Abstract
Cytokinesis in Saccharomyces cerevisiae involves coordination between actomyosin ring contraction and septum formation and/or targeted membrane deposition. We show that Mlc1p, a light chain for Myo2p (type V myosin) and Iqg1p (IQGAP), is the essential light chain for Myo1p, the only type II myosin in S. cerevisiae. However, disruption or reduction of Mlc1p–Myo1p interaction by deleting the Mlc1p binding site on Myo1p or by a point mutation in MLC1, mlc1-93, did not cause any obvious defect in cytokinesis. In contrast, a different point mutation, mlc1-11, displayed defects in cytokinesis and in interactions with Myo2p and Iqg1p. These data suggest that the major function of the Mlc1p–Myo1p interaction is not to regulate Myo1p activity but that Mlc1p may interact with Myo1p, Iqg1p, and Myo2p to coordinate actin ring formation and targeted membrane deposition during cytokinesis. We also identify Mlc2p as the regulatory light chain for Myo1p and demonstrate its role in Myo1p ring disassembly, a function likely conserved among eukaryotes.
Keywords: septins; Myo2p; Myo4p; cytokinesis; actomyosin ring
Introduction
Cytokinesis is the last event in the cell division cycle and likely occurs by the contraction of a cortical actomyosin ring, which consists of type II myosin and F-actin. This contraction mechanism must be coordinated with membrane deposition at the cleavage site to ensure efficient cytokinesis and cell separation. Analysis of cytokinesis in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe has demonstrated that the basic mechanisms underlying cytokinesis are conserved between yeast and animal cells.
Each heavy chain of the type II myosin binds to two light chains, an essential light chain (ELC) proximal to the myosin head and a regulatory light chain (RLC) downstream of the ELC. These interactions are mediated by two tandem IQ motifs in the heavy chain. The ELC in Dictyostelium discoideum is required for cytokinesis and may regulate the actin-activated ATPase of the heavy chain (Pollenz et al., 1992), but it is not clear whether the role of the ELC in cytokinesis is performed primarily through its regulation of the heavy chain or another IQ-containing protein. In S. pombe, Cdc4p is the ELC for the two type II myosins. Cdc4p also interacts with a type V myosin, Myo51p, and an IQGAP-related protein, Rng2p, and it appears that at least one essential function of Cdc4p is mediated through these proteins as deletion of the IQ domains of the essential type II myosin, Myo2, does not suppress the defects caused by mutations in Cdc4p (D 'souza et al., 2001). The major role of RLC may be to regulate the actin-activated Mg2+-ATPase activity and stability of myosin filaments (Sellers, 1991; Trybus, 1991; Matsumura et al., 1998). Studies in D. discoideum and S. pombe suggest the binding of RLC to the heavy chain relieves an auto-inhibitory function of the RLC binding site as deletion of the relevant IQ site on the heavy chain suppresses defects associated with mutations in the RLC (Uyeda and Spudich, 1993; Naqvi et al., 2000). Myosin II molecules purified from RLC null cells in D. discoideum assemble into thick filaments with normal kinetics, but display a defect in filament disassembly in vitro (Chen et al., 1994). This defect has not been confirmed by in vivo studies.
Unlike the case in most animal cells and S. pombe, deletion of MYO1, which encodes the only type II myosin in S. cerevisiae, causes a defect in cytokinesis and cell separation, but not lethality, suggesting that actomyosin ring-independent mechanisms can carry out cytokinesis, albeit less efficiently (Bi et al., 1998; Vallen et al., 2000; Tolliday et al., 2003). Further studies suggest that this alternative mechanism may involve septum formation and/or targeted secretion. In contrast to MYO1, deletion of IQG1/CYK1, which encodes the only IQGAP in S. cerevisiae, or deletion of MLC1, which encodes a light chain for the type V myosin Myo2p and for Iqg1p, causes lethality with cells arrested in cytokinesis (Epp and Chant, 1997; Stevens and Davis, 1998; Lippincott and Li, 1998a; Shannon and Li, 2000). Mlc1p is required for the recruitment of Iqg1p to the bud neck (Shannon and Li, 2000), which, in turn, is required for actin ring formation (Epp and Chant, 1997; Lippincott and Li, 1998a). Because Iqg1p and Mlc1p are essential, they must play a role in the alternative mechanism in cytokinesis in addition to a role in actomyosin ring function.
Myo1p in S. cerevisiae contains two noncanonical IQ motifs. The light chains for Myo1p have not previously been identified, although Mlc1p is coimmunoprecipitated with Myo1p (Boyne et al., 2000). We demonstrate here that Mlc1p is the ELC for Myo1p. However, binding of Mlc1p to Myo1p does not appear to play a major role in regulating Myo1p, but instead Mlc1p interacts with Myo1p, Iqg1p, and Myo2p to regulate actin ring formation and targeted secretion. In addition, we identify and demonstrate that Mlc2p is the RLC for Myo1p and that Mlc2p most likely plays a role in the disassembly of the Myo1p ring in vivo.
Results
Identification of Mlc2p
To identify the light chains for Myo1p, the head domain of Myo1p containing two putative IQ motifs (Fig. 1 A) was used in a two-hybrid screen against the yeast ORF-Gal4p activation domain fusion array to identify interacting proteins. Only one protein, encoded by YPR188C, was a positive interactor in this screen (Fig. 1 B). For reasons described later, YPR188C is named MLC2 (Myo1p light chain 2). Mlc2p consists of 163 amino acids and displays significant sequence homology to calmodulin (or myosin light chain)-related proteins, with identities in the range of 25–30% and similarities ∼50% (Fig. 1 C). Like other light chains, Mlc2p contains an EF hand (amino acids 28–40) and a serine at residue 6 that corresponds to one of the activating phosphorylation sites, Ser19, in the human type II myosin RLC (Uchimura et al., 2002; Fig. 1 C; MRLC2).
Figure 1.
Identification of Mlc2p. (A) The positions and sequences of the IQ1 and IQ2 motifs in Myo1p. The bold letters indicate generally conserved sequences in IQ motifs of type II myosins. (B) Myo1p-N (the head domain) interacts with Mlc2p by two-hybrid assay. Strain PJ69-4α carrying the bait plasmid pOBD (DBD vector) or pOBD-MYO1-N (DBD-MYO1-N) was crossed with strain PJ69-4a carrying the prey plasmid pOAD alone or carrying one of its derivatives that contain one of the six calmodulin-related genes in S. cerevisiae, MLC2, MLC1, CMD1, FRQ1, CDC31, and CNB1. Diploids from the mating reactions were selected on SC-Trp-Leu and then replica-plated onto SC-His for detecting positive interactions. (C) Alignment of Mlc2p with other myosin II RLCs. The putative phosphorylation site, Ser6, is indicated by an asterisk and the EF hand is marked with a top line. Mlc2p (Scer) from S. cerevisiae; MLC (Scas) from S. castelli (Contig 631.6, http://genome-www4.stanford.edu/cgi-bin/FUNGI/showAlign?locus=YPR188C&source=WashU); Rlc1p (Sp) from S. pombe; and MRLC2 (Hs) from Homo sapiens. Sequences are aligned with MacVector software.
Mlc1p, a light chain for the type V myosin Myo2p (Stevens and Davis, 1998) and also for the IQGAP-like protein Iqg1p/Cyk1p (Shannon and Li, 2000), can be coimmunoprecipitated with Myo1p (Boyne et al., 2000), suggesting that Mlc1p might also be a light chain for Myo1p. However, an interaction between Mlc1p and the Myo1p head (Fig. 1 B) was not detected by two-hybrid assays, even in the absence of Mlc2p (not depicted). Similarly, no interaction was detected between the Myo1p head and other calmodulin-related proteins.
Mlc2p localizes to the bud neck in a Myo1p-dependent manner
If Mlc2p were a light chain for Myo1p, Mlc2p should show similarity in its localization to Myo1p. Indeed, Mlc2p is the only known protein that displays an identical localization profile in the cell cycle as Myo1p. Mlc2p first localized to the presumptive bud site as a cortical ring (Fig. 2 A, cell 1). Once the bud emerged, Mlc2p formed a ring at the bud neck (Fig. 2 A, cells 2 and 3). The Mlc2p ring maintained its diameter at the neck until late anaphase when the Mlc2p ring started to contract. The contraction process, which took 8–10 min (Fig. 2 B, bottom), was followed closely by the centripetal septum formation (Fig. 2 B, top), a behavior very similar to that of Myo1p (Bi et al., 1998; Lippincott and Li, 1998a). The neck localization of Mlc2p was completely abolished in myo1Δ cells (Fig. 2 C, middle). In contrast, the neck localization of Myo1p was not affected by deletion of MLC2 (analyzed in detail later; Fig. 2 C, right). These results are consistent with the possibility that Mlc2p is a light chain for Myo1p and Mlc2p localizes to the neck through an interaction with Myo1p.
Figure 2.

Localization pf Mlc2p to the bud neck and its dependency on Myo1p. (A) Localization of Mlc2p in the cell cycle. Cells of YEF2474 (MLC2-GFP/ MLC2-GFP) grown exponentially in YM-P were fixed with formaldehyde and visualized by DIC and fluorescence microscopy. (B) Contraction of Mlc2p-GFP ring during late anaphase of the cell cycle. Cells of YEF2474 grown exponentially in SC medium were analyzed by time-lapse microscopy at 20°C. (C) Localization of Mlc2p to the bud neck depends on Myo1p. Localization of Mlc2p in YEF2565 (myo1Δ/myo1Δ MLC2-GFP/MLC2-GFP) with (left) or without (middle) the plasmid YCp50-MYO1 and localization of Myo1p in YEF2616 (mlc2Δ/mlc2Δ MYO1-GFP/MYO1-GFP; right) were examined after streaking the strains onto YPD plates and incubating the plates at 24°C for 12–16 h.
Mlc2p binds to IQ2 of Myo1p and thus defines an RLC for Myo1p
To determine the binding site of Mlc2p on Myo1p, we generated Myo1p mutants with a precise deletion of IQ1 (amino acids 795–815), IQ2 (amino acids 827–850), or both (amino acids 795–850; Fig. 3 A). All these variants were under the MYO1 promoter control and were tagged with GFP immediately after their start codon. Centromere-based plasmids carrying these myo1 alleles along with control plasmids were transformed into a myo1Δ MLC2:MYC strain, individually, for coimmunoprecipitation (coIP) experiments. As shown in Fig. 3 B, MYC-tagged Mlc2p coimmunoprecipitated effectively with GFP-tagged full-length Myo1p, Myo1p without IQ1 (IQ1Δ), and Myo1p with a point mutation at residue 806 [R806A (IQ1)] that changes the highly conserved arginine among all known type II myosins to alanine (Fig. 1 A). In contrast, the interaction between Mlc2p and Myo1p was abolished completely when IQ2 was deleted (Fig. 3 B). As expected, Mlc2p did not interact with Myo1p deleted for both IQ1 and IQ2 (Fig. 3 B).
Figure 3.

Mlc2p binds to IQ2 of Myo1p exclusively. (A) Diagram of IQ mutants of Myo1p. Asterisk indicates R806A change in IQ1. (B) Mlc2p binds to Myo1p through IQ2. Strain YEF3175 (a myo1Δ::Kan MLC2:MYC) was transformed individually with plasmid pRS316NoNot (Vector), pRS316-MYO1 (untagged Myo1p), and pRS316-N-MYO1-GFP or its IQ mutant derivatives. Transformants were grown exponentially in SC-Ura media at 24°C and processed for coIP experiments. Mlc2p-MYC: Mlc2p-MYC in the immunoprecipitates brought down by anti-MYC–conjugated agarose beads from different cell lysates; Myo1p-GFP-input: Myo1p-GFP in different cell lysates before immunoprecipitation; Myo1p-GFP-bound: Myo1p-GFP in the immunoprecipitates from different cell lysates. White lines indicate that intervening lanes have been spliced out. (C) Mlc2p fails to localize to the bud neck in cells carrying myo1-IQ2Δ. YEF2603 (myo1Δ/myo1Δ MLC2:GFP/MLC2:GFP) was transformed individually with plasmid pRS316-N-MYO1-R806A-GFP (R806A), pRS316-N-MYO1-IQ1Δ-GFP (IQ1Δ), pRS316-N-MYO1-IQ2Δ-GFP (IQ2Δ), or pRS316-N-MYO1-(IQ1Δ+IQ2Δ)-GFP (IQ1Δ+IQ2Δ). Transformants were grown exponentially in SC-Ura media, fixed with formaldehyde, and observed with DIC and fluorescence microscopy.
The coIP results correlate perfectly with the localization studies of Mlc2p in various IQ mutants of Myo1p. Mlc2p localized normally in cells containing Myo1p-R806A or Myo1p-IQ1Δ as the sole source of Myo1p (Fig. 3 C). In contrast, the neck localization of Mlc2p was abolished completely in cells containing Myo1p-IQ2Δ or Myo1p-(IQ1Δ+IQ2Δ) (Fig. 3 C), even though these mutant forms of MYO1 were expressed at a similar level as the corresponding wild type, and the Myo1p mutants themselves localized normally to the bud neck (Fig. 3 B and not depicted). This result is similar to those seen in S. pombe where deletion of the RLC binding site on type II myosins abolishes the localization of RLC to the division site (Naqvi et al., 2000). Together, the coIP results and the localization studies indicate that Mlc2p binds to Myo1p exclusively through IQ2. Based on the fact that RLCs of all known type II myosins bind to their heavy chains through IQ2, Mlc2p is likely a bona fide RLC for Myo1p.
Mlc2p plays a role in the disassembly of the Myo1p ring in vivo
Surprisingly, deletion of MLC2 did not produce any obvious defects in growth rate, cytokinesis, and cell separation at temperatures ranging from 18 to 37°C on plates containing synthetic complete (SC) media or YPD media in the presence or absence of 1 M sorbitol or 0.9 M KCl, except that occasionally mlc2Δ cells formed cell clusters containing more than three cell bodies (unpublished data). Consistent with the hypothesis that Mlc2p functions through Myo1p, deletion of MLC2 and MYO1 together did not produce any additive effect on cytokinesis and cell separation. In addition, deletion of MLC2 together with each of the known cytokinesis genes including BNI1, BNR1, HOF1, and CYK3 did not significantly enhance the phenotypes of the single mutants (unpublished data). Furthermore, screens against the ordered array of yeast deletion mutants at 30 and 37°C failed to identify any mutants that displayed synthetic–lethal or synthetic–sick interactions with mlc2Δ cells. These data suggest that there might be multiple genes sharing a role with Mlc2p in cytokinesis; or more likely, that Mlc2p plays a subtle role in cytokinesis under laboratory conditions.
Upon detailed analyses, we found that the rate of Myo1p ring contraction in mlc2Δ diploid cells was approximately the same as that of an isogenic wild-type strain, but that there was a defect in ring disassembly. In wild-type cells at 20°C, the Myo1p “dot” disappeared within 1 min at the end of contraction (n = 7). However, ∼46% of the mlc2Δ diploid cells (n = 13) showed a 2 to 8 min delay in the disassembly of the Myo1p dot at the end of its contraction at 20°C. We also performed time-lapse analyses on the same strains at 37°C. Unfortunately, the Myo1p-GFP signal at 37°C was not strong enough to produce interpretable time-lapse series. To pursue the mutant phenotype further, we reasoned that if the Myo1p ring were enlarged, the effect of mlc2 deletion on the disassembly of the Myo1p dot might be enhanced. Therefore, we constructed a tetraploid strain homozygous for MYO1-GFP and mlc2Δ. The diameter of the bud neck of tetraploid cells was increased ∼27% over that of isogenic diploid cells (n = 40; the width and the length of the tetraploid cells were increased ∼22% and ∼28% over those of the diploid cells, respectively). At 20°C, ∼50% of the mlc2Δ tetraploid cells (n = 10) showed persistence of the Myo1p dot for at least 2–8 min in comparison to ∼11% of the control cells (n = 9). In contrast to the diploid cells, the Myo1p-GFP signal at 37°C in tetraploid cells was strong enough to permit time-lapse analyses. ∼80% of the mlc2Δ tetraploid cells (n = 10; Fig. 4 B) showed a delay in the disassembly of the Myo1p dot in comparison to 0% of the control cells (n = 10; Fig. 4 A). The Myo1p-GFP contraction rates for these tetraploid cells at 37°C are similar for wild-type and for mlc2Δ cells (0.344 ± 0.014 μm/min, n = 6, and 0.347 ± 0.044 μm/min, n = 5, respectively). These data strongly suggest that Mlc2p might regulate the disassembly of Myo1p ring, at least at the end of its contraction.
Figure 4.
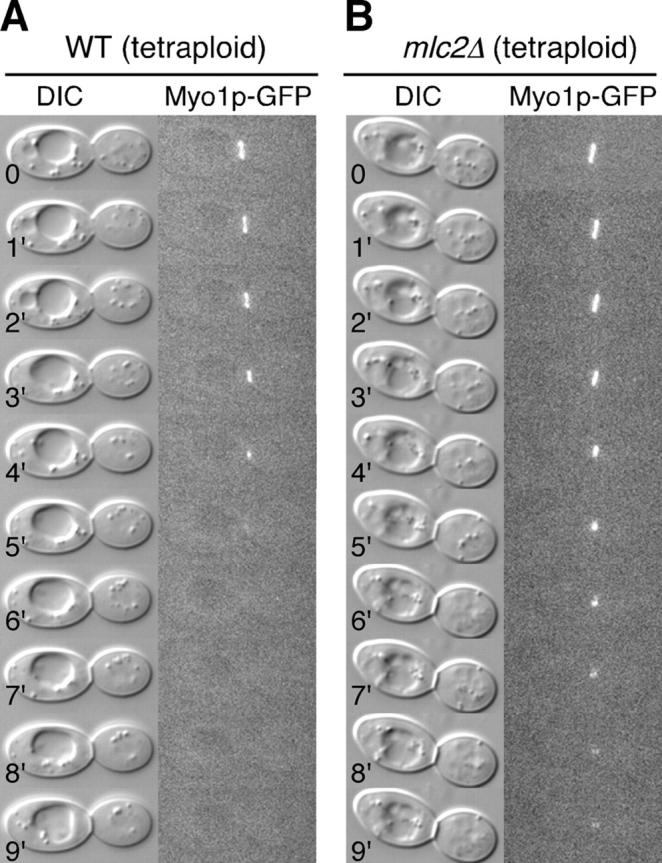
Myo1p ring disassembly is defective in mlc2Δ cells. Time-lapse analyses of tetraploid wild-type (YEF3323; A) and mlc2Δ (YEF3324; B) cells carrying MYO1-GFP by DIC and fluorescence microscopy at 37°C.
Because Mlc2p binds exclusively to IQ2 of Myo1p, we wondered if deletion of IQ2 would cause a similar defect in Myo1p disassembly. We performed time-lapse analyses on myo1Δ diploid cells carrying plasmid pRS316-MYO1-IQ2Δ-GFP, a centromeric plasmid carrying the myo1-IQ2Δ allele. Perhaps due to a slightly higher expression of Myo1p-IQ2Δ from the plasmid, this strain allowed time-lapse analyses at 37°C. Myo1p ring contraction and the disassembly of Myo1p dot occurred normally in ∼80% of cells carrying Myo1p-IQ2Δ as the sole source of Myo1p (n = 5) in comparison to 100% of the control cells carrying wild-type MYO1 (n = 5). These results suggest that IQ2 might be inhibitory to the disassembly of the Myo1p dot and that Mlc2p binding to IQ2 is normally involved in relieving this inhibitory effect, similar to what has been observed in S. pombe (Naqvi et al., 2000) and D. discoideum (Uyeda and Spudich, 1993).
Mlc1p is an ELC for Myo1p but deletion of its binding site on Myo1p does not appear to affect Myo1p function
Mlc1p is known to coimmunoprecipitate with Myo1p (Boyne et al., 2000), suggesting that it might be a light chain for Myo1p. To examine this possibility, we mapped the binding site of Mlc1p on Myo1p by coIP experiments with the various IQ mutants of Myo1p. In contrast to Mlc2p, Mlc1p bound to Myo1p-IQ2Δ very efficiently, but failed to bind to Myo1p-IQ1Δ or Myo1p-(IQ1Δ+IQ2Δ) (Fig. 5 A). These data indicate that Mlc1p binds to Myo1p through IQ1 and thus likely defines an “ELC” for Myo1p.
Figure 5.

Mlc1p–Myo1p interaction is disposable for Myo1p function. (A) Mlc1p binds to Myo1p through IQ1. Strain YEF3176 (a myo1Δ::Kan MLC1:3HA) was transformed individually with plasmid pRS316-N-MYO1-GFP or its IQ mutant derivatives. Transformants were used for coIP experiments as shown in Fig. 3. Mlc1p-HA: Mlc1p-HA in the immunoprecipitates brought down by anti-HA–conjugated agarose beads from different cell lysates; Myo1p-GFP-input and Myo1p-GFP-bound: as in Fig. 3. (B) Myo1p deficient in Mlc1p binding contracts normally. Time-lapse analysis of a cell from YEF1820 (myo1Δ/myo1Δ) carrying plasmid pRS316-N-MYO1-(IQ1Δ+IQ2Δ)-GFP at 20°C.
Interestingly, deletion of IQ1 alone or together with IQ2 did not affect the rate of the Myo1p ring contraction (n = 8) or the disassembly of the Myo1p dot at the end of Myo1p contraction (only one out of eight cells showed a slight delay in the disassembly of the Myo1p dot, similar to the behavior of Myo1p-IQ2Δ cells described previously; Fig. 5 B). These data suggest that binding of Mlc1p to Myo1p does not appear to play a major role in regulating Myo1p function and that the neck region of Myo1p including both IQ motifs is not essential for Myo1p function. A similar relationship between the type II myosins and their ELC Cdc4p has been observed in the fission yeast S. pombe (D 'souza et al., 2001).
Mlc1p targets to the bud neck in the absence of its interactions with Myo1p and Myo2p
Because Mlc1p localizes to the bud neck and plays an essential role in cytokinesis (Stevens and Davis, 1998; Shannon and Li, 2000; Wagner et al., 2002), understanding the mechanisms underlying its neck localization might facilitate understanding its function in cytokinesis in general. Previous studies showed that the neck localization of Mlc1p occurs in strains deleted for Myo1p and in strains deleted for the IQ motifs of Myo2p (Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002). In addition, although Mlc1p binds to Iqg1p, Mlc1p localization to the bud neck occurs before and independently of Iqg1p (Boyne et al., 2000). In contrast, bud tip localization of Mlc1p has been demonstrated to largely depend on its interaction with the IQ motifs of Myo2p (Shannon and Li, 2000). Some of these results are confirmed here (Fig. 6). We hypothesized that the IQ motifs in Myo1p and Myo2 might function redundantly in recruiting Mlc1p to the bud neck. Therefore, we examined whether or not simultaneous elimination of Mlc1p interactions with Myo1p and Myo2p would abolish the localization of Mlc1p to the bud neck. If so, this might cause cells to arrest at cytokinesis and/or cell separation, which would explain the essential role of Mlc1p in cytokinesis. However, the myo1Δ myo2IQ6Δ double mutant was viable and Mlc1p localized to the bud neck in the double mutant, albeit less efficiently than in either single mutant (Fig. 6 A). These results suggest that some Mlc1p molecules can localize to the bud neck independently of their interaction with either Myo1p or Myo2p. Surprisingly, the percentage of cells showing bud tip localization of Mlc1p in the myo1Δ myo2IQ6Δ double mutant was increased in comparison to the myo2IQ6Δ single mutant (Fig. 6 A), suggesting that Myo1p might normally trap some Mlc1p molecules at the bud neck and prevent them from reaching the bud tip. The tip, but not the neck, localization of Mlc1p was completely eliminated in the myo1Δ myo2IQ6Δ myo4Δ triple mutant (Fig. 6 A). Because budding was normal in the triple mutant, which presumably reflects normal targeting of secretory vesicles to the daughter cell, these results suggest that Mlc1p is normally targeted to the bud tip solely through its association with the two type V myosins, Myo2p and Myo4p, which have been shown to play a similar role in the bud tip localization of calmodulin (Stevens and Davis, 1998). These results also suggest that Mlc1p is not directly associated with secretory vesicles in contrast to a recent suggestion (Wagner et al., 2002).
Figure 6.
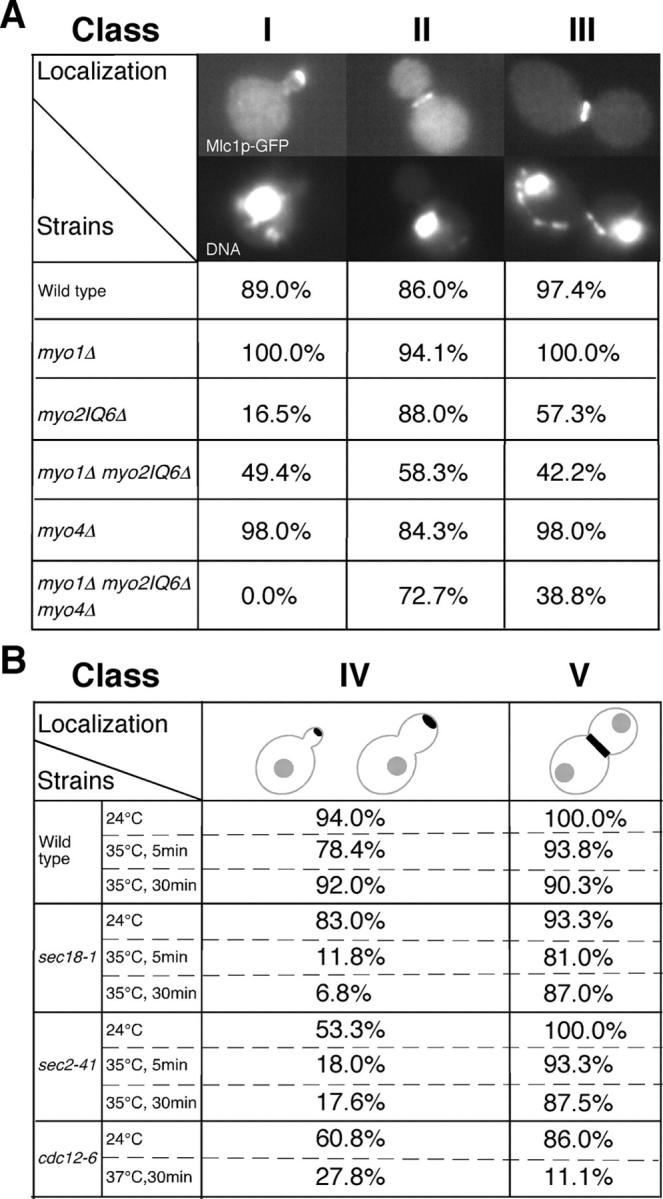
Mlc1p localization in different mutants. (A) Strains carrying pUG34-MLC1 were grown exponentially in SC-His media at 24°C and fixed for 10 min with 3.7% formaldehyde before visualization of GFP-tagged Mlc1p and DNA by fluorescence microscopy. Wild type, SSC1; myo1Δ, SSC350; myo2IQ6Δ, RSY21; myo1Δ myo2IQ6Δ, YEF3380; myo4Δ, YJL126; and myo1Δ myo2IQ6Δ myo4Δ, YJL114A. Class I, small-budded cells with a single nucleus. Class II, large-budded cells with a single nucleus. Class III, large-budded cells with two separated nuclei. (B) Indicated strains carrying pUG34-MLC1 were grown and processed for visualization of GFP-tagged Mlc1p as in A except some strains were shifted to 35 or 37°C for 5 or 30 min before sample collection. Wild type, SEY6210; sec18-1, SEY5188; sec2-41, JGY28B; and cdc12-6, M-17. (Class IV) Due to the strong influence on bud size and shape by secretory and septin mutants, budded cells (from small to large buds) with a single nucleus were grouped together and counted for the tip localization of Mlc1p. Class V was as Class III in A. At least 50 cells were counted for each sample.
What targets Mlc1p to the bud neck in the absence of its interactions with Myo1p, Myo2p, and Myo4p? Previous studies suggest that intact secretory pathway and septins may be involved in the bud neck localization of Mlc1p (Boyne et al., 2000; Shannon and Li, 2000; Wagner et al., 2002). However, a previous work does not distinguish tip localization of Mlc1p from its neck localization in scoring the percentage of cells with a polarized Mlc1p signal (Wagner et al. 2002). The role of septins in targeting Mlc1p to the bud neck remains controversial. One paper (Shannon and Li, 2000) indicates that the neck localization of Mlc1p depends on septins before anaphase, but is independent of the septins in cells with separated nuclei (late anaphase or telophase). Another paper indicates that the neck localization of Mlc1p depends on septins for the entire population of cells (Boyne et al., 2000). For these reasons, we reexamined the role of secretory pathway and the septins in the neck localization of Mlc1p. Our work showed that the neck localization of Mlc1p was largely unaffected by sec18-1, which blocks vesicle fusion at all stages of the secretory pathway or by sec2-41 and sec4-8, which block secretion from the Golgi to plasma membrane (Fig. 6 B). However, the bud tip localization of Mlc1p in these mutants was severely affected (Fig. 6 B), suggesting that the sec mutations demonstrated the expected defect. Further, these data suggest that the neck localization of Mlc1p is independent of the secretory pathway, which is consistent with the conclusion reached with the myo1Δ myo2IQ6Δ myo4Δ triple mutant. Our work also showed that the neck localization of Mlc1p strongly depended on the septins, as only 2% of the budded cells with a single nucleus and 11.1% of cells with separated nuclei had Mlc1p at the bud neck after shifting the temperature-sensitive septin mutant, cdc12-6, to 37°C for 30 min (Fig. 6 B). This result is in good agreement with one of the previous studies (Boyne et al., 2000).
In summary, there are at least three ways by which Mlc1p is targeted to the bud neck: by interacting with Myo1p or Myo2p directly and by interacting with the septins through an undefined mechanism.
Specific alleles of MLC1 reveal the functions of different Mlc1p interactions
In a screen for mutations that displayed synthetic–lethal interactions with the deletion of HOF1, a gene involved in coupling actomyosin-ring function to septum formation (details of the screen will be described elsewhere; Kamei et al., 1998; Lippincott and Li, 1998b; Vallen et al., 2000), we identified two mutant alleles of MLC1, which contain a single point mutation that causes either a glycine-to-aspartic acid change at residue 114 (G114D; mlc1-11) or a glycine-to-glutamic acid change at residue 135 (G135E; mlc1-93). Both changes occurred on amino acids that are highly conserved among the calmodulin (or light chain) superfamily (Fig. 7 A). The mlc1-11 cells were temperature-sensitive for growth (unpublished data) and showed a severe defect in cytokinesis even at 24°C, as the undivided mlc1-11 cells still shared a cytoplasm indicated by the decoration of the plasma membrane by GFP-tagged Ras2p (Fig. 7, B and C). In contrast, the mlc1-93 cells did not show any obvious defect in cytokinesis at temperatures ranging from 20 to 37°C (Fig. 7 B and not depicted). To determine the molecular basis for the behavior of the mlc1 mutant alleles, we did the following experiments. First, we determined the localization of the mlc1 mutant proteins and their interaction with Myo2p. We found that the protein encoded by mlc1-11 failed to localize efficiently to the presumptive bud site and to the bud tip (Fig. 7 D), which is very similar to the localization of wild-type Mlc1p in a myo2-IQ6Δ strain (Shannon and Li, 2000; Fig. 6 and Fig. 7 D). These data suggest that mlc1-11p may be defective in interaction with Myo2p, which was confirmed by coIP experiments (Fig. 8 A). We also found that mlc1-11p localized to the bud tip less efficiently than wild-type Mlc1p in myo2-IQ6Δ cells (Fig. 7 D, asterisks), suggesting that there must be another mechanism for targeting Mlc1p to the bud tip independently of the IQ motifs of Myo2p, in which mlc1-11p is deficient. This conclusion is supported by the fact that the bud tip localization of Mlc1p also depends on Myo4p (Fig. 6 A). However, the localization of mlc1-11p to the bud neck was similar to that of the wild-type Mlc1p (Fig. 7 D). Previous analysis indicates that the neck localization of Mlc1p largely depends on septin function, although a direct interaction has not been demonstrated (see previous section; Boyne et al. 2000; Shannon and Li, 2000). In contrast to mlc1-11p, mlc1-93p localized normally to the incipient bud site, the bud tip, and the bud neck (Fig. 7 D) and interacted normally with Myo2p (Fig. 8 A). Next, we determined the effect of the mlc1 mutations on the neck localization of Iqg1p, which depends on Mlc1p (Shannon and Li, 2000). Iqg1p completely failed to localize to the bud neck in mlc1-11 cells, indicating a defect in the Iqg1p–Mlc1p interaction, which was again confirmed by coIP experiments (Fig. 8 B). In contrast, Iqg1p was able to localize in mlc1-93 cells, albeit less efficiently (Fig. 7 B). CoIP experiments showed that mlc1-93p interacted with Iqg1p to a similar extent as the wild-type Mlc1p did (Fig. 8 B). Finally, we determined the interaction between the mlc1 mutant proteins with Myo1p. Both mlc1-11p and mlc1-93p displayed a similar reduced interaction with Myo1p (Fig. 8 C).
Figure 7.
Properties of mlc1 alleles. (A) Sequence alignment of Mlc1p with other calmodulin-related molecules. Mlc1p (Sc), Mlc1p in S. cerevisiae; Cdc4p (Sp), the ELC for type II myosins in S. pombe; Cmd1p (Sc), calmodulin in S. cerevisiae; and Calm1 (Hs), calmodulin 1 in H. sapiens (gi:31377794). (asterisks) Positions of the indicated mutations. (B) The effects of mlc1 mutations on cytokinesis and Iqg1p localization. “Abnormal cytokinesis” is defined as cells with four or more connected cell bodies. At least 200 cells were scored for each strain. MLC1: wild type, YEF473A; mlc1-11: Y5005-8D, and mlc1-93: Y5119-20A. For Iqg1p localization, anaphase cells with two separated two nuclei from the indicated strains carrying the plasmid pUG35-IQG1 that contains GFP-IQG1 were counted for the neck localization of Iqg1p. At least 50 cells were counted for each strain. (C) mlc1-11 cells display cytokinesis defect at 24°C. Strain Y5005-8D (mlc1-11) carrying plasmid pRS315-GFP-RAS2 was grown exponentially in liquid SC-Leu media at 24°C before the visualization of GFP-tagged Ras2p by fluorescence microscopy. (D) mlc1-11 is defective in bud tip localization. Plasmid pUG34 carrying wild-type MLC1 or mlc1-11 or mlc1-93 was transformed into YEF473A and RSY21. Transformants were grown exponentially in SC-His media at 30°C. Cells were fixed with formaldehyde, and GFP-tagged Mlc1p and DNA were visualized by fluorescence microscopy. Asterisks highlight the low percentage of myo2-IQ6Δ cells with the bud tip localization of mlc1-11p.
Figure 8.
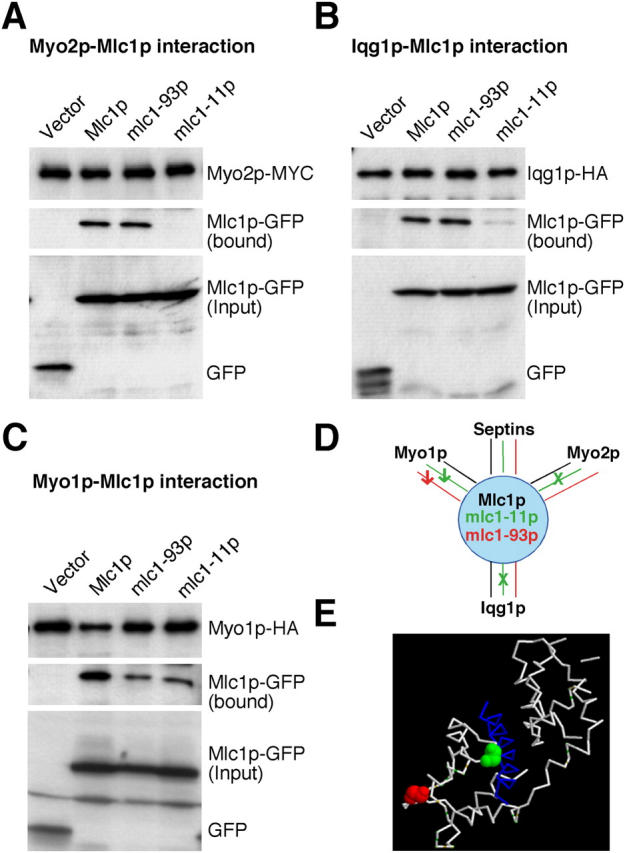
The interactions of mlc1 mutant proteins with Myo2p, Iqg1p, and Myo1p. (A) Myo2p-MYC was precipitated with anti-MYC-agarose beads from cell lysates of strains YJL176A (MYO1:HA MYO2:MYC) carrying pUG34 (vector), pUG34-MLC1 (Mlc1p), pUG34-MLC1-93 (mlc1-93p), or pUG34-MLC1-11 (mlc1-11p), respectively. Myo2p-MYC, Mlc1p-GFP, and its derivatives in the bound and in the input fractions were detected by Western-blot analysis with antibodies against MYC or GFP as indicated in A. (B) As in A except that strain YJL175A (myo1Δ IQG1:HA MYO2:MYC) carrying the different plasmids and the anti-HA-agarose beads were used to do the pull down. (C) As in A except that anti-HA-agarose beads were used to do the pull down. (D) Summary of the interactions between Mlc1p derivatives and its binding partners. (E) Position of the mlc1-11 and mlc1-93 mutations on the three-dimensional structure of the IQ2 (from Myo2p)–Mlc1p complex (Terrak et al., 2003).
In summary, mlc1-11p is defective in its interaction with Myo2p and Iqg1p; and both mlc1-11p and mlc1-93p display a reduced interaction with Myo1p (Fig. 8 D). The fact that both mutant proteins localized normally to the bud neck suggests that the mechanism underlying the septin-dependent neck localization of Mlc1p is still largely intact and that mlc1-11p is unable to recruit or maintain Iqg1p at the neck. Consistent with this separation of functions for Mlc1p is the observation that a mutation at the corresponding position as mlc1-11 in Cdc4p, the ELC for type II myosins in S. pombe, also causes a temperature-sensitive cytokinesis defect without affecting the overall conformation of the protein (Slupsky et al., 2001). Because mlc1-93 cells did not show any obvious defect in cytokinesis in an otherwise wild-type background, these results lend further support to our previous notion that the Myo1p–Mlc1p interaction may not play a major role in regulating Myo1p function or cytokinesis. Because elimination of the Mlc1p–Myo2p interaction by deletion of the IQ motifs of Myo2p (Stevens and Davis, 1998) also does not cause any obvious defect in cytokinesis, the phenotype of mlc1-11 cells suggests that the cytokinesis defect is mainly due to its defective interaction with Iqg1p or due to a combinatorial effect of its decreased interactions with all the binding partners. Strikingly, the mlc1-11 mutation (Fig. 8 E, green) mapped on the three-dimensional structure of IQ2 (from Myo2p)-Mlc1p complex (Terrak et al., 2003) at the interface between the IQ motif and the COOH-terminal domain of Mlc1p, whereas the mlc1-93 mutation (Fig. 8 E, red) was mapped at a site quite distal to the IQ2-Mlc1p interface. Previous data has demonstrated that alleles of CDC4 in S. pombe display intragenic complementation (Nurse and Nasmyth, 1976). Our data, taken with the structural studies (Slupsky et al., 2001; Terrak et al., 2003) suggest that Mlc1p interactions with the IQ regions of Myo1p and Myo2p and/or Iqg1p may be mediated, at least in part, through distinct domains and thereby explains the intragenic complementation of some ELC alleles.
Discussion
Mlc2p regulates the disassembly of the Myo1p ring
We have shown that Mlc2p shares a localization and contraction profile in the cell cycle with Myo1p and that the neck localization of Mlc2p completely depends on Myo1p. In addition, coIP and localization experiments indicate that Mlc2p binds to Myo1p exclusively through the IQ2 motif of Myo1p; thus defining Mlc2p as the RLC for Myo1p.
Deletion of MLC2 produced a mild but consistent defect in the disassembly of the Myo1p ring, at least at the end of its contraction. Because the Myo1p ring is small, even in tetraploid cells, we cannot rule out the possibility that there also might be a subtle change in the contraction kinetics in mlc2Δ cells that we could not detect. In fact, a defect in contraction could certainly arise from a defect in disassembly because these processes are normally coupled (Schroeder, 1972). The biochemical basis for the Myo1p disassembly defect is not clear. However, the type II myosin purified from D. discoideum RLC null cells formed thick filaments comparable to wild-type myosin, but displayed a defect in the disassembly properties in vitro (Chen et al., 1994). These data suggest that the basic function of an RLC may be conserved through evolution. Further biochemical studies on different pairs of RLC–heavy chain interactions are required to determine whether or not a common molecular mechanism underlies the similar function for the RLCs.
The family of RLCs for nonmuscle type II myosins appears to have three features. First, they share very limited sequence homology. For example, Mlc2p displays only 19% identity and 38% similarity in amino acid sequence with Rlc1p, the RLC in S. pombe. Second, the requirement for the function of the RLC in modulating the heavy-chain activity and thus in cytokinesis appears to increase with organismal complexity. In Drosophila melanogaster, mutations in the RLC gene spaghetti-squash cause embryonic lethality and a severe defect in cytokinesis (Karess et al., 1991). In D. discoideum, deletion of the RLC gene does not cause cell lethality, but produces a cytokinesis defect similar to that caused by deletion of the heavy chain (Chen et al., 1994). In S. pombe, rlc1 is only needed for cell viability and cytokinesis at low temperature (Le Goff et al., 2000; Naqvi et al., 2000). In S. cerevisiae, deletion of MLC2 produces a mild defect in the disassembly of the Myo1p ring that is only detectable with a sensitive approach such as time-lapse microscopy. Finally, RLCs appear to act on their respective type II myosins only, which may explain why these light chains are so evolutionarily divergent.
Mlc1p may interact with Myo1p, Iqg1p, and Myo2p to coordinate the formation and contraction of the actomyosin ring with targeted membrane deposition
We show here that Mlc1p binds to Myo1p through the IQ1 motif of Myo1p, suggesting that Mlc1p is an ELC for Myo1p. Mlc1p is also a light chain for Myo2p (the type V myosin) and Iqg1p (IQGAP; Stevens and Davis, 1998; Shannon and Li, 2000). It is not clear how Mlc1p interacts with different proteins in a temporally and spatially regulated manner to carry out its essential function in cytokinesis. The relative contribution of each Mlc1p interaction to its role in cytokinesis can be assessed by analyzing the consequences of disrupting each specific interaction by mutations in Mlc1p or its binding partners.
First, the Mlc1p–Myo1p interaction does not appear to play a significant role in regulating Myo1p function, because deletion of IQ1 alone or together with IQ2 in Myo1p, which abolished the binding of Mlc1p to Myo1p, did not cause an obvious defect in cytokinesis. The phenotype of myo1-(IQ1Δ+IQ2Δ) is much less severe than complete deletion of MYO1 in this strain background, suggesting that Myo1p can perform some functions in cytokinesis in the absence of Mlc1p binding. This conclusion is further supported by the fact that a specific mutation in MLC1 (mlc1-93) that decreased the Mlc1p–Myo1p interaction, but maintained the Mlc1p–Iqg1p and the Mlc1p–Myo2p interactions, did not show any obvious defect in cytokinesis either. Second, the Mlc1p–Myo2p interaction is not responsible for the major role of Mlc1p in cytokinesis because a myo2-IQ6Δ strain, in which Mlc1p does not interact with Myo2p directly (Stevens and Davis, 1998), is able to carry out cytokinesis to a large degree, although it may be slightly defective in cytokinesis and/or cell separation. Even simultaneous disruption of the Mlc1p–Myo1p and the Mlc1p–Myo2p interactions in a myo1-(IQ1Δ+IQ2Δ) myo2-IQ6Δ strain did not cause a defect in cytokinesis nearly as severe as the depletion of Mlc1p, although the double mutant showed a slight additive defect in cytokinesis (unpublished data). These data suggest that the interaction of Mlc1p with Myo1p and Myo2p plays a somewhat minor role in cytokinesis.
In contrast, the Mlc1p–Iqg1p interaction appears to play a major role in cytokinesis. A specific mutation in MLC1 (mlc1-11), which causes a defect in its interactions with Myo1p, Myo2p, and Iqg1p, is defective in cytokinesis. Because the Mlc1p–Myo1p and Mlc1p–Myo2p interactions play only a fine-tuning role in cytokinesis, the major role of Mlc1p in cytokinesis is likely performed through its interaction with Iqg1p. Mlc1p is required for the recruitment of Iqg1p to the bud neck (Boyne et al., 2000; Shannon and Li, 2000), which, in turn, is required for actin ring formation (Epp and Chant, 1997; Lippincott and Li, 1998a); thus, Mlc1p–Iqg1p interaction plays an essential role in the assembly of the actomyosin ring. Both Mlc1p and Iqg1p must play an additional role in cytokinesis independent of their role in actomyosin ring assembly because deletion of MLC1 or IQG1 causes cell lethality with cells accumulating in chains, whereas deletion of MYO1, which abolishes the actomyosin ring function (Bi et al., 1998), in the same strain background does not (Boyne et al., 2000; unpublished data). Indeed, multicopy HOF1 or CYK3 can suppress an iqg1 deletion and its associated cytokinesis defect without restoring the actomyosin ring function (Korinek et al., 2000). The actomyosin ring-independent function of Iqg1p in cytokinesis could involve targeted secretion to the bud neck and/or septum formation (Korinek et al., 2000; Bi, 2001; Osman et al., 2002).
In contrast to RLCs, the sequences and functions of the ELCs of the type II myosins appear to be better conserved through evolution. For example, Mlc1p shares 42% identity and 61% similarity in amino acid sequence with Cdc4p, the ELC of type II myosins in S. pombe. In addition, all known ELCs including Cdc4p in S. pombe, Mlc1p in S. cerevisiae, and ELC in D. discoideum play an essential role or as important a role as their respective heavy chains in cytokinesis (Pollenz et al., 1992; McCollum et al., 1995; Stevens and Davis, 1998; D 'souza et al., 2001). Like Mlc1p, Cdc4p in S. pombe also appears to interact with the IQGAP and a type V myosin to execute its function in cytokinesis (D 'souza et al., 2001; Win et al., 2001). The interactions of the ELCs with other conserved molecules may act as an evolutionary constraint and explain the higher conservation among the ELCs from different organisms.
An integrated view of cytokinesis in S. cerevisiae
Increasing evidence suggests that cytokinesis in animal cells and in fungi involves interplay between actomyosin ring function and targeted secretion to the division site (Hales et al., 1999; Shuster and Burgess, 2002; Wang et al., 2002). In S. cerevisiae, cytokinesis involves the coordinated action of the actomyosin ring and septum formation (Fig. 9 A), which probably requires targeted secretion (Vallen et al., 2000; Bi, 2001; Schmidt et al., 2002). The function of the actomyosin ring might be to guide septum formation by targeting secretion to the precise location at the precise time (Vallen et al., 2000; Bi, 2001). At the molecular level (Fig. 9 B), septins, a family of GTP-binding, filament-forming proteins in eukaryotes (Gladfelter et al., 2001; Longtine and Bi, 2003), form a collar at the bud neck that provides an anchoring site for the actomyosin ring components such as Myo1p and proteins involved in septum formation such as Chs2p, Bni4p, Chs3p, Chs4p, and Mlc1p (Shaw et al., 1991; DeMarini et al., 1997; Bi et al., 1998; Lippincott and Li, 1998a; Shannon and Li, 2000; Schmidt et al., 2002; Wagner et al., 2002). Mlc1p interacts with both Myo1p and Iqg1p to promote the formation of a functional actomyosin ring. Mlc2p may regulate the disassembly of the Myo1p ring during and/or at the end of its contraction. In contrast, Mlc1p also interacts with Myo2p and perhaps Iqg1p to promote targeted membrane deposition at the bud neck. Through these interactions, Mlc1p may effectively coordinate targeted secretion with the actomyosin ring. This coordination mechanism may ensure the maximal efficiency of the cytokinesis process.
Figure 9.
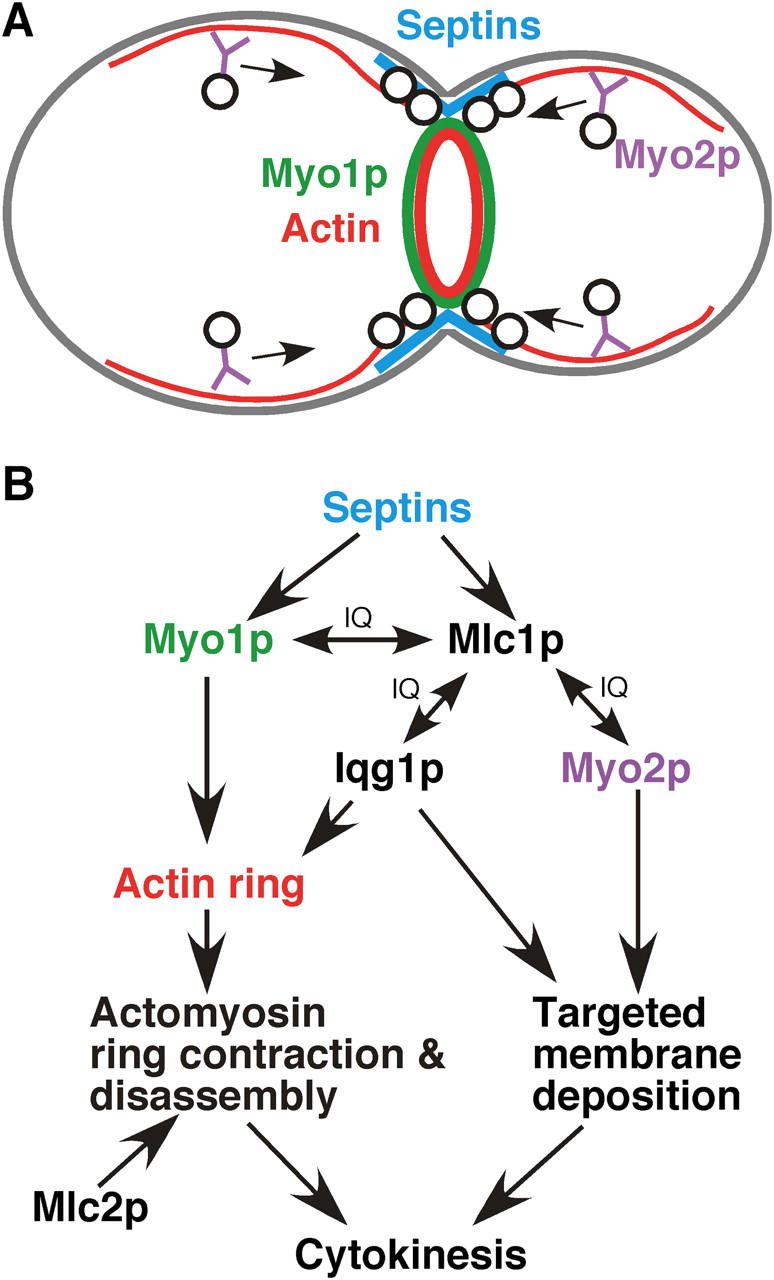
An integrated view of cytokinesis in S. cerevisiae. (A) Cytokinesis in S. cerevisiae. Both the actomyosin ring (Myo1p ring in green and actin ring in red) and the secretory machinery (vesicles in black and motors in purple) are targeted to the bud neck in a septin (blue)-dependent manner in late anaphase of the cell cycle to promote efficient cytokinesis. (B) Coordinating different elements of cytokinesis through the ELC Mlc1p. Mlc1p interacts with Myo1p and Iqg1p to promote actomyosin ring formation at the bud neck. Mlc1p interacts with Myo2p and Iqg1p to promote targeted membrane deposition at the bud neck. Mlc2p affects the disassembly of the Myo1p ring during and/or after the ring contraction.
The role of the ELC in promoting actomyosin ring formation through type II myosin and IQGAP and in coordinating the actomyosin ring contraction with targeted secretion through type V myosin, and perhaps IQGAP, appears to be a conserved feature in cytokinesis between S. cerevisiae and S. pombe, two distantly related fungi, although many details of biochemical interactions require further investigation. Because all the major molecules involved in these processes are conserved through evolution, it is tempting to speculate that similar mechanisms involving the ELC may exist in animal cells.
Materials and methods
Strains, growth conditions, and genetic methods
Yeast strains are listed in Table I. Standard culture media and genetic methods were used (Guthrie and Fink, 1991). In some cases, 1 mg/ml 5-fluoroorotic acid (Angus Buffers & Biochemicals) was added to media to select for loss of URA3-containing plasmids.
Table I. Yeast strains used in this study.
| Strain | Genotype | Reference or source |
|---|---|---|
| JGY28B | a sec2-41 his3 leu2 trp1 ura3 | Gao et al., 2003 |
| M-17 | a cdc12-6 leu2 ura3 | Caviston et al., 2003 |
| PJ69-4α | α trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| PJ69-4a | a trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al., 1996 |
| RSY21 | a myo2IQ6Δ ade2 can1-100 his3 leu2 trp1 ura3 | Stevens and Davis, 1998 |
| RSY22 | α myo2IQ6Δ ade2 can1-100 his3 leu2 trp1 ura3 | Stevens and Davis, 1998 |
| SEY6210 | α his3 leu2 lys2 suc2 trp1 ura3 | Burd et al., 1997 |
| SEY5188 | α sec18-1 leu2 suc2 ura3 | Burd et al., 1997 |
| SSC1 | a ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | Boyne et al., 2000 |
| SSC349 | a myo1Δ::URA3 MLC1:3HA-TRP1 ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | Boyne et al., 2000 |
| SSC350 | a myo1Δ::URA3 IQG1:3HA-TRP1 ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | Boyne et al., 2000 |
| Y3656 | α can1Δ::MFA1pr-HIS3-Mfa1pr-LEU2 | Tong et al., 2004 |
| YEF473 | a/α his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Bi and Pringle, 1996 |
| YEF473A | a his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF1804 | a myo1Δ::Kan his3 leu2 lys2 trp1 ura3 | Bi et al., 1998 |
| YEF1820 | As YEF473 except myo1Δ::Kan/myo1Δ::HIS3 | Bi et al., 1998 |
| YEF2056 | a myo1Δ::HIS3 his3 leu2 lys2 trp1 ura3 (YCp50-MYO1) | This studya |
| YEF2293 | a MYO1:GFP-Kan his3 leu2 lys2 trp1 ura3 | See text |
| YEF2294 | α MYO1:GFP-Kan his3 leu2 lys2 trp1 ura3 | See text |
| YEF2311 | As YEF473 except MYO1:GFP-Kan/ MYO1:GFP-Kan | YEF2293 X YEF2294 |
| YEF2455 | As YEF473 except MLC2::GFP-Kan/MLC2 | See text |
| YEF2473 | α MLC2:GFP-Kan his3 leu2 lys2 trp1 ura3 | Segregant of YEF2455 |
| YEF2565 | As YEF473 except myo1Δ::HIS3/myo1Δ::HIS3 MLC2:GFP-Kan/MLC2:GFP-Kan (YCp50-MYO1) | This studyb |
| YEF2598 | As YEF473 except mlc2Δ::HIS3/MLC2 | See text |
| YEF2603 | As YEF473 except myo1Δ::HIS3/myo1Δ::HIS3 MLC2:GFP-Kan/MLC2:GFP-Kan | This studyc |
| YEF2612 | a mlc2Δ::HIS3 MYO1:GFP-Kan his3 leu2 lys2 trp1 ura3 | This studyd |
| YEF2616 | As YEF473 except mlc2D::HIS3/mlc2D::HIS3 MYO1:GFP-Kan/ MYO1:GFP-Kan | This studyd |
| YEF2661 | As YEF473 except MLC2:MYC-HIS3/MLC2 | See text |
| YEF3175 | a myo1Δ::Kan MLC2:GFP-HIS3 his3 leu2 lys2 trp1 ura3 | This studye |
| YEF3176 | a myo1Δ::Kan MLC1:3HA-TRP1 ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | This studyf |
| YEF3233 | YEF1820 (pRS316-N-MYO1-GFP) | This study |
| YEF3235 | YEF1820 (pRS316-MYO1-IQ2Δ-GFP) | This study |
| YEF3302 | a mlc2Δ::TRP1 MYO1:GFP-Kan his3 leu2 lys2 trp1 ura3 | This studyg |
| YEF3303 | α mlc2Δ::TRP1 MYO1:GFP-Kan his3 leu2 lys2 trp1 ura3 | This studyg |
| YEF3323 | aa/αα (MYO1:GFP-Kan)×4(can1Δ::MFA1pr-HIS3-Mfa1pr-LEU2)×2 (his3 leu2 lys2 trp1 ura3)×4 | See text |
| YEF3324 | As YEF3323 except (mlc2Δ::TRP1)×4 | See text |
| YEF3380 | a myo2IQ6Δ myo1Δ::Kan ade2 can1-100 his3 leu2 trp1 ura3 | This studyh |
| Y5005-8D | a mlc1-11 his3 leu2 lys2 trp1 ura3 | This studyi |
| Y5119-20A | a mlc1-93 his3 leu2 lys2 trp1 ura3 | This studyi |
| YJL114A | a myo2IQ6Δ myo1Δ::NatR myo4Δ::Kan ade2 can1-100 his3 leu2 trp1 ura3 | This studyj |
| YJL126 | a myo4Δ::Kan ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | This studyk |
| YJL175A | a myo1Δ::URA3 IQG1:3HA-TRP1 MYO2:MYC-Kan ade2-1 ura3-52 his3 leu2-3, 112, trp1-1 can1-100 | See text |
| YJL176A | a MYO1:HA-TRP1 MYO2:MYC-Kan his3 leu2 lys2 trp1 ura3 | See text |
YEF1751 (myo1Δ::HIS3/MYO1) carrying YCp50-MYO1 (Vallen et al., 2000) was sporulated to generate YEF2056.
YEF2056 was crossed to YEF2473. Segregants with appropriate genotypes and opposite mating types were crossed to form YEF2565.
YEF2603 was obtained after curing YCp50-MYO1 from YEF2565 through 5FOA selection.
A PCR fragment carrying mlc2Δ::HIS3 was amplified from YEF2598 and transformed into YEF2293 and YEF2294, yielding YEF2612 and YEF2613, respectively, which were then crossed to generate YEF2616.
A PCR fragment carrying myo1Δ::Kan was amplified from YEF1804 and transformed into a segregant of YEF2661 to generate YEF3175.
A PCR fragment carrying myo1Δ::Kan was amplified from YEF1804 and transformed into SSC349, selecting for Kanr Ura− to generate YEF3176.
Constructed by transforming a PCR fragment carrying mlc2Δ::TRP1, generated as described previously (Longtine et al., 1998), into YEF2612, selecting for Trp+ His− colonies.
RSY21 carrying YEplac181 (2 μm LEU2; Gietz and Sugino, 1988) was crossed to RSY22 carrying pRS316 (CEN URA3; Sikorski and Hieter, 1989) to generate YEF3291. After curing both plasmids, the resulting diploid was transformed with a PCR fragment carrying myo1Δ::Kan amplified from YEF1804, generating YEF3304, sporulation of which gave rise to YEF3380.
Segregants from the second backcrosses between the mlc1 mutants (from the hof1Δ synthetic-lethal screen) and the wild-type strain YEF473A.
The myo1Δ::Kan in YEF3380 was converted to myo1Δ::NatR through homologous recombination (the NatR-carrying plasmid was provided by C. Boone). The resulting strain was then transformed with a PCR-amplified fragment carrying myo4Δ::Kan, generating strain YJL114A. The template for the PCR was the chromosomal DNA from a myo4Δ::Kan strain, which was provided by C. Burd (University of Pennsylvania School of Medicine, Philadelphia, PA).
A PCR-amplified fragment carrying myo4Δ::Kan was generated as in j and then transformed into SCC1 to yield YJL126.
Plasmids
Plasmid pOBD-MYO1-N was constructed by cloning the Myo1p-head (amino acids 1–855) coding sequence into pOBD by a gap repair-mediated method (Drees et al., 2001). Plasmids pOAD carrying different light chain-related genes used in Fig. 1 B were supplied by S. Fields' group (University of Washington, Seattle, WA). Plasmid YCp50-MYO1 (CEN URA3; supplied by S. Brown, University of Michigan, Ann Arbor, MI) carries wild-type MYO1 under its own promoter control. Plasmid pRS316NoNot is a derivative of pRS316 (CEN URA3; Sikorski and Hieter, 1989), in which the unique NotI site has been destroyed (Caviston et al., 2003). Plasmid pRS316-MYO1 carries wild-type MYO1 (Caviston et al., 2003). Plasmid pRS316-N-MYO1-GFP carries MYO1 with a GFP cassette inserted in-fame after the start codon of MYO1 (Caviston et al., 2003). Different IQ mutations (R806A, IQ1Δ, IQ2Δ, and IQ1Δ+IQ2Δ) of MYO1 were introduced into pRS316-N-MYO1-GFP using a PCR-based method with appropriate mutagenic primers. Plasmid pRS315-GFP-RAS2 contains an ∼2.5-kb HindIII-XbaI fragment carrying GFP-RAS2 from plasmid pIW192 (Philips and Herskowitz, 1997) that was cloned into the corresponding sites in pRS315 (CEN LEU2; Sikorski and Hieter, 1989). Plasmids pUG34-MLC1 (CEN HIS3) and pUG35-IQG1 (CEN URA3; supplied by A. Ragnini-Wilson, University of “Tor Vergata” Rome, Rome, Italy) carrying MET25p-yEGFP-MLC1 and MET25p-yEGFP-IQG1, respectively, were described previously (Wagner et al., 2002). Plasmid pUG34 (supplied by J.H. Hedgemann, Heinrich-Heine-Universität, Düsseldorf, Germany) carrying mlc1-11 or mlc-93 was constructed similarly as pUG34-MLC1. A PCR fragment flanking the MLC1 locus was amplified from the mutant strain carrying mlc1-11 or mlc1-93. PCR fragments were mixed with XmaI-EcoRI–digested pUG34 and transformed into YEF473A for gap-repair to generate the desired plasmids. All mutants were confirmed by DNA sequencing at the Sequencing Facility of the University of Pennsylvania. All oligonucleotide primers were purchased from Integrated DNA technologies.
Construction of yeast strains
Strains carrying MYO1:GFP-Kan (YEF2293 and YEF2294), MLC2:GFP-Kan (YEF2455), MLC2:MYC-HIS3 (YEF2661,) and mlc2Δ::HIS3 (YEF2598) were constructed using a PCR-based method (Longtine et al., 1998), except that the gfp allele in the template plasmid pFA6a-GFP (S65T, F64L)-KanMX6 carries two mutations (Caviston et al., 2003). Strains YJL175A (IQG1:3HA-TRP1 MYO2:MYC-Kan) and YJL176A (MYO1:3HA-TRP1 MYO2:MYC-Kan) were constructed by introducing an affinity-tagged allele of MYO1 and MYO2 by a PCR-based method (Longtine et al., 1998) into strains SSC350 and YEF473A, respectively.
To construct the tetraploid strains, an ∼3.9-kb PCR fragment carrying the dual reporter can1Δ::MFA1pr-HIS3-Mfα1pr-LEU2 was amplified from strain Y3656 (supplied by C. Boone, University of Toronto, Toronto, Canada) and transformed intoYEF2293 (a MYO1:GFP-Kan) and YEF3302 (a mlc2Δ::TRP1 MYO1:GFP-Kan), selecting for His+, Canavanine-resistant colonies, to generate YEF3305 and YEF3306, respectively. These two strains were crossed with YEF2294 and YEF3303, respectively, to form a/α strains YEF3316 and YEF3317 that are heterozygous for the dual reporter locus. YEF3316 and YEF3317 were streaked onto SC-His and SC-Leu plates, respectively, to generate aa and αα versions of strains carrying MYO1:GFP-Kan (YEF3318 and YEF3319) or mlc2Δ::TRP1 MYO1:GFP-Kan (YEF3320 and YEF3321). The mating types were confirmed by monitoring pheromone secretion. YEF3318 and YEF3319 or YEF3320 and YEF3321 were crossed to generate the tetraploid YEF3323 or YEF3324, which was confirmed by sporulation.
CoIP
50 OD600 of yeast cells grown exponentially in SC-Ura media were disrupted with glass beads by vortexing at 4°C in 500 μl of 1× PBS buffer, containing 0.1% NP-40 and a mixture of protease inhibitors. After centrifugation to remove cell debris, the supernatant was used for immunoprecipitation with 100 μl of slurry of agarose beads conjugated with either anti-HA mAb or anti-MYC mAb (Covance Research Products Inc.). After incubation at 4°C for 12–16 h, beads were washed five times with lysis buffer, and proteins bound to the beads were eluted with SDS sample buffer. The precipitated Mlc2p-Myc or Mlc1p-HA, the total Myo1p-GFP variants in supernatants, and the bound fractions of Myo1p-GFP variants to the anti-MYC or anti-HA beads were separated by SDS-PAGE and blotted with anti-MYC, anti-HA, and anti-GFP mAbs, respectively (Covance Research Products Inc.). The secondary antibodies were peroxidase-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories).
Microscopy
Cell morphologies and GFP-tagged proteins were visualized by differential interference contrast (DIC) and fluorescence microscopy, respectively. DNA was stained with 1 μg/ml bisBenzimide (Sigma-Aldrich).
For time-lapse microscopy, cells growing exponentially in liquid SC-Ura medium were spotted onto a microscope slide spread with a thin layer of SC-Ura medium solidified with 25% gelatin (Yeh et al., 1995). Individual cells were followed at 1- or 2-min intervals using a computer-controlled microscope (model Eclipse E800; Nikon) with motorized focus and a high-resolution CCD camera (model C4742-95; Hamamatsu Photonics). For each time point, one DIC image and one GFP image were acquired and analyzed using Image-Pro Plus software (Media Cybernetics). The contrast of the images was enhanced using Image-Pro Plus and/or Adobe PhotoShop Version 7.0 (Adobe Systems). For time-lapse microscopy at 37°C, the objective lens was heated and maintained at 37°C using an objective heater (Bioptechs) during the course of the experiments. The rate of Myo1p ring contraction was determined as follows: the diameter of Myo1p ring before shrinkage minus the diameter of the first Myo1p dot divided by the time required to undergo this change in diameter.
Acknowledgments
We thank S. Brown, A. Ragnini-Wilson, J.H. Hedgemann, T.N. Davis, C. Price, and C. Burd for plasmids and strains; M. Lippincott, K. Bass, P. Martin, and E. Falls for excellent technical assistance; A. Tong and C. Boone for the synthetic–lethal screens with an mlc2Δ strain; and M. Ostap for mapping the mlc1 mutations on the three-dimensional structure of Mlc1p.
This work was supported by grant RSG02039CSM from the American Cancer Society to E. Bi and grant 1R15GM065883-01 from the National Institutes of Health to E. Vallen.
Abbreviations used in this paper: coIP, coimmunoprecipitation; DIC, differential interference contrast; ELC, essential light chain; RLC, regulatory light chain; SC, synthetic complete.
References
- Bi, E. 2001. Cytokinesis in budding yeast: the relationship between actomyosin ring function and septum formation. Cell Struct. Funct. 26:529–537. [DOI] [PubMed] [Google Scholar]
- Bi, E., and J.R. Pringle. 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., P. Maddox, D.J. Lew, E.D. Salmon, J.N. McMillan, E. Yeh, and J.R. Pringle. 1998. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 142:1301–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyne, J.R., H.M. Yosuf, P. Bieganowski, C. Brenner, and C. Price. 2000. Yeast myosin light chain, Mlc1p, interacts with both IQGAP and class II myosin to effect cytokinesis. J. Cell Sci. 113:4533–4543. [DOI] [PubMed] [Google Scholar]
- Burd, C.G., M. Peterson, C.R. Cowles, and S.D. Emr. 1997. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell. 8:1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston, J.P., M. Longtine, J.R. Pringle, and E. Bi. 2003. The role of Cdc42p GTPase-activating proteins (GAPs) in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., B.D. Ostrow, S.R. Tafuri, and R.L. Chisholm. 1994. Targeted disruption of the Dictyostelium RMLC gene produces cells defective in cytokinesis and development. J. Cell Biol. 127:1933–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D.J., A.E.M. Adams, H. Fares, C. De Virgilio, G. Valle, J.S. Chuang, and J.R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L., B. Sundin, E. Brazeau, J.P. Caviston, G.-C. Chen, W. Guo, K.G. Kozminski, M.W. Lau, J.J. Moskow, A. Tong, et al. 2001. A protein interaction map for cell polarity development. J. Cell Biol. 154:549–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D 'souza, V.M., N.I. Naqvi, H. Wang, and M.K. Balasubramanian. 2001. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct. Funct. 26:555-565. [DOI] [PubMed] [Google Scholar]
- Epp, J.A., and J. Chant. 1997. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7:921–929. [DOI] [PubMed] [Google Scholar]
- Gao, X.–D., S. Alberts, S.E. Tcheperegine, C.G. Burd, D. Gallwitz, and E. Bi. 2003. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J. Cell Biol. 162:635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 74:527–534. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., J.R. Pringle, and D.J. Lew. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681–689. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink, eds. 1991. Guide to yeast genetics and molecular biology. Methods in Enzymolology, Vol. 194. San Diego, CA: Academic Press.
- Hales, K.G., E. Bi, J.Q. Wu, J.C. Adam, I.C. Yu, and J.R. Pringle. 1999. Cytokinesis: an emerging unified theory for eukaryotes? Curr. Opin. Cell Biol. 11:717–725. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei, T., K. Tanaka, T. Hihara, M. Umikawa, H. Imamura, M. Kikyo, K. Ozaki, and Y. Takai. 1998. Interaction of Bnr1p with a novel Src Homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J. Biol. Chem. 273:28341–28345. [DOI] [PubMed] [Google Scholar]
- Karess, R.E., X.-J. Chang, K.A. Edwards, S. Kulkarni, I. Aguilera, and D.P. Kiehart. 1991. The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell. 65:1177–1189. [DOI] [PubMed] [Google Scholar]
- Korinek, W.S., E. Bi, J.A. Epp, L. Wang, J. Ho, and J. Chant. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10:947–950. [DOI] [PubMed] [Google Scholar]
- Le Goff, X., F. Motegi, E. Salimova, I. Mabuchi, and V. Simanis. 2000. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J. Cell Sci. 113:4157–4163. [DOI] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 1998. a. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140:355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott, J., and R. Li. 1998. b. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J. Cell Biol. 143:1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M., and E. Bi. 2003. Regulation of septin organization and function in yeast. Trends Cell Biol. 13:403–409. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. DeMarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Matsumura, F., S. Ono, Y. Yamakita, G. Totsukawa, and S. Yamashiro. 1998. Specific localization of serine 19 phosphorylated myosin II during cell locomotion and mitosis of cultured cells. J. Cell Biol. 140:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., M.K. Balasubramanian, L.E. Pelcher, S.M. Hemmingsen, and K.L. Gould. 1995. Schizosaccharomyces pombe cdc4 + gene encodes a novel EF-hand protein essential for cytokinesis. J. Cell Biol. 130:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi, N., K.C.Y. Wong, X. Tang, and M.K. Balasubramanian. 2000. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat. Cell Biol. 2:855–858. [DOI] [PubMed] [Google Scholar]
- Nurse, P., and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. [DOI] [PubMed] [Google Scholar]
- Osman, M.A., J.B. Konopka, and R.A. Cerione. 2002. Iqg1p links spatial and secretion landmarks to polarity and cytokinesis. J. Cell Biol. 159:601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, J., and I. Herskowitz. 1997. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 138:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollenz, R.S., T.L. Chen, L. Trivinos-Lagos, and R.L. Chisholm. 1992. The Dictyostelium essential light chain is required for myosin function. Cell. 69:951–962. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., B. Bowers, A. Varma, D.-H. Roh, and E. Cabib. 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115:293–302. [DOI] [PubMed] [Google Scholar]
- Schroeder, T.E. 1972. The contractile ring. II. determining its brief existence, volumetric changes, and vital role in cleavage Arbacia eggs. J. Cell Biol. 53:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers, J.R. 1991. Regulation of cytoplasmic and smooth muscle myosin. Curr. Opin. Cell Biol. 3:98–104. [DOI] [PubMed] [Google Scholar]
- Shannon, K.B., and R. Li. 2000. A myosin light chain mediates the localization of the budding yeast IQGAP-like protein during contractile ring formation. Curr. Biol. 10:727–730. [DOI] [PubMed] [Google Scholar]
- Shaw, J.A., P.C. Mol, B. Bowers, S.J. Silverman, M.H. Valdivieso, A. Duran, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster, C.B., and D.R. Burgess. 2002. Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc. Natl. Acad. Sci. USA. 99:3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupsky, C.M., M. Desautels, T. Huebert, R. Zhao, S.M. Hemmingsen, and L.P. McIntosh. 2001. Structure of Cdc4p, a contractile ring protein essential for cytokinesis in Schizosaccharomyces pombe. J. Biol. Chem. 276:5943–5951. [DOI] [PubMed] [Google Scholar]
- Stevens, R.C., and T.N. Davis. 1998. Mlc1p is a light chain for the unconventional myosin Myo2p in Saccharomyces cerevisiae. J. Cell Biol. 142:711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak, M., G. Wu, W.F. Stafford, R.C. Lu, and R. Dominguez. 2003. Two distinct myosin light chain structures are induced by specific variations within the bound IQ motifs-functional implications. EMBO J. 22:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolliday, N., M. Pitcher, and R. Li. 2003. Direct evidence for a critical role of Myosin II in budding yeast cytokinesis and the evolvability of new cytokinetic mechanisms in the absence of Myosin II. Mol. Biol. Cell. 14:798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H., G. Lesage, G.D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G.F. Berriz, R.L. Brost, M. Chang, et al. 2004. Global mapping of the yeast genetic interaction network. Science. 303:808–813. [DOI] [PubMed] [Google Scholar]
- Trybus, K.M. 1991. Regulation of smooth muscle myosin. Cell Motil. Cytoskeleton. 18:81–85. [DOI] [PubMed] [Google Scholar]
- Uchimura, T., K. Fumoto, Y. Yamamoto, K. Ueda, and H. Hosoya. 2002. Spatial localization of mono- and diphosphorylated myosin II regulatory light chain at the leading edge of motile HeLa cells. Cell Struct. Funct. 27:479–486. [DOI] [PubMed] [Google Scholar]
- Uyeda, T.Q., and J.A. Spudich. 1993. A functional recombinant myosin II lacking a regulatory light chain-binding site. Science. 262:1867–1870. [DOI] [PubMed] [Google Scholar]
- Vallen, E.A., J. Caviston, and E. Bi. 2000. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, W., P. Bielli, S. Wacha, and A. Ragnini-Wilson. 2002. Mlc1p promotes septum closure during cytokinesis via the IQ motifs of the vesicle motor Myo2p. EMBO J. 21:6397–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., X. Tang, J. Liu, S. Trautmann, D. Balasundaram, D. McCollum, and M.K. Balasubramanian. 2002. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol. Biol. Cell. 13:515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win, T.Z., Y. Gachet, D.P. Mulvihill, K.M. May, and J.S. Hyams. 2001. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J. Cell Sci. 114:69–79. [DOI] [PubMed] [Google Scholar]
- Yeh, E., R.V. Skibbens, J.W. Cheng, E.D. Salmon, and K. Bloom. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]




