Abstract
Components of the DNA replication machinery localize into discrete subnuclear foci after DNA damage, where they play requisite functions in repair processes. Here, we find that the replication factors proliferating cell nuclear antigen (PCNA) and RPAp34 dynamically exchange at these repair foci with discrete kinetics, and this behavior is distinct from kinetics during DNA replication. Posttranslational modification is hypothesized to target specific proteins for repair, and we find that accumulation and stability of PCNA at sites of damage requires monoubiquitination. Contrary to the popular notion that phosphorylation on the NH2 terminus of RPAp34 directs the protein for repair, we demonstrate that phosphorylation by DNA-dependent protein kinase enhances RPAp34 turnover at repair foci. Together, these findings support a dynamic exchange model in which multiple repair factors regulated by specific modifications have access to and rapidly turn over at sites of DNA damage.
Keywords: proliferating cell nuclear antigen; replication protein A; cisplatin; FRAP; foci
Introduction
The cellular response to DNA damage requires a wide range of protein factors to ensure the fidelity of genetic material passed on to daughter cells. These factors include detection and signaling proteins to warn that damage has occurred, checkpoint response proteins to inhibit cell division allowing time for repair, and the plethora of proteins that function in the repair of genetic lesions (Rouse and Jackson, 2002). Apart from its function in the replication of the genome, the DNA replication machinery plays an essential role in the repair of damaged DNA. The heterotrimeric replication protein A (RPA) complex has been implicated in the damage detection process, whereas factors including proliferating cell nuclear antigen (PCNA), DNA polymerases, and DNA ligases are known to be important in the resynthesis of excised nucleotide sequences (Shivji et al., 1992; He et al., 1995; Zou and Elledge, 2003). Though DNA replication and repair both involve DNA synthesis, we speculated that classical replication proteins might behave distinctly during these two processes and that discrete signaling pathways might underlie these differences.
Results and discussion
To investigate the action of specific replication factors during DNA replication and repair in living cells, we generated Rat-1 and U2OS cell lines stably expressing GFP fused to the replication proteins PCNA and RPAp34 (the 34-kD subunit of the replication protein A complex, also termed RPA2). The GFP-PCNA fusion protein colocalized with sites of BrdU incorporation in S-phase cells and was homogeneously distributed throughout the nucleoplasm during other phases of the cell cycle (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200312048/DC1). GFP-RPAp34 displayed similar localization but was more difficult to detect at sites of replication until the formation of large replication clusters during late S-phase (Fig. S1). This finding is likely due to the transient role of RPA in DNA replication and rapid turnover at replication sites (Sporbert et al., 2002). Similar localization and behavior of these GFP-fusion proteins during replication in other cell systems has been documented (Leonhardt et al., 2000; Sporbert et al., 2002). As such, these cell lines provide an effective model for studying the action of the replication machinery in the DNA damage response.
Numerous proteins have been identified to relocalize within the nucleus in response to genotoxic insult. For example, ionizing radiation induces the formation of nuclear foci enriched in proteins including BRCA1, 53BP1, and the Rad50/Mre11/Nbs1 complex (Maser et al., 1997; Wang et al., 2002). Therefore, the localization of PCNA and RPAp34 in response to DNA damage was examined (Fig. 1). Upon treatment of the stable lines with cis-diamminedichloroplatinum II (CDDP), a chemotherapeutic agent that induces platinum-DNA (Pt-DNA) adducts (Siddik, 2003), the GFP-fusion proteins accumulated into discrete subnuclear foci (Fig. 1 A). GFP-RPAp34 and GFP-PCNA also accumulated into numerous subnuclear foci in response to acute UV irradiation (Fig. 1 B and not depicted), consistent with reports on the function of these replication factors in the nucleotide excision repair of pyrimidine dimers (He et al., 1995; Riedl et al., 2003). Focus formation of GFP-RPAp34 and GFP-PCNA was restricted to microdomains of the nucleus that were irradiated through pores in a polycarbonate filter, indicating that the nucleation of these factors in response to UV irradiation is not a pan-nuclear event (Fig. 1 C and Fig. S1). Although GFP-RPAp34 was largely diffuse in replicating cells, it assembled into discrete foci when DNA replication was stalled with hydroxyurea (HU) or aphidicolin (APH; Fig. 1 D). These foci presumably represent the accumulation of RPA at regions of single-stranded DNA occurring at stalled or collapsed replication forks, as appearance of these foci preceded the development of strand breaks (Fig. 1 E) indicated by the accumulation of H2AX phosphorylation (γ-H2AX; Ward and Chen, 2001). The topoisomerase inhibitor camptothecin also induced the formation of GFP-RPAp34 foci that colocalized with γ-H2AX foci (Fig. 1 F). Insult with CDDP or UV irradiation induced the formation of GFP-RPAp34 foci irrespective of ongoing cell cycle progression (Fig. 1 G). In contrast, challenge with HU only induced GFP-RPAp34 and GFP-PCNA foci formation in cells progressing into S-phase (Fig. 1 G and not depicted). Together, these results demonstrate that replication factors are recruited to sites of genetic insult, independent of ongoing DNA replication.
Figure 1.
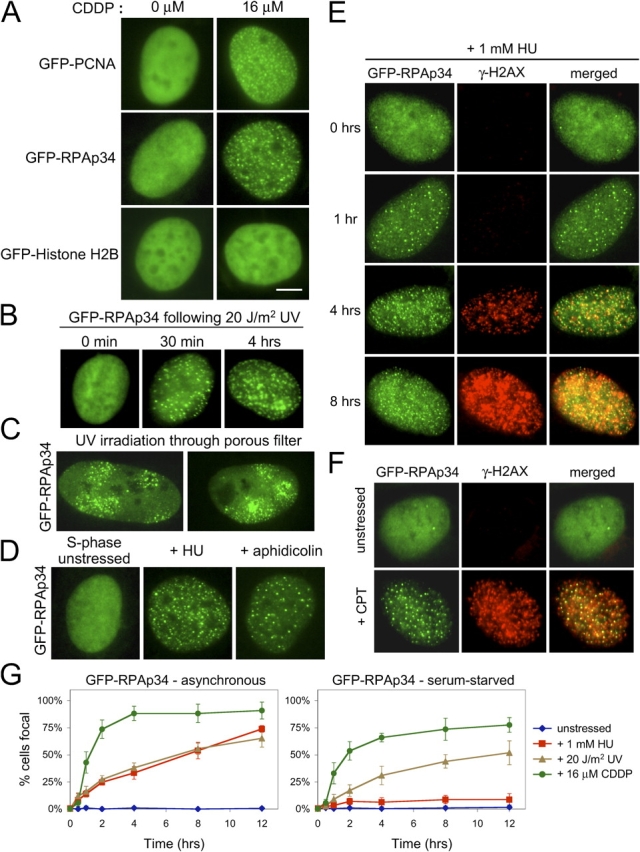
Visualization of replication factors in the DNA damage response. (A) CDDP induces the relocalization of GFP-PCNA and GFP-RPAp34, but not GFP-histone H2B, into discrete foci within the nucleus of stably expressing Rat-1 cells. Bar, 5 μm. (B) RPAp34 rapidly accumulates into foci in response to UV irradiation. (C) Cells expressing GFP-RPAp34 were exposed to UV irradiation through a porous polycarbonate filter. GFP-RPAp34 only accumulates into foci within the irradiated microdomains 3 μm in diameter. (D) RPAp34 accumulates into foci in response to the stalling of DNA replication with HU or APH. (E) RPAp34 foci appear before the accumulation of H2AX phosphorylation after HU treatment. (F) Camptothecin (CPT) induces the formation of RPAp34 foci that colocalize with γ-H2AX foci. (G) Quantitation of the number of cells with punctate versus diffuse localization of GFP-RPAp34 in asynchronous (left) or serum-starved (right) cultures after various stresses over time. 200 cells were counted at each time point from each of three independent experiments. After 48 h serum-starvation, 4% of the unstressed cells labeled BrdU positive during the 12-h time course of the experiment.
Given its clinical relevance and complexity of action, we extended the analyses of PCNA and RPA recruitment in the response to CDDP. Initially, we determined the kinetics of formation of Pt-DNA adducts. Using an antibody specific for Pt-DNA (Tilby et al., 1991), we found that Pt-DNA adducts were detectable within 2 h of exposure to CDDP (Fig. 2 A). Corresponding analyses of PCNA and RPA indicated that peak focal recruitment coincided with maximal levels of Pt-DNA (Fig. 2 A). By microscopic analyses we found that Pt-DNA was relatively dispersed throughout the nucleus, whereas both GFP-PCNA and GFP-RPAp34 accumulated at discrete subnuclear foci after treatment with CDDP (Fig. 2 B). This result led us to speculate that the RPA/PCNA foci represented discrete sites of active repair. It has been previously shown that the repair of Pt-DNA adducts involves DNA strand breaks (Siddik, 2003), and we found that RPA/PCNA foci temporally colocalized with γ-H2AX (Fig. 2, A and C). To further determine the nature of these foci, the coordinate localization of GFP-PCNA and GFP-RPAp34 with other factors known to contribute to the repair of Pt-DNA adducts was analyzed. The repair of Pt-DNA adducts is a complex process that likely involves both nucleotide excision repair and mismatch repair (Siddik, 2003). We found that the mismatch repair factor MSH2 colocalized with GFP-PCNA at replication foci in S-phase cells and similarly colocalized with GFP-PCNA and GFP-RPAp34 at foci induced by CDDP damage (Fig. 2 D and not depicted). The nucleotide excision repair factor ERCC1 was largely diffuse in unstressed cells, including those in S-phase, but accumulated into numerous foci after CDDP damage that colocalized with GFP-PCNA and GFP-RPAp34 foci (Fig. 2 E and not depicted). These data indicate that the foci represent repair factories and led us to question their persistence as a function of repair. CDDP damage is difficult to efficiently repair, and the presence of Pt-DNA was greatly diminished yet clearly persisted above background levels by 48 h (Fig. 2 F). Concurrent with the decreased levels of Pt-DNA, we observed a decline in the number of cells with focal RPA/PCNA and γ-H2AX reactivity (Fig. 2 F), suggesting the completion of repair was being achieved.
Figure 2.
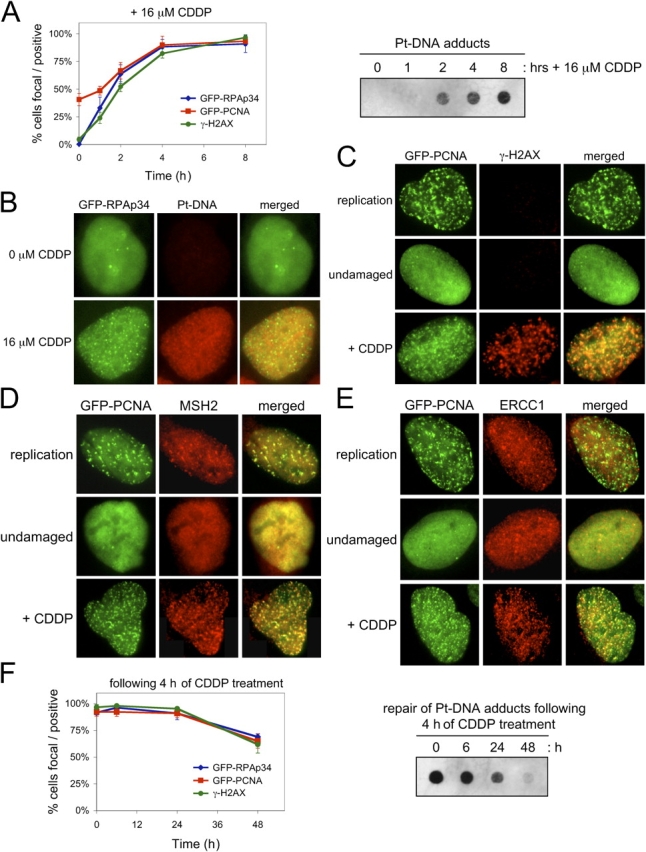
Kinetic induction of repair foci by cisplatin. (A) Cells were treated with 16 μM CDDP, and total DNA was analyzed for Pt-DNA levels at the indicated times by dot blotting (right). Cells were concurrently scored for PCNA/RPAp34 foci and γ-H2AX reactivity (left). (B) Representative images of GFP-RPAp34 and Pt-DNA adducts after 4 h of CDDP damage. (C–E) Representative images of GFP-PCNA and γ-H2AX (C), MSH2 (D), or ERCC1 (E) localization in replicating, undamaged, and CDDP-treated cells. (F) Cells were treated with 16 μM CDDP for 4 h and then cultured in the absence of CDDP for the indicated times. DNA was analyzed for Pt-DNA levels by dot blotting (right), and cells were concurrently scored for PCNA/RPAp34 foci and γ-H2AX reactivity (left). The error bars are the SD in the cell, counting from three independent experiments.
Numerous studies have demonstrated the localization of repair factors to foci in fixed cells after damage. However, very little is known about the kinetic behavior of replication factors during DNA repair processes in living cells. To study this dynamic behavior, we performed FRAP analysis. After irreversible bleaching of the GFP signal within a defined region of the nucleus, the recovery of fluorescence intensity into the bleached region was assessed (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200312048/DC1). These analyses revealed discrete dynamic properties of replication factors dependent on their biological action (Fig. 3). In cycling cells not in S-phase, GFP-PCNA was diffuse and exhibited highly mobile behavior with recovery reaching 50% in 260 ms. In contrast, PCNA assembled into replication foci readily bleached and exhibited recovery over longer intervals (t 1/2 of ∼25 s). Surprisingly, GFP-PCNA at repair foci after insult with 16 μM CDDP was highly immobile and turned over only on the order of minutes (Fig. 3, A and B). Thus, the kinetic behavior of subnuclear PCNA foci is distinct dependent on the biological process, demonstrating a separation of function between replication and repair. In interphase nuclei, GFP-RPAp34 was diffuse and highly mobile with recovery reaching 50% in 170 ms. To enrich for S-phase cells and render GFP-RPAp34 focal, cells were synchronized with APH for 8 h and then released into a replicative state before imaging. Under these conditions, GFP-RPAp34 was focal but still highly mobile, as indicated by rapid recovery after bleaching (t 1/2 of 220 ms). Treatment with CDDP increased GFP-RPAp34 retention, but turnover was still seen with 50% recovery in 660 ms (Fig. 3 C). Thus, like PCNA, RPA is more stably associated at repair than replicative foci. Interestingly, these studies also provided the opportunity to directly compare the kinetic behavior of these replication factors in the repair process. From the data it is clear that RPAp34 is a highly dynamic participant in the DNA damage detection and repair process, whereas PCNA is a more static component of the repair machinery.
Figure 3.
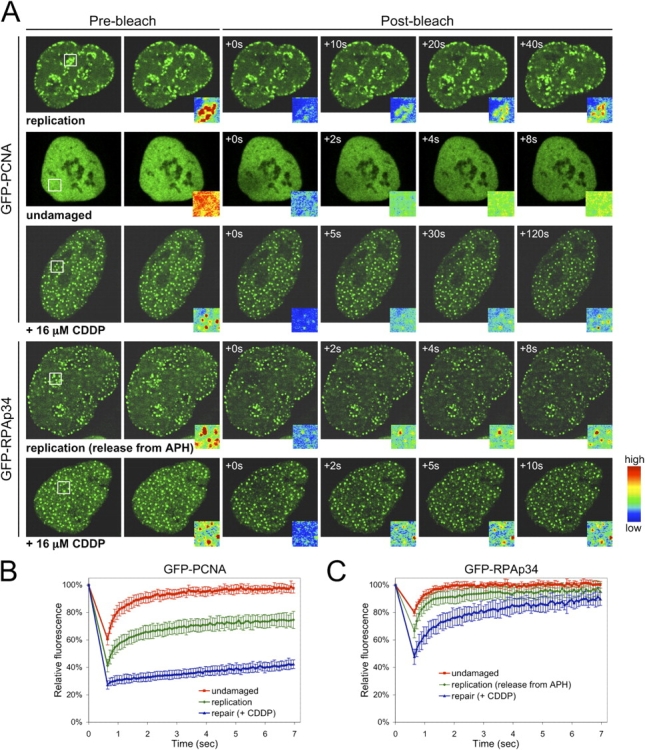
Components of the replication machinery turn over at repair foci with distinct kinetics. (A) FRAP on undamaged, replicating, and CDDP-treated Rat-1 cells expressing GFP-PCNA and GFP-RPAp34. Pseudocolored insets display the intensity profile of the photobleached region boxed in white, 2.9 μm × 2.9 μm. (B and C) Quantitative FRAP analysis of GFP-PCNA (B) and GFP-RPAp34 (C) in undamaged, replicating, and CDDP-treated (4 h) Rat-1 cells. These replication proteins exhibit distinct dynamic properties and recover more slowly during repair than replication. The error bars are the SD in the FRAP measurements.
To understand the molecular basis associated with the recruitment of replication factors to repair foci, we investigated the impact of specific posttranslational modifications on their dynamic behavior. Subunits of the RPA complex are known to be phosphorylated by kinases activated in the DNA damage response (e.g., ATM, ATR, and DNA-dependent protein kinase [DNA-PK]) or during progression into S-phase (e.g., CDK/cyclins) (Dutta and Stillman, 1992; Niu et al., 1997; Shao et al., 1999; Wang et al., 2001; Barr et al., 2003). It has been hypothesized that such modifications may direct RPA association with repair versus replication complexes (Shao et al., 1999; Wang et al., 2001; Barr et al., 2003; Vassin et al., 2004). As an initial screen for phospho-modifications of RPA behavior, we used an array of pharmacological kinase inhibitors. Specifically, we used caffeine (an inhibitor of PI3 kinases with highest specificity for ATM and ATR), PD98059 (a MAPK kinase inhibitor), roscovitine (a CDK inhibitor), and LY294002 (another PI3 kinase inhibitor with highest specificity for DNA-PK; Meijer et al., 1997; Izzard et al., 1999; Sarkaria et al., 1999; Walker et al., 2000). Roscovitine and PD98059 did not qualitatively or quantitatively influence the behavior of RPAp34 after CDDP damage (Fig. 4 A). However, caffeine blocked foci assembly after CDDP damage, as has recently been reported after IR irradiation (Barr et al., 2003), and resulted in a corresponding increase in protein mobility as assessed by FRAP (Fig. 4 A). In contrast, we found that LY294002 had little effect on foci formation but markedly impaired exchange after CDDP damage (Fig. 4 A). Although the use of such pharmacological agents does not preclude indirect effects, these results indicate that specific modifications may have discrete impacts on RPA dynamics. Thus, point mutations in GFP-RPAp34 were constructed in which either the two CDK phosphorylation sites (Ser-23 and Ser-29) or the defined DNA-PK phosphorylation sites (Thr-21 and Ser-33) were mutated to alanine (Dutta and Stillman, 1992; Niu et al., 1997). Interestingly, the assembly of these mutants into repair foci after DNA damage was not qualitatively impacted, nor was there a significant impact on their behavior at replication foci (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200312048/DC1). However, when examined by FRAP analysis the DNA-PK phosphorylation site mutant failed to exhibit rapid turnover at repair foci induced by CDDP insult (t 1/2 of 1.8 s), unlike the wild-type or CDK phosphorylation site mutant with t 1/2 values of 540 and 590 ms, respectively (Fig. 4, B and C). To further test the action of DNA-PK in regulating RPA behavior, wild-type GFP-RPAp34 was transfected into the glioblastoma cell line M059J deficient in DNA-PK activity and also into the cell line M059K restored with the DNA-PK catalytic subunit. We observed no qualitative difference in focus formation induced by various stresses (Fig. 4 D and not depicted), which is consistent with recent reports (Barr et al., 2003). However, when analyzed by photobleaching analysis a significant difference in the exchange at repair foci was detected (Fig. 4 D). GFP-RPAp34 exhibited rapid turnover in the DNA-PK–proficient cell line M059K (t 1/2 of 680 ms), whereas turnover was drastically impaired in M059J cells (t 1/2 of 3.1 s). Together, these data clearly demonstrate that DNA-PK plays a direct role in promoting RPAp34 turnover at repair foci, presumably by altering its binding affinity for substrates such as single-stranded DNA or other repair factors. These results provide a dynamic framework for RPA modification in the regulation of repair and led us to speculate that similar modifications may dictate the behavior of PCNA.
Figure 4.
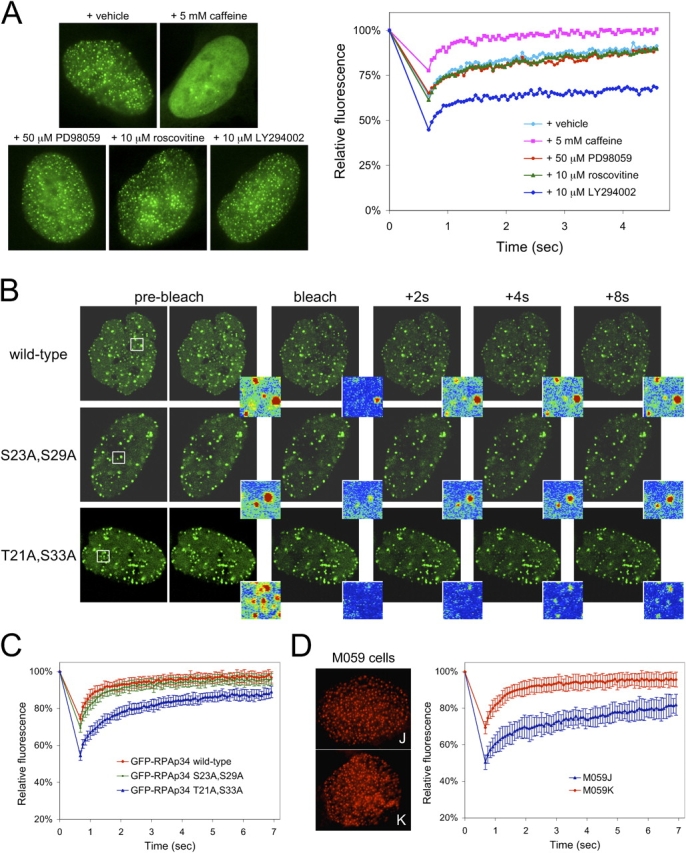
Phosphorylation by DNA-PK enhances turnover of RPAp34 at repair foci. (A) Localization and quantitative FRAP analysis of U2OS cells stably expressing GFP-RPAp34 after damage with 16 μM CDDP in combination with the indicated drug for 4 h. (B) FRAP on U2OS cells stably expressing GFP-RPAp34 wild type, a CDK phosphorylation site mutant (S23A and S29A), or a DNA-PK phosphorylation site mutant (T21A and S33A) after damage with 16 μM CDDP for 4 h. Pseudocolored insets display the intensity profile of the photobleached region boxed in white, 2.9 μm × 2.9 μm. (C) Quantitative FRAP analysis of cells in B. (D) Representative images of endogenous RPA (left) and quantitative FRAP analysis of GFP-RPAp34 (right) expressed in M059J and M059K cells after damage with 16 μM CDDP for 4 h. The error bars are the SD in the FRAP measurements.
In contrast to RPA, which is highly phosphorylated after DNA damage, there is little evidence that PCNA is regulated via direct phosphorylation. However, we used kinase inhibitors to examine the consequence of attenuating signaling pathways that may have indirect influence on PCNA dynamics (e.g., via RPA modulation). As with RPAp34, caffeine treatment inhibited the recruitment of PCNA to subnuclear foci and resulted in highly mobile protein behavior (Fig. 5 A). Similarly, roscovitine and PD98059 had little influence on PCNA retention (Fig. 5 A). Surprisingly, LY294002 significantly enhanced PCNA turnover at repair foci (Fig. 5 A), suggesting that the action of DNA-PK has opposing effects on PCNA versus RPA dynamics. To determine if direct influences on PCNA could mimic the effect of either caffeine or LY294002, we sought to examine if any posttranslational modification is responsible for dictating PCNA function in repair. It has been recently shown in Saccharomyces cerevisiae that a lysine residue (K164) on the surface of PCNA is ubiquitinated by RAD6 after DNA damage (Hoege et al., 2002). The critical role of this residue in the DNA damage response was highlighted by the finding that yeast strains unable to modify the residue (K164R substitution) are viable yet sensitive to DNA damage (Hoege et al., 2002). As this residue is highly conserved, we made an analogous substitution in the human PCNA (K164R) and studied its behavior. As shown in Fig. 5 (B and E), the K164R allele colocalized with sites of active DNA replication and exhibited fluorescence recovery comparable to wild-type PCNA in S-phase cells. Consistent with the results of Hoege et al. (2002), we found that wild-type PCNA was monoubiquitinated after damage, whereas no modification of the K164R mutant allele could be detected (Fig. 5 C). When treated with CDDP, the K164R allele of PCNA failed to efficiently accumulate into repair foci (Fig. 5 D). Additionally, we failed to observe localization of the K164R allele to γ-H2AX foci after CDDP damage (Fig. S3). By FRAP analysis, the K164R allele remained mobile after insult with CDDP, unlike the highly immobile wild-type allele (Fig. 5 E). Thus, loss of the K164 modification site precludes the stable assembly of PCNA at repair foci.
Figure 5.
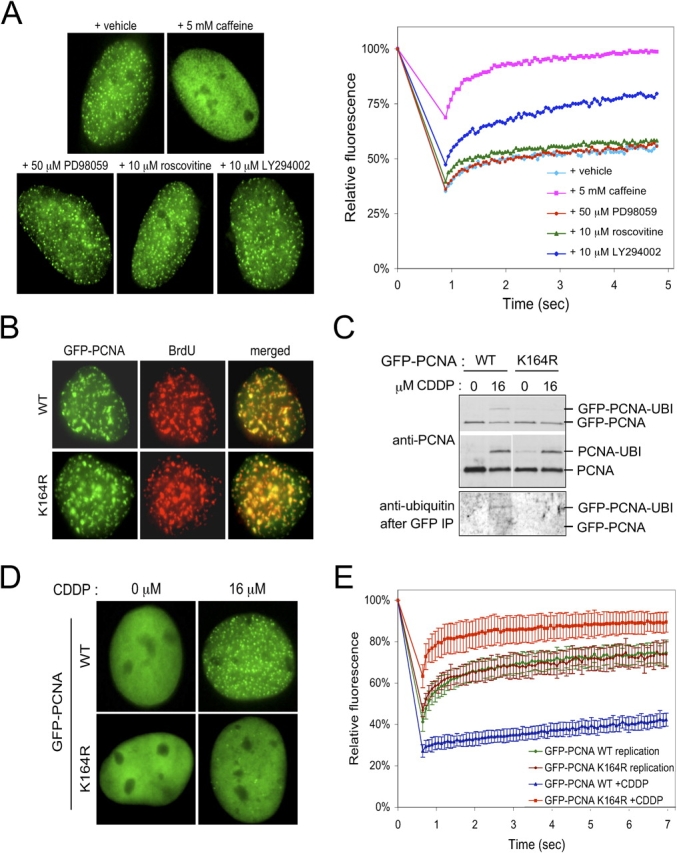
Monoubiquitination of Lys-164 targets PCNA for DNA damage repair. (A) Localization and quantitative FRAP analysis of U2OS cells stably expressing GFP-PCNA after damage with 16 μM CDDP in combination with the indicated drug for 4 h. (B) Colocalization of GFP-PCNA wild type and K164R mutant with sites of active BrdU incorporation. (C) K164R mutation renders PCNA refractory to ubiquitination after genotoxic insult. Total cell lysate (top) and protein immunoprecipitated with GFP antibody (bottom) from U2OS cells stably expressing GFP-PCNA wild type or K164R mutant after treatment with vehicle or CDDP were resolved by SDS-PAGE, and the indicated protein species were detected by immunoblot. (D) Localization of PCNA wild type and K164R mutant after insult with CDDP for 4 h. (E) Quantitative FRAP analysis of GFP-PCNA wild-type and K164R mutant in U2OS cells during replication and after damage with 16 μM CDDP for 4 h. The error bars are the SD in the FRAP measurements.
Our study presents the first dynamic analyses of the replication factors PCNA and RPAp34 in the DNA damage response in living cells. It has been largely viewed that the role of replication factors after genotoxic insult mirrors their respective roles in DNA replication. The RPA complex is believed to stabilize regions of single-stranded DNA that are uncovered during the unwinding of replication origins and ongoing replication processes at the fork (Waga and Stillman, 1998). In the context of DNA repair, it has been hypothesized that RPA plays a similar role during resynthesis reactions and signaling that damage is present (Riedl et al., 2003; Zou and Elledge, 2003). In contrast, PCNA would be viewed as functioning as an accessory subunit for polymerases involved either in synthesis of the genome or resynthesis in regions of damage (Waga and Stillman, 1998; Riedl et al., 2003). However, our analyses illustrate that the behavior of both RPAp34 and PCNA in replication versus repair is discrete. A dual-function protein, TFIIH, has previously been shown to display distinct residence times depending on its action in transcription versus repair (Hoogstraten et al., 2002). Interestingly, TFIIH displayed longer residence times during repair than transcription, similar to our observations with PCNA and RPAp34 during repair versus replication. However, in the case of TFIIH there is a well documented biochemical distinction between the repair and transcriptional complex (Svejstrup et al., 1995). It is less clear what might underlie the differences in PCNA and RPAp34 behavior between replication and repair. In the case of PCNA, we speculate that the types of DNA polymerases associated with the sliding clamp, or alternatively association with other repair proteins (e.g., MSH2 or Rad9), may be a primary determinant.
Several studies have described specific signaling pathways that regulate the biological activity of replication factors. Specifically, phosphorylation of RPA subunits after genotoxic insult has been observed (Shao et al., 1999; Wang et al., 2001; Barr et al., 2003). These reports have speculated that phosphorylation enhances RPA affinity for damaged or single-stranded DNA, effectively stabilizing a denatured DNA structure that RPA has been predicted to play a static role in maintaining. Our results strongly question this model. RPAp34 was observed to rapidly exchange at repair foci with significantly faster kinetics than other repair factors (i.e., PCNA). Contrary to expectation, we found that phosphorylation of RPAp34 by DNA-PK was not required for focus formation. Rather, this phosphorylation event destabilized RPAp34 at sites of damage. The use of increasing the turnover of RPAp34 at repair foci is at present unknown. However, several tantalizing possibilities exist, such as a role for phosphorylated RPA in signaling for the recruitment of additional factors from the nucleoplasm or to facilitate accessibility of repair factors to the region of damage. Alternatively, phosphorylation by DNA-PK may be required for efficient functioning of RPAp34 in repair processes. Cells deficient in DNA-PK activity have defects in DNA repair and V(D)J recombination, and an inability to rapidly modulate RPA–DNA interactions may explain these defects. Additionally, we have delineated a posttranslational mechanism by which PCNA is targeted to or stabilized at repair foci. Although there appears to be no significant modification of human PCNA during replication or other phases of the cell cycle, modification by ubiquitin after DNA damage is essential for the stable assembly of PCNA at repair foci. This result provides an explanation for the lack of viability in yeast harboring the K164R allele after DNA damage (Hoege et al., 2002). Notably, ubiquitination also plays a role in targeting the Fanconi anemia protein FANCD2 to foci after DNA damage (Garcia-Higuera et al., 2001). It will be interesting to further examine the critical role of ubiquitination in the DNA damage response.
The dynamic behavior of repair factors in living cells was first demonstrated by Houtsmuller et al. (1999) who observed that the endonuclease complex ERCC1-XPF rapidly exchanges at sites of UV-induced damage. However, not all proteins behave identically at sites of damage, as Nbs1 is largely tethered whereas Chk2 remains highly mobile (comparable to our observations with PCNA and RPAp34, respectively; Lukas et al., 2003). In this context, our data support a dynamic exchange model in which multiple repair factors have access to and rapidly turn over at sites of genetic insult. Further analysis of the DNA damage response in living cells will continue to provide powerful insight into the complex process of repair essential for the maintenance of genomic stability.
Materials and methods
Plasmids and mutagenesis
Expression plasmids encoding GFP-PCNA, GFP-RPAp34, and GFP-histone H2B have been described previously (Leonhardt et al., 2000; Phair and Misteli, 2000; Sporbert et al., 2002). Lys-164 of GFP-PCNA was mutated to Arg, and multiple NH2-terminal phosphorylation sites of GFP-RPAp34 (Thr-21, Ser-23, Ser-29, and Ser-33) were mutated to Ala and confirmed by sequencing.
Cell culture and generation of stable cell lines
Rat-1, U2OS, and M059 cells were grown in DME supplemented with 10% FBS. Rat-1 and U2OS cells were transfected with GFP-PCNA, GFP-RPAp34, and GFP-histone H2B along with pBABE-Puro using FuGene 6 (Roche), and multiple independent stable cell lines were established after puromycin selection. Transient transfection of M059 cells was performed using FuGene 6. Clinical grade CDDP was obtained from Bristol Oncology. All other drugs were obtained from commercially available sources.
Immunoprecipitation and immunoblotting
Cells were lysed in NET-N supplemented with protease inhibitors, and clarified lysate was immunoprecipitated with a monoclonal GFP antibody (B-2; Santa Cruz Biotechnology, Inc.). Proteins were detected with the following antibodies: PCNA (FL-261; Santa Cruz Biotechnology, Inc.), RPA (Ab-3; Oncogene Research Products), and ubiquitin (U5379; Sigma-Aldrich).
BrdU labeling and immunofluorescence
Asynchronously growing cells were pulsed with BrdU for 15 min, and sites of BrdU incorporation were visualized as described previously (Angus et al., 2003). For detection of specific DNA lesions, cells were fixed in 3.7% formaldehyde and probed with antibodies to cyclobutane pyrimidine dimers (T. Matsunaga, Osaka University, Osaka, Japan) or Pt-DNA adducts (M. Tilby, University of Newcastle upon Tyne, Newcastle, UK). For all other immunofluorescence, cells were fixed in cold methanol for 20 min and probed with antibodies to RPA (Ab-3; Oncogene Research Products), MSH2 (N-20; Santa Cruz Biotechnology, Inc.), ERCC1 (FL-297; Santa Cruz Biotechnology, Inc.), or phospho-histone H2A.X Ser-139 (JBW301; Upstate Biotechnology). Imaging was performed on a microscope (model Microphot-FX; Nikon) using a plan-apochromat 60× 1.4 NA objective.
UV microirradiation
UVC radiation was delivered using a low-pressure mercury lamp with maximal output at 254 nm. Polycarbonate filters containing either 3- or 5-μm diameter pores were gently laid over the cells before UV irradiation.
Dot-blot assay of Pt-DNA adduct levels
4 μg of genomic DNA isolated from U2OS cells after CDDP damage was extensively boiled and sheared by sonication, transferred onto a nylon membrane, and probed with the Pt-DNA adduct antibody.
Live-cell imaging and FRAP
Live-cell imaging and photobleaching analysis was performed at 37°C on a confocal microscope (model LSM510 Axiovert; Carl Zeiss MicroImaging, Inc.) using the 488-nm line of an argon laser (25 mW nominal output, photomultiplier detection through a pinhole diameter of 96 μm and a 505-nm long-pass filter) as described previously (Phair and Misteli, 2000; Angus et al., 2003).
Online supplemental material
Three supplemental figures and accompanying figure legends are available online at http:www.jcb.org/cgi/conent/full/jcb.200312048/DC1.
Acknowledgments
We thank Nancy Kleene, Robert Hennigan, Michael Tilby, Tsukasa Matsunaga, Yoli Sanchez, and Kathleen Dixon for kindly providing reagents and technical assistance. We thank Andre Nussenzweig, Yoli Sanchez, Peter Stambrook, and Karen Knudsen for critical reading of the manuscript.
M.C. Cardoso is funded by the Deutsche Forschungsgemeinschaft, and E.S. Knudsen is funded by the American Cancer Society (grant RSG-01-254-ECG) and National Cancer Institute (grant CA-106471).
D.A. Solomon's present address is Dept. of Cell Biology, Lombardi Cancer Center, Georgetown University School of Medicine, Washington, DC 20057.
Abbreviations used in this paper: APH, aphidicolin; CDDP, cisplatin or cis-diamminedichloroplatinum II; DNA-PK, DNA-dependent protein kinase; HU, hydroxyurea; PCNA, proliferating cell nuclear antigen.
References
- Angus, S.P., D.A. Solomon, L. Kuschel, R.F. Hennigan, and E.S. Knudsen. 2003. Retinoblastoma tumor suppressor: analyses of dynamic behavior in living cells reveal multiple modes of regulation. Mol. Cell. Biol. 23:8172–8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, S.M., C.G. Leung, E.E. Chang, and K.A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047–1051. [DOI] [PubMed] [Google Scholar]
- Dutta, A., and B. Stillman. 1992. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 11:2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera, I., T. Taniguchi, S. Ganesan, M.S. Meyn, C. Timmers, J. Hejna, M. Grompe, and A.D. D'Andrea. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 7:249–262. [DOI] [PubMed] [Google Scholar]
- He, Z., L.A. Henricksen, M.S. Wold, and C.J. Ingles. 1995. RPA involvement in the damage-recognition and incision steps of nucleotide excision repair. Nature. 374:566–569. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G.L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141. [DOI] [PubMed] [Google Scholar]
- Hoogstraten, D., A.L. Nigg, H. Heath, L.H. Mullenders, R. van Driel, J.H. Hoeijmakers, W. Vermeulen, and A.B. Houtsmuller. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell. 10:1163–1174. [DOI] [PubMed] [Google Scholar]
- Houtsmuller, A.B., S. Rademakers, A.L. Nigg, D. Hoogstraten, J.H. Hoeijmakers, and W. Vermeulen. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science. 284:958–961. [DOI] [PubMed] [Google Scholar]
- Izzard, R.A., S.P. Jackson, and G.C. Smith. 1999. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 59:2581–2586. [PubMed] [Google Scholar]
- Leonhardt, H., H.P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M.C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas, C., J. Falck, J. Bartkova, J. Bartek, and J. Lukas. 2003. Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat. Cell Biol. 5:255–260. [DOI] [PubMed] [Google Scholar]
- Maser, R.S., K.J. Monsen, B.E. Nelms, and J.H. Petrini. 1997. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol. Cell. Biol. 17:6087–6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer, L., A. Borgne, O. Mulner, J.P. Chong, J.J. Blow, N. Inagaki, M. Inagaki, J.G. Delcros, and J.P. Moulinoux. 1997. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 243:527–536. [DOI] [PubMed] [Google Scholar]
- Niu, H., H. Erdjument-Bromage, Z.Q. Pan, S.H. Lee, P. Tempst, and J. Hurwitz. 1997. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J. Biol. Chem. 272:12634–12641. [DOI] [PubMed] [Google Scholar]
- Phair, R.D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature. 404:604–609. [DOI] [PubMed] [Google Scholar]
- Riedl, T., F. Hanaoka, and J.M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse, J., and S.P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science. 297:547–551. [DOI] [PubMed] [Google Scholar]
- Sarkaria, J.N., E.C. Busby, R.S. Tibbetts, P. Roos, Y. Taya, L.M. Karnitz, and R.T. Abraham. 1999. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59:4375–4382. [PubMed] [Google Scholar]
- Shao, R.G., C.X. Cao, H. Zhang, K.W. Kohn, M.S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji, K.K., M.K. Kenny, and R.D. Wood. 1992. Proliferating cell nuclear antigen is required for DNA excision repair. Cell. 69:367–374. [DOI] [PubMed] [Google Scholar]
- Siddik, Z.H. 2003. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 22:7265–7279. [DOI] [PubMed] [Google Scholar]
- Sporbert, A., A. Gahl, R. Ankerhold, H. Leonhardt, and M.C. Cardoso. 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol. Cell. 10:1355–1365. [DOI] [PubMed] [Google Scholar]
- Svejstrup, J.Q., Z. Wang, W.J. Feaver, X. Wu, D.A. Bushnell, T.F. Donahue, E.C. Friedberg, and R.D. Kornberg. 1995. Different forms of TFIIH for transcription and DNA repair: holo-TFIIH and a nucleotide excision repairosome. Cell. 80:21–28. [DOI] [PubMed] [Google Scholar]
- Tilby, M.J., C. Johnson, R.J. Knox, J. Cordell, J.J. Roberts, and C.J. Dean. 1991. Sensitive detection of DNA modifications induced by cisplatin and carboplatin in vitro and in vivo using a monoclonal antibody. Cancer Res. 51:123–129. [PubMed] [Google Scholar]
- Vassin, V.M., M.S. Wold, and J.A. Borowiec. 2004. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 24:1930–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67:721–751. [DOI] [PubMed] [Google Scholar]
- Walker, E.H., M.E. Pacold, O. Perisic, L. Stephens, P.T. Hawkins, M.P. Wymann, and R.L. Williams. 2000. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell. 6:909–919. [DOI] [PubMed] [Google Scholar]
- Wang, B., S. Matsuoka, P.B. Carpenter, and S.J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science. 298:1435–1438. [DOI] [PubMed] [Google Scholar]
- Wang, H., J. Guan, A.R. Perrault, Y. Wang, and G. Iliakis. 2001. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 61:8554–8563. [PubMed] [Google Scholar]
- Ward, I.M., and J. Chen. 2001. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. J. Biol. Chem. 276:47759–47762. [DOI] [PubMed] [Google Scholar]
- Zou, L., and S.J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 300:1542–1548. [DOI] [PubMed] [Google Scholar]


