Abstract
PEX19 is a chaperone and import receptor for newly synthesized, class I peroxisomal membrane proteins (PMPs). PEX19 binds these PMPs in the cytoplasm and delivers them to the peroxisome for subsequent insertion into the peroxisome membrane, indicating that there may be a PEX19 docking factor in the peroxisome membrane. Here we show that PEX3 is required for PEX19 to dock at peroxisomes, interacts specifically with the docking domain of PEX19, and is required for recruitment of the PEX19 docking domain to peroxisomes. PEX3 is also sufficient to dock PEX19 at heterologous organelles and binds PEX19 via a conserved motif that is essential for this docking activity and for PEX3 function in general. Not surprisingly, transient inhibition of PEX3 abrogates class I PMP import but has no effect on class II PMP import or peroxisomal matrix protein import. Taken together, these results suggest that PEX3 plays a selective, essential, and direct role in PMP import as a docking factor for PEX19.
Keywords: PMP import; temperature-sensitive mutants; Zellweger syndrome; organelle biogenesis; protein import
Introduction
Peroxisomes are single membrane organelles present in nearly all free-living eukaryotes. The biogenesis of peroxisomes requires a group of protein factors referred to as peroxins and the PEX genes that encode them (Distel et al., 1996; Sacksteder and Gould, 2000). A variety of studies have demonstrated that all, or nearly all, peroxisomal proteins are synthesized on free polyribosomes and imported posttranslationally (Lazarow and Fujiki, 1985). Peroxisomal matrix enzymes typically possess a peroxisomal targeting signal (PTS1 or PTS2; Gould et al., 1989; Swinkels et al., 1991) and their import is dependent upon the predominantly cytoplasmic PTS1 and PTS2 receptors (PEX5 and PEX7, respectively; McCollum et al., 1993; Marzioch et al., 1994; Dodt et al., 1995; Wiemer et al., 1995), receptor docking factors on the peroxisome membrane (PEX14 and PEX13; Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997; Girzalsky et al., 1999; Stein et al., 2002) and a host of other peroxins of ill-defined function (Sacksteder and Gould, 2000; Gould and Collins, 2002). Like their cousin enzymes, PMPs are also imported by a direct cytoplasm-to-peroxisome mechanism, as suggested by kinetic studies in vivo (Fujiki et al., 1984; Imanaka et al., 1996) and the ability of peroxisomes to import newly synthesized PMPs directly from cytosolic lysates (Diestelkotter and Just, 1993; Pause et al., 1997). However, PMPs use different types of signals (PMP targeting signals [mPTSs]; Dyer et al., 1996; Jones et al., 2001, 2004) and are imported independently of the peroxins involved in matrix protein import (Chang et al., 1999a; Hettema et al., 2000).
Two pathways of PMP import have been described (Jones et al., 2004). The class I pathway of PMP import is dependent upon PEX19, which binds a wide variety of PMPs (Gloeckner et al., 2000; Sacksteder et al., 2000; Snyder et al., 2000; Fransen et al., 2001; Brosius et al., 2002), and then binds their transmembrane domains, stabilizes them during their transit through the cytoplasm, and releases them before or during their insertion into the peroxisome membrane (Jones et al., 2004). PEX19 also binds to the targeting signals (mPTSs) of numerous class I PMPs (Jones et al., 2001, 2004; Brosius et al., 2002), is required for class I mPTS function in vivo, and is essential for the import of class I PMPs, though it is not required for import of peroxisomal matrix enzymes or a class II PMP (Jones et al., 2004). Taken together, these results indicate that PEX19 is a cytoplasmic chaperone and import receptor for newly synthesized class I PMPs.
The class I PMPs are a diverse group and include metabolite transporters such as PMP22, PMP34, and PMP70, peroxins involved in peroxisome division such as PEX11, peroxins involved in peroxisomal matrix protein import such as PEX2 and PEX13, and even one peroxin involved in peroxisome membrane synthesis, PEX16 (Sacksteder et al., 2000; Jones et al., 2001, 2004; Brosius et al., 2002). In fact, the only PMP that is known to be imported independently of PEX19 is PEX3, which currently defines the class II PMP import pathway (Jones et al., 2004).
The observation that PEX19 binds newly synthesized class I PMPs in the cytoplasm and then transports them to the peroxisome indicates that there is at least one docking factor for PEX19 on the peroxisome membrane. A docking factor for PEX19 can be expected to have certain properties. For example, it must bind to PEX19 and it must reside in the peroxisome membrane. However, these properties are also shared by all integral PMPs that depend on PEX19 for their import (Sacksteder et al., 2000; Jones et al., 2004). More stringent criteria for the PEX19 docking factor are necessary and some are as follows. First, loss of a putative docking factor must block the recruitment of PEX19 to peroxisomes. Second, a docking factor should interact with a region of PEX19 that (a) differs from its PMP-binding domain, (b) is sufficient for docking, and (c) requires the docking factor for its recruitment to peroxisomes. Third, mislocalization of a PEX19 docking factor to another organelle should be sufficient to dock PEX19 at the heterologous organelle. Fourth, the docking factor for PEX19 should be required for class I PMP import but dispensable for class II PMP import and peroxisomal matrix protein import. Here we show that PEX3 meets all of these criteria.
Results
PEX3 is required for PEX19 docking
Aside from PEX19, only one conserved peroxin, PEX3, is known to be required for peroxisome membrane biogenesis (PEX16 is required for peroxisome membrane biogenesis in mammalian cells [Honsho et al., 1998; South and Gould, 1999], but is absent from the yeast Saccharomyces cerevisiae and serves a different function in the yeast Yarrowia lipolytica [Eitzen et al., 1997]). Human and yeast cells lacking the PEX3 gene are devoid of detectable peroxisomes, rapidly degrade most PMPs, and mislocalize other PMPs to the mitochondria and other organelles (Chang et al., 1999a; Hettema et al., 2000; South et al., 2000). Thus, of the known peroxins, PEX3 represents an obvious candidate for a PEX19 docking factor.
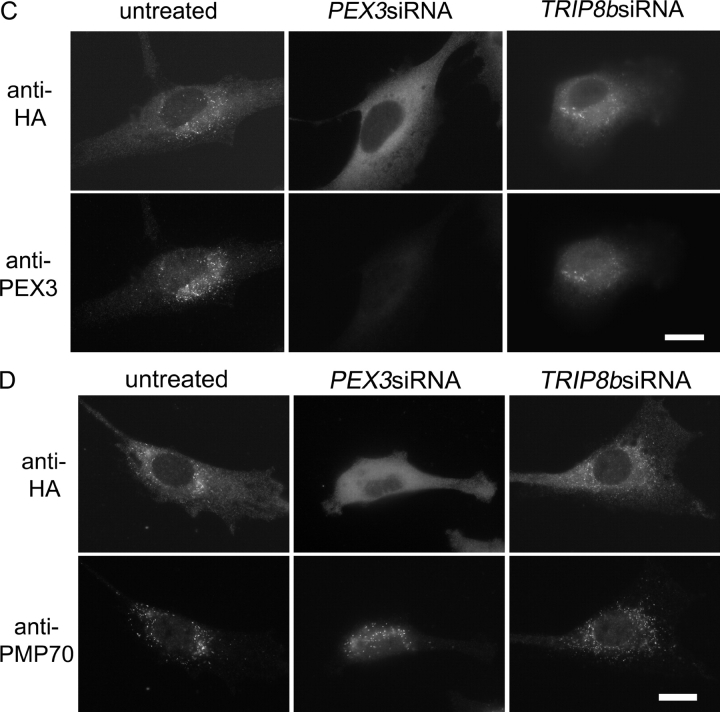
To determine whether PEX3 is required for PEX19 to dock at the peroxisome membrane we tested whether transient depletion of PEX3, using RNA interference (RNAi) technology, affected PEX19's association with peroxisomes. Wild-type (WT) human fibroblasts transfected with small interfering RNAs (siRNAs) specific for the PEX3 gene show a significant reduction in PEX3 protein levels at 2–3 d after transfection, with levels reduced ∼90% on day 2 (Fig. 1 A). At this time point, PEX3 can no longer be detected on peroxisomes by immunofluorescence (Fig. 1 B). Control RNAi directed against TRIP8b (a gene that has no role in peroxisome biogenesis; Chen et al., 2001) or PEX5 (a gene that functions only in peroxisomal matrix protein import; Dodt et al., 1995; Chang et al., 1999a) had no effect on PEX3 abundance within the cell.
Figure 1.
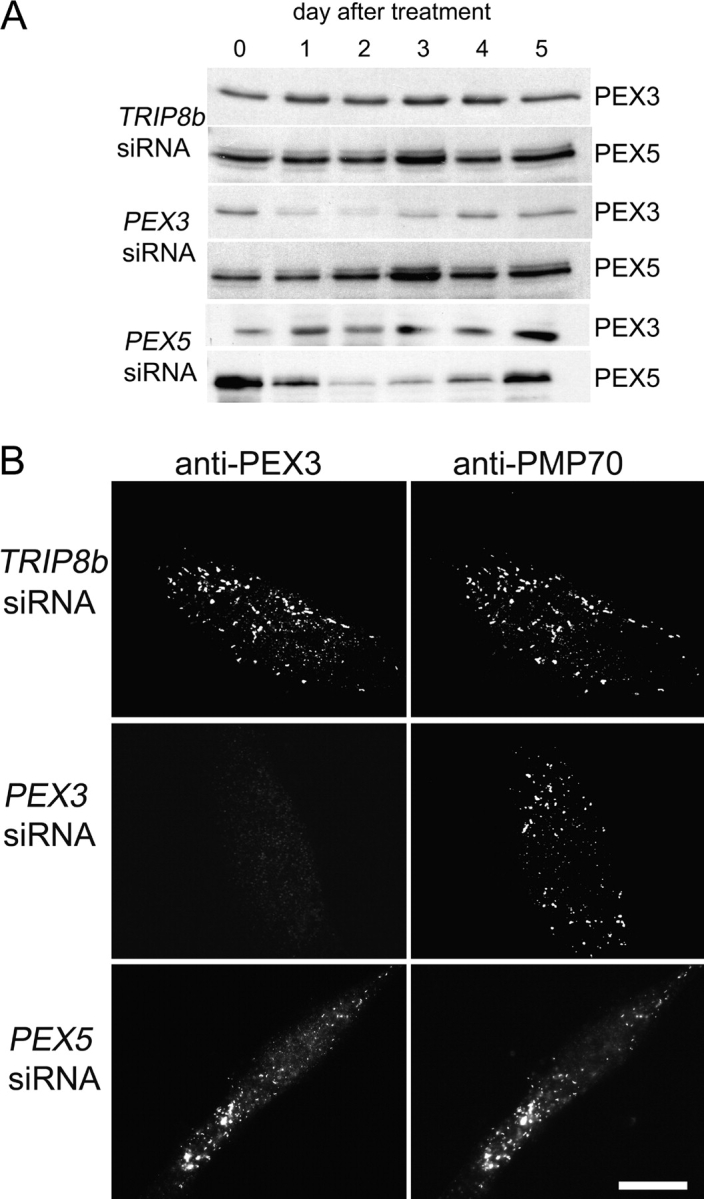
The effect of PEX3 on the localization of 3xHA-PEX19. (A) GM5756-TI cells were transfected with TRIP8bsiRNAs, PEX3siRNAs, or PEX5siRNAs, respectively. Cells were lysed each day following the siRNA treatment, and equivalent amounts of protein from each sample were processed for immunoblot with antibodies against PEX3 and PEX5. (B) On day 2 after the siRNA treatment, cells were processed for double indirect immunofluorescence microscopy with anti-PEX3 and anti-PMP70 antibodies. (C and D) 3xHA-PEX19/PBD399-TI cells (untreated) and 3xHA-PEX19/PBD399-TI cells transfected with TRIP8bsiRNAs or PEX3siRNAs were grown for 2 d and processed for double indirect immunofluorescence microscopy with antibodies against (C) the HA epitope and PEX3, or (D) the HA epitope and PMP70. Bars, 20 μm.
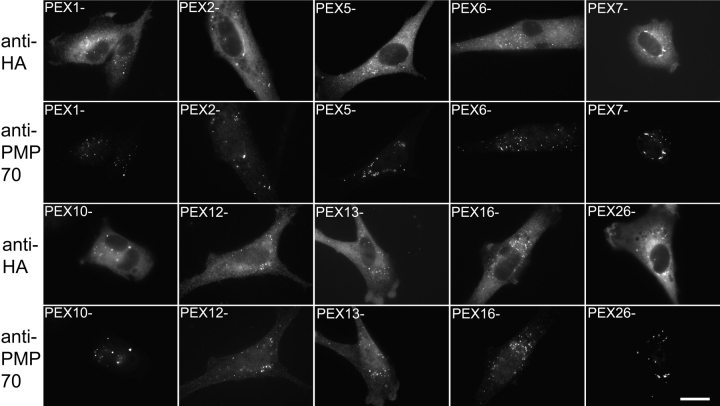
Although PEX19 is partly peroxisomal, the vast majority of PEX19 is located in the cytoplasm and we have been unable to detect appreciable levels of WT PEX19 on peroxisomes either by subcellular fractionation or by immunofluorescence microscopy (Sacksteder et al., 2000). However, the Fujiki lab has reported that an NH2-terminally tagged form of PEX19 can be detected on peroxisome membranes (Matsuzono et al., 1999) and we therefore used a 3xHA-PEX19 fusion protein to assess PEX19 docking in the presence or absence of PEX3. 3xHA-PEX19 is detected easily on peroxisomes though the cytoplasmic pool of the protein is also significant (Fig. 1 C). In contrast, 3xHA-PEX19 cannot be detected on peroxisomes in cells that were cotransfected with the PEX3 siRNAs and lack PEX3 protein. The absence of peroxisome-associated 3xHA-PEX19 is not due to a complete absence of peroxisomes from PEX3-depleted cells, since they still contain peroxisome-associated PMP70 (Fig. 1 D). The requirement for PEX3 also appears specific, as 3xHA-PEX19 colocalizes with peroxisomes in human fibroblasts from peroxisome biogenesis disorder patients (Gould and Valle, 2000) that carry inactivating mutations in the PEX1, PEX2, PEX5, PEX6, PEX7, PEX10, PEX12, PEX13, and PEX26 genes, as well as in WT cells that are depleted for PEX16 by RNAi (Fig. 2).
Figure 2.
The localization of 3xHA-PEX19 in other human pex mutants. Human fibroblasts from peroxisome biogenesis disorder patients that carry inactivating mutations in the PEX1, PEX2, PEX5, PEX6, PEX7, PEX10, PEX12, PEX13, and PEX26 genes were transfected with 3xHA-PEX19 expression plasmid, grown overnight, and processed for indirect immunofluorescence microscopy with anti-HA and anti-PMP70 antibodies. GM5756-TI cells were also transfected with PEX16siRNAs and the 3xHA-PEX19 expression plasmid. 2 d after the transfection, at a time when PEX16 activity is impaired (unpublished data), these cells (PEX16-) were subjected to double indirect immunofluorescence microscopy using anti-HA and anti-PMP70 antibodies. Bar, 20 μm.
The docking domain of PEX19 binds to PEX3
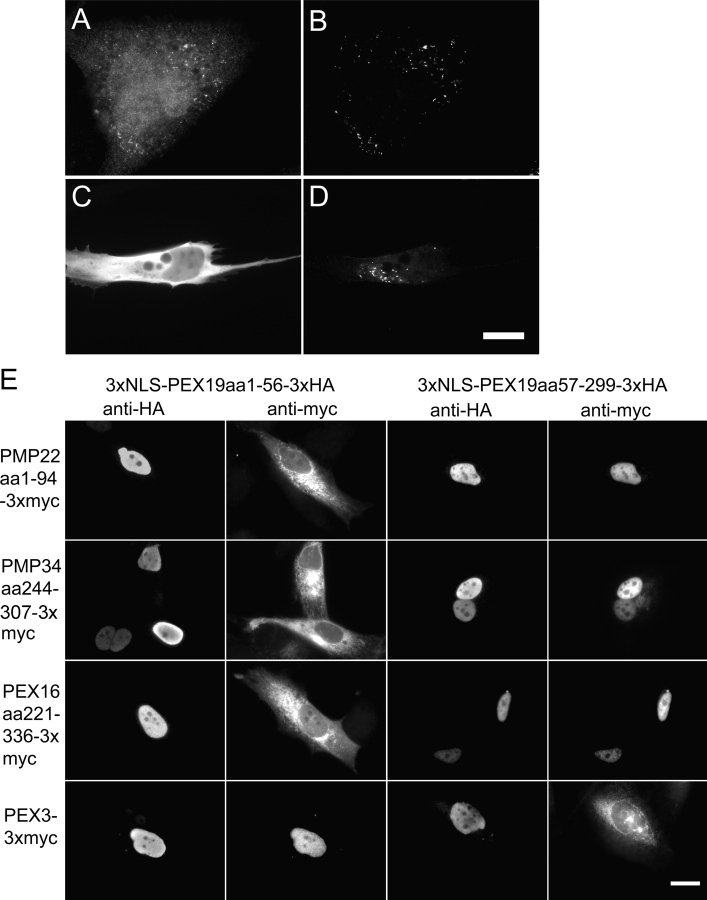
As a chaperone and import receptor for newly synthesized class I PMPs, PEX19 is likely to use distinct domains for binding these ligands and for docking with the peroxisome membrane. We constructed a series of deletion mutants in PEX19 and assayed these for docking to peroxisomes. A fragment consisting of just the NH2-terminal 56 amino acids of PEX19 is sufficient for docking to peroxisomes but is unable to bind class I PMPs, whereas the remainder of the protein, amino acids 57–299, retains the ability to bind numerous class I PMPs but is unable to dock at peroxisomes (Fig. 3). Previous studies have established that this same segment of PEX19 is sufficient to bind PEX3 (Snyder et al., 2000; Fransen et al., 2001), which we show again here (Fig. 3, bottom four panels). Thus, the same short region of PEX19 that binds to PEX3 is also sufficient for docking PEX19 at the peroxisome membrane.
Figure 3.
Characterization of the docking domain of PEX19. (A–D) GM5756-TI cells were transfected with (A and B) the 3xHA-PEX19aa1–56 or (C and D) 3xHA-PEX19aa57–299 expression plasmids, respectively, grown overnight and processed for double indirect immunofluorescence using (A and C) anti-HA and (B and D) anti-PMP70 antibodies. (E) PBD399-TI cells were cotransfected with the plasmids designed to express either 3xNLS-PEX19aa1–56–3xHA or 3xNLS-PEX19aa57–299–3xHA, together with plasmids designed to express PMP22aa1–94–3xmyc, PMP34aa244–307–3xmyc, PEX16aa221–336–3xmyc, and PEX3–3xmyc. 24 h after the transfection, cells were processed for double indirect immunofluorescence using anti-HA and anti-myc antibodies. Bars, 20 μm.
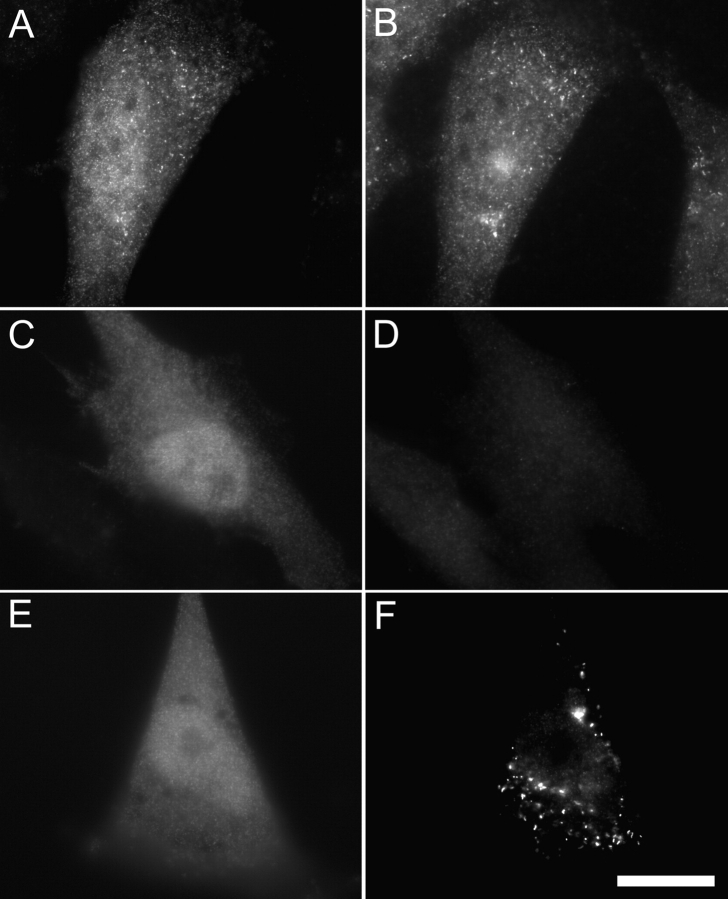
The preceding results indicate that PEX3 might function as a docking factor for PEX19. If this hypothesis is correct, transient reduction of PEX3 levels should block recruitment of the PEX19 docking domain to peroxisomes. To test this we examined the subcellular distribution of PEX19aa1–56 in human fibroblasts transfected with control and PEX3 siRNAs. Control siRNA had no affect on PEX3 abundance and did not affect the association of the PEX19 docking domain with peroxisomes (Fig. 4, A and B). In contrast, cells that had reduced levels of PEX3 protein (after transfection with PEX3 siRNAs) were unable to recruit the PEX19 docking domain to peroxisomal membranes (Fig. 4, C–F).
Figure 4.
The effect of PEX3 depletion on the localization of the PEX19 docking domain. GM5756-TI cells were cotransfected with a plasmid designed to express 3xHA-PEX19aa1–56 and siRNAs specific for either (A and B) TRIP8b or (C–F) PEX3. On day 2 after transfection, cells were processed for double indirect immunofluorescence microscopy with (A and C) anti-HA antibody and (B and D) anti-PEX3 antibodies. The same samples were also processed for double indirect immunofluorescence assay with (E) anti-HA and (F) anti-PMP70 antibodies. Bar, 20 μm.
A conserved peptide motif of PEX3 binds PEX19 and is required for PEX19 docking
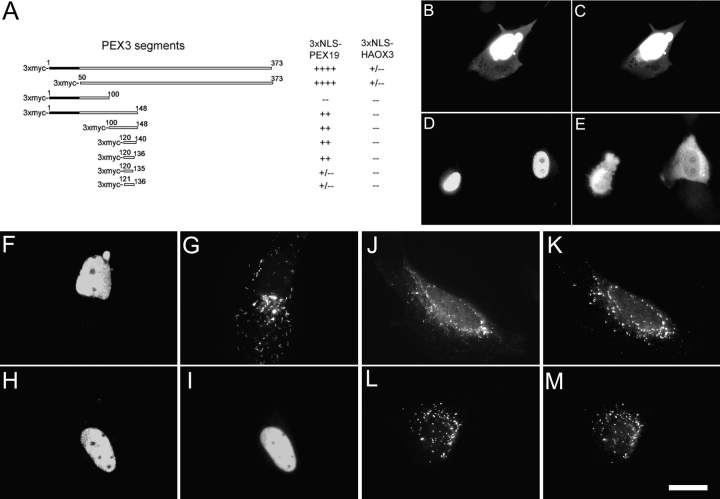
Another way to assess the possibility that PEX3 is a PEX19 docking factor is to identify a region of PEX3 that binds to PEX19 and then determine whether it is important for PEX19 docking. To identify a PEX19-binding site within PEX3 we assayed a series of PEX3 protein fragments for their ability to interact with PEX19 in vivo. Segments of the PEX3 protein were coexpressed with 3xNLS-PEX19 (Jones et al., 2001) in human fibroblasts, followed by immunofluorescence microscopy to determine whether 3xNLS-PEX19 bound the fragments well enough to concentrate them in the nucleus (Fig. 5, A–E). These studies revealed the existence of a conserved, 17–amino acid-long segment of human PEX3, amino acids 120–136 (NH2-YSTCMLVVLLRVQLNII-COOH), which is sufficient for binding to PEX19 in vivo.
Figure 5.
Characterization of one PEX19-interacting motif of PEX3. (A) Selected segments of PEX3 tagged with 3xmyc were coexpressed in PBD399-TI cells with either 3xNLS-PEX19 or 3xNLS-HAOX3 followed by double indirect immunofluorescence microscopy. The interaction strength was calculated by counting cells in which PEX3 segments accumulated in the nucleus (++++, >80% cells concentrated the PEX3 segment in the nucleus; +++, 60–79%; ++, 40–59%; +, 15–39%; +/−, <15%; −, 0%). (B–E) PBD399-TI cells were cotransfected with plasmids to express (B, C) 3xmyc-PEX3aa120–136 and 3xNLS-PEX19, or (D, E) 3xmyc-PEX3aa120–136 and 3xNLS-HAOX3. 24 h after the transfection, cells were processed for double indirect immunofluorescence microscopy with (B) anti-PEX19 and (C) anti-myc antibodies, or (D) anti-HAOX3 and (E) anti-myc antibodies. (F–I) PBD399-TI cells were cotransfected with plasmids designed to express (F and G) 3xNLS-PEX19 and PEX3L125P/N134D-3xmyc, or (H and I) 3xNLS-PEX19 and PEX3–3xmyc, grown overnight, and processed for double indirect immunofluoresence microscopy with (F and H) anti-PEX19 and (G and I) anti-myc antibodies. (J–M) GM5756-TI cells were transfected with (J and K) pcDNA3-PEX3L125P/N134D-3xmyc or (L and M) pcDNA3-PEX3–3xmyc, respectively, and grown overnight. The localization of PEX3L125P/N134D-3xmyc and PEX3–3xmyc was examined by double indirect immunofluoresence with antibodies against (J and L) the myc epitope and (K and M) endogenously expressed PMP70. Bar, 20 μm.
To determine whether this region of PEX3 is important for PEX3 function and for PEX19 docking, we first created a mutant version of PEX3 altered in this region. This motif is predicted to have a strong α-helical propensity. To disrupt this motif we replaced a conserved leucine near the NH2 terminus of the motif with a proline (L125P), as well as a conserved asparagine with an aspartate (N134D). This mutant PEX3 protein no longer interacted with PEX19 (as evident from its lack of colocalization with 3xNLS-PEX19; Fig. 5, F–I), no longer complemented PEX3-deficient human cells (unpublished data), but was still properly targeted to peroxisomes in WT human fibroblasts (Fig. 5, J–M).
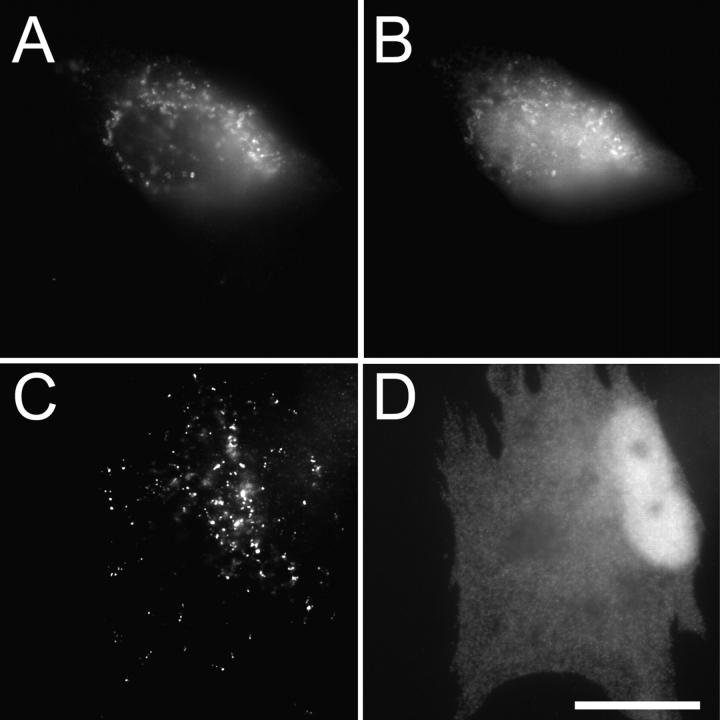
The mislocalization of PEX3 to other subcellular organelles in cells that lack PEX19 (and therefore lack peroxisomes) makes it possible to ask whether PEX3 is sufficient to recruit PEX19 to organelle surfaces. PEX19-deficient cells were cotransfected with plasmids designed to express the docking domain of PEX19 (3xHA-PEX19aa1–56) and either WT PEX3–3xmyc or the L125P/N134D mutant form of PEX3–3xmyc. Both PEX3 proteins are imported into heterologous organelles, including mitochondria and small unidentifiable small vesicles. WT PEX3 is able to recruit the docking domain of PEX19 to these other organelles (Fig. 6, A and B) but the L125P/N134D mutant form of PEX3myc is unable to recruit the docking domain of PEX19 to these organelles (Fig. 6, C and D).
Figure 6.
The effect of PEX3L125P/N134D on PEX19 docking. PBD399-TI cells were cotransfected with (A and B) pcDNA3–3xHA-PEX19aa1–56 and pcDNA3-PEX3–3xmyc or (C and D) pcDNA3–3xHA-PEX19aa1–56 and pcDNA3-PEX3L125P/N134D-3xmyc, grown overnight, and processed for double indirect immunofluorescence microscopy using (A and C) anti-myc and (B and D) anti-HA antibodies. Bar, 20 μm.
PEX3 is required for PMP import
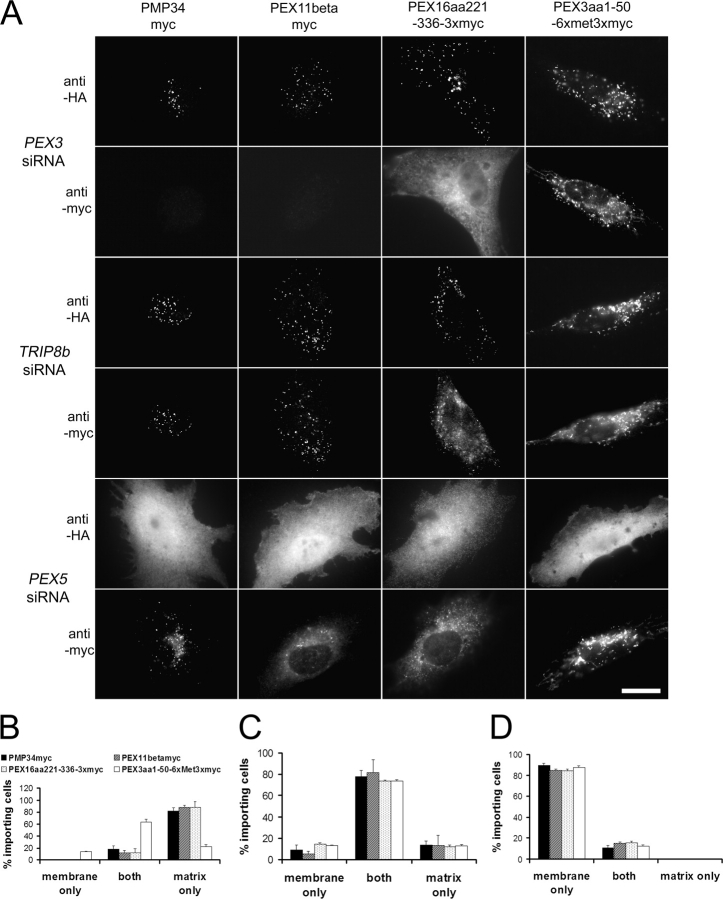
The observation that PEX3 is both necessary and sufficient to mediate PEX19 docking indicates that it should also be essential for class I PMP import. To test this we again used PEX3 RNAi to generate a transient depletion of PEX3 protein in cells that still possessed peroxisomes. We also used control RNAi specific for the TRIP8b and PEX5 transcripts (please refer to Fig. 1 for control immunoblots). 3 d after transfection with the RNAi, the cells from each population were retransfected with a pair of plasmids designed to express (a) an HA-tagged peroxisomal matrix protein (HA-PTE1; Jones et al., 1999) and (b) any of several myc-tagged PMPs. These included the class I PMPs PMP34 (PMP34myc), PEX11β (PEX11βmyc) and a PEX16 mPTS, as well as a class II PMP, the mPTS of PEX3 (Jones et al., 2004). At 2 h after the DNA transfection, the cells were fixed and processed for immunofluorescence microscopy using antibodies specific for the matrix and membrane marker proteins.
These experiments revealed that inhibition of PEX3 induced a selective defect in import of class I PMPs but had no detectable effect on the import of a class II PMP or a peroxisomal matrix protein (Fig. 7). In addition to the immunofluorescence microscopy images, which provide a visual representation of these results (Fig. 7 A), we assessed import capabilities of hundreds of randomly selected cells from each population. The statistical analysis of this data confirms that inhibition of PEX3 resulted in a specific defect in class I PMP import (Fig. 7, B–D).
Figure 7.
The effect of PEX3 depletion on class I and II PMP import. (A) GM5756-TI cells were electroporated with PEX3, TRIP8b, or PEX5 siRNAs, respectively, grown for 2–3 d, retransfected with a pair of plasmids designed to express (a) HA-PTE1 and (b) each of the following myc-tagged proteins: PMP34myc, PEX11βmyc, the mPTS of PEX16 (PEX16aa221–336–3xmyc), and PEX3(PEX3aa1–50–6xmet3xmyc). 2 h later, cells were processed for double indirect immunofluoresence microscopy with antibodies against the HA and myc epitopes. Bar, 20 μm. (B–D) Cells showing peroxisomal staining (import of either marker) were counted as to whether they imported HA-PTE1 only (matrix only), PMPs-myc only (membrane only), or both (both) in the populations transfected with (B) PEX3siRNAs, (C) TRIP8bsiRNAs, and (D) PEX5siRNAs. Each experiment was done in triplicate and the mean and standard deviation are presented.
Yeast PEX3 also plays a selective role in PMP import
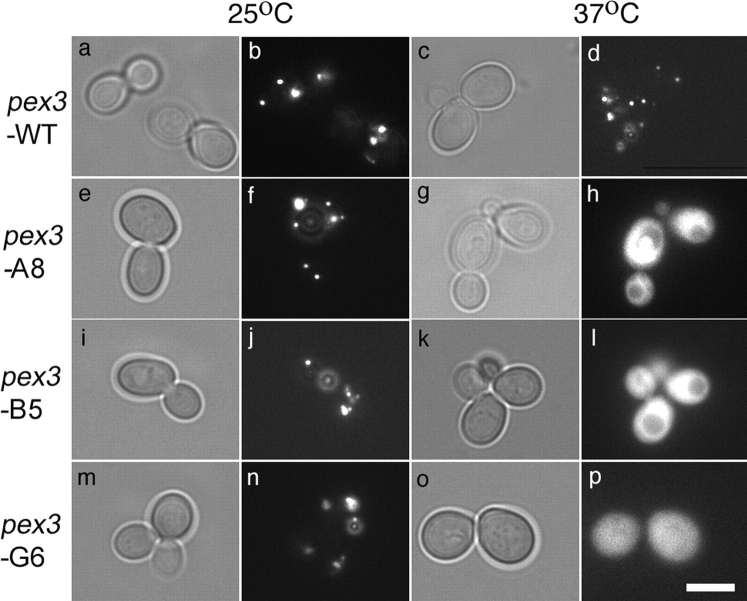
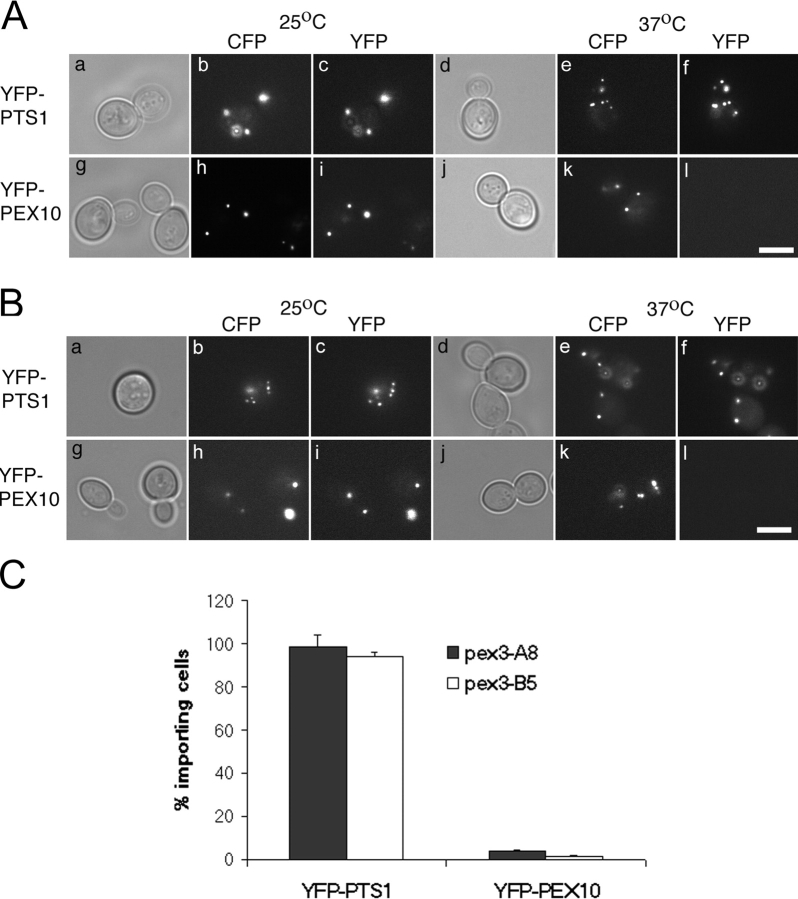
PEX3 is also required for peroxisome membrane synthesis in the yeast S. cerevisiae, as evident from the rapid destruction of PMPs and lack of peroxisomal membranes in pex3-null mutants of this yeast (Hettema et al., 2000). To test whether yeast PEX3 also plays a selective and essential role in PMP import we first sought to generate temperature-sensitive pex3 mutants of this yeast. These experiments used a pex3Δ derivative of the strain BY4733 (Baker-Brachmann et al., 1998), YFY1, that also carries a LEU2-containing plasmid that constitutively expresses CFP-PTS1 (a cyan fluorescing form of green fluorescent protein (CFP) that contains a type 1 peroxisomal-targeting signal at its COOH terminus and is imported into the peroxisome lumen (Kalish et al., 1996)). In YFY1, CFP-PTS1 accumulates in the cytoplasm instead of being targeted to peroxisomes, though it is imported into peroxisomes in YFY1 cells that carry the WT PEX3 gene on a plasmid (unpublished results). To obtain temperature-sensitive pex3 mutants, YFY1 cells were cotransformed with (a) a PEX3 gap repair plasmid (pRS314-scPEX3UTR) that contained the PEX3 promoter and 3′-flanking regions but lacked all PEX3 coding sequences and (b) a PCR-generated PEX3 DNA fragment that contained the entire PEX3 gene, including about 100 bps of flanking sequences on either side of the PEX3 ORF. 1,000 transformants were spotted onto duplicate plates, incubated at 25° and 37°C for 3 d, and then examined by epifluorescence microscopy to determine the subcellular distribution of CFP-PTS1. Three conditional pex3 mutants were identified, each of which imported CFP-PTS1 at the permissive temperature (25°C) but accumulated CFP-PTS1 in the cytoplasm at the restrictive temperature (37°C): pex3-A8, pex3-B5, and pex3-G6 (Fig. 8).
Figure 8.
Characterization of conditional pex3 mutants. A pex3-null strain that constitutively expresses CFP-PTS1 was transformed with plasmids that express the WT PEX3 allele (a–d), pex3-A8 allele (e–h), pex3-B5 allele (i–l), or pex3-G6 allele (m–p). The resulting strains were grown at either (a, b, e, f, i, j, m, and n) 25°C or (c, d, g, h, k, l, o, and p) 37°C for 3 d, fixed, and examined by phase contrast (a, c, e, g, i, k, m, and o) and epifluorescence microscopy specific for CFP (b, d, f, h, j, l, n, and p). Bar, 5 μm.
The PEX3-expression plasmids were recovered from each of the three yeast strains, transformed into bacteria, and amplified. After their reintroduction into YFY1 cells, all three plasmids again conferred a temperature-sensitive pex3 phenotype, demonstrating that the conditional phenotypes of the pex3-A8, pex3-B5, and pex3-G6 strains mapped to their plasmid-borne pex3 genes. The sequences of the pex3 alleles carried on these plasmids were determined and each contained several amino acid substitutions: Y144N, T161S, T261A, and N430Y for pex3-A8; E132G, K369N, T398S for pex3-B5; L11Q, Q248P, F275Y, L305F, and D415V for pex3-G6.
To determine whether PEX3 played essential roles in PMP import, peroxisomal matrix protein import, or both, we assayed both processes in YFY1 cells carrying either a WT PEX3 expression plasmid or the pex3-G6 plasmid, which appeared to confer the “tightest” conditional phenotype of the three mutant plasmids we obtained. These two strains, which constitutively express CFP-PTS1, were then transformed, individually, with plasmids that carried the galactose-inducible GAL1 promoter driving the expression of four peroxisomal marker proteins: (a) YFP-PTS1, a peroxisomal matrix protein (Kalish et al., 1996), (b) YFP-PEX10, a YFP-tagged form of a PMP that is involved in peroxisome biogenesis (Kalish et al., 1995), (c) YFP-PEX15, another YFP-tagged form of a PMP that is involved in peroxisome biogenesis (Elgersma et al., 1997), and (d) ANT1-YFP, a YFP-tagged form of a PMP that is involved in ATP translocation across the peroxisome membrane and plays no role in peroxisome biogenesis (Palmieri et al., 2001). The resulting strains were grown at the permissive temperature in minimal glucose medium to repress expression of the YFP marker proteins. Each strain was then transferred to galactose medium to induce expression of the YFP marker proteins and either maintained at the permissive temperature or shifted to the restrictive temperature. 4 h after the temperature shift the cells were fixed and examined by epifluorescence microscopy to determine the subcellular distribution of the proteins as marked by CFP and YFP. As expected, YFY1 cells containing the WT PEX3 gene imported both YFP-PTS1 and the YFP-PMP fusion proteins into peroxisomes at both the permissive and restrictive temperatures (Fig. 9 A). The temperature-sensitive pex3-G6 strain also imported all four peroxisomal marker proteins at the permissive temperature. However, when the pex3-G6 strain was shifted to 37°C, it was unable to import YFP-PEX10, ANT1-YFP, or YFP-PEX15 proteins into peroxisomes, even though it continued to import the peroxisomal matrix protein marker YFP-PTS1 (Fig. 9 B). Similar results were obtained in the pex3-A8 and pex3-B5 strains (Fig. 10). Thus, transient inhibition of yeast PEX3 results in a specific defect in PMP import with no apparent defect in peroxisomal matrix protein import. Previous studies have also reported variable and aberrant fates for PMPs in cells that lack PEX3 activity (Hettema et al., 2000; South et al., 2000), and these are reflected in our inability to detect YFP-PEX10 and YFP-PEX15 and the mislocalization of ANT1-YFP to nonperoxisomal structures in cells that lack PEX3 activity.
Figure 9.
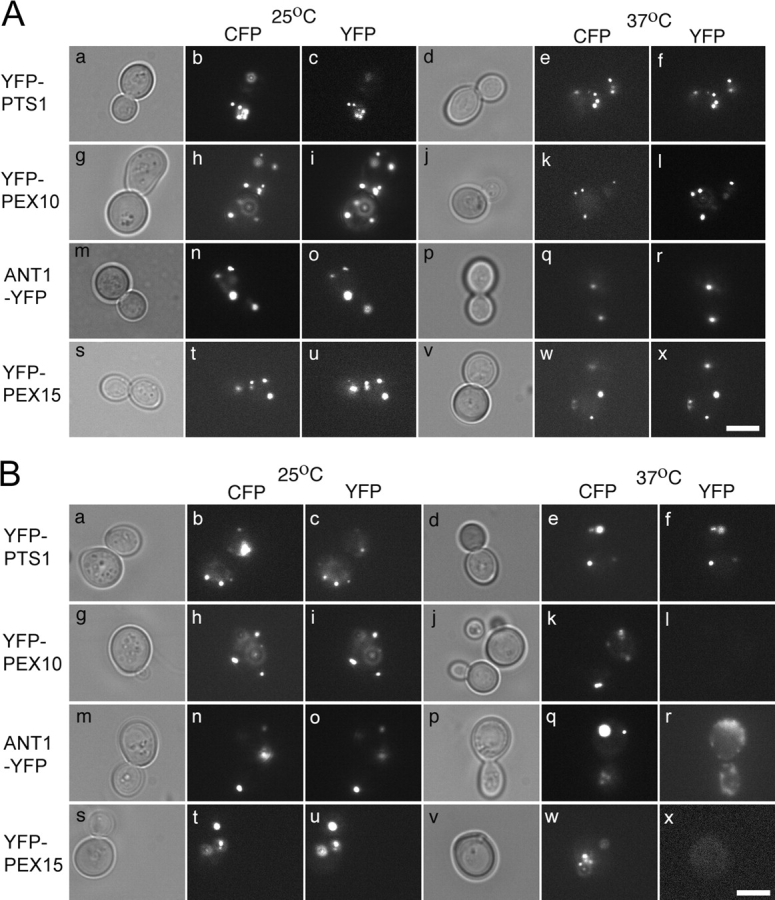
The effect of PEX3 activity on PMP import. A pex3-null mutant that constitutively expresses CFP-PTS1 and (A) WT PEX3 allele or (B) pex3-G6 allele was transformed with plasmids designed to express (a–f) YFP-PTS1, (g–l) YFP-PEX10, (m–r) ANT1-YFP, or (s–x) YFP-PEX15 under the control of galactose. The resulting strains were grown on minimal glucose medium at 25°C, transferred to minimal galactose medium, and maintained at (a–c, g–i, m–o, and s–u) 25°C, or shifted to (d–f, j–l, p–r, and v–x) 37°C for 2.5–4 h, followed by examination by phase contrast (a, d, g, j, m, p, s, and v), and epifluorescence microscopy specific for (b, e, h, k, n, q, t, and w) CFP and (c, f, i, l, o, r, u, and x) YFP. Bars, 5 μm.
Figure 10.
The effect of other two conditional pex3 mutants on PMP import. A pex3-null mutant, which constitutively expresses CFP-PTS1 and (A) pex3-A8 allele or (B) pex3-B5 allele, was transformed with plasmids designed to express (a–f) YFP-PTS1, or (g–l) YFP-PEX10 under the control of galactose. After being transferred to minimal galactose medium for 3 h at 25°C, the resultant strains were either maintained at (a–c and g–i) 25°C, or shifted to (d–f and j–l) 37°C for four more hours, and examined by phase contrast (a, d, g, and j) and epifluorescence microscope specific for (b, e, h, and k) CFP and (c, f, i, and l) YFP. Bars, 5 μm. (C) All cells with peroxisomal CFP-PTS1 were scored as to whether YFP was imported or not. Then the calculated percentage of import of each protein at 37°C was normalized to that at 25°C, respectively. Each experiment was done in triplicate and the mean and standard deviation are shown.
Discussion
In a recent study we established that there are at least two mechanistically distinct pathways of PMP import, a class I pathway that is dependent upon PEX19 and a class II pathway that is independent of PEX19 (Jones et al., 2004). Here we observed that PEX3 is essential for PEX19 docking at peroxisomes and sufficient to dock PEX19 at heterologous membranes. The PEX3-binding domain of PEX19 is sufficient for docking PEX19 at the peroxisome surface, but only in the presence of PEX3 on the peroxisome surface. Disruption of the PEX19-binding site on PEX3 disrupts its ability to recruit PEX19 to membranes and eliminates PEX3 function. Finally, we show that PEX3 is essential for import of class I PMPs but is not required for either class II PMP import or peroxisomal matrix protein import. These observations support the hypothesis that PEX3 plays an essential and selective role in the import of class I PMPs and that this role includes being a docking factor for PEX19, the class I PMP import receptor. It is also consistent with the recent observation that PEX3 and PEX19 interact primarily at the surface of the peroxisome membrane (Muntau et al., 2003).
The most widely accepted model of peroxisome biogenesis is that peroxisomes are formed by the growth and division of preexisting peroxisomes (Lazarow and Fujiki, 1985). Under this model, any significant defect in PMP import would be expected to cause the loss of peroxisomes from the cell. This is precisely the phenotype of pex3- and pex19-null mutants in both humans and S. cerevisiae (Matsuzono et al., 1999; Hettema et al., 2000; Sacksteder et al., 2000; South et al., 2000). Although class I PMP import might require more protein factors than PEX19 and PEX3, it is also possible that it does not. Therefore, we should not exclude additional roles for PEX19 and PEX3 in PMP import. In fact, some studies have detected interactions between class I PMPs and both PEX19 and PEX3 at the peroxisome membrane (Snyder et al., 2000; Hazra et al., 2002) and these observations might reflect a role for PEX3 and/or PEX19 in the insertion of class I PMPs.
Neither PEX3 nor PEX19 are required for class II PMP import (Jones et al., 2004). Of the PMPs that have been tested, PEX3 is the only class II PMP yet identified. Given the essential role for PEX3 in PMP import and the phenotypes associated with the loss of PEX3 activity (loss of peroxisomes from the cell, rapid destruction of PMPs, and cytosolic accumulation of matrix enzymes in the cytosol), it is expected that loss of any factor involved in PEX3 import would result in phenotypes that are at least as severe as the pex3-null mutant. No such mutants are known in yeast but the pex16 mutant of humans does meet these criteria. Future studies that test whether PEX16 acts as a PEX3 import factor, as another class I PMP import factor, or as a critical player in some other aspect of peroxisome membrane biogenesis (lipid import, peroxisome division, etc.) are a high priority. In addition to searching for factors that might mediate PEX3 import, it might be possible that PEX3 does not require any specific protein factors for import into peroxisomes, perhaps autocatalyzing its import into peroxisomes. Interestingly, Haan et al. (2002) have observed that an engineered epitope appears exposed to the cytosol regardless of where it is inserted in PEX3, and on this basis have argued that PEX3 might not even be an integral PMP, even though it has the biochemical properties of being embedded in the membrane.
The unique targeting pathway for PEX3 and its conserved role in the import of most PMPs can also be interpreted in the context of more speculative models of peroxisome biogenesis. For example, several researchers believe that peroxisomes arise, directly or indirectly, from the ER (Mullen et al., 2001; Titorenko and Rachubinski, 2001a,b; Tabak et al., 2003). A critical requirement of such models is the existence of a peroxin that is essential for the formation of peroxisome membranes but traffics to peroxisomes by a mechanism that is fundamentally distinct from that used by the vast majority of PMPs. Moreover, Faber et al. (2002) have claimed that the overexpression of nonfunctional forms of PEX3 in a pex3-null mutant can stimulate the formation of “preperoxisomal” vesicles from the ER and that these vesicles subsequently mature into fully functional peroxisomes upon the expression of WT PEX3. Our observation that PEX3 is imported differently than most other PMPs might be interpreted by some as indirect support for both an ER-to-peroxisome route for PEX3 biogenesis and a direct role for the ER in the genesis of peroxisomal vesicles.
However, we do not feel that such a conclusion is warranted at this time, and for several reasons. First, the kinetics of PEX3-mediated peroxisome synthesis during the complementation of pex3-null mutants is one to two orders of magnitude slower than the kinetics of PEX3 import into peroxisomes of WT cells (South et al., 2000). This result means that virtually all PEX3 is imported into preexisting peroxisomes long before it even has the chance to mediate “de novo” peroxisome synthesis. These considerations raise serious doubts as to whether any observations of peroxisome synthesis in the absence of preexisting peroxisomes are physiologically relevant. Second, the one study that has attempted to follow the biogenesis of PEX3 in vivo found no evidence for PEX3 migration through the ER to the peroxisome in WT cells or in cells that lack peroxisomes and instead detected PEX3 first in the peroxisome (South et al., 2000). Third, PEX3-mediated peroxisome synthesis occurs independently of the SEC61 and SSH1 protein translocons in the ER membrane, independently of the COPII coat complex, which is essential for vesicular traffic from the ER, and independently of the COPI coat complex, which is required for other vesicle-mediated trafficking events in the early secretory pathway (South et al., 2001, 2000). These concerns and observations, coupled with the unique function of PEX3 in peroxisome biogenesis and its enigmatic route of import, make it more important than ever to resolve the details of PEX3 biogenesis through direct analysis of endogenously expressed PEX3 in normal cells.
Materials and methods
Plasmids
The plasmids pcDNA3-PMP34myc (Sacksteder et al., 2000), pcDNA3-PEX11βmyc (Schrader et al., 1998), pNHA-PTE1, pcDNA3-PMP34aa244–307–3xmyc, pcDNA3–3xNLS-PEX19 and pcDNA3–3xNLS-HAOX3 (Jones et al., 1999, 2001), pcDNA3-PEX3aa1–50–6xmet3xmyc, pcDNA3-PEX16aa221–336–3xmyc, and pcDNA3-PMP22aa1–94–3xmyc (Jones et al., 2004) have been described previously. pcDNA3-PEX3L125P/N134D-3xmyc was generated by PCR PEX3aa83–139 (with CAGCAACTGAATTCCGAGAGCCTC, ATATCCACCAATTATGTCTAACTGGACCCGCAAAAGAACAACCGGCATAC) and PEX3aa135–232 (with CATAATTGGTGGATATATTTAC, AGGTTTGGATCCATCTTTATTAATC). Then, the two PCR products were mixed and used as templates to do a second PCR (with CAGCAACTGAATTCCGAGAGCCTC, AGGTTTGGATCCATCTTTATTAATC). The resultant PCR products were then digested with EcoRI and BamHI and inserted into pcDNA3-PEX3–3xmyc digested with the same enzymes. The rest of plasmids were created by amplifying DNA fragments with the indicated primers (Table I), cleaving the fragments with restriction enzymes able to cut the terminal palindromes on the fragments, and inserting the resulting DNAs into the specified vectors. The plasmids pRS313-GAL1-YFP-SKL, pRS313-GAL1-YFP-PEX10, pRS313-GAL1-YFP-PEX15, and pRS313-GAL1-ANT1-YFP were generated by insertion of the GAL1 promoter upstream of the YFP-tagged genes, and pRS315-PGK1-CFP-SKL was generated by insertion of the PGK1 promoter upstream of CFP-SKL. All PCR-generated segments were sequenced in their entirety and only clones that possessed the correct sequence were used in the study.
Table I. Primers used.
| Plasmid | Primers | Vectors |
|---|---|---|
| PEX3-3xmyc | 5′CCGGTACCATGCTGAGGTCTGTATGGAATTTTCTG 3′CCAGATCTTTTCTCCAGTTGCTGAGGGGTAC |
pcDNA-(C)3xmyc |
| 3xmyc-PEX3aa50-373 | 5′CCGTCGACCGCCCAAGCACGACGACAATATCATTTTG 3′CCGCGGCCGCTCATTTCTCCAGTTGCTGAGGGGTAC |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa1-100 | 5′CCCTCGAGCCTGAGGTCTGTATGGAATTTTCTG 3′CCGCGGCCGCTCACTTGTTTGAAGGCCTGTTTTTTAGCAG |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa1-148 | 5′CCCTCGAGCCTGAGGTCTGTATGGAATTTTCTG 3′CCGCGGCCGCTCAGCCAACTGCTGCATTATC |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa100-148 | 5′CCCTCGAGCAAGCTAGAAATATGGGAGGATC 3′CCGCGGCCGCTCAGCCAACTGCTGCATTATC |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa120-140 | 5′CCCTCGAGCTACAGTACCTGTATGCTGGTTG 3′CCGCGGCCGCTCAAATATATCCACCAATTATGTTTAACTG |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa120-136 | 5′ CCCTCGAGCTACAGTACCTGTATGCTGGTTG 3′CCGCGGCCGCTCAAATTATGTTTAACTGGACCCGC |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa121-136 | 5′CCCTCGAGCAGTACCTGTATGCTGGTTGTTC 3′CCGCGGCCGCTCAAATTATGTTTAACTGGACCCGC |
pcDNA3-(N)3xmyc |
| 3xmyc-PEX3aa120-135 | 5′CCCTCGAGCTACAGTACCTGTATGCTGGTTG 3′CCGCGGCCGCTCATATGTTTAACTGGACCCGCAAAAG |
pcDNA3-(N)3xmyc |
| pcDNA3-3xNLS | 5′CCCAAGCTTGCCATGGCATGTCCAAAG 3′CCGGTACCATCACCGACTTTCCGTTTCTTC |
pcDNA3 |
| 3xNLS-PEX19aa1-56-3xHA | 5′CCGGTACCGCCGCCGCTGAGGAAGGCTGTAG 3′CCGGATCCTCCTGGCGATCTCTTCTGGGG |
pcDNA3- (N)3xNLS |
| 3xNLS-PEX19aa57-299-3xHA | 5′CCGGTACCGACACTGCCAAAGATGCCCTCTTC 3′CCGGATCCCATGATCAGAACACTGTTCACCAC |
pcDNA3- (N)3xNLS |
| 3XHA-PEX19 | 5′CCCTCGAGCGCCGCCGCTGAGGAAGGCTGTAG 3′CCGCGGCCGCTCACATGATCAGACACTGTTCACC |
pcDNA3- (N)3xHA |
| 3xHA-PEX19aa1-56 | 5′CCCTCGAGCGCCGCCGCTGAGGAAGGCTGTAG 3′CCGCGGCCGCTCATCCTGGCGATCTCTTCTG |
pcDNA3- (N)3xHA |
| 3xHA-PEX19aa57-299 | 5′CCCTCGAGCGACACTGCCAAAGATGCCCTCTTC 3′CCGCGGCCGCTCACATGATCAGACACTGTTCACC |
pcDNA3-(N)3xHA |
| scPEX3 | 5′CGCCTCGAGCGGCATCTGAATCTACAGAGGTGG 3′CGCCCGCGGTCACAATCTCGTCGTCATCCTCATC |
pRS314 |
| scPEX3UTR | 5′CGCCTCGAGCGGCATCTGAATCTACAGAGGTGG 3′CGCGGATCCGTCGACATCCTTTTAGTGGTTTGCCTCCTTG 5′CGCGGATCCGCGGCCGCTCTCTGAATAAGTACTGACACTCACAC 3′CGCCCGCGGTCACAATCTCGTCGTCATCCTCATC |
pRS314 |
| pRS313-(N)YFP | 5′CCAAGGATCCATGGTGAGCAAGGGCGAGGAG 3′CCCCTCGAGTTCTTGTACAGCTCGTCCATGCC |
pRS313 |
| YFP-scPEX10 | 5′CAAGTCGACGAAGAATGATAATAAGTTGCAAAAGGAAGC 3′CCAAGCGGCCGCTATTGCCGCAGGACCAGAATTTC |
pRS313-(N)YFP |
| YFP-scPEX15 | 5′CCAAGCGGCCGCATTGGTTGGACACGGTTAC 3′CAAGTCGACGGCTGCAAGTGAGATAATG |
pRS313-(N)YFP |
| pRS313-(C)YFP | 5′CCCGCGGCCGCAGTGAGCAAGGGCGAGGAGCTGTTC 3′CCCCCGCGGTTACTTGTACAGCTCGTCCATGCC |
pRS313 |
| ANT1-YFP | 5′GGGGGATCCCATGTTAACTCTAGAGTCTGCATTAAC 3′CCTGCGGCCGCAGTGGAAGCCAGCTTGCGTTGTCCGTG |
pRS313- (C)YFP |
| YFP-SKL and CFP-SKL |
5′CCAAGGATCCATGGTGAGCAAGGGCGAGGAG 3′CCCTCTAGATCACAGCTTCGACTTGTACAGCTCGTCCATGCC |
pRS313 pRS315 |
Cell lines, immunofluorescence, and antibodies
The immortalized WT human skin fibroblast cell line (GM5756-TI) has been described (Jones et al., 2004). The 3xHA-PEX19/PBD399-TI stable cell line was generated by stably transfecting the immortalized PEX19-deficient human cell line (PBD399-TI) (Jones et al., 2004) with NH2-terminally tagged form of PEX19 (3xHA-PEX19), followed by selection with 300 μg/ml G418 (GIBCO BRL). All transfections were performed by electroporation (Chang et al., 1997). For immunofluorescence, cells on cover glasses were fixed with 3% formadehyde, permealized with 1% Triton X-100, and incubated with primary antibodies for 30 min followed by extensive wash with Dulbecco's-modified PBS (GIBCO BRL), and then incubated with FITC or Texas red-labeled secondary antibodies (Jackson ImmunoResearch Laboratories) for 10 min followed by extensive washes, and mounted on slides. The anti–c-myc monoclonal antibody, sheep anti-PMP70 antibodies, rabbit anti-PEX3 antibodies (South et al., 2000), rabbit anti-PEX5 (Chang et al., 1999b), rabbit anti-PEX19, and anti-HAOX3 antibodies (Jones et al., 2001) have been described previously. The polyclonal and monoclonal antibodies against the HA epitope (Santa Cruz Biotechnology, Inc.) and the HRP-labeled secondary antibodies (Jackson ImmunoResearch Laboratories) were from commercial sources.
Immunofluorescence images were captured on a BH2-RFCA microscope (Olympus) with an Olympus SplanApo 60x 0.40 oil objective and a Sensicam QE (Cooke) digital camera using IPLab 3.6 software (Scanalytics) at room temperature, and processed with Adobe Photoshop 7.0 software (Adobe Systems, Inc.). Contrast, brightness, and gamma values were adjusted to approximate the original IPLab image.
SiRNA treatment and immunoblots
RNA oligonucleotides were obtained from Dharmacon Research, Inc. Four pairs of RNA oligonucleotides were used in this study: PEX5: 5′-AGAAGCUACUCCCAAAGGCdTdT-3′, 5′-GCCUUUGGGAGUAGCUUCUdTdT-3′; TRIP8b: 5'-GCAGGGAAAAGGCUCUAGGdTdT-3′, 5′-CCUAGAGCCUUUUCCCUGCdTdT-3′; PEX3: 5′-CGGACAGAUCCAUUCAGUUdTdT-3′, 5′-AACUGAAUGGAUCUGUCCGdTdT-3′; PEX16: 5′-UGACGGGAUCCUACGGAAGdTdT-3′, 5′-CUUCCGUAGGAUCCCGUCAdTdT-3′. To test the effect of siRNA on protein levels, GM 5756-TI cells from each confluent 75-cm2 flask were suspended in 500 μl Hepes-buffered saline (21 mM Hepes, pH 7.15, 137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 6 mM dextrose), mixed with 50 μl siRNA (20 μM in annealing buffer: 100 mM KAc, 30 mM Hepes-KOH, pH 7.4, 2 mM MgAc), and electroporated in a BTX ECM600 Electroporation System at 230 V, 1,500 μF, 129Ω. After transfection, cells were grown for 5 d. Every 24 h, cells were fixed with 3% formaldehyde for indirect immunofluoresence study or lysed with SDS-loading buffer for immunoblot study, respectively. An equal amount of total protein from each sample was subjected to SDS-PAGE gel. The proteins were then transferred to PVDF membrane (Millipore), followed by incubation with primary antibodies and HRP-conjugated secondary antibodies sequentially, and visualized by using ECL detection reagents (Amersham Biosciences).
SiRNA treatment and peroxisomal protein import assay
GM5756-TI cells were transfected with PEX3, PEX5, and TRIP8b siRNAs three times at 24-h intervals, respectively. At 72 h after the initial treatment, cells were cotransfected with (a) pNHA-PTE1 and (b) pcDNA3-PMP34myc, pcDNA3-PEX11βmyc, pcDNA3-PEX16aa221–336–3xmyc, or pcDNA3-PEX3aa1–50–6xmet3xmyc. 2 h after the DNA transfection, cells were processed for double indirect immunofluoresence microscopy using anti-myc and anti-HA antibodies. To quantify the import assay, all cells showing peroxisomal staining (import of either marker) were counted as to whether they imported HA-PTE1, PMP34myc (PEX11βmyc, PEX3aa1–50-6xmet3xmyc, PEX16aa221–336-3xmyc), or both. Each experiment was done in triplicate and the mean and standard deviation were presented.
Yeast strains and screening for temperature-sensitive mutants of PEX3
The genotype of the S. cerevisiae strain BY4733 used in this study is MATa, his3Δ200, leu2Δ0, met15Δ0, trp1Δ63, ura3Δ0. The YFY1 strain was generated by one-step PCR-mediated disruption of PEX3 in BY4733 with URA3 as the selectable marker (Baker-Brachmann et al., 1998), followed by transforming the cells with pRS315-PGK1-CFP-PTS1. To screen for the temperature-sensitive mutants of PEX3, YFY1 cells were cotransformed with the linearized gap repair plasmid pRS314-scPEX3UTR (digested by SalI and NotI) and the error-prone PCR product of PEX3 flanked by 5′ 102 bps and 3′ 120 bps. Transformants were selected on minimal S medium (0.17% yeast nitrogen base without ammonium sulfate [Sigma-Aldrich], 0.5% ammonium sulfate) with 2% glucose lacking uracil, leucine, and tryptophan. Each transformant was spotted onto duplicate plates, which were then incubated at 25° and 37°C for 3 d, and then examined by epifluorescence microscopy to determine the subcellular distribution of CFP-PTS1. Those that imported CFP-PTS1 at 25°C but accumulated CFP-PTS1 in the cytoplasm at 37°C were considered temperature-sensitive mutants of PEX3.
PMP import assay in WT and pex3ts strains
The YFY1 strain was transformed with pRS314-PEX3 as WT control for pex3 ts strains. Next, the WT and pex3 ts strains, which constitutively expressed CFP-PTS1, were retransformed with the plasmids: pRS313-GAL1-YFP-PTS1, pRS313-GAL1-YFP-PEX10, pRS313-GAL1-ANT1-YFP, pRS313-GAL1-YFP-PEX15. The YFP-tagged peroxisomal proteins were expressed under the control of the galactose-inducible GAL1 promoter. Cells were grown at 25°C on minimal glucose medium lacking uracil, leucine, histidine, and tryptophan to repress expression of the YFP marker proteins. For the import assay of YFP-PTS1, YFP-PEX10, YFP-PEX15, pex3-WT, and pex3-G6 cells were transferred to minimal medium with 2% galactose at 25°C for 2–3 h, and then either maintained at 25°C or shifted to 37°C for four more hours. For the import assay of ANT1-YFP, pex3-WT and pex3-G6 cells grown in glucose minimal medium were shifted to 37°C for 2.5 h, and then transferred to minimal galactose (2%) medium for two and a half more hours. For the import assay of YFP-PTS1 and YFP-PEX10 in pex3-A8 and pex3-B5 strains, cells were transferred to minimal galactose medium at 25°C for 2–3 h, and then either maintained at 25°C or shifted to 37°C for four more hours. Cells with CFP-PTS1 imported to peroxisome were scored as to whether YFP imported or not. Then the calculated percentage of import of each protein at 37°C was normalized to that at 25°C, respectively. Each experiment was performed in triplicate and the mean and standard deviation are presented.
Acknowledgments
This study was supported by a grant (DK59479) from the National Institutes of Health to S.J. Gould.
Abbreviations used in this paper: mPTS, PMP targeting signal; PMP, peroxisomal membrane protein; PTS, peroxisomal targeting signal; RNAi, RNA interference; siRNA, small interfering RNAs; WT, wild type.
References
- Albertini, M., P. Rehling, R. Erdmann, W. Girzalsky, J.A.K.W. Kiel, M. Veenhuis, and W.-H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 89:83–92. [DOI] [PubMed] [Google Scholar]
- Baker-Brachmann, C., A. Davies, G.J. Cost, E. Caputo, J. Li, P. Hieter, and J.D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132. [DOI] [PubMed] [Google Scholar]
- Brosius, U., T. Dehmel, and J. Gartner. 2002. Two different targeting signals direct human peroxisomal membrane protein 22 to peroxisomes. J. Biol. Chem. 277:774–784. [DOI] [PubMed] [Google Scholar]
- Chang, C.C., W.H. Lee, H.W. Moser, D. Valle, and S.J. Gould. 1997. Isolation of the human PEX12 gene, mutated in group 3 of the peroxisome biogenesis disorders. Nat. Genet. 15:385–388. [DOI] [PubMed] [Google Scholar]
- Chang, C.C., S. South, D. Warren, J. Jones, A.B. Moser, H.W. Moser, and S.J. Gould. 1999. a. Metabolic control of peroxisome abundance. J. Cell Sci. 112:1579–1590. [DOI] [PubMed] [Google Scholar]
- Chang, C.C., D.S. Warren, K.A. Sacksteder, and S.J. Gould. 1999. b. PEX12 binds PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J. Cell Biol. 147:761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., M.C. Liang, J.N. Chia, J.K. Ngsee, and A.E. Ting. 2001. Rab8b and its interacting partner TRIP8b are involved in regulated secretion in AtT20 cells. J. Biol. Chem. 276:13209–13216. [DOI] [PubMed] [Google Scholar]
- Diestelkotter, P., and W.W. Just. 1993. In vitro insertion of the 22 kD peroxisomal membrane protein into isolated rat liver peroxisomes. J. Cell Biol. 123:1717–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel, B., R. Erdmann, S.J. Gould, G. Blobel, D.I. Crane, J.M. Cregg, G. Dodt, Y. Fujiki, J.M. Goodman, W.W. Just, et al. 1996. A unified nomenclature for peroxisome biogenesis. J. Cell Biol. 135:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt, G., N. Braverman, C. Wong, A. Moser, H.W. Moser, P. Watkins, D. Valle, and S.J. Gould. 1995. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9:115–124. [DOI] [PubMed] [Google Scholar]
- Dyer, J.M., J.A. McNew, and J.M. Goodman. 1996. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J. Cell Biol. 133:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen, G.A., R.K. Szilard, and R.A. Rachubinski. 1997. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane protein. J. Cell Biol. 137:1265–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma, Y., L. Kwast, A. Klein, T. Voorn-Brouwer, M. van den Berg, B. Metzig, T. America, H.F. Tabak, and B. Distel. 1996. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1 containing proteins. J. Cell Biol. 135:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma, Y., L. Kwast, M. van den Berg, W.B. Snyder, B. Distel, S. Subramani, and H.F. Tabak. 1997. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 16:7326–7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., and G. Blobel. 1996. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, K.N., G.J. Haan, R.J. Baerends, A.M. Kram, and M. Veenhuis. 2002. Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J. Biol. Chem. 277:11026–11033. [DOI] [PubMed] [Google Scholar]
- Fransen, M., T. Wylin, C. Brees, G.P. Mannaerts, and P.P. Van Veldhoven. 2001. Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol. Cell. Biol. 21:4413–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki, Y., R.A. Rachubinski, and P.B. Lazarow. 1984. Synthesis of a major integral membrane polypeptide of rat liver peroxisomes on free polysomes. Proc. Natl. Acad. Sci. USA. 81:7127–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girzalsky, W., P. Rehling, K. Stein, J. Kipper, L. Blank, W.H. Kunau, and R. Erdmann. 1999. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2–dependent protein import into peroxisomes. J. Cell Biol. 144:1151–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner, C.J., P.U. Mayerhofer, P. Landgraf, A.C. Muntau, A. Holzinger, J.K. Gerber, S. Kammerer, J. Adamski, and A.A. Roscher. 2000. Human adrenoleukodystrophy protein and related peroxisomal ABC transporters interact with the peroxisomal assembly protein PEX19p. Biochem. Biophys. Res. Commun. 271:144–150. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., and C.S. Collins. 2002. Opinion: peroxisomal-protein import: is it really that complex? Nat. Rev. Mol. Cell Biol. 3:382–389. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., and D. Valle. 2000. The genetics and cell biology of the peroxisome biogenesis disorders. Trends Genet. 16:340–344. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., G.A. Keller, N. Hosken, J. Wilkinson, and S. Subramani. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S.J., J.E. Kalish, J.C. Morrell, J. Bjorkman, A.J. Urquhart, and D.I. Crane. 1996. PEX13p is an SH3 protein in the peroxisome membrane and a docking factor for the PTS1 receptor. J. Cell Biol. 135:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haan, G.J., K.N. Faber, R.J. Baerends, A. Koek, A. Krikken, J.A. Kiel, I.J. van der Klei, and M. Veenhuis. 2002. Hansenula polymorpha Pex3p is a peripheral component of the peroxisomal membrane. J. Biol. Chem. 277:26609–26617. [DOI] [PubMed] [Google Scholar]
- Hazra, P.P., I. Suriapranata, W.B. Snyder, and S. Subramani. 2002. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 3:560–574. [DOI] [PubMed] [Google Scholar]
- Hettema, E.H., W. Girzalsky, M. van Den Berg, R. Erdmann, and B. Distel. 2000. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honsho, M., S. Tamura, N. Shimozawa, Y. Suzuki, N. Kondo, Y. Fujiki. 1998. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am. J. Hum. Genet. 63:1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka, T., Y. Shiina, T. Hashimoto, and T. Osumi. 1996. Insertion of the 70-kDa peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J. Biol. Chem. 271:3706–3713. [DOI] [PubMed] [Google Scholar]
- Jones, J.M., K. Nau, M.T. Geraghty, R. Erdmann, and S.J. Gould. 1999. Identification of peroxisomal acyl-CoA thioesterases in yeast and humans. J. Biol. Chem. 274:9216–9223. [DOI] [PubMed] [Google Scholar]
- Jones, J.M., J.C. Morrell, and S.J. Gould. 2001. Multiple distinct targeting signals in integral peroxisomal membrane proteins. J. Cell Biol. 153:1141–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.M., J.C. Morrell, and S.J. Gould. 2004. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J. Cell Biol. 164:57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish, J.E., C. Theda, J.C. Morrell, J.M. Berg, and S.J. Gould. 1995. Formation of the peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol. Cell. Biol. 15:6406–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalish, J.E., G.A. Keller, J.C. Morrell, S.J. Mihalik, B. Smith, J.M. Cregg, and S.J. Gould. 1996. Characterization of a novel component of the peroxisomal protein import apparatus using fluorescent peroxisomal proteins. EMBO J. 15:3275–3285. [PMC free article] [PubMed] [Google Scholar]
- Lazarow, P.B., and Y. Fujiki. 1985. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1:489–530. [DOI] [PubMed] [Google Scholar]
- Marzioch, M., R. Erdmann, M. Veenhuis, and W.-H. Kunau. 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13:4908–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono, Y., N. Kinoshita, S. Tamura, N. Shimozawa, M. Hamasaki, K. Ghaedi, R.J. Wanders, Y. Suzuki, N. Kondo, and Y. Fujiki. 1999. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc. Natl. Acad. Sci. USA. 96:2116–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., E. Monosov, and S. Subramani. 1993. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells. The PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal and is a member of the TPR protein family. J. Cell Biol. 121:761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen, R.T., C.R. Flynn, and R.N. Trelease. 2001. How are peroxisomes formed? The role of the endoplasmic reticulum and peroxins. Trends Plant Sci. 6:256–261. [DOI] [PubMed] [Google Scholar]
- Muntau, A.C., A.A. Roscher, W.H. Kunau, and G. Dodt. 2003. The interaction between human PEX3 and PEX19 characterized by fluorescence resonance energy transfer (FRET) analysis. Eur. J. Cell Biol. 82:333–342. [DOI] [PubMed] [Google Scholar]
- Palmieri, L., H. Rottensteiner, W. Girzalsky, P. Scarcia, F. Palmieri, and R. Erdmann. 2001. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 20:5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, B., P. Diestelkotter, H. Heid, and W.W. Just. 1997. Cytosolic factors mediate protein insertion into the peroxisomal membrane. FEBS Lett. 414:95–98. [DOI] [PubMed] [Google Scholar]
- Sacksteder, K.A., and S.J. Gould. 2000. The genetics of peroxisome biogenesis. Annu. Rev. Genet. 34:623–652. [DOI] [PubMed] [Google Scholar]
- Sacksteder, K.A., J.M. Jones, S.T. South, X. Li, Y. Liu, and S.J. Gould. 2000. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J. Cell Biol. 148:931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader, M., B.E. Reuber, J.C. Morrell, G. Jimenez-Sanchez, C. Obie, T. Stroh, D. Valle, T.A. Schroer, and S.J. Gould. 1998. Expression of PEX11b mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273:29607–29614. [DOI] [PubMed] [Google Scholar]
- Snyder, W.B., A. Koller, A.J. Choy, and S. Subramani. 2000. The peroxin Pex19p interacts with multiple, integral membrane proteins at the peroxisomal membrane. J. Cell Biol. 149:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., and S.J. Gould. 1999. Peroxisome synthesis in the absence of pre-existing peroxisomes. J. Cell Biol. 144:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., K.A. Sacksteder, X. Li, Y. Liu, and S.J. Gould. 2000. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149:1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., E. Baumgart, and S.J. Gould. 2001. Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc. Natl. Acad. Sci. USA. 98:12027–12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, K., A. Schell-Steven, R. Erdmann, and H. Rottensteiner. 2002. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 22:6056–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinkels, B.W., S.J. Gould, A.G. Bodnar, R.A. Rachubinski, and S. Subramani. 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10:3255–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak, H.F., J.L. Murk, I. Braakman, and H.J. Geuze. 2003. Peroxisomes start their life in the endoplasmic reticulum. Traffic. 4:512–518. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., and R.A. Rachubinski. 2001. a. Dynamics of peroxisome assembly and function. Trends Cell Biol. 11:22–29. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., and R.A. Rachubinski. 2001. b. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357–368. [DOI] [PubMed] [Google Scholar]
- Wiemer, E.A.C., W.M. Nuttley, B.L. Bertolaet, X. Li, U. Franke, M.J. Wheelock, W.K. Anne, K.R. Johnson, and S. Subramani. 1995. Human peroxisomal targeting signal-1 receptor restores peroxisomal protein import in cells from patients with fatal peroxisomal disorders. J. Cell Biol. 130:51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]