Abstract
Growth, repair, and regeneration of adult skeletal muscle depends on the persistence of satellite cells: muscle stem cells resident beneath the basal lamina that surrounds each myofiber. However, how the satellite cell compartment is maintained is unclear. Here, we use cultured myofibers to model muscle regeneration and show that satellite cells adopt divergent fates. Quiescent satellite cells are synchronously activated to coexpress the transcription factors Pax7 and MyoD. Most then proliferate, down-regulate Pax7, and differentiate. In contrast, other proliferating cells maintain Pax7 but lose MyoD and withdraw from immediate differentiation. These cells are typically located in clusters, together with Pax7−ve progeny destined for differentiation. Some of the Pax7+ve/MyoD−ve cells then leave the cell cycle, thus regaining the quiescent satellite cell phenotype. Significantly, noncycling cells contained within a cluster can be stimulated to proliferate again. These observations suggest that satellite cells either differentiate or switch from terminal myogenesis to maintain the satellite cell pool.
Keywords: stem; skeletal muscle regeneration; Pax7; MyoD; myogenin
Introduction
Precise voluntary movement in vertebrates is achieved by the fine coordinated control of skeletal muscles. The cellular unit of adult skeletal muscle is the myofiber, a highly specialized syncytium sustained by hundreds of post-mitotic myonuclei. Repair and maintenance of this syncytium is conventionally assigned to a pool of undifferentiated myogenic precursors, satellite cells, located beneath the basal lamina that surrounds the myofiber. During postnatal development, satellite cells divide to provide new myonuclei to the growing muscle fibers (Moss and Leblond, 1971) before becoming quiescent in normal muscle as it matures (Schultz et al., 1978). However, satellite cells retain the ability to proliferate and differentiate in response to the needs for myonuclear turnover and myofiber hypertrophy (Snow, 1977, 1978; Bischoff, 1986; Schmalbruch and Lewis, 2000). This activation and proliferation of satellite cells is thought to be largely, if not entirely, responsible for the remarkable capacity for efficient repair and regeneration of damaged skeletal muscle, with the generation of large numbers of new myotubes within as little as 3–4 d after severe acute damage (Whalen et al., 1990). The rate and extent of this repair is particularly striking considering the paucity of satellite cells associated with each myofiber (∼2.5–6% of fiber nuclei), implying a rapid activation and expansion of precursors to restore the full complement of myofibers (Zammit et al., 2002).
The activation of satellite cells from a state of quiescence and their subsequent progression along the myogenic lineage are controlled by various transcription factors, chief among which are the myogenic regulatory factors (MRFs) Myf5, MyoD, myogenin, and MRF4 (for review see Zammit and Beauchamp, 2001). Myf5 and MyoD determine the myogenic lineage (Rudnicki et al., 1993; Tajbakhsh et al., 1996), whereas myogenin is essential for muscle cell differentiation (Hasty et al., 1993). In adult muscle, the Myf5 locus is active in quiescent satellite cells, with MyoD appearing during activation and myogenin following as differentiation begins (Fuchtbauer and Westphal, 1992; Grounds et al., 1992; Yablonka-Reuveni and Rivera, 1994; Cooper et al., 1999; Beauchamp et al., 2000). The absence of MyoD adversely affects muscle regeneration (Megeney et al., 1996), delaying the transition of satellite cell–derived myoblasts from proliferation to differentiation (Sabourin et al., 1999; Yablonka-Reuveni et al., 1999). The role of Myf5 and myogenin during muscle regeneration has not been fully explored due to the perinatal mortality of the relevant null mice (Braun et al., 1992; Tajbakhsh et al., 1996). The expression of these MRFs in satellite cells provides a series of molecular landmarks for the transition from quiescence to activation and subsequent differentiation (Yablonka-Reuveni and Rivera, 1994; Beauchamp et al., 2000). Pax3 and Pax7, members of the paired box transcription factor family, have also been shown to be integral to muscle biology. Pax3 is essential for the migration of muscle precursors from the somites during development (Tajbakhsh et al., 1996) and is expressed in a small population of satellite cells (Buckingham et al., 2003), whereas Pax7 is required for satellite cell specification (Seale et al., 2000). However, the role of Pax7 during satellite cell activation and muscle regeneration has not yet been fully investigated.
An important question is: how is the satellite cell compartment maintained? For effective restoration of structure and function in the face of repeated injury (Sadeh et al., 1985; Luz et al., 2002), the pool of quiescent satellite cells must be replenished. There is evidence to support three scenarios that might achieve this. First, it has been suggested that satellite cells are a heterogeneous population, with some differentiating rapidly, whereas others are responsible for maintaining the pool (Rantanen et al., 1995). Second, there is a view that satellite cells are intrinsically homogenous and simultaneously activate but then adopt different fates to provide both new myonuclei and maintain the satellite cell pool (Moss and Leblond, 1971). More recently, it has been proposed that satellite cells may be part of a hierarchical system and represent a committed myogenic precursor that is restricted to providing myonuclei with satellite cell replacement occurring from a stem cell located within the muscle interstitium (Gussoni et al., 1999; Asakura et al., 2002) and/or outside muscle tissue (Fukada et al., 2002; LaBarge and Blau, 2002).
To explore the relative contribution of these three mechanisms to the maintenance of the satellite cell pool, we have used cultured myofibers, isolated complete with their retinue of satellite cells. When these myofibers are maintained in suspension culture, the associated satellite cells become activated, proliferate, and differentiate, while still exposed to signals from the myofiber (Beauchamp et al., 2000). This permits us to follow the fate of an entire cohort of satellite cells without any bias of selection. More importantly in the present context, the myofiber is isolated from potential exogenous sources of myogenic cells such as connective tissue and blood supply (Ferrari et al., 1998; LaBarge and Blau, 2002; Tamaki et al., 2002).
Here, we show that satellite cells can adopt divergent fates. Quiescent satellite cells become synchronously activated to coexpress both Pax7 and MyoD. Most satellite cells then undergo limited proliferation before down-regulating Pax7 and differentiating. Alternatively, satellite cell progeny can maintain Pax7 but lose MyoD. These Pax7+ve/MyoD−ve cells are typically located in clusters together with Pax7−ve cells destined for differentiation. Pax7+ve/MyoD−ve cells persist and eventually divide slowly or not at all. Significantly, although most cells within a cluster express myogenin and differentiate, some retain the ability to be reactivated and reenter the cell cycle. Thus, our observations show that dividing satellite cells can either enter terminal differentiation or regain characteristics of quiescence. This finding suggests that the satellite cell pool is maintained via self-renewal, involving withdrawal from the terminal myogenic program, and may not require a contribution from elsewhere.
Results
Satellite cells activate to coexpress Pax7 and MyoD before division
Isolated myofibres provide an accessible means to study the activation, proliferation, and differentiation of satellite cells in their native position beneath the basal lamina that surrounds each muscle fiber. This model preserves potentially important interactions between satellite cells and/or myofibers. Myofibers were isolated from the extensor digitorum longus (EDL) muscle of adult mice and cultured in suspension. After 24 h in growth medium, the majority (98.5 ± 1.0%, 20 myofibers from mouse 2) had become activated, as shown by the presence of MyoD, in accord with our previous observations (Zammit et al., 2002). By 48 h, the majority (97.7 ± 1.1%, 20 myofibers from mouse 2) of satellite cells remained Pax7+ve/MyoD+ve while proliferating, as confirmed by BrdU incorporation in 98.2 ± 0.8% (20 myofibers) of such cells. Thus, most of these Pax7+ve/MyoD+ve satellite cells had entered S phase or divided and were distributed on the myofiber as either single cells or small groups of two to six cells. Over this time, only rare Pax7+ve cells remained BrdU−ve (1.8 ± 0.8% distributed over 20% of the myofibers). Therefore, during the initial 48 h in culture almost all satellite cells coexpress Pax7 and MyoD before, and during, division.
Satellite cells can adopt divergent fates
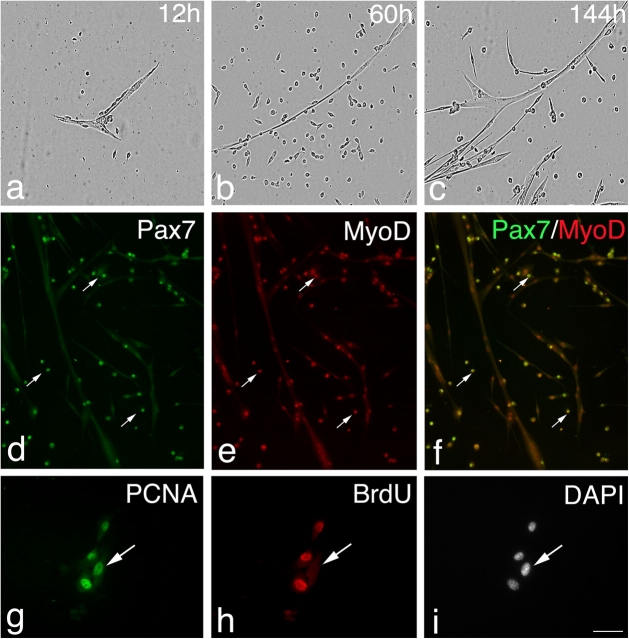
However, beyond 48 h, satellite cells exhibited a variety of Pax7, MyoD, and myogenin expression profiles (Table I). Immunostaining of the satellite cells present on wild-type myofibers after 72 h in culture (Fig. 1) showed that the majority contained MyoD (76.8%) and were divided into those that also coexpressed Pax7 (17.3%) and those that did not (59.5%; Fig. 1). At this time, a significant proportion of satellite cell progeny (23.2%; Table I) expressed Pax7 without MyoD (Fig. 1). The Pax7+ve/MyoD−ve cells were typically located in clusters (defined as four or more intimately associated cells; Fig. 1, e and f). Myofiber-associated satellite cells were also coimmunostained for MyoD and myogenin after 72 h in culture (Fig. 2) to determine how many cells were in the earliest phase of differentiation (Andres and Walsh, 1996). Of the myofiber-associated satellite cells that were immunostained, only 6% were myogenin+ve/MyoD−ve and 21.4% MyoD+ve/myogenin−ve, whereas the remaining 72.7% contained both MyoD and myogenin protein (Table I). Together, these observations show that virtually all satellite cells initially coexpress Pax7 and MyoD before proliferating and that most then commit to differentiation. Importantly though, at the same time some satellite cells begin to adopt a different fate, maintaining Pax7 but not MyoD. Furthermore, these Pax7+ve/MyoD−ve cells were still found when the growth medium was replaced each day, arguing that these cells had not simply withdrawn from myogenesis in response to mitogen depletion (unpublished data).
Table I. Pax7+ve/MyoD−ve satellite cell progeny appear by 72 h in culture.
| Pax7 | MyoD | Pax7/ MyoD |
MyoD | Myogenin | MyoD/ Myogenin |
|
|---|---|---|---|---|---|---|
| Mouse 1 | 19.1% | 66.8% | 14.0% | 16.7% | 4.5% | 78.8% |
| (± 1.4) | (± 4.0) | (± 3.5) | (± 4.9) | (± 2.6) | (± 3.6) | |
| Mouse 2 | 27.3% | 52.2% | 20.5% | 26.0% | 7.4% | 66.5% |
| (± 4.5) | (± 4.4) | (± 2.1) | (± 1.8) | (± 1.5) | (± 3.0) | |
| Mean | 23.2% | 59.5% | 17.3% | 21.4% | 6.0% | 72.7% |
Batches of myofibers from the same mouse were coimmunostained for either Pax7 and MyoD (first three columns) or MyoD and myogenin (last three columns). Values are population means from the pooled data from T72 myofibers from each of two age-matched wild-type adult mice. The number of satellite cells in each category is expressed as a mean percentage of the total immunostained cells present on the myofiber (± SEM).
Figure 1.
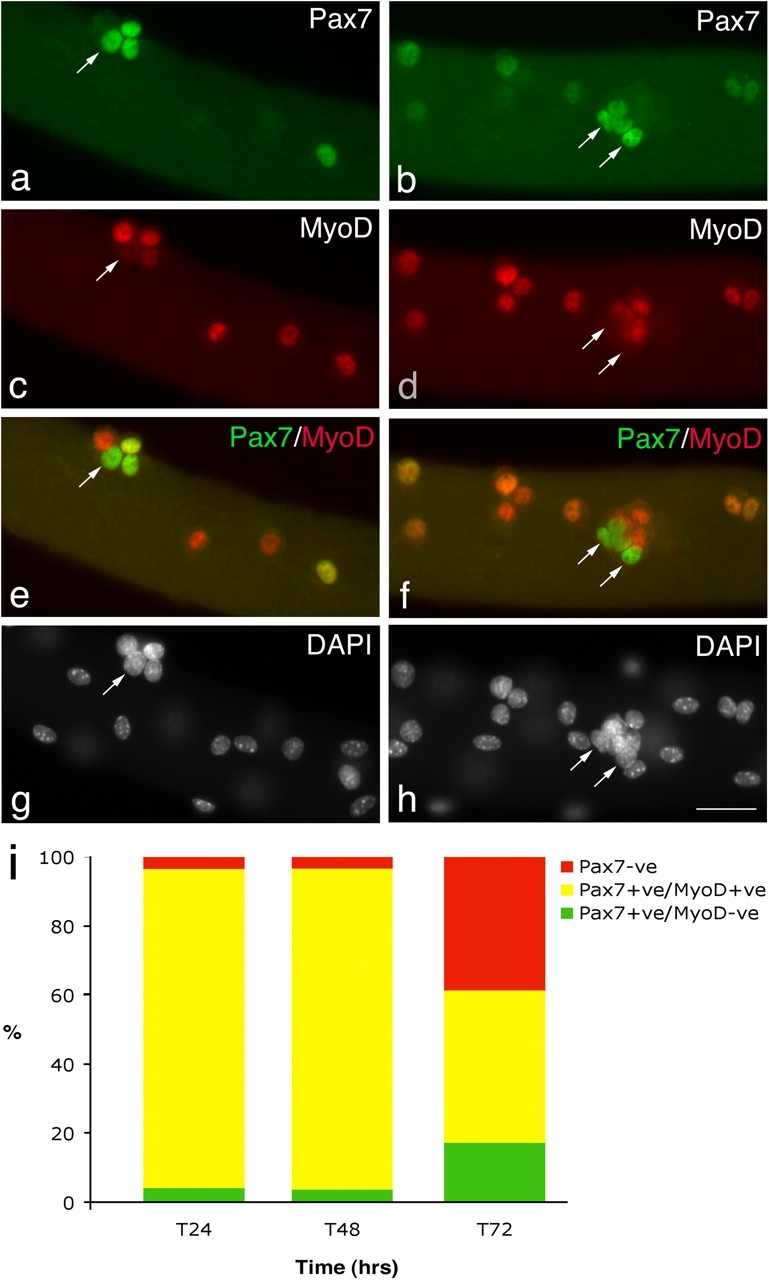
Satellite cells adopt divergent fates. After 72 h in culture, coimmunostaining of EDL myofibers demonstrates that in the majority of satellite cells, Pax7 (a and b) is becoming down-regulated whereas MyoD (c and d) remains strongly expressed. However, combining the Pax7 (green) and MyoD (red) fluorescent immunosignals (e and f) clearly shows that some (∼23%) MyoD−ve daughters (arrows) maintain robust Pax7 expression. Pax7+ve/MyoD−ve satellite cell (arrows) are typically located (85%) in clusters (defined as four or more overlying/touching cells) together with both Pax7+ve/MyoD+ve and Pax7−ve/MyoD+ve cells. Counterstaining with DAPI was used to identify all nuclei present on the myofiber (g and h). Bar, 30 μm. The number of satellite cells from 3F-nlacZ-E mouse 3 and 4 (Table II) that were either Pax7+ve/MyoD−ve, Pax7+ve/MyoD+ve, or Pax7−ve at T24, T48, and T72 were counted. Expressing this data as the mean percentage in each category of the total number of satellite cells per myofiber (i) illustrates that the Pax7+ve/MyoD+ve cells present at T24 and T48 give rise to the majority of Pax7+ve/MyoD−ve and Pax7−ve populations at T72.
Figure 2.
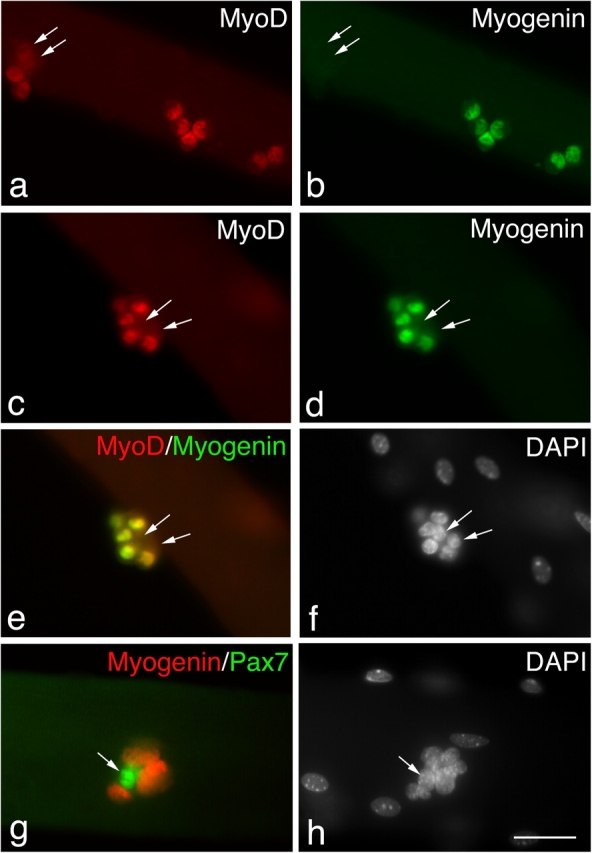
Not all satellite cells are committed to differentiation. Coimmunostaining of EDL myofibers that had been in culture for 72 h showed that most (∼73%) MyoD+ve satellite cells (a and c) were committed to differentiation, as shown by the presence of myogenin (b and d). The yellow color resulting from combining the MyoD (c) and myogenin (d) fluorescent immunosignals (e) clearly shows that most MyoD+ve daughters within this cluster also contain myogenin. However, it is significant that the MyoD−ve daughters in this cluster were also myogenin−ve (arrows), and at least some of these are presumably Pax7+ve (Fig. 1). Coimmunostaining for Pax7 (green) and myogenin (red) showed that these two antigens were essentially mutually exclusive (g) with only 6.8 ± 1.6% of Pax7+ve cells also expressing myogenin. Counterstaining with DAPI was used to identify all nuclei present (f and h). Bar, 30 μm.
Pax7+ve/MyoD−ve satellite cells arise from the Pax7+ve/MyoD+ve–activated satellite cell pool
The aforementioned data do not unequivocally determine the origin of Pax7+ve/MyoD−ve cells after 72 h in culture. Although almost all (∼98%) satellite cells at both 24 and 48 h coexpress Pax7 and MyoD, we could not be certain whether these cells had lost MyoD or whether they arose by the rapid proliferation (which would require a cell cycle time of <6 h) of ∼2% of Pax7+ve/MyoD−ve cells seen at these early time points. It was also possible that the Pax7+ve/MyoD−ve cells had arisen from satellite cells that did not initially express either Pax7 or MyoD, but which began to express Pax7 by 72 h. To address these points, we used the 3F-nlacZ-E transgenic mouse in which the myonuclei of fast myofibers have robust β-galactosidase activity (Kelly et al., 1995), permitting us to identify all satellite cells by their failure to stain with X-gal (Beauchamp et al., 2000). EDL myofibers isolated from this mouse were cultured in suspension and fixed at 24-h intervals from T0 to T72. They were then coimmunostained for either Pax7 and MyoD, MyoD and myogenin, or Pax7 and myogenin. Antibody binding was revealed with fluorescently labeled secondary antibodies before myofibers were briefly incubated in X-gal and all nuclei counterstained with DAPI. Using this procedure, the myonuclei are identified by the deposition of a Prussian blue X-gal reaction product, which obscures the fluorescent DAPI nuclear counterstain. Therefore, clearly visible DAPI staining identifies the nuclei of all satellite cells associated with the isolated myofiber whether or not they are expressing any of the antigens under investigation. Each category of cell, defined by its pattern of immunoreactivity for Pax7, MyoD, and myogenin, including totally negative cells, can thus be related to the total population of satellite cells. The mean number of cells per myofiber in each category at each time point studied is given in Table II. The data from different mice was also pooled and expressed as the mean percentage of the total number of satellite cells (defined as nonquenched DAPI) per myofiber in each category (Fig. 1 i).
Table II. Most Pax7+ve/MyoD−ve satellite cell progeny arise from Pax7+ve/MyoD+ve parents.
| Pax7 | MyoD | Both | Neither | Myogenin | MyoD | Both | Neither | Pax7 | Myogenin | Both | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3F-nlacZ-E mouse 3 | |||||||||||
| T0 | 3.5 | n/d | n/d | 0.1 | |||||||
| (± 0.7) | (± 0.7) | (± 0.1) | |||||||||
| T24 | 0.4 | 0 | 5.1 | 0.2 | |||||||
| (± 0.2) | (± 0.7) | (± 0.1) | |||||||||
| T48 | 0.7 | 0.1 | 9.5 | 0.3 | 0 | 10.4 | 0 | 0.6 | |||
| (± 0.3) | (± 0.1) | (± 1.2) | (± 0.1) | (± 1.6) | (± 0.3) | ||||||
| T72 | 15.3 | 30.2 | 31.3 | 8.0 | 2.2 | 30.5 | 21.4 | 9.5 | 48.2 | 25.0 | 3.9 |
| (± 3.0) | (± 7.5) | (± 5.1) | (± 2.2) | (± 0.8) | (± 5.8) | (± 4.8) | (± 2.8) | (± 5.8) | (± 6.6) | (± 1.2) | |
| 3F-nlacZ-E mouse 4 | |||||||||||
| T0 | 5.3 | n/d | n/d | 0.3 | |||||||
| (± 0.7) | (± 0.1) | ||||||||||
| T24 | 0.2 | 0 | 5.45 | 0.3 | |||||||
| (± 0.1) | (± 0.8) | (± 0.2) | |||||||||
| T48 | 0.7 | 0.5 | 19.3 | 0.3 | 0 | 11.5 | 0 | 0.6 | |||
| (± 0.2) | (± 0.2) | (± 2.6) | (± 0.2) | (± 2.0) | (± 1.8) | ||||||
| T72 | 14.3 | 28.7 | 40.0 | 4.0 | 1.7 | 58.9 | 28.1 | 21.2 | 32.2 | 41.4 | 3.0 |
| (± 3.3) | (± 6.3) | (± 6.4) | (± 0.9) | (± 0.8) | (± 3.8) | (± 3.5) | (± 3.5) | (± 5.6) | (± 6.6) | (± 1.0) |
Batches of myofibers from the same mouse were coimmunostained for either Pax7 and MyoD (first four columns), MyoD and myogenin (middle four columns), or Pax7 and myogenin (last three columns) before myonuclear β-galactosidase activity was revealed with X-gal. Values are population means of the number of satellite cells in each category (± SEM) per myofiber isolated from each of two age-matched 3F-nlacZ-E mice. n/d, not determined.
Like the 24- and 48-h data from wild-type mice in which 97–98% of satellite cells coexpressed Pax7 and MyoD, the 3F-nlacZ-E transgenic myofibers showed again that the vast majority of the total satellite cell pool was Pax7+ve/MyoD+ve (92.4% at 24 h and 92.9% at 48 h), the remainder being either Pax7+ve/MyoD−ve (4.1% and 3.7%, respectively) or negative for both Pax7 and MyoD (3.5% and 2.2%, respectively). Only very rare cells expressed myogenin at T48, but by 72 h in culture many cells were differentiating, as shown by the presence of myogenin (∼30%). At this time, a significant number of Pax7+ve/MyoD−ve cells were also present (17.2%; Fig. 1 i). Coimmunostaining for Pax7 and myogenin revealed an essentially mutually exclusive relationship between these two proteins (Fig. 2, g and h) with only rare Pax7+ve cells (6.8 ± 1.6% of total Pax7+ve cells) found to coexpress myogenin, usually at a low level. The Pax7+ve/MyoD−ve cells were mainly (85%) located in clusters of four or more cells. These clusters contained a mean of 7.9 ± 0.4 cells consisting of 1.9 ± 0.2 Pax7+ve/MyoD−ve cells, 2.5 ± 0.2 Pax7−ve/MyoD+ve cells, 2.8 ± 0.2 Pax7+ve/MyoD+ve cells, and 0.7 ± 0.1 unlabeled cells (133 clusters analyzed from mouse 3 and 4). The overriding presence of Pax7+ve/MyoD−ve cells in clusters indicates that interactions within the cluster may be an important feature of this diversification of phenotype.
Satellite cells can down-regulate MyoD without committing to differentiation
Where do the Pax7+ve/MyoD−ve cells come from? As mentioned above, based on the pooled data from wild-type mice, one possible explanation could be the rapid expansion of a Pax7+ve/MyoD−ve population. However, the distribution of Pax7+ve/MyoD−ve cells at 24 h excludes this possibility. At 24 h, only 15% of myofibers contained Pax7+ve/MyoD−ve satellite cells, whereas by 72 h, virtually every myofiber (97%) did. Therefore, analysis of the myofiber distribution in the 3F-nlacZ-E study unequivocally shows that most of the Pax7+ve/MyoD−ve cells must have arisen from satellite cells that were initially Pax7+ve/MyoD+ve.
When is MyoD lost? Satellite cells on T0 myofibers are typically located as single cells distributed along the length of the entire myofiber; only 4 pairs were found among 59 satellite cells on 16 myofibers from mouse 3 and 5 pairs among 106 satellite cells on 19 myofibers from mouse 4. Therefore, the pairs and small groups of satellite cells observed at 48 h are probably clonal in origin. Rare instances of clones of satellite cells could be observed that contained both Pax7+ve/MyoD+ve and Pax7+ve/MyoD−ve cells. An example is shown in Fig. 3, where the cell surface protein CD34 (Beauchamp et al., 2000) marks four satellite cells, only three of which coexpress MyoD (Fig. 3, a and b). In addition, MyoD−ve cells could incorporate BrdU, showing that they were the progeny of an activated satellite cell, and not rare quiescent MyoD−ve cells that had failed to activate (Fig. 3, c–e). To assess the frequency of this event, myofibers were cultured in the presence of BrdU for 48 h and then immunostained. Of 50 pairs of touching BrdU+ve cells, presumed to have recently divided, 43 pairs were symmetrically MyoD+ve, whereas 7 pairs (14%) had conspicuously higher levels of MyoD in one daughter than the other. If we recalculate, taking single and groups of satellite cells into account, the 14% that down-regulate MyoD in the touching satellite cell pairs becomes ∼2.2% of the total satellite cells present.
Figure 3.
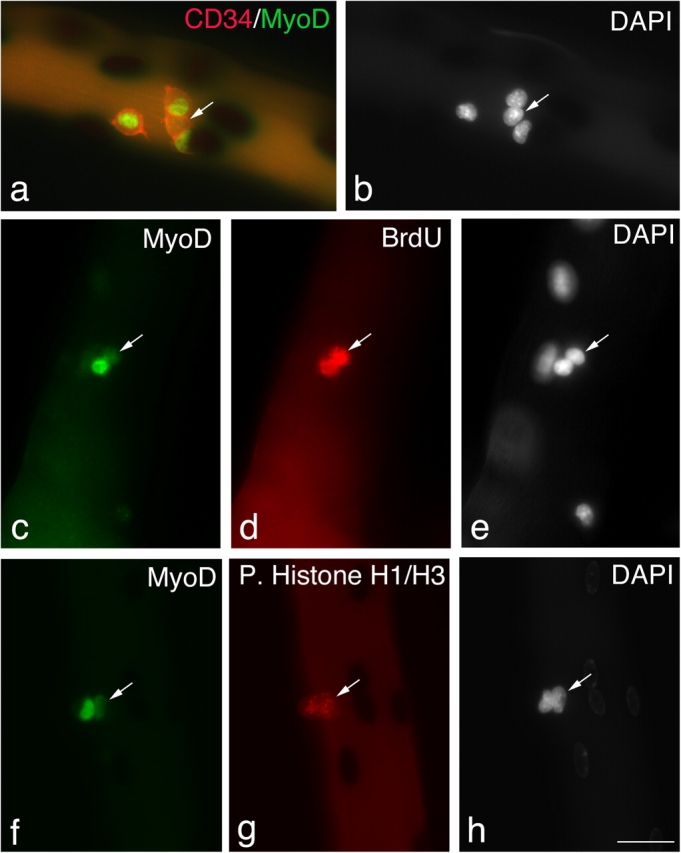
Satellite cells can maintain Pax7, but lose MyoD. Quiescent satellite cells are generally distributed as single cells along the length of the myofiber. Between 24–48 h in culture, almost all satellite cells express both Pax7 and MyoD and begin to divide; therefore, it can be assumed that most pairs/small groups of cells present at these times are derived from a single parent satellite cell. Immunostaining followed by X-gal incubation of EDL myofibers from a 3F-nlacZ-E mouse after 48 h in culture shows that of four CD34+ve satellite cells (a, red-cell surface), three contained MyoD protein (a, green nuclear), but significantly one did not (a and b, arrows). These MyoD−ve cells (c–e, arrows) had undergone division, as shown by the incorporation of BrdU into the daughter cells (d), and were not merely quiescent satellite cells. Whether MyoD protein levels vary with cell cycle is unresolved (Kitzmann et al., 1998; Lindon et al., 1998). However, it is important that both the MyoD+ve and MyoD−ve progeny of a single satellite cell could be in the same phase of the cell cycle (f–h). An example is shown where immunostaining followed by X-gal incubation of an EDL myofiber from a 3F-nlacZ-E mouse after 48 h in culture shows that MyoD+ve (f) and MyoD−ve (f–h, arrows) daughters are both in the same phase of the cell cycle as shown by phosphorylated Histone H1/H3 immunostaining (g). Counterstaining with DAPI was used to identify all nuclei present (b, e, and h). Bar, 30 μm.
Whether or not MyoD protein levels vary with cell cycle is a matter of debate (Kitzmann et al., 1998; Lindon et al., 1998). The characteristic pattern of phosphorylated Histone H1 and H3 immunostaining can be used as a guide to the phase of the cell cycle. To explore whether or not the low level of MyoD observed in some satellite cells reflected cell cycle variation, coimmunostaining for phosphorylated Histone H1 and H3 and MyoD was performed and showed that cells with radically different levels of MyoD could be in the same phase of the cell cycle (Fig. 3, f–g). The doublet of cells shown in Fig. 3 (f–h) with different MyoD levels contained the only MyoD−ve cell observed in a total of 126 satellite cells on 20 myofibers derived from a 3F-nlacZ-E mouse. MyoD+ve and MyoD−ve satellite cell progeny still contained Pax7 (Fig. 1), and Pax7 without MyoD is characteristic of quiescent satellite cells (Zammit and Beauchamp, 2001).
Some Pax7+ve/ MyoD−ve cells eventually divide slowly or stop cycling
Until ∼72 h, the majority of satellite cells were cycling rapidly, as shown by the presence of PCNA in their nuclei (Fig. 4, a–c). However, from 72 h onwards the number of PCNA+ve cells dropped markedly, as differentiation ensued. The low level of BrdU incorporation after a 24-h pulse from 96–120 h confirmed that very few satellite cells were still cycling. Interestingly, 42.5% of the Pax7+ve cells (∼20 myofibers from each of three mice) also failed to incorporate BrdU during this time, indicating that they were either cycling slowly (i.e., not entering S phase during the 24-h period of the pulse) or were quiescent (Fig. 4, d–f). Crucially, Pax7+ve cells were almost all negative for MyoD at 120 h (Fig. 4, g–i), showing that the Pax7+ve/MyoD−ve phenotype was not transient but persisted until at least 120 h, the latest time point examined in this study. In addition, virtually no expression of MyoD (Fig. 4, g–i) or myogenin (not depicted) was detectable in any satellite cells at this time, demonstrating that the expression of these two MRFs is transient. Thus, some satellite cells cultured in association with their myofiber maintain expression of Pax7, down-regulate MyoD, and begin to cycle slowly or not at all; all characteristics of quiescent satellite cells.
Figure 4.
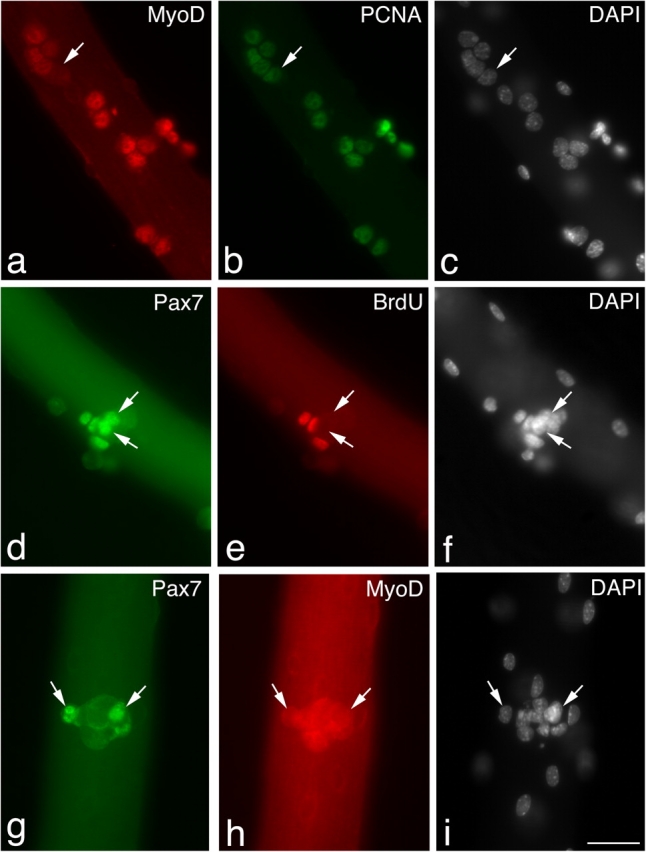
Pax7+ve/MyoD−ve satellite cells eventually cycle slowly, or not at all. Immunostaining of EDL myofibers after 72 h in culture showed that the majority of MyoD+ve cells (a) were also expressing PCNA (b), indicating that they were still cycling. Rare MyoD−ve cells (a–c, arrows) were also PCNA+ve (b, arrow) and would presumably be either Pax7+ve or myogenin+ve. However, a pulse of BrdU between 96–120 h in culture (d–f) followed by immunostaining showed that by 120 h, 42.5% of Pax7+ve cells (d–f, arrows) had failed to incorporate BrdU (e, arrows) over the previous 24 h. At 120 h, the Pax7+ve cells (g–i, arrow) were negative for MyoD (h, arrow), with MyoD being undetectable in almost all satellite cells at this time. Counterstaining with DAPI was used to identify all nuclei present (c, f, and i). Bar, 30 μm.
Clusters contain cells that can reactivate and proliferate
To investigate whether or not the satellite cells contained in clusters on a myofiber retain the ability to reactivate and proliferate, individual clusters of satellite cells were removed with a pipette and plated on a Matrigel substrate in fresh growth medium. These conditions stimulate satellite cells to divide and migrate over the Matrigel substrate. The clusters rapidly adhered to the substrate, and both proliferating cells and small myotubes were observed within 24 h of plating (Fig. 5 a). Cells continued to proliferate with a second wave of myotube formation occurring several days later (Fig. 5, b and c). Satellite cells associated with myofibers that have been cultured in suspension for 120 h are almost all completely MyoD negative (Fig. 4, g–i), whereas many cells derived from plated clusters became Pax7+ve/MyoD+ve (Fig. 5, d–f), showing that MyoD expression had been reinduced.
Figure 5.
Isolated clusters of satellite cell progeny contain cells able to reactivate, proliferate, and differentiate. EDL myofibers were maintained in suspension for 120 h, at which time clusters were removed with a pipette, plated onto Matrigel, and photographed over the next 144 h (a–c). Clusters quickly adhered to the Matrigel substrate, and both single cells and small myotubes rapidly appeared (a). Cells proliferated over the subsequent days (b) until a second wave of myotube formation began around 96 h (c). In separate experiments, myofibers were maintained in suspension for 120 h, at which time MyoD protein was virtually absent. Clusters were then removed, plated, and fixed after a further 120 h and immunostained (d–f). The proliferating cells (d–f, arrows) expressed both Pax7 (d, green) and MyoD (e, red), seen as yellow when the red and green immunosignals are combined (f) interspersed with myocytes and myotubes. To determine if satellite cells that had stopped cycling while associated with a myofiber were able to still undergo division, a BrdU pulse was administered between 96–120 h. Clusters were then plated and 24 h later coimmunostained for PCNA and BrdU (g–i, arrows). Proliferating cells were identified by PCNA (g) expression that were BrdU−ve (h), indicating that they had been refractory to BrdU incorporation while associated with the myofiber, but were able to reenter cell cycle when dissociated/stimulated. Counterstaining with DAPI was used to identify all nuclei present (i). Bars: (d–f) 100 μm; (g–i) 30 μm.
Because some satellite cells were not cycling after 96/120 h in culture, we next sought to determine if these cells retained the ability to proliferate again or if they had merely become senescent. EDL myofibers were maintained as nonadherent cultures for 120 h in growth medium, with BrdU present for the final 24 h. Clusters were then removed, plated on Matrigel for 24 h, fixed, and coimmunostained for PCNA and BrdU. PCNA+ve/BrdU+ve cells were observed, indicating that cells that had been cycling while on the myofiber remained in cycle when moved onto a Matrigel substrate. Crucially, we also found PCNA+ve/BrdU−ve cells, indicating that some cells that had become refractory to BrdU incorporation while in clusters on the myofiber were able to reenter the cell cycle when explanted from this communal environment.
Discussion
Although the remarkable ability of skeletal muscle to recover from injury has been much studied in vivo, attempts to model the process in tissue culture systems have been able to provide only crude parallels. After severe muscle damage, many myotubes appear in as little as 3–4 d (Whalen et al., 1990), a process that entails a rapid and efficient mobilization of the satellite cell compartment, with a phase of intense proliferation over a period of some 3–4 d (McGeachie and Grounds, 1987). The capacity of muscle to cope successfully with serial damage (Sadeh et al., 1985; Luz et al., 2002) and chronic degenerative muscle disorders, such as that observed in mdx mice, demonstrates that the satellite cell pool is effectively replenished during each episode of regeneration. To better understand this process, we investigated the dynamics of satellite cell activation, proliferation, and differentiation on isolated myofibers maintained in culture. We observed a sequence of events that is strongly reminiscent of that which occurs during muscle regeneration in vivo. We found that satellite cell progeny adopted divergent fates: although most differentiate, others regain a phenotype characteristic of quiescent satellite cells. Our observations support the general hypothesis that self-renewal is a major mechanism used to maintain the satellite cell pool, as suggested by the work of Moss and Leblond (1971) over 30 yr ago, without need for a significant input from outside the satellite cell compartment. Further evidence for self-renewal has come from studies of myoblast transplantation into adult muscle, where neonatal myoblasts/satellite cell–derived myoblasts gave rise to both differentiated myonuclei and functional satellite cells (Blaveri et al., 1999; Heslop et al., 2001).
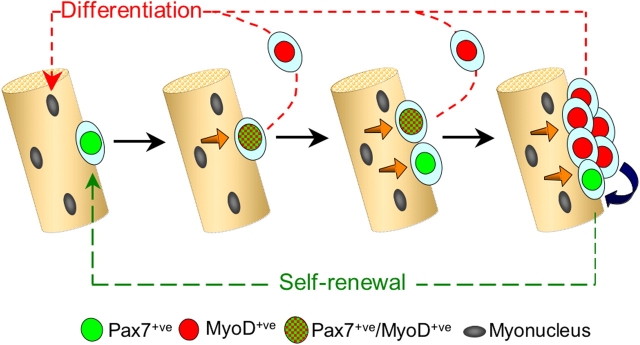
On the basis of our observations on the alternate fates adopted by satellite cells, we propose a model for satellite cell self-renewal (Fig. 6). Virtually all quiescent satellite cells coexpress Pax7 and MyoD within a few hours of activation. After this point, phenotypic and behavioral diversification becomes apparent. Most cells undergo rapid but limited proliferation and down-regulate Pax7 as they begin to differentiate, consistent with previous descriptions of MyoD and myogenin expression during satellite cell activation (Grounds et al., 1992; Yablonka-Reuveni and Rivera, 1994; Cooper et al., 1999). This behavior shows close parallels to the transit-amplifying population resident in skin (Watt, 2002). However, not all Pax7+ve/MyoD+ve satellite cell progeny follow this highly coordinated program of gene expression toward a differentiated state. Others adopt an alternative fate in which Pax7 is maintained but MyoD is lost as the cells withdraw from differentiation. Later, Pax7+ve/MyoD−ve cells are observed in clusters, together with Pax7−ve cells that are destined to differentiate to replace lost myonuclei. The Pax7+ve/MyoD−ve cells then become quiescent, thus maintaining the satellite cell pool. This model is consistent with observations of satellite cells during regeneration in vivo where most rapidly differentiate after only a limited number of divisions, whereas a minority proliferate for an extended period (Grounds and McGeachie, 1987; McGeachie and Grounds, 1987; Rantanen et al., 1995).
Figure 6.
Model of satellite cell self-renewal. Quiescent satellite cells (green) activate to coexpress Pax7 and MyoD (green and red tartan), and then most proliferate, down-regulate Pax7, maintain MyoD (red), and differentiate (red pathway). However, activated Pax7+ve/MyoD+ve (green and red tartan) satellite cells can also divide to give rise to cells that adopt a different fate. These give rise to clusters of cells containing both Pax7−ve/MyoD+ve (red) progeny, whereas others down-regulate MyoD expression and cycle while maintaining only Pax7 (green). These clusters may grow by the further generation of cells with divergent fates. Pax7+ve/MyoD−ve cells (green) become quiescent, thus renewing the satellite cell pool (green pathway), whereas the MyoD+ve cells (red) differentiate to produce myonuclei (red pathway). Signaling from the myofiber (orange arrows) and/or between cells within the clusters (blue arrow) may dictate which fate the satellite cell adopts.
Once expressed, MyoD tends to initiate a self-reinforcing cascade of myogenic commitment by virtue of its auto- and cross-activation driving the production of all MRFs. Indeed, the ability of MyoD to efficiently direct many nonmuscle cells down the myogenic lineage testifies to its potency in this respect (Weintraub et al., 1991). In light of this, our finding that most MyoD−ve satellite cell progeny must be derived from MyoD+ve parents is unexpected, for it implies that expression of MyoD does not necessarily result in differentiation. A mechanism must exist to suppress MyoD, so as to divert cells away from the differentiation pathway into a precursor state. This contradicts the general dogma that expression of key lineage commitment genes in a stem cell enforces an irreversible linear progression toward differentiation. This concept has also been challenged in the adult central nervous system, where committed oligodendrocyte precursors can revert to a multipotent neural stem cell phenotype (Kondo and Raff, 2000).
What dictates the divergent fates of satellite cells? Is it an innate, lineage based, heterogeneity of the satellite cell population, or differences within the microenvironment on the myofiber and/or within the cell cluster? The satellite cell pool is heterogeneous by several criteria. For example, although the majority of quiescent satellite cells in limb muscle transcribe Myf5 and contain M-cadherin and CD34 protein, a minority are negative for any of these markers (Beauchamp et al., 2000). On a larger scale, the majority of muscles in the Pax3 nLacZ/+mouse contain only rare Pax3+ve satellite cells, but some show more widespread expression (Buckingham et al., 2003). Functional heterogeneity within muscle is also evident, with at least two populations identified by their differential sensitivity to irradiation (Heslop et al., 2000). Clonal studies also suggest heterogeneity as defined by both proliferation rate and clonogenic capacity (Schultz and Lipton, 1982; Molnar et al., 1996). Furthermore, during both growth and regeneration, satellite cells can clearly be divided with respect to their growth factor requirements for proliferation (Yablonka-Reuveni and Rivera, 1994), by their rate of cell division (Grounds and McGeachie, 1987; McGeachie and Grounds, 1987; Rantanen et al., 1995; Schultz, 1996), and by the myosin heavy chain isoforms they express upon differentiation (Hoh et al., 1988; Hoh and Hughes, 1991; Rosenblatt et al., 1996).
That divergence in fate of the Pax7+ve/MyoD+ve satellite cells occurs without the synchronizing signal of mitogen reduction indicates that this is a normal part of the program of satellite cell replacement. However, it remains to be shown whether satellite cell heterogeneity reflects the stochaistic generation of diversity within a dynamic system or a distinct lineage-based subpopulation. We have shown that this divergence in fate of satellite cells occurs reproducibly in cultures on suspended myofibers. This finding contrasts with the behavior of satellite cells that are allowed to migrate from a myofiber onto the tissue culture substrate, where Pax7+ve/MyoD−ve cells are rarely observed and three-dimensional clusters do not form (unpublished data). This suggests that the culture of satellite cells on isolated myofibers in suspension is an apposite model to mimic the self-regulatory events of myogenesis during muscle regeneration in vivo. The potential importance of environmental regulation is also suggested by the behavior of primary myoblasts or myogenic cell lines induced to differentiate by mitogen withdrawal. Although most differentiate, a population of “reserve cells” has been described that stop dividing yet retain the ability to reenter cell cycle and differentiate when passaged (Kitzmann et al., 1998; Yoshida et al., 1998). Significantly, reserve cells can be generated from clones, supporting the notion that this is not an intrinsic property of a permanent subset but that any myoblast can acquire this behavioral phenotype (Baroffio et al., 1996).
Therefore, we suggest that generation of Pax7+ve/MyoD−ve cells is not a cell autonomous event, but instead arises from signaling between the myofiber and satellite cell, or the microenvironment within cell clusters, as indicated in our model (Fig. 6). Maintenance in vitro of satellite cells in their native position under the basal lamina and adjacent to the myofiber preserves such contact-dependent interactions and signaling. It may be that an instructive signal from the myofiber plays some part in directing the cell to become quiescent, much as IL-4 secreted by myotubes controls the recruitment of further myoblasts (Horsley et al., 2003). It is also possible that the differentiation of some satellite cells may signal others to become quiescent, possibly using systems used to direct cell fate elsewhere, such as the Notch signaling pathway (Artavanis-Tsakonas et al., 1999), components of which are expressed by satellite cells (Conboy and Rando, 2002).
The single fiber culture model used here also has the advantage that the source of cells is defined as satellite cells, i.e., those resident under the basal lamina, thus isolating the process of satellite cell activation and renewal from potential contributions from elsewhere. This finding is important in light of several recent studies that have shown that other cells isolated from muscle and from a diverse range of tissues are able to adopt a myogenic phenotype, albeit at a very low frequency (Ferrari et al., 1998; Gussoni et al., 1999; LaBarge and Blau, 2002).
Side population stem cells, characterized by their ability to exclude Hoescht dye, can be isolated from muscle tissue and have been shown to be distinct from satellite cells (Gussoni et al., 1999; Jackson et al., 1999; Asakura et al., 2002). Side population cells can contribute to both the hematopoietic and myogenic lineages (Gussoni et al., 1999; Asakura et al., 2002), leading to the suggestion that they are a normal, routine source of satellite cells. Myogenic precursors also reside in the interstitium of muscle (Tamaki et al., 2002), and blood vessel–derived mesoangioblasts too have myogenic potential (De Angelis et al., 1999; Cusella De Angelis et al., 2003; Sampaolesi et al., 2003). Moreover, transplantation studies show that cells derived from various nonmuscle tissues can contribute to muscle (for review see Grounds et al., 2002). The most intriguing of these nonmuscle sources is bone marrow, which produces cells that are able to move into the satellite cell niche, arriving via the circulation following bone marrow grafts (Fukada et al., 2002; LaBarge and Blau, 2002), suggesting that this could also be a usual source of myogenic cells. However, the number of myofibers with a donor contribution is very low and does not increase significantly with time in either mouse (Ferrari et al., 2001; Brazelton et al., 2003) or man (Gussoni et al., 2002), implying that either bone marrow cells are unable to contribute to any great extent to the stem cell compartment within muscle or to be continually recruited.
The results presented here, showing that satellite cells can adopt divergent fates, suggest that the satellite cell pool is maintained by self-renewal and that this is independent of a contribution from outside. Although this finding does not exclude participation by other cells to myogenesis in vivo, possibly to augment the number of myoblasts during severe muscle regeneration, no evidence has been produced of more than a minor contribution from these other sources. Indeed, the ablation of the satellite cell pool by irradiation suggests that cells from outside of muscle are not able to restore functional regeneration (Heslop et al., 2000). Certainly, no such contribution appears to be required because satellite cells associated with a muscle fiber are capable of producing enough myoblasts to entirely replace that myofiber in a time scale consistent with in vivo regeneration (Zammit et al., 2002).
Materials and methods
Animal models
Adult (>6 wk of age) wild-type or 3F-nlacZ-E transgenic mice (generated by R. Kelly and M. Buckingham, Pasteur Institute, Paris, France) were used in this study. 3F-nlacZ-E mice contain seven copies of a construct consisting of a 2-kb myosin light chain (MLC) 3F promoter, nlacZ-SV40 poly (A) in frame in the second MLC3F specific exon, 1 kb of MLC3F sequence 3′ of nlacZ, and a 260bp 3′ MLC1F/3F enhancer (Kelly et al., 1995).
Tissue preparation and single fiber isolation
Mice were killed by cervical dislocation before the EDL muscles were carefully removed. Myofibers were isolated as described previously (Rosenblatt et al., 1995) and either fixed for 5–20 min in 4% PFA/PBS or cultured.
Myofiber culture
For suspension culture, myofibers were incubated in growth medium (DME with 10% [vol/vol] horse serum [PAA Laboratories] and 0.5% [vol/vol] chick embryo extract [ICN Biomedicals]) at 37°C in 5% CO2. For adherent cultures, isolated clusters were placed in 24-well Primaria plates (Marathon) coated with 1 mg/ml of Matrigel (Collaborative Research Inc.). Growth medium was added and the cultures maintained at 37°C in 5% CO2. Where used, BrdU was added to the medium at a final concentration of 10 μM. Myofibers and cells were fixed in 4% PFA/PBS for 5–20 min.
Immunostaining
Fixed myofibers were permeabilized with 0.5% (vol/vol) Triton X-100/PBS and blocked using 20% (vol/vol) goat serum/PBS, as described previously (Beauchamp et al., 2000). Primary antibodies used were monoclonal rat anti-BrdU (clone BU1/75; Abcam), monoclonal mouse antimyogenin (clone F5D; DakoCytomation or Developmental Studies Hybridoma Bank [DSHB]), anti-MyoD1 (clone 5.8a; DakoCytomation), anti-Pax7 (DSHB), anti-PCNA (clone PC10; DakoCytomation), rabbit polyclonal anti-MyoD (Santa Cruz Biotechnology, Inc.), antimyogenin (Santa Cruz Biotechnology, Inc.), antiphosphorylated Histone H1 (Upstate Biotechnology), and antiphosphorylated Histone H3 (Upstate Biotechnology). Primary antibodies were visualized with fluorochrome-conjugated secondary antibodies (Molecular Probes) before mounting in Faramount fluorescent mounting medium (DakoCytomation) containing 100 ng/ml DAPI.
Histology
To visualize β-galactosidase activity, immunostained myofibers were incubated in X-gal solution (4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, 400 μg/ml X-gal, and 0.02% NP-40 in PBS) for ∼15 min at RT. Myofibers were rinsed several times in PBS and mounted in Faramount aqueous mounting medium containing 100 ng/ml DAPI.
Quantification
For wild-type mice, the number of cells in each category (i.e., Pax7+ve/MyoD−ve, Pax7+ve/MyoD+ve, or Pax7−ve/MyoD+ve) were counted and expressed as a percentage of the total immunostained cells on the myofiber, and the data from multiple myofibers were pooled to give a population mean (± SEM). For 3F-nlacZ-E mice, the absolute number of satellite cells per myofiber was determined using unquenched DAPI fluorescence after X-gal incubation. Each cell was categorized by immunostaining, and the data from multiple myofibers were pooled to give a population mean (± SEM) for cells in each category and also expressed as a percentage of the total satellite cell pool.
Image capture
Immunostained myofibers were viewed on an epifluorescence microscope (model Axiophot; Carl Zeiss MicroImaging, Inc.) using a 40×/0.75 Ph2-Neofluar lens. Digital images were acquired with a Charge-Coupled Device (model RTE/CCD-1300-Y; Princeton Instruments Inc.) at −10°C using Metamorph software version 4.5r5 (Universal Imaging Corp). Live cells were viewed on a microscope (model Axiovert 100; Carl Zeiss MicroImaging, Inc.) with Achrostigmat lenses (Carl Zeiss MicroImaging, Inc.) and digital images acquired on a three-color Charge-Coupled Device (model DXC-930P; Sony) using Sirius VI software version 2.0c (Optivision). Images were optimized globally for contrast and brightness and assembled into figures using Adobe Photoshop 6.0.1.
Acknowledgments
We would like to thank Robert Kelly and Margaret Buckingham for providing the 3F-nlacZ-E mouse and the colleagues who made their antibodies available through the Developmental Studies Hybridoma Bank.
This work was supported by the Medical Research Council, European Community (EC) Biotechnology grant BIO4 CT 95-0228, EC Framework 5 QLRT-99-00020, EC Framework 5QLK6-1999-02034, and the British Council/EGIDE Alliance 2000/2001 grant PN 00.172. P.S. Zammit was funded by the Muscular Dystrophy Association and the Medical Research Council. Y. Nagata was funded by the Japan Scholarship Foundation. J.P. Golding was funded by EC Framework 5QLK6-1999-02034 and a Medical Research Council Collaborative Career Development Fellowship in Stem Cell Research. T. Partridge was partially funded by a Winter Fuel Payment from the UK Department for Work and Pensions. The Muscle Cell Biology Group also received support from EC Biotechnology grants BIO4 CT95-0284 and BMH4 CT97-2767 and the Leopold Muller Foundation.
Note added in proof. Recent work by Yablonka-Reuveni and associates (Halevy, O., Y. Piestun, M. Allouh, B. Rosser, Y. Rinkevich, R. Reshef, I. Rozenboim, M. Wleklinski-Lee, and Z. Yablonka-Reuveni. 2004. Dev. Dynam. In Press) is in accordance with our model of satellite cell self-renewal. These authors show that Pax7+ve/MyoD−ve cells can be observed in cultures initiated from wing chicken muscle and that a single cell can give rise to both Pax7+ve/MyoD−ve and Pax7+ve/MyoD+ve progeny.
Y. Nagata's present address is Dept. of Life Sciences, The University of Tokyo, 3-8-1 Komaba, Meguro-ku, Tokyo 153-8902, Japan.
V. Hudon's present address is McGill Cancer Center, 3655 Promenade Sir William Osler, Montreal, Quebec, H3G 1Y6, Canada.
Abbreviations used in this paper: EDL, extensor digitorum longus; MLC, myosin light chain; MRF, myogenic regulatory factor.
References
- Andres, V., and K. Walsh. 1996. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 132:657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., M.D. Rand, and R.J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science. 284:770–776. [DOI] [PubMed] [Google Scholar]
- Asakura, A., P. Seale, A. Girgis-Gabardo, and M.A. Rudnicki. 2002. Myogenic specification of side population cells in skeletal muscle. J. Cell Biol. 159:123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroffio, A., M. Hamann, L. Bernheim, M.L. Bochaton-Piallat, G. Gabbiani, and C.R. Bader. 1996. Identification of self-renewing myoblasts in the progeny of single human muscle satellite cells. Differentiation. 60:47–57. [DOI] [PubMed] [Google Scholar]
- Beauchamp, J.R., L. Heslop, D.S. Yu, S. Tajbakhsh, R.G. Kelly, A. Wernig, M.E. Buckingham, T.A. Partridge, and P.S. Zammit. 2000. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151:1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, R. 1986. Proliferation of muscle satellite cells on intact myofibers in culture. Dev. Biol. 115:129–139. [DOI] [PubMed] [Google Scholar]
- Blaveri, K., L. Heslop, D.S. Yu, J.D. Rosenblatt, J.G. Gross, T.A. Partridge, and J.E. Morgan. 1999. Patterns of repair of dystrophic mouse muscle: studies on isolated fibers. Dev. Dyn. 216:244–256. [DOI] [PubMed] [Google Scholar]
- Braun, T., M.A. Rudnicki, H.H. Arnold, and R. Jaenisch. 1992. Targeted inactivation of the muscle regulatory gene Myf-5 results in abnormal rib development and perinatal death. Cell. 71:369–382. [DOI] [PubMed] [Google Scholar]
- Brazelton, T.R., M. Nystrom, and H.M. Blau. 2003. Significant differences among skeletal muscles in the incorporation of bone marrow-derived cells. Dev. Biol. 262:64–74. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy, I.M., and T.A. Rando. 2002. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 3:397–409. [DOI] [PubMed] [Google Scholar]
- Cooper, R.N., S. Tajbakhsh, V. Mouly, G. Cossu, M. Buckingham, and G.S. Butler-Browne. 1999. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 112:2895–2901. [DOI] [PubMed] [Google Scholar]
- Cusella De Angelis, M.G., G. Balconi, S. Bernasconi, L. Zanetta, R. Boratto, D. Galli, E. Dejana, and G. Cossu. 2003. Skeletal myogenic progenitors in the endothelium of lung and yolk sac. Exp. Cell Res. 290:207–216. [DOI] [PubMed] [Google Scholar]
- De Angelis, L., L. Berghella, M. Coletta, L. Lattanzi, M. Zanchi, M.G. Cusella-De Angelis, C. Ponzetto, and G. Cossu. 1999. Skeletal myogenic progenitors originating from embryonic dorsal aorta coexpress endothelial and myogenic markers and contribute to postnatal muscle growth and regeneration. J. Cell Biol. 147:869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari, G., G. Cusella-De Angelis, M. Coletta, E. Paolucci, A. Stornaiuolo, G. Cossu, and F. Mavilio. 1998. Muscle regeneration by bone marrow-derived myogenic progenitors. Science. 279:1528–1530. [DOI] [PubMed] [Google Scholar]
- Ferrari, G., A. Stornaiuolo, and F. Mavilio. 2001. Failure to correct murine muscular dystrophy. Nature. 411:1014–1015. [DOI] [PubMed] [Google Scholar]
- Fuchtbauer, E.M., and H. Westphal. 1992. MyoD and myogenin are coexpressed in regenerating skeletal muscle of the mouse. Dev. Dyn. 193:34–39. [DOI] [PubMed] [Google Scholar]
- Fukada, S., Y. Miyagoe-Suzuki, H. Tsukihara, K. Yuasa, S. Higuchi, S. Ono, K. Tsujikawa, S. Takeda, and H. Yamamoto. 2002. Muscle regeneration by reconstitution with bone marrow or fetal liver cells from green fluorescent protein-gene transgenic mice. J. Cell Sci. 115:1285–1293. [DOI] [PubMed] [Google Scholar]
- Grounds, M.D., and J.K. McGeachie. 1987. A model of myogenesis in vivo, derived from detailed autoradiographic studies of regenerating skeletal muscle, challenges the concept of quantal mitosis. Cell Tissue Res. 250:563–569. [DOI] [PubMed] [Google Scholar]
- Grounds, M.D., K.L. Garrett, M.C. Lai, W.E. Wright, and M.W. Beilharz. 1992. Identification of skeletal muscle precursor cells in vivo by use of MyoD1 and myogenin probes. Cell Tissue Res. 267:99–104. [DOI] [PubMed] [Google Scholar]
- Grounds, M.D., J.D. White, N. Rosenthal, and M.A. Bogoyevitch. 2002. The role of stem cells in skeletal and cardiac muscle repair. J. Histochem. Cytochem. 50:589–610. [DOI] [PubMed] [Google Scholar]
- Gussoni, E., Y. Soneoka, C.D. Strickland, E.A. Buzney, M.K. Khan, A.F. Flint, L.M. Kunkel, and R.C. Mulligan. 1999. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 401:390–394. [DOI] [PubMed] [Google Scholar]
- Gussoni, E., R.R. Bennett, K.R. Muskiewicz, T. Meyerrose, J.A. Nolta, I. Gilgoff, J. Stein, Y.M. Chan, H.G. Lidov, C.G. Bonnemann, et al. 2002. Long-term persistence of donor nuclei in a Duchenne muscular dystrophy patient receiving bone marrow transplantation. J. Clin. Invest. 110:807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty, P., A. Bradley, J.H. Morris, D.G. Edmondson, J.M. Venuti, E.N. Olson, and W.H. Klein. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 364:501–506. [DOI] [PubMed] [Google Scholar]
- Heslop, L., J.E. Morgan, and T.A. Partridge. 2000. Evidence for a myogenic stem cell that is exhausted in dystrophic muscle. J. Cell Sci. 113:2299–2308. [DOI] [PubMed] [Google Scholar]
- Heslop, L., J.R. Beauchamp, S. Tajbakhsh, M.E. Buckingham, T.A. Partridge, and P.S. Zammit. 2001. Transplanted primary neonatal myoblasts can give rise to functional satellite cells as identified using the Myf5nlacZ/+ mouse. Gene Ther. 8:778–783. [DOI] [PubMed] [Google Scholar]
- Hoh, J.F., and S. Hughes. 1991. Basal lamina and superfast myosin expression in regenerating cat jaw muscle. Muscle Nerve. 14:398–406. [DOI] [PubMed] [Google Scholar]
- Hoh, J.F., S. Hughes, and J.F. Hoy. 1988. Myogenic and neurogenic regulation of myosin gene expression in cat jaw-closing muscles regenerating in fast and slow limb muscle beds. J. Muscle Res. Cell Motil. 9:59–72. [DOI] [PubMed] [Google Scholar]
- Horsley, V., K.M. Jansen, S.T. Mills, and G.K. Pavlath. 2003. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 113:483–494. [DOI] [PubMed] [Google Scholar]
- Jackson, K.A., T. Mi, and M.A. Goodell. 1999. Hematopoietic potential of stem cells isolated from murine skeletal muscle. Proc. Natl. Acad. Sci. USA. 96:14482–14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, R., S. Alonso, S. Tajbakhsh, G. Cossu, and M. Buckingham. 1995. Myosin light chain 3F regulatory sequences confer regionalized cardiac and skeletal muscle expression in transgenic mice. J. Cell Biol. 129:383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann, M., G. Carnac, M. Vandromme, M. Primig, N.J. Lamb, and A. Fernandez. 1998. The muscle regulatory factors MyoD and myf-5 undergo distinct cell cycle–specific expression in muscle cells. J. Cell Biol. 142:1447–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, T., and M. Raff. 2000. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 289:1754–1757. [DOI] [PubMed] [Google Scholar]
- LaBarge, M.A., and H.M. Blau. 2002. Biological progression from adult bone marrow to mononucleate muscle stem cell to multinucleate muscle fiber in response to injury. Cell. 111:589–601. [DOI] [PubMed] [Google Scholar]
- Lindon, C., D. Montarras, and C. Pinset. 1998. Cell cycle–regulated expression of the muscle determination factor Myf5 in proliferating myoblasts. J. Cell Biol. 140:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz, M.A., M.J. Marques, and H. Santo Neto. 2002. Impaired regeneration of dystrophin-deficient muscle fibers is caused by exhaustion of myogenic cells. Braz. J. Med. Biol. Res. 35:691–695. [DOI] [PubMed] [Google Scholar]
- McGeachie, J.K., and M.D. Grounds. 1987. Initiation and duration of muscle precursor replication after mild and severe injury to skeletal muscle of mice. An autoradiographic study. Cell Tissue Res. 248:125–130. [DOI] [PubMed] [Google Scholar]
- Megeney, L.A., B. Kablar, K. Garrett, J.E. Anderson, and M.A. Rudnicki. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173–1183. [DOI] [PubMed] [Google Scholar]
- Molnar, G., M.L. Ho, and N.A. Schroedl. 1996. Evidence for multiple satellite cell populations and a non-myogenic cell type that is regulated differently in regenerating and growing skeletal muscle. Tissue Cell. 28:547–556. [DOI] [PubMed] [Google Scholar]
- Moss, F.P., and C.P. Leblond. 1971. Satellite cells as the source of nuclei in muscles of growing rats. Anat. Rec. 170:421–435. [DOI] [PubMed] [Google Scholar]
- Rantanen, J., T. Hurme, R. Lukka, J. Heino, and H. Kalimo. 1995. Satellite cell proliferation and the expression of myogenin and desmin in regenerating skeletal muscle: evidence for two different populations of satellite cells. Lab. Invest. 72:341–347. [PubMed] [Google Scholar]
- Rosenblatt, J.D., A.I. Lunt, D.J. Parry, and T.A. Partridge. 1995. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim. 31:773–779. [DOI] [PubMed] [Google Scholar]
- Rosenblatt, J.D., D.J. Parry, and T.A. Partridge. 1996. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 60:39–45. [DOI] [PubMed] [Google Scholar]
- Rudnicki, M.A., P.N. Schnegelsberg, R.H. Stead, T. Braun, H.H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 75:1351–1359. [DOI] [PubMed] [Google Scholar]
- Sabourin, L.A., A. Girgis-Gabardo, P. Seale, A. Asakura, and M.A. Rudnicki. 1999. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh, M., K. Czyewski, and L.Z. Stern. 1985. Chronic myopathy induced by repeated bupivacaine injections. J. Neurol. Sci. 67:229–238. [DOI] [PubMed] [Google Scholar]
- Sampaolesi, M., Y. Torrente, A. Innocenzi, R. Tonlorenzi, G. D'Antona, M.A. Pellegrino, R. Barresi, N. Bresolin, M.G. De Angelis, K.P. Campbell, et al. 2003. Cell therapy of α-sarcoglycan null dystrophic mice through intra-arterial delivery of mesoangioblasts. Science. 301:487–492. [DOI] [PubMed] [Google Scholar]
- Schmalbruch, H., and D.M. Lewis. 2000. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle Nerve. 23:617–626. [DOI] [PubMed] [Google Scholar]
- Schultz, E. 1996. Satellite cell proliferative compartments in growing skeletal muscles. Dev. Biol. 175:84–94. [DOI] [PubMed] [Google Scholar]
- Schultz, E., and B.H. Lipton. 1982. Skeletal muscle satellite cells: changes in proliferation potential as a function of age. Mech. Ageing Dev. 20:377–383. [DOI] [PubMed] [Google Scholar]
- Schultz, E., M.C. Gibson, and T. Champion. 1978. Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J. Exp. Zool. 206:451–456. [DOI] [PubMed] [Google Scholar]
- Seale, P., L.A. Sabourin, A. Girgis-Gabardo, A. Mansouri, P. Gruss, and M.A. Rudnicki. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786. [DOI] [PubMed] [Google Scholar]
- Snow, M.H. 1977. Myogenic cell formation in regenerating rat skeletal muscle injured by mincing. II. An autoradiographic study. Anat. Rec. 188:201–217. [DOI] [PubMed] [Google Scholar]
- Snow, M.H. 1978. An autoradiographic study of satellite cell differentiation into regenerating myotubes following transplantation of muscles in young rats. Cell Tissue Res. 186:535–540. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh, S., D. Rocancourt, and M. Buckingham. 1996. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice. Nature. 384:266–270. [DOI] [PubMed] [Google Scholar]
- Tamaki, T., A. Akatsuka, K. Ando, Y. Nakamura, H. Matsuzawa, T. Hotta, R.R. Roy, and V.R. Edgerton. 2002. Identification of myogenic-endothelial progenitor cells in the interstitial spaces of skeletal muscle. J. Cell Biol. 157:571–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt, F.M. 2002. The stem cell compartment in human interfollicular epidermis. J. Dermatol. Sci. 28:173–180. [DOI] [PubMed] [Google Scholar]
- Weintraub, H., R. Davis, S. Tapscott, M. Thayer, M. Krause, R. Benezra, T.K. Blackwell, D. Turner, R. Rupp, S. Hollenberg, et al. 1991. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 251:761–766. [DOI] [PubMed] [Google Scholar]
- Whalen, R.G., J.B. Harris, G.S. Butler-Browne, and S. Sesodia. 1990. Expression of myosin isoforms during notexin-induced regeneration of rat soleus muscles. Dev. Biol. 141:24–40. [DOI] [PubMed] [Google Scholar]
- Yablonka-Reuveni, Z., and A.J. Rivera. 1994. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 164:588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonka-Reuveni, Z., M.A. Rudnicki, A.J. Rivera, M. Primig, J.E. Anderson, and P. Natanson. 1999. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev. Biol. 210:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, N., S. Yoshida, K. Koishi, K. Masuda, and Y. Nabeshima. 1998. Cell heterogeneity upon myogenic differentiation: down-regulation of MyoD and Myf-5 generates ‘reserve cells’. J. Cell Sci. 111:769–779. [DOI] [PubMed] [Google Scholar]
- Zammit, P., and J. Beauchamp. 2001. The skeletal muscle satellite cell: stem cell or son of stem cell? Differentiation. 68:193–204. [DOI] [PubMed] [Google Scholar]
- Zammit, P.S., L. Heslop, V. Hudon, J.D. Rosenblatt, S. Tajbakhsh, M.E. Buckingham, J.R. Beauchamp, and T.A. Partridge. 2002. Kinetics of myoblast proliferation show that resident satellite cells are competent to fully regenerate skeletal muscle fibers. Exp. Cell Res. 281:39–49. [DOI] [PubMed] [Google Scholar]