Abstract
Heterochromatin is thought to play a critical role for centromeric function. However, the respective contributions of the distinct repetitive sequences found in these regions, such as minor and major satellites in the mouse, have remained largely unsolved. We show that these centric and pericentric repeats on the chromosomes have distinct heterochromatic characteristics in the nucleus. Major satellites from different chromosomes form clusters associated with heterochromatin protein 1α, whereas minor satellites are individual entities associated with centromeric proteins. Both regions contain methylated histone H3 (Me-K9 H3) but show different micrococcal nuclease sensitivities. A dinucleosome repeating unit is found specifically associated with major satellites. These domains replicate asynchronously, and chromatid cohesion is sustained for a longer time in major satellites compared with minor satellites. Such prolonged cohesion in major satellites is lost in the absence of Suv39h histone methyltransferases. Thus, we define functionally independent centromeric subdomains, which spatio-temporal isolation is proposed to be important for centromeric cohesion and dissociation during chromosome segregation.
Keywords: centromere; cohesion; replication; nuclear organization; cluster
Introduction
In eukaryotic cells, centromeres ensure that during cellular division each daughter cell receives one copy of each chromosome. Therefore, these chromosomal regions are key elements for the correct segregation and inheritance of genetic information (Pidoux and Allshire, 2000). In the budding yeast Saccharomyces cerevisiae, a defined 125-bp DNA element is sufficient to confer such activity (Fitzgerald-Hayes et al., 1982). In contrast, in most other eukaryotes, including mammals, centromere identity and function cannot be transmitted simply by a specific DNA sequence. This finding has led to the concept of an “epigenetic” component in centromere function that can be inherited throughout multiple divisions, yet is not encoded genetically (Karpen and Allshire, 1997). Importantly, an increasing number of epigenetic marks have been uncovered that are associated with the heterochromatic state (Maison and Almouzni, 2004). Such a state has been considered a hallmark of centromeric regions, which remain condensed during interphase, thus by definition is constitutively heterochromatic (Heitz, 1928). Furthermore, the heterochromatic nature of these domains is thought to contribute to centromere identity and function (Perrod and Gasser, 2003).
In the mouse, Mus musculus domesticus, two types of repetitive DNA sequences are associated with centromeres. These are the major satellite repeats (6 megabases of 234 bp units) and minor satellite repeats (∼600 kb of 120 bp units; Choo, 1997). In situ hybridization on metaphase chromosomes has shown that major satellite sequences are located pericentrically, whereas minor satellite sequences coincide with the centric constriction (Wong and Rattner, 1988; Joseph et al., 1989). In interphase nuclei, the association of centromeres of different chromosomes results in an organization in clusters. These highly condensed clusters are easily detectable cytologically (Hsu et al., 1971). It is believed that ectopic pairing of repetitive sequences and/or association of heterochromatin components produces this organization (Comings, 1980; Manuelidis, 1990). However, it has yet to be elucidated whether this association occurs through major and/or minor satellite domains, and whether the nature of the heterochromatin in these domains differs in any way. To date, several typical marks have been associated with centromeric heterochromatin: histones are generally hypoacetylated (Jeppesen et al., 1992) and specifically methylated on lysine 9 in the NH2-terminal tail of histone H3 (Me-K9 H3; Peters et al., 2001). In mouse cells, both of these characteristics (Peters et al., 2001; Taddei et al., 2001), in conjunction with an RNA component (Maison et al., 2002; Muchardt et al., 2002), are necessary for the maintenance of heterochromatin protein 1 (HP1) within centromeric regions. The presence of the latter is thought to contribute to centromere function (Eissenberg and Elgin, 2000). Most importantly, how these heterochromatic marks distribute between the two subdomains in interphase to contribute to a mitotic function is still unknown. This is particularly critical given that in mitosis a clear specialization is already evidenced by the fact that kinetochore-associated proteins can be found at centric regions, whereas heterochromatin-associated proteins are associated mainly with pericentric domains (Craig et al., 2003). These findings prompted us to explore the (three-dimensional) organization of the minor and major satellite domains in the interphase nucleus and to compare this to their organization on mitotic chromosomes to gain insights into their spatio-temporal dynamics and potential importance for centromere function.
We find that in mouse nuclei, centromeric heterochromatin clusters (“chromocenters”) are formed by the coalescence of the major satellites, whereas the corresponding minor satellites are located in a surrounding domain as several separate entities. By combining immunofluorescence and DNA FISH, we observed that HP1α specifically accumulates on the major satellites, whereas centromeric protein (CENP) distribution is associated only with the minor satellites. Although both of these regions contain Me-K9 H3, these domains display different micrococcal nuclease (Mnase) sensitivity. Furthermore, chromatin immunoprecipitation reveals the existence of distinct Me-K9 H3–containing dinucleosomes within major satellite regions. Thus, each of these regions appears to be associated with a distinct higher order chromatin organization. In addition, we find that these domains replicate asynchronously: major satellites replicate in the middle of S-phase, and minor satellites replicate later during S-phase. Another important feature is detected in mitosis when we find that chromatid cohesion is sustained for a longer time in major satellites compared with minor satellites. Remarkably, in cells lacking the Suv39h histone methyltransferases, which is important for HP1 localization at centromeric regions, such prolonged cohesion in major satellite regions is lost while minor satellite regions remain unaffected.
Thus, we conclude that two spatially distinct domains can be defined with specific marks and differential timing in both replication and chromatid separation. Such spatio-temporal organization is proposed to be critical to ensure a proper kinetochore function with coordination between centromeric cohesion and dissociation necessary during chromosome segregation.
Results
Major and minor satellites form polar clusters within interphase nuclei
In mouse acrocentric chromosomes, the minor satellites are centric, whereas the major satellites are pericentric (Wong and Rattner, 1988; Joseph et al., 1989; Fig. 1 A, top). These sequences are well conserved between chromosomes, except for the Y chromosome, so that a single probe can stain the centromeres of all chromosomes by in situ hybridization (Choo, 1997). We analyzed the spatial organization of these regions using two-color DNA FISH with appropriate probes (Lehnertz et al., 2003), both on metaphase chromosomes (Fig. 1 A, bottom) and interphase nuclei (Fig. 1 B). On metaphase chromosomes, major satellites display a single large signal, whereas sister chromatids of minor satellites appear as a doublet signal (Fig. 1 A, bottom). In interphase, major satellites were detected as large spots, which colocalize with the DAPI-dense clusters (Fig. 1 B, a; Matsuda and Chapman, 1991). In contrast, the signal corresponding to minor satellites appeared at the periphery of the clusters as several individual spots (Fig. 1 B, b). Therefore, the two types of sequences clearly appear as distinct entities that are spatially segregated. This result was reproducibly obtained with several mouse cell lines (unpublished data). The three-dimensional organization of major and minor satellites, as shown by confocal analysis (Fig. 1 C, top), reveals that even at this higher resolution, minor and major satellites are juxtaposed but segregated. Confocal images were used for three-dimensional reconstruction (Fig. 1 C, middle). Minor satellites were always detected at one side of the cluster (Fig. 1 C, bottom), indicating a polar association of the clusters. Together, our results suggest that major satellites from different chromosomes are associated to form clusters, whereas minor satellites are located at the periphery of the cluster and form individual entities.
Figure 1.

Major and minor satellite DNA define three-dimensional domains in mouse interphase nuclei. (A, top) Scheme of a typical acrocentric mouse chromosome with the primary constriction corresponding to the centromeric region close to telomere. (a) The location of telomeres (black), major satellites (green), minor satellites (red), and the long arm of the chromosome (blue) are indicated. (b) A close-up view of the centromeric region is shown. (bottom) Localization of major and minor satellites on metaphase chromosomes by FISH analysis. (a–c) Minor satellite (red), major satellite (green) probes, and DNA (DAPI staining; blue) are shown. (d) Close-up view of the centromeric region. (B) Major and minor satellites in interphase. (a) Triple color image. (b) Two-color image of DAPI and minor satellites. (c) Two-color image of minor and major satellites. (d) A close-up view of the cluster. (C, top) Mid-zone confocal sections of an individual interphase nucleus showing major (green) and minor (red) satellites by two-color DNA FISH. Bars, 2 μm. (bottom) Three-dimensional reconstruction of the major (green) and minor (red) satellite DNA in mouse interphase nuclei. (top) Reconstructed model of a single nucleus. (middle) Close-up of a single cluster, around the Y-axis at 90° intervals from left to right.
Major and minor satellite regions present distinct timing of chromatid separation during the cell cycle
Accurate chromosome segregation requires that sister chromatids remain attached until chromosomes have bilaterally attached to the spindle, only then can anaphase ensue. Given that mammalian and fly cohesin is retained only at centromeric regions until anaphase (Waizenegger et al., 2000; Warren et al., 2000), a specialized role in holding sister chromatids together has been attributed to centromeric regions. To address whether or not specific parts within the centromeric region could be responsible for such sustained cohesion, we followed the dynamic association of minor and major satellites during this process by DNA FISH. The number of major satellite spots was used as a measure of chromosome dissociation. We observed that these spots in interphase nuclei (on average 16) increase significantly to reach a maximum in metaphase chromosomes (80 spots associated to each chromosome; Fig. 2 A). This number fits with the average number of chromosomes estimated to be present in this cell line. Prophase stage was identified using Phospho-Ser 10 histone H3 immunostaining (Hendzel et al., 1997). At this stage, major satellite spots increase to 40–80 spots in 80% of the cells, indicating that dissociation between major satellites from different chromosomes had already begun (Fig. 2 A). Subsequently, when this chromosome separation is achieved (prophase with 80 major spots), we observe one minor satellite spot per major satellite spot (Fig. 2 A, close-up images); at this stage our resolution does not allow us to distinguish if minor satellite chromatids already separate. However, timing of sister chromatid resolution during mitosis for both minor and major satellites showed that, in metaphase, for each chromosome one single spot of major satellite corresponds to two spots of minor satellites (Fig. 2 A, close-up). At this stage, when cohesins from chromosome arms have already dissociated, centromeres represent the only connecting part between chromatids before anaphase (Waizenegger et al., 2000). Our data support the fact that the major satellites are key to ensure that sister chromatids remain attached. It is only later in anaphase, when the two chromatids of each chromosome migrate to the opposite spindle pole, that we can observe one major satellite spot per minor satellite spot. Thus, major satellite chromatids are the last centromeric part to separate during mitosis. A precise choreography of these events is summarized in our model (Fig. 2 B).
Figure 2.

Major and minor satellite organization during the cell cycle. (A) DNA FISH of major (green) and minor (red) satellite DNA on 3T3 cells throughout the cell cycle. DNA was visualized with DAPI. A close-up of selected foci or chromosome (insets, bottom panels). Bars, 5 μm. (B) Scheme for major and minor satellite dynamics of association during the cell cycle. Minor (small dots) and major (large dots) satellites from three individual chromosomes are presented in different color. A dark color is used for replicated chromatids. (1) Major satellites from different chromosomes associate in clusters in interphase. (2) Major satellites from different chromosomes dissociate in prophase. (3) Minor satellites from sister chromatids dissociate, whereas the major satellite sister chromatids still cohere. (4) Finally, during anaphase major satellite sister chromatids separate.
HP1α marks major satellite domains, whereas CENPs characterize minor satellite domains
We analyzed the relative distribution of specific centromeric heterochromatin marks in interphase nuclei and metaphase chromosomes, using combined protein immunolabeling and DNA FISH. The HP1α variant of HP1 has been shown recently to be a conserved mark of constitutive heterochromatin in several higher eukaryotes (Gilbert et al., 2003). In mouse cells, HP1α accumulates at DAPI dense heterochromatin clusters in interphase nuclei (Wreggett et al., 1994; Minc et al., 1999). We found that HP1α colocalized with major satellite domains, whereas minor satellites are juxtaposed but segregated from HP1α (Fig. 3, top, left). Using the anticentromere antibody (ACA) that recognizes the three intrinsic CENPs CENP-A, CENP-B, and CENP-C (Earnshaw and Rothfield, 1985), we found that the staining with ACA marks the blocks of minor satellites in interphase nucleus (Fig. 3, top, middle), revealing a specific association of centromeric core components with minor satellite domains. Coimmunostaining further confirmed the distinct labeling of separate domains by HP1α and ACA. Under these conditions, we failed to reveal any colocalization of ACA and HP1α antibodies. Indeed, the two signals are juxtaposed but clearly segregated (Fig. 3, top, right). Thus, the vast majority of CENP-A, B, and C proteins are mostly associated with the minor satellites but not with the major satellites. Our results indicate that major and minor satellite domains, besides their contrasting organization in the nucleus, carry different marks. During mitosis, we were unable to detect HP1α on fixed metaphase chromosomes (unpublished data), although a clear signal was obtained without fixation of the material, to ensure maximal accessibility of the antibodies (Jeppesen et al., 1992). Therefore, the fixation method used appears to be critical for the detection of components that are present in condensed chromatin. We found that HP1α colocalizes with DAPI-dense spots, which colocalize with major satellites on metaphase chromosome (Fig. 3, bottom). This result is consistent with previous data (Minc et al., 1999; Craig et al., 2003) and comforts the view of an association of HP1α with major satellites (found at DAPI-dense spots; Fig. 1) that is at least partly maintained during mitosis. In addition, ACA staining on metaphase chromosomes confirm that the CENPs display the same pattern as minor satellites at the centric region of the chromosome during metaphase (Fig. 3, bottom). Thus, HP1α and ACA marked distinct domains in interphase that could be stably maintained during mitosis.
Figure 3.
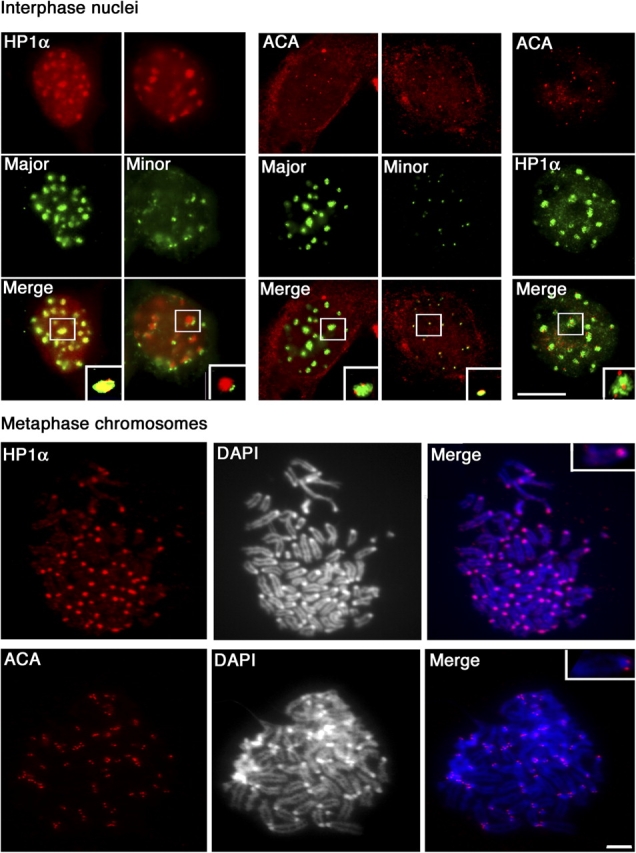
Major and minor satellite-specific marks in interphase and metaphase nuclei. Interphase: (left) HP1α (red) and major and minor DNA (green). (middle) ACA (CENP-A, B, C) is presented in red, major and minor DNA in green. (right) Costaining with ACA serum (red) combined with anti-HP1α antibodies (green). A close-up of selected foci (inset in the merged images). Metaphase: DNA was visualized with DAPI and protein staining in red. Double labeling is shown in merged images. A close-up of a chromosome selected (inset in the merged images). Bars, 5 μm.
Major and minor satellite domains marked by Me-K9 H3 harbor a distinct spatial organization
Another epigenetic mark thought to be typical of heterochromatin is histone H3 specifically methylated at lysine 9 (Me-K9 H3; Lachner et al., 2003). However, lysine residues can accept multiple methyl groups and, thus, can be mono-, di-, or tri-methylated (mono-Me-K9, di-Me-K9, tri-Me-K9; Rice and Allis, 2001; Peters et al., 2003; Rice et al., 2003). To investigate the H3-K9 methylation status of centromeric regions, we used two distinct antibodies raised against linear synthetic H3 peptides with either a di- or tri-Me-K9. The anti-di-Me-K9 antibody already used in several studies proved highly specific (Nakayama et al., 2001; Maison et al., 2002). The anti-tri-Me-K9 antibody was characterized by slot-blot using various H3 peptides corresponding to the amino terminal tail region, which were either unmodified or mono-, di-, or tri-methylated at the K4, K9, and K27 residues to test possible cross-reactions (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200403109/DC1). In addition, peptide competition experiments showed that the staining of interphase mouse nucleus obtained with the antibody was specifically competed away only by the tri-Me-K9 linear peptide.
We then assessed the nuclear locations of K9 methylation of H3 by immunofluorescence. The anti-di-Me-K9 antibody revealed a broad granular staining through the nucleoplasm (Maison et al., 2002). In contrast, the anti-tri-Me-K9 stained nuclear spots that colocalized with HP1α at pericentric heterochromatin (Fig. 4 A, left). This staining was maintained on metaphase chromosomes (Fig. 4 D). Immunofluorescence followed by DNA FISH showed that tri-Me-K9 labeling colocalized clearly with major satellites and to a lesser extent with minor satellites (Fig. S2 for three-dimensional analysis, available at http://www.jcb.org/cgi/content/full/jcb.200403109/DC1). In contrast, we could not detect any specific enrichment for di-Me-K9 (Fig. 4 A, middle and right). These data suggest that H3-K9 tri-methylation is generally enriched across the whole centromere.
Figure 4.
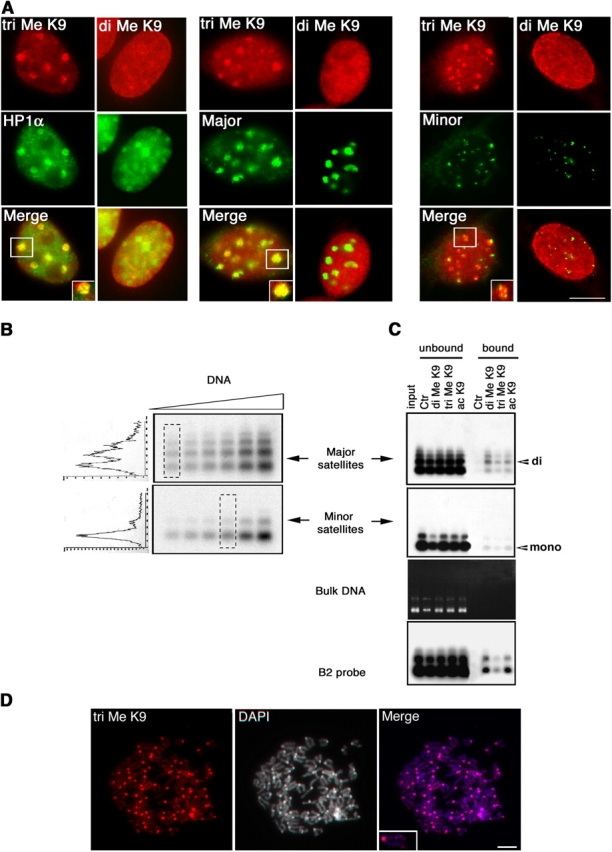
Major and minor satellite domains differ in their higher chromatin organization. (A) Staining of interphase nuclei with di-Me-K9 and tri-Me-K9 antibodies (red), combined with either anti-HP1α (left, green) or FISH for major satellites (middle, green) or minor satellites (right, green). Insets correspond to close-ups of selected foci. (B) A range of concentration corresponding to native oligonucleosomes isolated by partial Mnase digestion were analyzed by gel electrophoresis followed by transfert and hybridization with indicated probes. Scans are presented for signals of comparable intensity as indicated. (C) NChIP with antibodies against di- and tri-Me-K9 H3. Autoradiographs of the membrane after hybridization are presented using labeled probes: mouse major satellite (top) or minor satellite (below) or B2 repeat (bottom), and ethidium bromide-stained gel (Bulk DNA) to show migration of bulk DNA in the nucleosome preparation. Positions of mononucleosomal (mono) and dinucleosomal (di) DNA are indicated. (D) Immunofluorescence on metaphase with tri-Me-K9 antibodies (red), DNA visualized with DAPI, and a close-up of a selected chromosome are shown (inset). Bars, 5 μm.
We also examined the H3-K9 methylation status of centromeric regions at the molecular level by native chromatin immunoprecipitation (NChIP). Compared with the classical formaldehyde cross-linked chromatin immunoprecipitation technique, this procedure has the advantage of avoiding the formaldehyde cross-linking step that can fix chromatin interactions that are transient or simply due to spatial proximity. For NChIP, native oligonucleosomes were prepared and purified after Mnase digestion of mouse cell nuclei (O'Neill and Turner, 1995) to obtain a high enrichment in mononucleosome on bulk DNA visualized by EtBr (Fig. 4 C, Bulk DNA). Consistently, with the supposedly heterochromatic nature of centromeric regions, we find that these regions were more resistant to digestion, as attested by the detection of larger fragments when probing for major and to a lesser extent for minor satellite repeats when compared with bulk DNA (Fig. 4 C, compare minor and major to bulk). Detection of the larger nucleosomal fragments within major satellite regions demonstrated that they constitute the most resistant part of centromeric DNA compared with minor satellites, which are mainly composed of dinucleosomes and mononucleosomes (Fig. 4 C). To verify that our interpretation was not biased by effects, due to hybridization efficiency with the different probes and higher representation of major satellites compared with minor satellites in the genome, we loaded different amounts of material. Reproducibly for all points we confirmed a differential sensitivity to Mnase digestion. Comparison of the scans performed for the points, which provided signal of comparable cumulated intensity by densitometry, further evidenced this difference in digestion profiles (Fig. 4 B). Next, we used these native oligonucleosomes for immunoprecipitation with antibodies against modified Me-K9 H3. These NChIP experiments indicated that major and minor satellite regions were pulled-down by both di-Me-K9 and tri-Me-K9 antibodies (Fig. 4 C) as observed by formaldehyde cross-linked chromatin immunoprecipitation (Peters et al., 2003). However, when we compared the nucleosomal ladder in the input and pulled-down material, we noted that for major satellite the immunoprecipitated material was enriched in a dinucleosome fraction (such enrichment was not seen in the input). In contrast, for minor satellites, the majority of the immunoprecipitated material appeared mainly as mononucleosomes. To confirm that such a dinucleosome unit was specific to major satellite repeat, we used another type of repeat sequence in the genome (B2 repeat; Boyle et al., 1990). Using this probe, no enrichment was found while we confirmed the general hypoacetylation of centromeric regions compared with B2 repeats. Given that it has been previously proposed that a distinct architecture is required for HP1 accumulation within pericentromeric regions (Maison et al., 2002), we suggest that such architecture is built using an asymmetric dinucleosomal Me-K9 H3 subunit to establish higher order folding. These results further support the idea that major and minor satellite chromatins have distinct higher order structures that depend on nucleosome folding.
Major and minor satellites replicate asynchronously in S-phase
Replication of DNA late in S-phase of the cell cycle has been considered a hallmark of heterochromatin (Goldman et al., 1984; Hatton et al., 1988; Schubeler et al., 2002), which could play an important role in centromere function (Csink and Henikoff, 1998). Data from most organisms, with the exception of S. cerevisiae, favor the notion of generally mid to late replication timing patterns for centromeric regions (Ten Hagen et al., 1990; O'Keefe et al., 1992; Shelby et al., 2000; Sullivan and Karpen, 2001). Given the aforementioned distinct properties of centric and pericentric heterochromatin, we wished to examine their relative timing of DNA replication. We decided to combine BrdU immunostaining with DNA FISH on synchronized cells to address this issue. Synchronization of NIH 3T3 cells at the G1/S border was achieved using aphidicolin, an inhibitor of DNA polymerase. Cells were released into S-phase by washing away the inhibitor at different time points (hourly between 1 to 8 h) and pulse labeled for 10 min with the nucleotide analogue BrdU. Immunostaining of BrdU incorporation combined with either major or minor satellite DNA FISH was performed (Fig. 5). S-phase usually takes 7–8 h to complete in NIH 3T3 cells. Different replication patterns could be distinguished corresponding to early, mid, and late S-phase, as revealed by detection of BrdU incorporation (Fox et al., 1991; Dimitrova and Berezney, 2002). Typical replication patterns (Fig. 5, red) are shown combined with specific probing (Fig. 5, green) for major (Fig. 5, top) or minor (Fig. 5, bottom). In early S-phase, replication begins with discrete punctuate sites distributed throughout the nucleus and no colocalization is found at this time point for any of the probes. In middle S, around 3 to 6 h after release, the cells show BrdU rings around the major satellite clusters (Fig. 5, top, Merged), possibly reflecting newly synthesized DNA occurring at the periphery of the domain (Quivy et al., 2004). At this stage, no colocalization between minor satellites and BrdU is observed (Fig. 5, bottom). Finally, in late S, 7–8 h after release, BrdU incorporation again became more granular with small foci in the interior of the nucleus. Only at this stage could we detect some minor satellite colocalization with BrdU (Fig. 5, bottom). These data indicated that major and minor satellite domains replicate at different times during S-phase, with replication of major satellites in middle S, followed by replication of minor satellites in later S-phase.
Figure 5.
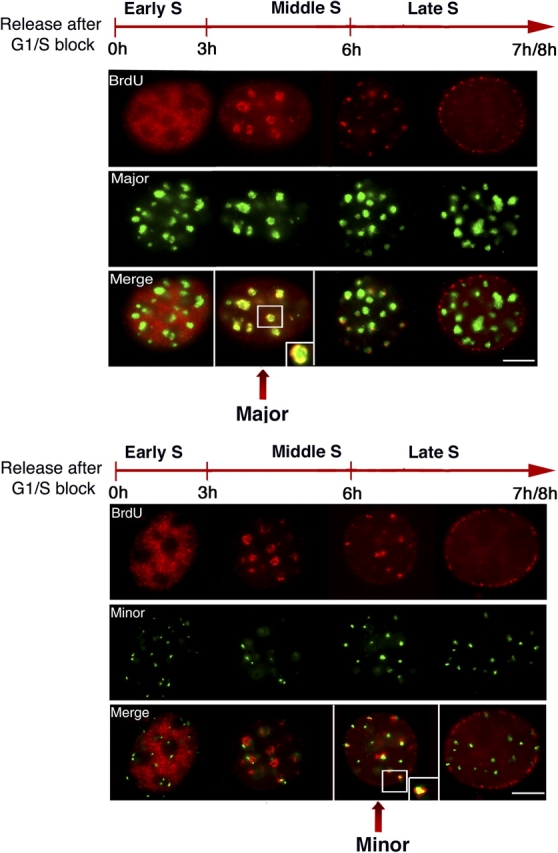
Replication timing of major and minor satellites during S-phase. Synchronized NIH 3T3 cells into S-phase were pulse labeled with BrdU for 10 min at the indicated times after release and stained for incorporation (BrdU, red) combined with DNA FISH (green) either for major (top) or minor satellites (bottom). Colocalization of BrdU staining with major or minor satellite DNA is presented in merged images. A close-up of a selected chromosome is shown (inset). Bars, 5 μm.
To further analyze this distinct timing for replication of minor and major, active sites of DNA synthesis were labeled by in situ elongation in the presence of Biotin-16-deoxyuridine (BiodU; Fig. 6, green), followed by DNA FISH of minor satellites (Fig. 6, red). Under these in vitro conditions, on permeabilized cells, no further replication initiation is permitted (Taddei et al., 1999), thus the BiodU enables detection of elongation alone, from origins that had fired before. Given that the time between the appearances of the BrdU in major satellite regions and that in the minor satellites is <3 h, we choose to perform the in situ elongation assay every 30 min between 0 to 3 h. After 90 min, the BiodU staining was found in large spots, which covered entirely the DAPI dense clusters (Fig. 6), indicating completion of the elongation reaction in major satellite clusters. Quantitatively this population represent 37.5% of labeled nuclei (population 1) without labeling minor satellites. This pattern arises most likely due to the prolonged labeling with BiodU of the major satellite domains, which replication initiation occurred previously. However, on the same slides for the same labeling time period, we could find in 13.2% of nuclei labeled (population 2) a clear colocalization between BiodU in small spots and minor satellites DNA, which can be assigned to elongation events for replication sites within minor satellite domains (Fig. 6, bottom). Although we cannot exclude that a minor fraction of labeled nuclei with both major and minor satellite regions colabeled, below the sensitivity of our detection method could exist, such a low frequency event argues against a common replication control. These data support the hypothesis whereby major and minor satellite domains replicate asynchronously possibly using independent initiation from different origins.
Figure 6.
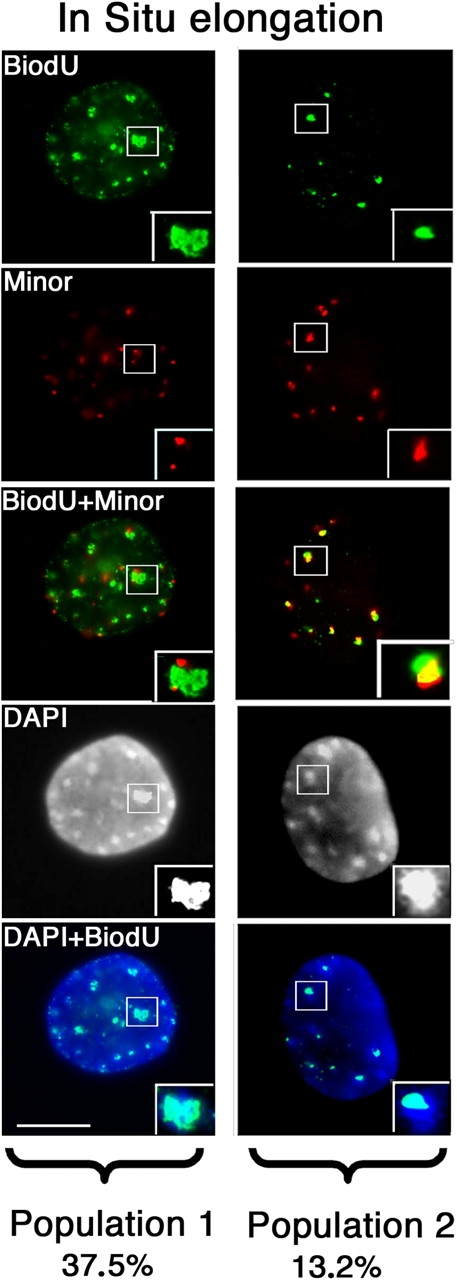
Major and minor satellites replication using in situ elongation with BiodU and minor satellite DNA FISH. Sites of DNA synthesis were labeled on MEFs by in situ elongation in the presence of Bio-16-dUTP for 90 min (green), followed by DNA FISH of minor satellites (red). Population 1, nuclei showing a complete staining of the DAPI dense clusters with BiodU. Population 2, nuclei showing a colocalization between BiodU and minor satellites DNA. Bar, 5 μm.
Sister chromatid cohesion is lost in major satellite region during metaphase in cells lacking the Suv39h methyltransferases
Given that in fission yeast, HP1-related proteins are required for association of cohesin with centromeres to ensure sister centromere cohesion and proper chromosome segregation (Bernard et al., 2001; Nonaka et al., 2002), we wondered if HP1 proteins would also be required for chromatid cohesion in major satellite region in mouse. We took advantage of the fact that in mouse the association of HP1α at pericentric heterochromatin is severely compromised in cells lacking the histone H3 Lys 9 methyltransferases Suv39h1 and Suv39h2, and we therefore investigate the HP1α localization and cohesion properties for minor and major satellites domains in such cells (Suv39h double null [dn]). Using coimmunostaining with HP1α and ACA in interphase nuclei, we observe that whereas HP1α staining is lacking in centromeric regions as previously described (Maison et al., 2002), the CENPs were still present as several separate entities in the Suv39h dn nuclei (Fig. 7 A). By combining ACA immunolabeling and DNA FISH, we observe a perfect colocalization between ACA signal and minor satellites (Fig. 7 A). Thus, the loss of histone H3 Lys 9 methylation at the centromeric regions seems to specifically affect the HP1α association with the major satellites. We analyzed the timing of chromatid segregation of minor and major satellites using DNA FISH on metaphase chromosomes. For each cell culture corresponding to wild type and Suv39h dn, over 50 metaphases were scored. The single spot of major satellites observed in the wild-type metaphase chromosome corresponds to the sister chromatids that still held together (Fig. 7 B, left). In the Suv39h dn metaphase chromosomes, 60–90% of the chromosomes per metaphase show a double spot evidenced both with the major satellite probe and DAPI staining (Fig. 7 B, right; indeed, these double spots are clearly observed when the chromosomes are well spread on the slide), indicating a defect in chromatid cohesion specific to major satellites. No difference was observed for the separation of minor satellite chromatids. These data indicate that the loss of H3 Lys 9 methylation and HP1α at pericentric major satellites leads to a defect in their chromatid cohesion and may cause a precocious segregation at metaphase stage.
Figure 7.
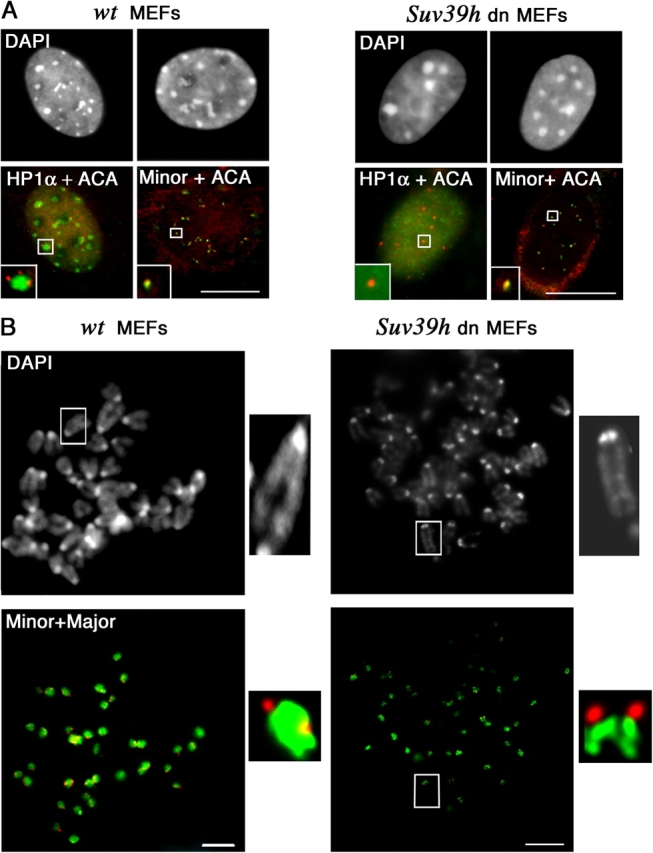
Major and minor satellite domains in Suv39h double mutant cells. (A) Major and minor satellite chromatin in MEFs Suv39h double mutant (Suv39h dn) compared with wild-type (wt) nuclei. Costaining with ACA serum (red) and anti-HP1α antibodies (green). ACA immunostaining (red) combined with minor satellite FISH (green). DNA was visualized with DAPI. Insets correspond to close-ups of selected foci. Bars, 5 μm. (B) Sister chromatid separation of minor and major satellites at metaphase stage in MEFs Suv39h double mutant (Suv39h dn) compared with wild type (wt). DNA FISH for major (green) and minor (red) satellites. DNA was visualized with DAPI. A close-up of a selected chromosome is shown along each panel (insets). Bars, 2 μm.
Discussion
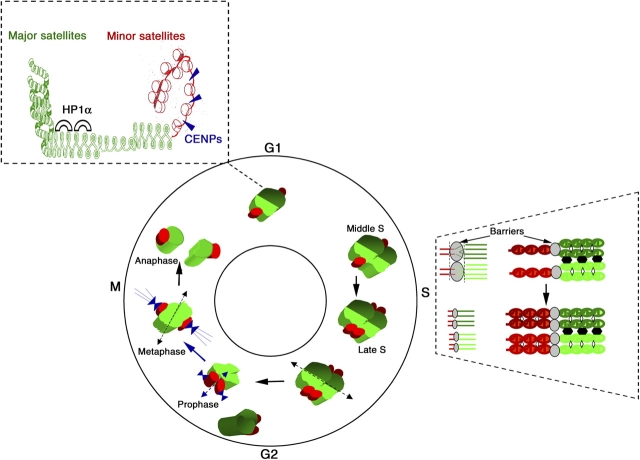
Centric and pericentric heterochromatin domains that are composed of minor and major satellite DNA, respectively, differ in their higher order nuclear organization, their chromatin content, and their replication. Our three-dimensional FISH analysis indicates that in interphase nuclei major satellites of different chromosomes are associated in clusters, whereas the corresponding minor satellites surround the clusters, but do not coalesce. Cluster organization within the nucleus is observed in different species and is thought to be important for a functional nuclear organization. In the yeast S. cerevisiae, their centromeric regions cluster at the nuclear periphery (Jin et al., 1998) as opposed to the telomeres, which themselves form multiple clusters at the opposite part of the nucleus (Gotta et al., 1996). However, the importance of this organization for centromere structure is still an open question. We show that clusters of major satellites from different chromosomes replicate together in the mid S-phase, and remain coalesced (Fig. 8, middle). In contrast, the corresponding minor satellites replicate later during S-phase and segregate earlier. Our data suggest that distinct replication timing and/or epigenetic barriers exist between major and minor satellite domains (Fig. 8, right). Interestingly, the fact that major and minor satellites replicate asynchronously provides an opportunity for them to be processed in an independent fashion to assemble different chromatin components that can permit either the maintenance or disruption of cohesion at different times after replication. Remarkably, whereas in mouse centromeric regions replicate from mid to late S-phase, in both Drosophila melanogaster and S. pombe they were found to replicate early (Ahmad and Henikoff, 2001; Kim et al., 2003). However, their centric and pericentric domains replicate at a different time, suggesting that a differential replication timing, rather than an absolute time in S-phase, may be the common key feature for centromere organization and function.
Figure 8.
Model for the spatio-temporal isolation of major and minor satellite domains in the nucleus. (top) Major satellites in green associated with HP1α (black semi-circle) form a stable 30-nm fiber with a dinucleosomal periodicity. Minor satellites (red), which are more accessible, are associated with CENPs (blue triangle). During the cell cycle, clusters of major satellites from two different chromosomes (dark and light green spots) with corresponding minor satellites surrounding them (red spots) are presented in G1. In S-phase, the major and minor satellites replicate asynchronously. The two distinct domains are depicted on the right for the major (green) and minor (red). Newly synthesized sister chromatids are held together by specific proteins (white diamond). The clustering of major satellites is maintained during replication (black bridges). In prophase, the clusters of major satellites from different chromosomes dissociate (dotted arrow). Sister chromatid cohesion of minor satellites is lost first (blue arrow) followed by the separation of major satellite sister chromatids as a last event during anaphase (black arrow).
Our data suggest that minor satellite domains contain both histone H3 and CENP-A. Such an interspersed organization may be a general property of centric heterochromatin, considering recent work showing that centric chromatin contain interspersed nucleosomes of histones H3 and CENP-A, both in D. melanogaster and human cells (Blower et al., 2002). Because histone tails were proposed to be critical for the targeting of HP1α (Lachner et al., 2001; Maison et al., 2002) and both major and minor satellites contain Me-K9 H3, the absence of HP1α accumulation in minor satellites might result from the differences in chromatin organization between minor and major satellites. Indeed, major satellite chromatin appears less accessible than the minor satellites, as revealed by Mnase digestion. Moreover, Me-K9 H3 tails in these domains display a specific organization with a dinucleosomal periodicity, whereas Me-K9 H3 in minor satellites are present in a simple mononucleosomal configuration (Fig. 8). Such a dinucleosome repeat is reminiscent of the repeating unit of the zig-zag nucleosomal organization, which stabilization in the 30-nm fiber state in vivo may occur through interaction with proteins such as HP1α (Richmond and Widom, 2000; Woodcock and Dimitrov, 2001; Fig. 8). The interspersed nucleosomes of histones H3 and CENP-A within the minor satellite chromatin might also contribute to the absence of accumulation of HP1α at the minor satellites (Fig. 8).
Importantly, minor and major satellite domains show distinct timing for chromatid separation during mitosis. Minor satellite chromatids, which are associated with CENPs, dissociate early at the metaphase stage. The major satellites associated with HP1α, which is also important for centromeric function in mouse (Peters et al., 2001; Taddei et al., 2001), constitute the latest centromeric part to separate. These data suggest that these domains possess distinct roles in centromere function. Minor satellite domains may provide a nucleation center for kinetochore assembly, which is required for the proper segregation of sister chromatids during mitosis, whereas major satellites may help to ensure cohesion between sister chromatids next to centromeres. Indeed, in yeast S. pombe outer centromere heterochromatin components associated to the outer centromeres such as the HP1 homologue Swi6, H3 Lys 9 methylation and more recently the RNA interference machinery have been involved in sister chromatid cohesion at centromere and proper chromosome segregation (Hall et al., 2003). In mammals, several cytological observations support a role of pericentric heterochromatin in centromere cohesion (Vig, 1982). However, it is the first time that a direct link is made between H3 Lys 9 methylation/HP1α and centromere cohesion in mammals. Indeed, our data suggest that the loss of HP1α at major satellites pericentric heterochromatin as observed in Suv39h dn cells leads to a defect in centromeric cohesion in mouse cells. Mechanistically, the maintenance of cohesion between major satellite sisters may be advantageous when tension is imposed at the most centric part, which nucleates kinetochore formation and microtubule anchoring.
In conclusion, we have characterized how centromeric domains are organized into distinct types of heterochromatin that can self-perpetuate. This functional organization may be conserved in human. The next challenge will be to address how these domains can be established de novo. Neocentromere formation in human cells, which is accompanied with the association of several CENPs and chromatin components (Warburton, 2001), is a challenge of particular interest. Given that they do not necessarily contain repeats found in pericentromeric regions, one may speculate that the cohesion property could be acquired “in trans,” especially because specific centromeric clusters are also observed in human (Alcobia et al., 2000). Alternatively, it is possible that other means could be used in human cells to ensure this cohesion property. Nevertheless, the conceptual advance in mouse cells in the present study should promote future work to elucidate how other organisms have solved the puzzling issue of constructing a centromeric region with dual properties: cohesion and segregation, and thus open avenues into the evolution of epigenetic identity of centromeres.
Materials and methods
Cell culture
Mouse cells: 3T3, L929 cells, and MEFs derived from wild type or from Suv39h1 Suv39h2 double null (Suv39h dn) embryos were grown in DME supplemented with 10% FCS, 10 μg/ml penicillin and streptomycin, and 2 mM l-glutamine (GIBCO BRL).
Two-color DNA FISH
Plasmids pCR4 Maj9-2 and pCR4 Min5-1 (Lehnertz et al., 2003) contain major and minor satellite DNA (provided by T. Jenuwein, Research Institute of Molecular Pathology, The Vienna Biocenter, Vienna, Austria). DNA fragments were labeled by nick translation (Life Technologies) with digoxigenin-11-dUTP or biotin-16-dUTP (Boehringer). Cells grown on coverslips were fixed and treated with 0.7% Triton X-100 and 0.1 M HCl for 10 min on ice followed by denaturation with 2× SSC/50% formamide 30 min at 80°C. Heat-denatured probes were hybridized overnight at 37°C. After hybridization and washes with 2× SSC/50% formamide and 2× SSC at 42°C, probe detection used a three-step procedure for amplification (Manuelidis et al., 1982). Biotin was revealed using Texas red–conjugated streptavidin and biotinylated antistreptavidin antibody (Vector Laboratories), followed by Texas red–conjugated streptavidin. Digoxigenin was detected using a sheep FITC-conjugated anti-digoxigenin serum (Roche), rabbit FITC-conjugated anti–sheep antibodies (Jackson ImmunoResearch Laboratories), and goat FITC-conjugated anti–rabbit antibodies (Jackson ImmunoResearch Laboratories). Coverslips were mounted in Vectashield containing 0.5 μg/ml DAPI (Vector Laboratories).
Combined protein immunolabeling and DNA FISH
Immunostaining was performed on Triton X-100 extracted cells (Taddei et al., 2001) using the following: anti-HP1α (Euromedex HP1α, 2HP-1H5-As, at a dilution of 1:400), anti-di-Me-K9 H3 (Upstate Biotechnology; 07–212, at 1:2,000), anti-tri-Me-K9 H3 (Abcam; ab1186 at 1:500), di-Me K4 antibody (Upstate Biotechnology; 07–030 at 1:200), ACA serum (provided by G. Steiner, Northwestern University, McGaw Medical Center, Chicago, IL; at 1:100), and anti-phospho H3-S10 (Upstate Biotechnology; 05–598, at 1:400) and secondary antibodies coupled to FITC or Texas red (Jackson ImmunoResearch Laboratories). DNA FISH was performed after PFA post-fixation.
Metaphase chromosome spreads, protein immunolabeling, and DNA FISH
Mitotic cells collected after growth for 30 min in medium containing colcemid (GIBCO BRL; 0.1 μg/ml) were washed in PBS. After swelling in 75 mM KCl, they were recovered onto glass slides by cytocentrifugation. Protein immunolabeling was performed without fixation (Jeppesen et al., 1992). For DNA FISH procedure, slides were fixed after cytocentrifugation.
Replication timing analysis combining BrdU immunolabeling and DNA FISH
NIH 3T3 cells were synchronized at the G1/S border by aphidicolin arrest, and maintained at 100% confluence for 1–2 d, and then reseeded to 60–80% confluence in media containing 3 μg/ml aphidicolin (Sigma-Aldrich) for 18 h. Aliquots were released from the block every hour (from 1 to 8 h) and pulse-labeled for 10 min with 40 μM BrdU (Sigma-Aldrich). BrdU was immunodetected after a denaturation step in 4 N HCl for 10 min using rat mAb (AbCys; OBT0030) at 1:200 dilution. Cells were subsequently post-fixed with 2% PFA and DNA FISH was performed.
Combined in situ elongation assay and DNA FISH
in situ elongation was performed on Triton X-100 permeabilized MEFs. Slides were incubated for different time points at 37°C in buffer containing 40 mM K-Hepes, pH 7.8, 7 mM MgCl2; 3 mM ATP; 0.1 mM each of GTP, CTP, and UTP; 0.1 mM each of dATP, dGTP, and dCTP; 40 μM BiodU; 20 mM of creatine phosphate; 0.5 mM DTT; and 2.5 μg of phosphocreatine kinase (Boehringer). Reactions were stopped in 40 mM K-Hepes, pH 7.8, 7 mM MgCl2, 3 mM CaCl2, 0.5 mM DTT and fixed in 2% PFA. BiodU was detected with Texas red–conjugated streptavidin (Vector Laboratories) and cells were subsequently refixed with 2% PFA. DNA FISH was performed using DNA probes labeled with digoxigenin-11-dUTP to avoid antibody cross-reactions. The assay was performed twice, in each independent experiment 200–300 nuclei were analyzed.
Microscopy and image analysis
Image acquisition was performed with an epifluorescence photomicroscope (DMR; Leica) equipped with a chilled charge-coupled-device camera (C5985; Hamamatsu Photonics), in which the resolution is 200 nm in x-y. Confocal sections were obtained with a confocal scanning microscope (model TCS-4D; Leica) equipped with an acousto-optical tunable filter and with 100× numerical aperture 1.4 plan-apochromat oil-immersion objective. For two-color images, 25–30 serial sections were collected at each imaging time point (256 × 256 pixels or 512 × 512; pixel size, 120–200 nm; z-step, 0.4–0.5 μm). Data in 8-bit TIFF format series for each color were analyzed using Metamorph software (Universal Imaging Corp.). Selected images were assembled using Adobe Photoshop. Three-dimensional reconstruction of confocal image stacks was performed using AmiraTM 2.3 (TGS).
Mnase digestion and NChIP
Native oligonucleosomes were isolated by partial Mnase digestion on L929 cell nuclei and sucrose gradient purification and analyzed by gel electrophoresis followed by transfert and hybridization with probes. For NChIP, 10 μg of oligonucleosomes were incubated with 4 μg of either purified IgG anti–mouse (Jackson ImmunoResearch Laboratories), anti-di-Me-K9 (Upstate Biotechnology; 07–212), anti-tri-Me-K9 (Abcam; ab1186), or anti-ac K9 (acetylated histone H3 on lysine 9; Upstate Biotechnology; 06–942) antibodies in 1 ml of 20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 5 mM EDTA at 4°C overnight. 100 μl of protein A–Sepharose slurry (50% wt/vol; Amersham Biosciences) was added for 3-h incubation at 4°C. Immunoprecipitated and nonimmunoprecipitated oligonucleosomes were recovered by centrifugation. After washes in 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, both unbound and bound fractions were adjusted to 0.4% SDS. DNA was purified and analyzed by agarose gel electrophoresis, stained with ethidium bromide, and transferred onto a Hybond N+ membrane (Amersham Biosciences). For hybridization, sequences of the probes used were minor satellite (947: 5′-CACATTCGTTGGAAACGGGATTTGTAGAAC-3′; Kipling et al., 1994), major satellite (204: 5′-GTGAAATATGGCGAGGAAAACT-3′; Nicol and Jeppesen, 1996), and B2 repeats (Krayev et al., 1982).
Online supplemental material
Fig. S1 shows characterization of the tri-Me-K9 H3 antibody by slot-blot. Fig. S2 shows three-dimensional analysis of tri-Me-K9 H3 staining at minor and major satellites. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200403109/DC1.
Acknowledgments
We thank P.A. Defossez, E. Heard, and J.-P. Quivy for helpful comments, T. Jenuwein for plasmids, and P. Le Baccon and S. Huart for help in image analysis.
M. Guenatri was supported by Ministère de l'Education Nationale de l'Enseignement Supérieur de la Recherche and la Ligue Nationale contre le Cancer. G. Almouzni benefited from grants from Programme Collaboratif Institut Curie/Commissariat à l'Energie Atomique, la Ligue Nationale contre le Cancer (Equipe labelisée la Ligue), Euratom (FIGH-CT-1999-00010 and FIGH-CT-2002-00207), the Commissariat à l'Energie Atomique (LRC no. 26), and Research Training Network (HPRN-CT-2000-00078 and HPRN-CT-2002-00238).
Abbreviations used in this paper: ACA, anticentromere antibody; BiodU, Biotin-16-deoxyuridine; CENP, centromeric protein; di-Me-K9 H3, histone H3 di-methylated at lysine 9; dn, double null; HP1, heterochromatin protein 1; Me-K9 H3, histone H3 methylated at lysine 9; Mnase, micrococcal nuclease; mono-Me-K9 H3, histone H3 mono-methylated at lysine 9; NChIP, native chromatin immunoprecipitation; tri-Me-K9 H3, histone H3 tri-methylated at lysine 9.
References
- Ahmad, K., and S. Henikoff. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcobia, I., R. Dilao, and L. Parreira. 2000. Spatial associations of centromeres in the nuclei of hematopoietic cells: evidence for cell-type-specific organizational patterns. Blood. 95:1608–1615. [PubMed] [Google Scholar]
- Bernard, P., J.F. Maure, J.F. Partridge, S. Genier, J.P. Javerzat, and R.C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science. 294:2539–2542. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., B.A. Sullivan, and G.H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2:319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, A.L., S.G. Ballard, and D.C. Ward. 1990. Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA. 87:7757–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo, K.H.A. 1997. The Centromere. Oxford University Press, Oxford, UK. 304 pp.
- Comings, D.E. 1980. Arrangement of chromatin in the nucleus. Hum. Genet. 53:131–143. [DOI] [PubMed] [Google Scholar]
- Craig, J.M., E. Earle, P. Canham, L.H. Wong, M. Anderson, and K.H. Choo. 2003. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 12:3109–3121. [DOI] [PubMed] [Google Scholar]
- Csink, A.K., and S. Henikoff. 1998. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 14:200–204. [DOI] [PubMed] [Google Scholar]
- Dimitrova, D.S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J. Cell Sci. 115:4037–4051. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., and N. Rothfield. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 91:313–321. [DOI] [PubMed] [Google Scholar]
- Eissenberg, J.C., and S.C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204–210. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes, M., L. Clarke, and J. Carbon. 1982. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 29:235–244. [DOI] [PubMed] [Google Scholar]
- Fox, M.H., D.J. Arndt-Jovin, T.M. Jovin, P.H. Baumann, and M. Robert-Nicoud. 1991. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J. Cell Sci. 99:247–253. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., S. Boyle, H. Sutherland, J. de Las Heras, J. Allan, T. Jenuwein, and W.A. Bickmore. 2003. Formation of facultative heterochromatin in the absence of HP1. EMBO J. 22:5540–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman, M.A., G.P. Holmquist, M.C. Gray, L.A. Caston, and A. Nag. 1984. Replication timing of genes and middle repetitive sequences. Science. 224:686–692. [DOI] [PubMed] [Google Scholar]
- Gotta, M., T. Laroche, A. Formenton, L. Maillet, H. Scherthan, and S.M. Gasser. 1996. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, I.M., K. Noma, and S.I. Grewal. 2003. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA. 100:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton, K.S., V. Dhar, E.H. Brown, M.A. Iqbal, S. Stuart, V.T. Didamo, and C.L. Schildkraut. 1988. Replication program of active and inactive multigene families in mammalian cells. Mol. Cell. Biol. 8:2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, E. 1928. Das heterochromatin der Moose. Jahrb. Wiss. Botanik. 69:728. [Google Scholar]
- Hendzel, M.J., Y. Wei, M.A. Mancini, A. Van Hooser, T. Ranalli, B.R. Brinkley, D.P. Bazett-Jones, and C.D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360. [DOI] [PubMed] [Google Scholar]
- Hsu, T.C., J.E. Cooper, M.L. Mace, Jr., and B.R. Brinkley. 1971. Arrangement of centromeres in mouse cells. Chromosoma. 34:73–87. [DOI] [PubMed] [Google Scholar]
- Jeppesen, P., A. Mitchell, B. Turner, and P. Perry. 1992. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 101:322–332. [DOI] [PubMed] [Google Scholar]
- Jin, Q., E. Trelles-Sticken, H. Scherthan, and J. Loidl. 1998. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J. Cell Biol. 141:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, A., A.R. Mitchell, and O.J. Miller. 1989. The organization of the mouse satellite DNA at centromeres. Exp. Cell Res. 183:494–500. [DOI] [PubMed] [Google Scholar]
- Karpen, G.H., and R.C. Allshire. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489–496. [DOI] [PubMed] [Google Scholar]
- Kim, S.M., D.D. Dubey, and J.A. Huberman. 2003. Early-replicating heterochromatin. Genes Dev. 17:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipling, D., H.E. Wilson, A.R. Mitchell, B.A. Taylor, and H.J. Cooke. 1994. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma. 103:46–55. [DOI] [PubMed] [Google Scholar]
- Krayev, A.S., T.V. Markusheva, D.A. Kramerov, A.P. Ryskov, K.G. Skryabin, A.A. Bayev, and G.P. Georgiev. 1982. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 10:7461–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 410:116–120. [DOI] [PubMed] [Google Scholar]
- Lachner, M., R.J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117–2124. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Y. Ueda, A.A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A.H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192–1200. [DOI] [PubMed] [Google Scholar]
- Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296–304. [DOI] [PubMed] [Google Scholar]
- Maison, C., D. Bailly, A.H. Peters, J.P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329–334. [DOI] [PubMed] [Google Scholar]
- Manuelidis, L. 1990. A view of interphase chromosomes. Science. 250:1533–1540. [DOI] [PubMed] [Google Scholar]
- Manuelidis, L., P.R. Langer-Safer, and D.C. Ward. 1982. High-resolution mapping of satellite DNA using biotin-labeled DNA probes. J. Cell Biol. 95:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, Y., and V.M. Chapman. 1991. In situ analysis of centromeric satellite DNA segregating in Mus species crosses. Mamm. Genome. 1:71–77. [DOI] [PubMed] [Google Scholar]
- Minc, E., Y. Allory, H.J. Worman, J.C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 108:220–234. [DOI] [PubMed] [Google Scholar]
- Muchardt, C., M. Guilleme, J.S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 3:975–981. 10.1093/embo-reports/kvf194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama, J., J.C. Rice, B.D. Strahl, C.D. Allis, and S.I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 292:110–113. [DOI] [PubMed] [Google Scholar]
- Nicol, L., and P. Jeppesen. 1996. Chromatin organization in the homogeneously staining regions of a methotrexate-resistant mouse cell line: interspersion of inactive and active chromatin domains distinguished by acetylation of histone H4. J. Cell Sci. 109:2221–2228. [DOI] [PubMed] [Google Scholar]
- Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S.I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89–93. [DOI] [PubMed] [Google Scholar]
- O'Keefe, R.T., S.C. Henderson, and D.L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific α-satellite DNA sequences. J. Cell Biol. 116:1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, L.P., and B.M. Turner. 1995. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 14:3946–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrod, S., and S.M. Gasser. 2003. Long-range silencing and position effects at telomeres and centromeres: parallels and differences. Cell. Mol. Life Sci. 60:2303–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A.H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, et al. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 107:323–337. [DOI] [PubMed] [Google Scholar]
- Peters, A.H., S. Kubicek, K. Mechtler, R.J. O'Sullivan, A.A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 12:1577–1589. [DOI] [PubMed] [Google Scholar]
- Pidoux, A.L., and R.C. Allshire. 2000. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 12:308–319. [DOI] [PubMed] [Google Scholar]
- Quivy, J.P., D. Roche, D. Kirschner, H. Tagami, Y. Nakatani, and G. Almouzni. 2004. A CAF-1 dependent pool of HP1 heterochromatin duplication. EMBO J. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, J.C., and C.D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263–273. [DOI] [PubMed] [Google Scholar]
- Rice, J.C., S.D. Briggs, B. Ueberheide, C.M. Barber, J. Shabanowitz, D.F. Hunt, Y. Shinkai, and C.D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 12:1591–1598. [DOI] [PubMed] [Google Scholar]
- Richmond, T., and J. Widom. 2000. Chromatin Structure and Gene Expression. Oxford University Press, Oxford, UK. 327 pp.
- Schubeler, D., D. Scalzo, C. Kooperberg, B. van Steensel, J. Delrow, and M. Groudine. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32:438–442. [DOI] [PubMed] [Google Scholar]
- Shelby, R.D., K. Monier, and K.F. Sullivan. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B., and G. Karpen. 2001. Centromere identity in Drosophila is not determined in vivo by replication timing. J. Cell Biol. 154:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., D. Roche, J. Sibarita, B. Turner, and G. Almouzni. 1999. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147:1153–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114–120. [DOI] [PubMed] [Google Scholar]
- Ten Hagen, K.G., D.M. Gilbert, H.F. Willard, and S.N. Cohen. 1990. Replication timing of DNA sequences associated with human centromeres and telomeres. Mol. Cell. Biol. 10:6348–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig, B.K. 1982. Sequence of centromere separation: role of centromeric heterochromatin. Genetics. 102:795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger, I.C., S. Hauf, A. Meinke, and J.M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103:399–410. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E. 2001. Epigenetic analysis of kinetochore assembly on variant human centromeres. Trends Genet. 17:243–247. [DOI] [PubMed] [Google Scholar]
- Warren, W.D., S. Steffensen, E. Lin, P. Coelho, M. Loupart, N. Cobbe, J.Y. Lee, M.J. McKay, T. Orr-Weaver, M.M. Heck, and C.E. Sunkel. 2000. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 10:1463–1466. [DOI] [PubMed] [Google Scholar]
- Wong, A.K., and J.B. Rattner. 1988. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 16:11645–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock, C.L., and S. Dimitrov. 2001. Higher-order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 11:130–135. [DOI] [PubMed] [Google Scholar]
- Wreggett, K.A., F. Hill, P.S. James, A. Hutchings, G.W. Butcher, and P.B. Singh. 1994. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 66:99–103. [DOI] [PubMed] [Google Scholar]