washes were done in TBS-BSA (10 mM Tris, pH 7.5, 150 mM NaCl, and 1% BSA). Primary antibodies were detected using species–specific fluorescein- or Texas red–conjugated secondary antibodies (Vector Laboratories). DNA was detected with 0.2 μg/ml DAPI (Sigma-Aldrich). Coverslips were mounted with ProLong Antifade mounting medium (Molecular Probes).
Fluorescent images were captured with a cooled CCD camera (model Orca II; Hamamatsu) mounted on a microscope (model Axioplan 2; Carl Zeiss MicroImaging, Inc.). A series of 0.5-μm optical sections were collected in the z plane for each channel (DAPI, fluorescein, and/or Texas red) using a 63× 1.4NA objective and deconvolved using Openlab software (Improvision Inc.) to eliminate extraneous fluorescence background.
RNA interference
Kif2a levels were reduced using the sequence 5′-GGCAAAGAGAUUGACCUGG-3′. Nuf2 and MCAK levels were reduced using published sequences (DeLuca et al., 2003; Cassimeris and Morabito, 2004). All siRNA duplexes were synthesized with 3′ dTdT overhangs and were annealed (Ambion). Approximately 50,000 U2OS cells were plated on coverslips in 35-mm dishes the day before transfection and grown without antibiotics. Double stranded RNAs were transfected into cells using OligofectamineTM reagent (Invitrogen) as described previously (Elbashir et al., 2001). Samples were analyzed 48 h after transfection by either indirect immunofluorescence or immunoblot analysis.
Online supplemental material
Fig. S1 shows centrosome position during both prophase and metaphase in control cells and cells lacking Kif2a. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200404012/DC1.
Acknowledgments
We thank J. Gaetz and T. Kapoor for sharing unpublished data and J.B. Rattner for providing human anticentrosome sera.
This work was supported by National Institutes of Health grants GM51542 (D.A. Compton) and GM008704 (N. Ganem).
References
- Cassimeris, L., and J. Morabito. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A. 2000. Spindle assembly in animal cells. Annu. Rev. Biochem. 69:95–114. [DOI] [PubMed] [Google Scholar]
- Cottingham, F.R., L. Gheber, D.L. Miller, and M.A. Hoyt. 1999. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J. Cell Biol. 147:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi, S., E. Fontanella, L. De Gregorio, M.A. Pierotti, and D. Delia. 1997. Identification of a novel human kinesin-related gene (HK2) by the cDNA differential display technique. Genomics. 42:67–73. [DOI] [PubMed] [Google Scholar]
- DeLuca, J.G., B.J. Howell, J.C. Canman, J.M. Hickey, G. Fang, and E.D. Salmon. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103–2109. [DOI] [PubMed] [Google Scholar]
- Desai, A., S. Verma, T.J. Mitchison, and C.E. Walczak. 1999. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 96:69–78. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Endow, S.A., R. Chandra, D.J. Domma, A.H. Yamamoto, and E.D. Salmon. 1994. Mutants of the Drosophila ncd microtubule motor protein cause centrosomal and spindle defects in mitosis. J. Cell Sci. 107:859–867. [DOI] [PubMed] [Google Scholar]
- Gaetz, J., and M. Kapoor. 2004. Dynein/dynactin regulate metaphase spindle length by targeting depolymerizing activities to spindle poles. J. Cell Biol. 166:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmfeldt, P., S. Stenmark, and M. Gullberg. 2004. Differential functional interplay of TOGp/XMAP215 and the KinI kinesin MCAK during interphase and mitosis. EMBO J. 23:627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma, N., Y. Takei, Y. Tanaka, T. Nakata, S. Terada, M. Kikkawa, Y. Noda, and N. Hirokawa. 2003. Kinesin superfamily protein 2A (KIF2A) functions in suppression of collateral branch extension. Cell. 114:229–239. [DOI] [PubMed] [Google Scholar]
- Hunter, A.W., M. Caplow, D.L. Coy, W.O. Hancock, S. Diez, L. Wordeman, and J. Howard. 2003. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol. Cell. 11:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A., and E. Karsenti. 1996. Morphogenetic properties of microtubules and mitotic spindle assembly. Cell. 84:401–410. [DOI] [PubMed] [Google Scholar]
- Inoue, S., and E.D. Salmon. 1995. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 6:1619–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M.A., D. Thrower, and L. Wilson. 1992. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J. Cell Sci. 102:401–416. [DOI] [PubMed] [Google Scholar]
- Kinoshita, K., I. Arnal, A. Desai, D.N. Drechsel, and A.A. Hyman. 2001. Reconstitution of physiological microtubule dynamics using purified components. Science. 294:1340–1342. [DOI] [PubMed] [Google Scholar]
- Kline-Smith, S.L., and C.E. Walczak. 2002. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol. Biol. Cell. 13:2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith, S.L., A. Khodjakov, P. Hergert, and C.E. Walczak. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque, A.A., and D.A. Compton. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack, G.J., and D.A. Compton. 2001. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc. Natl. Acad. Sci. USA. 98:14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P., A. Straight, P. Coughlin, T.J. Mitchison, and E.D. Salmon. 2003. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J. Cell Biol. 162:377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., A.W. Hunter, M. Wagenbach, and L. Wordeman. 1998. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney, T., M. Wagenbach, and L. Wordeman. 2001. Molecular dissection of the microtubule depolymerizing activity of mitotic centromere-associated kinesin. J. Biol. Chem. 276:34753–34758. [DOI] [PubMed] [Google Scholar]
- Mayer, T.U., T.M. Kapoor, S.J. Haggarty, R.W. King, S.L. Schreiber, and T.J. Mitchison. 1999. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 286:971–974. [DOI] [PubMed] [Google Scholar]
- McIntosh, J.R., and M.P. Koonce. 1989. Mitosis. Science. 246:622–628. [DOI] [PubMed] [Google Scholar]
- Moores, C.A., M. Yu, J. Guo, C. Beraud, R. Sakowicz, and R.A. Milligan. 2002. A mechanism for microtubule depolymerization by KinI kinesins. Mol. Cell. 9:903–909. [DOI] [PubMed] [Google Scholar]
- Morfini, G., S. Quiroga, A. Rosa, K. Kosik, and A. Caceres. 1997. Suppression of KIF2 in PC12 cells alters the distribution of a growth cone nonsynaptic membrane receptor and inhibits neurite extension. J. Cell Biol. 138:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountain, V., C. Simerly, L. Howard, A. Ando, G. Schatten, and D.A. Compton. 1999. The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, Y., R. Sato-Yoshitake, S. Kondo, M. Nangaku, and N. Hirokawa. 1995. KIF2 is a new microtubule-based anterograde motor that transports membranous organelles distinct from those carried by kinesin heavy chain or KIF3A/B. J. Cell Biol. 129:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, T., R. Nitta, Y. Okada, and N. Hirokawa. 2004. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 116:591–602. [DOI] [PubMed] [Google Scholar]
- Ohi, R., M.L. Coughlin, W.S. Lane, and T.J. Mitchison. 2003. An inner centromere protein that stimulates the microtubule depolymerizing activity of a KinI kinesin. Dev. Cell. 5:309–321. [DOI] [PubMed] [Google Scholar]
- Rogers, G.C., S.L. Rogers, T.A. Schwimmer, S.C. Ems-McClung, C.E. Walczak, R.D. Vale, J.M. Scholey, and D.J. Sharp. 2003. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., K. LeFuellec, M. Philippe, and T.J. Mitchison. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., G.C. Rogers, and J.M. Scholey. 2000. Microtubule motors in mitosis. Nature. 407:41–47. [DOI] [PubMed] [Google Scholar]
- Tournebize, R., A. Popov, K. Kinoshita, A.J. Ashford, S. Rybina, A. Pozniakovsky, T.U. Mayer, C.E. Walczak, E. Karsenti, and A.A. Hyman. 2000. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat. Cell Biol. 2:13–19. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., and R.J. Fletterick. 1997. The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13:745–777. [DOI] [PubMed] [Google Scholar]
- Vasquez, R.J., B. Howell, A.M. Yvon, P. Wadsworth, and L. Cassimeris. 1997. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell. 8:973–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak, C.E., T.J. Mitchison, and A. Desai. 1996. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 84:37–47. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., E.C. Gan, A. Desai, T.J. Mitchison, and S.L. Kline-Smith. 2002. The microtubule-destabilizing kinesin XKCM1 is required for proper chromosome positioning during spindle assembly. Curr. Biol. 12:1885–1889. [DOI] [PubMed] [Google Scholar]
- Wordeman, L., and T.J. Mitchison. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X., K.L. Anderson, and D.W. Cleveland. 1997. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 139:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]