Abstract
The transcription factor Elk-1 is a nuclear target of mitogen-activated protein kinases and regulates immediate early gene activation by extracellular signals. We show that Elk-1 is also conjugated to SUMO on either lysines 230, 249, or 254. Mutation of all three sites is necessary to fully block SUMOylation in vitro and in vivo. This Elk-1 mutant, Elk-1(3R), shuttles more rapidly to nuclei of Balb/C cells fused to transfected HeLa cells. Coexpression of SUMO-1 or -2 strongly reduces shuttling by Elk-1 without affecting that of Elk-1(3R), indicating that SUMOylation regulates nuclear retention of Elk-1. Accordingly, overexpression of Elk-1(3R) in PC12 cells, where cytoplasmic relocalization of Elk-1 has been linked to differentiation, enhances neurite extension relative to Elk-1. The effect of Elk-1, but not of the 3R mutant, was blocked upon cotransfection with SUMO-1 or -2 and enhanced by coexpression with mutant Ubc-9. Thus, SUMO conjugation is a novel regulator of Elk-1 function through the control of its nuclear-cytoplasmic shuttling.
Keywords: Elk-1; SUMO; neuronal differentiation; nuclear localization
Introduction
MAPK cascades play a central role in the cellular response to diverse extracellular stimuli and therefore regulate numerous processes, such as cell proliferation, fate, and differentiation. Upon their activation, MAPKs translocate to the nucleus, where they target transcription factors, either directly or indirectly via the activation of other kinases, to induce the appropriate genetic response. A primary nuclear target of activated MAPKs is the ternary complex factor family of ETS-domain transcription factors, namely the proteins Elk-1, SAP-1, and NET/ERP/SAP-2/Elk-3 (Wasylyk et al., 1998). Ternary complex factor phosphorylation drives the rapid induction of immediate early gene transcription through a ternary complex formed on serum response elements together with serum response factor (SRF). This activity is mediated by several regions of sequence and functional homology: the NH2-terminal ETS DNA-binding domain, a motif that mediates protein–protein interactions with SRF, and the COOH-terminal transcription activation domain induced by MAPK phosphorylation on multiple sites. Elk-1 also contains the R motif, which represses basal transcriptional activation, particularly in the context of fusion proteins containing the Gal4 DNA-binding domain (Yang et al., 2002). Recently, Yang et al. (2003) reported that this repression is dependent on SUMOylation.
SUMO modification is an important mechanism for posttranslational regulation of protein function (Seeler and Dejean, 2003). Three SUMO isoforms exist. SUMO-1 shows 47% homology at the protein level with SUMO-2 and -3, whereas SUMO-2 and -3 are 95% homologous (Kim et al., 2002). SUMOylation of target proteins is a multi-step process involving the E1-activating complex SAE1/2 and the E2-conjugating enzyme Ubc-9 in mammalian cells (Tatham et al., 2001). These two suffice to drive SUMO conjugation in vitro; in vivo, this may also involve E3 factors (Seeler and Dejean, 2003). SUMO proteases have also been identified, underscoring the reversible, transient nature of SUMO modification. Many transcription regulatory proteins are SUMOylated, which is linked to numerous nuclear processes, including subnuclear localization, transcriptional activation or repression, and nucleo-cytoplasmic trafficking (Seeler and Dejean, 2003). Importantly, SUMO addition appears to occur upon nuclear import, and mutation of the NLS in certain proteins blocks their SUMOylation (Seeler and Dejean, 2003).
Elk-1, like many transcriptional regulators, is localized to the nucleus in cultured cells (Janknecht et al., 1994). This is mediated through an NLS, present within the ETS domain, that is independent of DNA binding (Janknecht et al., 1994; Vanhoutte et al., 2001). In the central nervous system, however, Elk-1 is found in both the nuclear and cytoplasmic compartments (Sgambato et al., 1998; Vanhoutte et al., 2001). This redistribution is found upon NGF-driven differentiation of PC12 cells, where neurite elongation is not observed when Elk-1 is restricted to the nucleus (Vanhoutte et al., 2001). Here, we report that SUMOylation of Elk-1 has an important role in its nuclear localization and thus its ability to induce neuritogenesis in PC12 cells. All SUMO isoforms can be conjugated to Elk-1 on any of three lysines that lie within or near the R motif. Elk-1 mutated in all three SUMOylation sites shuttles more rapidly than wild-type (WT) Elk between nuclei in HeLa-Balb/C heterokaryons, and enhances neurite extension in PC12 cells. Shuttling by WT Elk-1 is virtually eliminated by coexpression with SUMO, which also blocks Elk-1–driven PC12 differentiation. Thus, SUMOylation is a novel regulator of Elk-1 function that acts by controlling Elk-1 nuclear presence.
Results and discussion
Elk-1 binds to Ubc9 and can be conjugated to all three SUMO isoforms in vitro
The motif ψKxE, where ψ is an aliphatic amino acid and x is any amino acid, is the binding site for the E2 conjugating enzyme Ubc9 and for transfer of SUMO to the ɛ amino group of lysine (Rodriguez et al., 2001; Sampson et al., 2001). Elk-1 contains three potential SUMOylation sites, surrounding lysines 230, 249, and 254, that lie outside the domains involved in DNA binding, interactions with SRF and MAPKs, and transcriptional activation (Fig. 1 A). K230 and K249 lie within the R motif, which mediates repression of basal transcriptional activity when fused to the Gal4 DNA-binding domain (Yang et al., 2002). This has recently been linked to their SUMOylation (Yang et al., 2003). We used several tests to evaluate SUMO conjugation to Elk-1 at these sites in our system.
Figure 1.
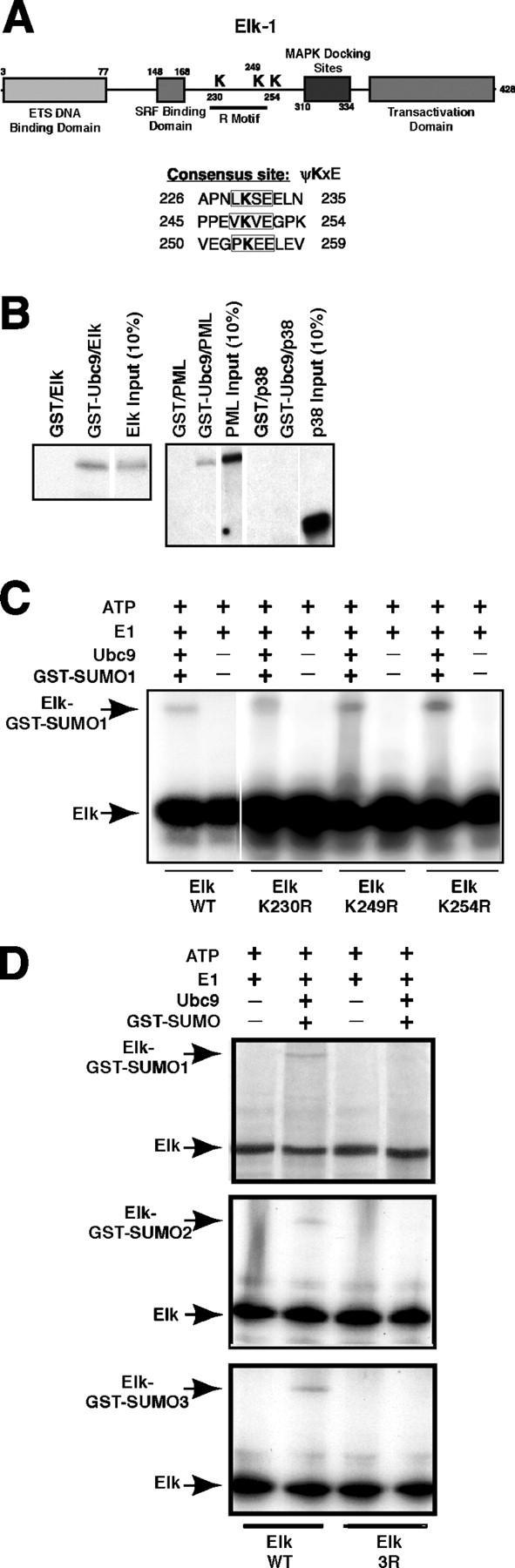
In vitro SUMOylation of Elk-1 is blocked by mutating all three consensus sites. (A) The major functional domains in human Elk-1 are indicated. Shown below is the consensus site for Ubc-9 binding and SUMOylation ψKxE, where ψ is an aliphatic amino acid and K is the lysine conjugated to SUMO, as are the sequences surrounding the three major consensus sites in Elk-1, centered at lysines 230, 249, and 254. (B) 35S-labeled Elk-1, PML, or p38α were incubated with GST or GST-Ubc9 bound to glutathione agarose. After washing, bound proteins were separated by SDS-PAGE and visualized by autoradiography of the dried gel. Input indicates 10% of the 35S-protein added to the incubation. White lines indicate that intervening lanes have been spliced out. (C) 35S-Elk-1, either WT or mutated in each consensus site (as shown below the panel) was incubated with the indicated components. The reactions were analyzed by SDS-PAGE as in B. Elk-1 and SUMOylated Elk-1 bands are indicated. The white line indicates that intervening lanes were spliced out. (D) Elk-1 WT and the triple mutant 3R were tested for conjugation to SUMO-1 (top), -2 (middle), or -3 (bottom) as in C.
GST pull-down assays analyzed solution interactions between recombinant Elk-1 and Ubc9 (Sampson et al., 2001) relative to PML, a known SUMO substrate, and the MAPK p38α, which has no Ubc9 binding site. 35S-labeled Elk-1 bound to purified GST-Ubc9 but not GST alone (Fig. 1 B). Binding to Elk-1 was more efficient than to 35S-PML and was specific because 35S-p38α did not interact with Ubc9 (Fig. 1 B). To test whether Elk is a substrate for the SUMO pathway, 35S-Elk-1 was incubated in an in vitro SUMO-conjugation assay system containing GST-SUMO-1, recombinant Ubc9, a HeLa cell fraction containing SUMO E1 activity and ATP (Rodriguez et al., 1999). A portion of 35S-Elk-1 was shifted to a slower migrating species conjugated to GST-SUMO-1 (Fig. 1 C); its appearance required the addition of Ubc9 and GST-SUMO-1. Elk-1 could also be modified with SUMO-2 and SUMO-3 in vitro (Fig. 1 D). To identify the residue involved, the three candidate lysines were mutated to arginine. No single Lys-Arg mutation blocked SUMO-1 conjugation (Fig. 1 C). Double mutation of the lysines reduced but did not fully block SUMOylation in vitro (not depicted), whereas mutation all three consensus sites eliminated conjugation of SUMO-1, -2, and -3 (Fig. 1 D).
Overexpressed Elk-1 is SUMOylated on lysines 230, 249, and 254 in cellula
To show that Elk-1 was modified by SUMO in cells, HeLa cells were transfected with expression vectors for either WT Elk or Elk3R bearing the HA epitope and one of the three forms of SUMO with a NH2-terminal oligohistidine tag. To avoid problems with SUMO peptidases, cells were lysed with 6 M Guanidinium-HCl and the histidine-tagged proteins, i.e. conjugated to SUMO, purified by metal affinity chromatography. The purified proteins were visualized by immunoblotting with an Elk-specific antiserum. A fraction of overexpressed WT Elk was conjugated to (His)SUMO-1 (Fig. 2 A), which was not observed with Elk3R (Fig. 2 A). SUMO conjugation to WT Elk, but not to Elk3R, was also apparent upon coexpression with either (His)SUMO-2 or -3 (Fig. 2, B and C). In addition, both SUMO-2 and -3 generated a ladder of SUMOylated Elk that could reflect modification on multiple sites and/or their oligomerization (Tatham et al., 2001). The same ladder was observed after cotransfection of expression vectors for the single mutants K230R, K249R, and K254R and (His)SUMO-2 (Fig. 2 D). Nevertheless, K249 appears to be preferentially modified, followed by K254 and finally K230, even though equivalent levels of each mutant were expressed (Fig. 2 D). Notably, combination of any two mutations lowered but did not abolish SUMOylation (unpublished data). Thus, the ladder reflects SUMO oligomerization and that any of the three sites is a target for SUMO conjugation in transfected cells.
Figure 2.
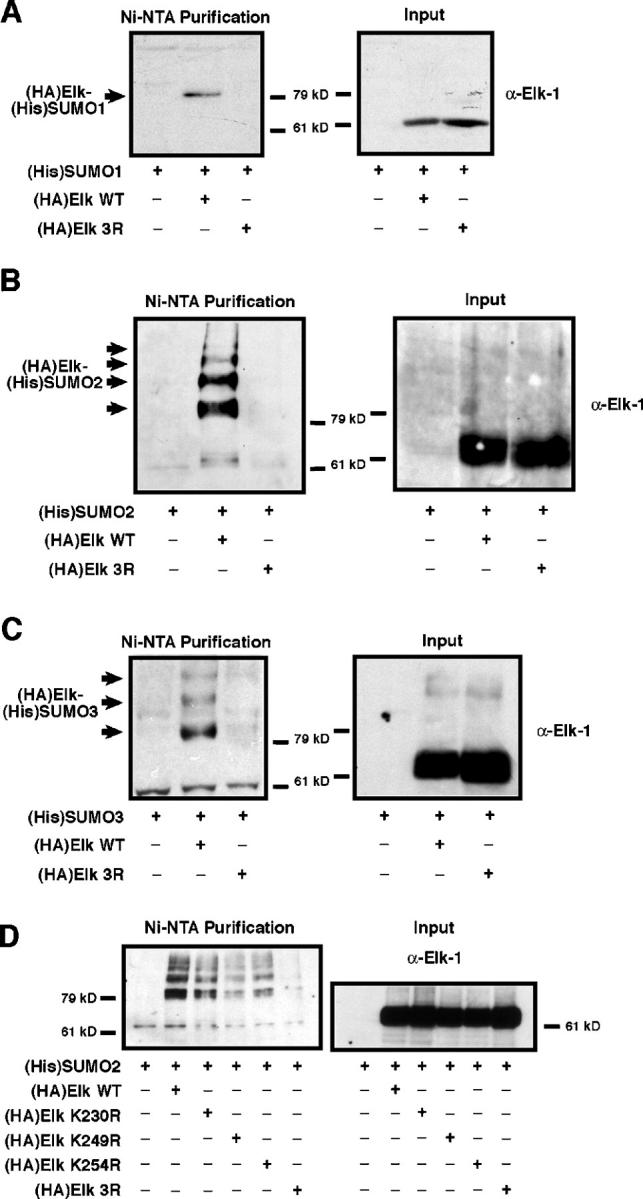
Conjugation of all three SUMO isoforms to Elk-1 in transfected cells. HeLa cells were transfected with expression vectors for Elk-1 WT or 3R, together with vectors for (His)SUMO-1 (A), -2 (B), or -3 (C) as indicated below the lanes. Total proteins (Input) and His-tagged proteins were purified from denaturing cell lysates, separated on SDS-PAGE and visualized by immunoblotting for Elk-1. Left, Ni-NTA bound proteins; right, total protein. (D) HeLa cells transfected with expression vectors for (His)SUMO-2 and either WT Elk-1 or the mutants K230R, K249R, K254R, or 3R were analyzed as in A–C.
SUMO conjugation to Elk plays a role in the dynamics of its nuclear-cytoplasmic shuttling
SUMO modification correlates with nuclear localization of certain transcriptional regulators (Seeler and Dejean, 2003), and mutation of the NLS compromises SUMOylation of the proteins SP100, HDAC4, Smad4, and MDM1 (Sternsdorf et al., 1999; Kirsh et al., 2002; Miyauchi et al., 2002; Lin et al., 2003). To analyze intracellular localization, we generated expression vectors for EGFP-Elk WT and 3R fusion proteins. In HeLa cells, these vectors encoded proteins of the expected molecular size recognized by an Elk-specific antibody (not depicted). The GFP-Elk3R fusion was predominantly nuclear, like WT protein, 24 h after transfection (Fig. 3 A). This indicates that the three K to R mutations do not disrupt the NLS, which lies mainly in the Ets domain.
Figure 3.
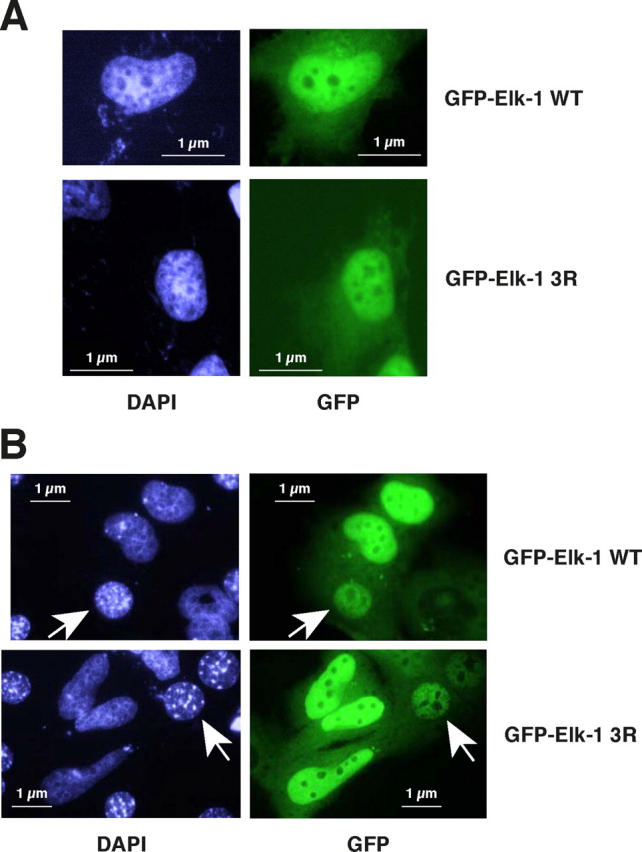
Shuttling of EGFP-Elk-1 WT and 3R in HeLa/Balb/C heterokaryons. (A) HeLa cells on coverslips were transfected with expression vectors for EGFP-Elk-1 WT or 3R, as indicated. Cells were fixed 36 h later and counterstained with DAPI. (B) Heterokaryons between Balb/C mouse fibroblasts and HeLa cells transfected as in A, along with pDsRed2 to visualize the heterokaryons (not depicted). Cells were fixed 60 min later and counterstained with DAPI. The arrows indicate mouse nuclei to which EGFP-Elk-1 WT (top) and 3R (bottom) have shuttled.
To test whether the mutant protein might show altered kinetics of nuclear export, we used heterokaryon analysis. HeLa cells transfected with the WT and mutant EGFP-Elk expression vectors were fused to Balb/C mouse fibroblasts using polyethylene glycol (Caceres et al., 1998). Nuclei of the two cell types are easily distinguished by fluorescence (Fig. 3 B). No EGFP signal could be detected in unfused mouse cells, whereas EGFP staining was readily visible in both the HeLa and mouse nuclei in the fused cells (Fig. 3 B, arrows indicate the mouse nucleus). Analysis of EGFP staining in mouse nuclei at different times after the induction of cell fusion revealed that EGFP-Elk3R shuttled more quickly to mouse nuclei (Table I). To test the effect of SUMO overexpression on shuttling, we used HeLa cells that stably overexpress SUMO-1 or -2. Transfection and fusion of HeLa-SUMO-1 or -2 cells enhanced the differential kinetics observed between WT and mutant Elk. SUMO strongly reduced shuttling of EGFP-Elk to the fused mouse nucleus, whereas it barely affected the kinetics of EGFP-Elk3R transfer (Table I). Thus, Elk localization to the nucleus actually reflects a balance between nuclear import and nuclear export. This balance is regulated by SUMOylation because it was sensitive to coexpression with SUMO and to mutation of the three SUMO acceptor sites in Elk, which led to a more rapid localization of Elk3R to the Balb/C nucleus in heterokaryons.
Table I. Quantification of EGFP-Elk-1 WT and 3R shuttling in HeLa-Balb/C heterokaryonsa .
| HeLa | HeLa-SUMO-1 | HeLa-SUMO-2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Min after fusion | wt | 3R | ratiob | wt | 3R | ratiob | wt | 3R | ratiob |
| 30 | 28% | 48% | 0.58 | 17% | 43% | 0.40 | 13% | 37% | 0.35 |
| 45 | 33% | 56% | 0.59 | 20% | 58% | 0.35 | 25% | 58% | 0.43 |
| 60 | 53% | 70% | 0.76 | 22% | 70% | 0.31 | 30% | 77% | 0.39 |
Kinetics of transfer of EGFP-Elk-1 WT and 3R to Balb/C nuclei as a function of stable overexpression of SUMO-1 or -2 in HeLa cells. The percentage of positive Balb/C nuclei in heterokaryons relative to total number of fused cells is shown. The values are representative of at least three independent experiments.
wt/3R.
Elk3R enhances neurite outgrowth by PC12 cells relative to the WT protein
NGF treatment of PC12 cells induces their neuronal differentiation, which can be easily visualized by neurite outgrowth. Transfection of an expression vector for WT Elk leads to increased PC12 differentiation (Fig. 4 A), an effect that is linked to the relocalization of Elk to the cytoplasm (Vanhoutte et al., 2001). Therefore, Elk3R would be expected to increase PC12 differentiation in the absence of NGF because it shuttles more rapidly than WT Elk from the nucleus. Thus, PC12 cells were transfected with expression vectors that encode HA-tagged versions of WT Elk or two mutants, Elk3R and Elk3A. Elk3A has Ser to Ala mutations in the major phosphorylation sites in the activation domain, which compromises signaling-driven reporter gene activation (Janknecht et al., 1993). 48 h after transfection, cells were fixed and evaluated microscopically for neurite outgrowth and by immunofluorescence for expression of GFP or HA-tagged Elk. Expression of WT Elk, Elk3A and Elk3R induced neurite extension relative to GFP-transfected cells (Fig. 4). WT Elk and Elk3A led to a twofold increase in the number of cells presenting neurites relative to GFP (Fig. 4 B), suggesting that Elk-1–driven gene expression is not important in inducing neurite extension under basal conditions. In contrast, shuttling is important because differentiation was increased another twofold by Elk3R (Fig. 4 B; Fig. 5, B–D). Thus, Elk expression is sufficient to increase PC12 differentiation in the absence of NGF and Elk3R enhances this phenotype.
Figure 4.
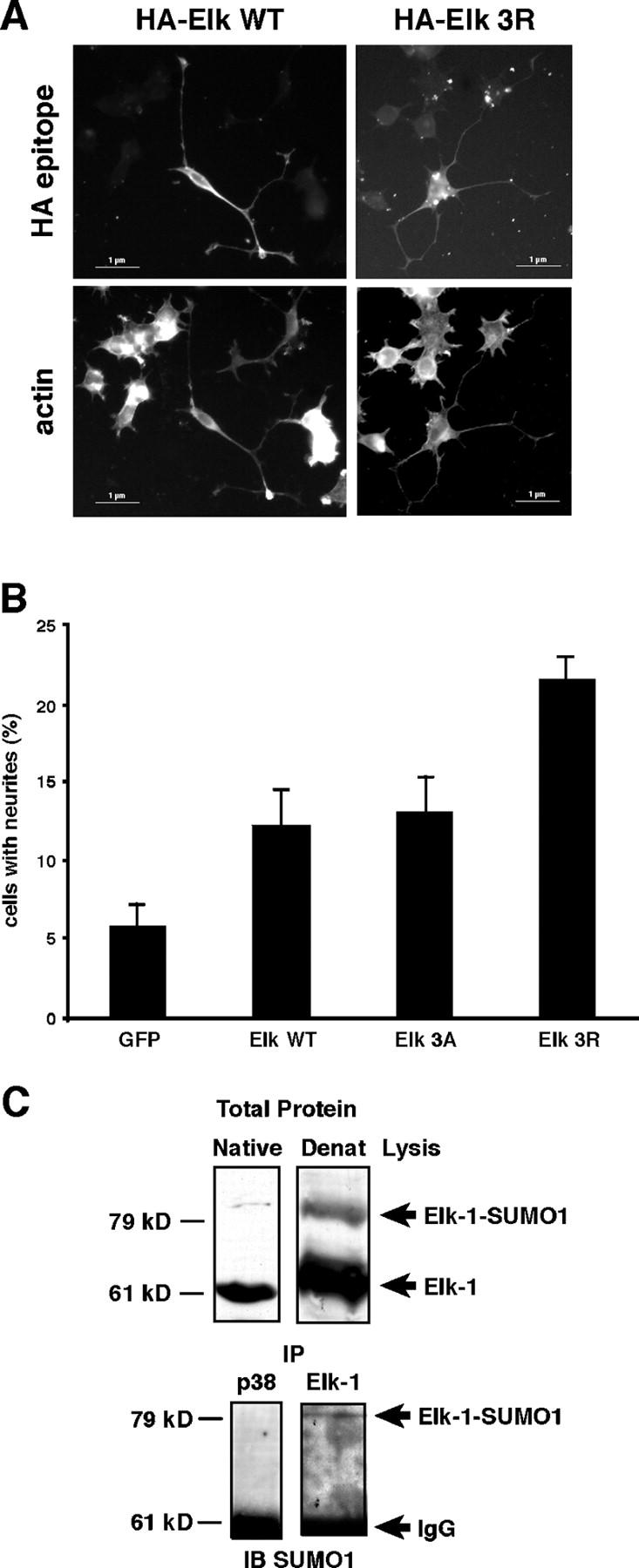
Elk3R enhances PC12 differentiation relative to Elk-1 WT. PC12 cells were transfected with expression vectors for (HA)Elk-1 WT, 3A or 3R, or EGFP as a control. Cells were processed 48 h later. (A) Elk-1 was visualized using an anti-HA antibody and F-actin with phalloidin-CPITC. (B) Percentage of HA-positive PC12 cells presenting neurites, defined by a length at least twice that of the cell body (Estrach et al., 2002). The numbers are representative of four experiments. The error bars indicate the SEM. (C) Immunoblot detection of endogenous Elk-1 in native and denaturing lysates of PC12 cells (top), and of SUMO-1–conjugated Elk-1 in Elk-1 but not p38MAPK immunoprecipitates from native lysates of PC12 cells (bottom). IgG denotes the signal from the antibody used for precipitation.
Importantly, SUMOylation of endogenous Elk-1 was detectable in PC12 cell extracts. A band migrating at the position expected for SUMOylated Elk-1 reacted with an Elk-1 antibody in immunoblots of denaturing lysates of PC12 cells (Fig. 4 C). A similar band was detected using a SUMO-1 antibody when Elk-1 was purified by immunoprecipitation from nondenaturing lysates (Fig. 4 C), indicating that a proportion of endogenous Elk-1 is conjugated to SUMO-1 in PC12 cells.
Cotransfection of SUMO suppresses and dominant negative (DN) Ubc9 enhances differentiation induced by WT Elk
In light of these results, enhancing or blocking SUMOylation of WT Elk would be expected to diminish or augment its ability to induce neurites, respectively. To test the former, PC12 cells were cotransfected with expression vectors encoding either GFP-SUMO-1 or -2 and those described above for WT and Elk3R. GFP-SUMO-1 and HA-Elk localized to the same cells, and their coexpression clearly affected WT Elk-driven differentiation (Fig. 5 A), lowering the percentage of cells with neurites to that observed with SUMO-1 alone (Fig. 5 B). SUMO-2 showed the same reduction in WT Elk-driven differentiation (Fig. 5 C). In contrast, SUMO-1 or -2 did not affect neurite induction by Elk3R (Fig. 5, B and C). To test the effect of blocking SUMOylation, we used a version of Ubc9 bearing a C35S mutation in the catalytic site that acts as a DN inhibitor of SUMO conjugation. Cotransfection with DN-Ubc9 increased neurite induction by WT Elk to the level observed with Elk3R (Fig. 5 D), which was unaltered in the presence of mutant Ubc9. These data show that PC12 differentiation, a physiological indicator of Elk intracellular localization, is strongly affected by altering the balance of SUMOylation of Elk and thereby the dynamics of its nucleo-cytoplasmic shuttling. This illustrates a novel functional role for Elk conjugation to SUMO of regulating its nuclear presence. This would have important consequences for the other role attributed to Elk SUMOylation, involving repression of Elk basal transcriptional activation, as well as for Elk function as a key mediator of intracellular signaling-driven transcriptional activation.
Figure 5.
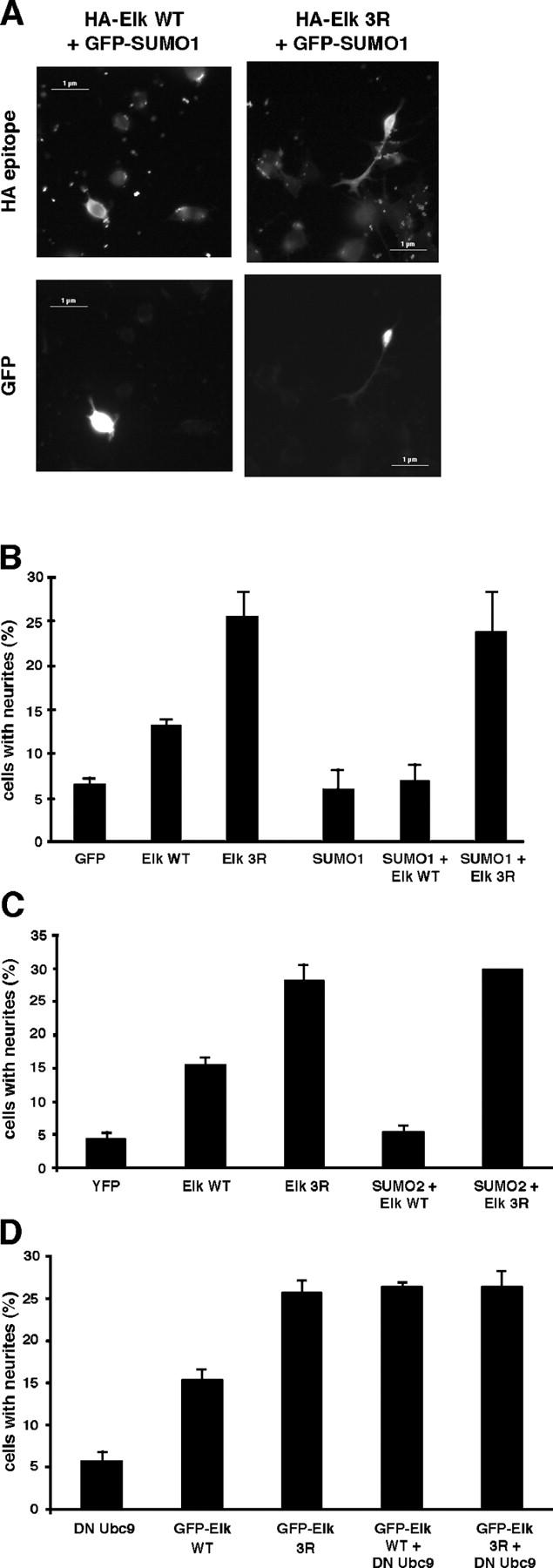
Coexpression of SUMO blocks and of DN-Ubc9 enhances PC12 differentiation driven by Elk-1 WT. (A) PC12 cells were transfected with expression vectors for pEGFP-SUMO-1 and (HA)Elk-1 WT or 3R. Cells were processed 48 h later. Elk-1 was visualized with an anti-HA antibody. (B and C) PC12 cells were transfected with expression vectors for pEGFP (B) or pEYFP (C), (HA)Elk-1 WT or 3R, pEGFP-SUMO-1 (B) or -2 (C) as indicated, and processed as in A. Percentage of HA- and GFP-positive PC12 cells presenting neurites, as defined in the legend to Fig. 4 B. The numbers are representative of three experiments. (D) PC12 cells were transfected with expression vectors for HA-DN-Ubc9 and pEGFP Elk-1 WT or 3R as indicated and processed as described above. The numbers are representative of three experiments. The error bars indicate the SEM.
Materials and methods
Reagents
Peroxidase-coupled secondary antibodies, glutathione agarose, protein A-Sepharose, oligonucleotides, and phalloidin coumarin phenyl isothiocyanate (CPITC) were obtained from Sigma-Aldrich. All other reagents were obtained from local suppliers.
Constructs and mutagenesis
pCDNA3-HA-Elk-1 contains the Elk-1 coding region inserted downstream of the HA epitope in pCDNA3.1. Lysines 230, 249, and 254 were mutated to arginine using a site-directed mutagenesis kit (Stratagene) and appropriate mutant primers. Double and triple mutants were generated by subsequent mutagenesis. All mutations were confirmed by sequence analysis. pGEX2T-Ubc9, pGEX2T-SUMO-1, -2, and -3, pCDNA3(His)SUMO-1, -2, and -3, and DN-Ubc9 were a gift from R. Hay (University of St. Andrews, St. Andrews, UK). DN-Ubc9 was subcloned in pcDNA3.1-HA. pEGFPL11-Elk was constructed by the insertion of Elk-1 sequence into the AccI and BamHI sites of pEGFPL11. For pEGFPL11-Elk3R, the BlpI–PflMI fragment of pEGFPL11-Elk was replaced with that from Elk3R.
Cell culture, heterokaryon analysis, and immunofluorescence microscopy
HeLa and Balb/C cells were grown in DME (Invitrogen) supplemented with 10% FBS (Invitrogen) and antibiotics in 5% CO2 in a humidified incubator. PC12-E2 cells were maintained in DME complemented with 5% FCS and 10% horse serum as described above. PC12-E2 cells were plated on rat tail collagen as described previously (Estrach et al., 2002), and transfected using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. HeLa cells were transfected using Fugene 6 (Roche) according to the manufacturer's instructions. HeLa cells stably expressing (His)SUMO-1 and -2 were provided by R. Hay.
To study localization of EGFP-Elk WT and 3R, transfected HeLa cells were fixed on coverslips with 3.7% formaldehyde in PBS for 10 min. After washes in PBS, cells were then permeabilized with 0.1% Triton X-100 in PBS, counterstained with DAPI and mounted in Mowiol. For heterokaryon analysis (Caceres et al., 1998), 2.5 × 105 HeLa cells were plated on coverslips the day before transfection. Cells were transfected with 1 μg EGFP-Elk WT or 3R and 1 μg pDsRed2 to allow visualization of heterokaryons. 24 h later, 106 Balb/C cells were plated on the coverslips in culture medium containing 50 μg/ml cycloheximide for 3 h. After a PBS wash, cells were fused by incubation in 50% (wt/vol) polyethylene glycol. After three PBS washes, cells were incubated for the indicated times in medium containing 100 μg/ml cycloheximide. Cells were processed for immunofluorescence as described above. PC12 cells were transfected with the vectors described in the figure legend. 48 h later cells were fixed and stained with phalloidin-CPITC and the anti-HA antibody as described previously (Estrach et al., 2002).
Coverslips were examined on a DMRA fluorescence microscope (Leica) with a Leica PL APO 40X/1.25 (PC12 cells) or 63X/1.32 (heterokaryons) oil lens. GFP, phalloidin-CPITC, DAPI, RFP, and fluorescently tagged secondary antibodies were visualized using appropriate filters. Images were captured with a Photometrics CoolSnapTM Fx digital camera operated via Metamorph software, version 4.6r6 (Universal Imaging Corp.), and further processed using Adobe Photoshop® 7.
Binding assays and in vitro SUMOylation
20 μg of GST or GST-Ubc9 were bound to 20 μl of glutathione agarose beads in PBS-1 mM DTT. Non-specific binding was blocked by incubation in 2% BSA in PBS for 1 h at 4°C. Elk-1 was produced using the T3-TNT reticulocyte lysate system (Promega) and 5 μCi L-[35S]methionine (43 TBq/mMol, 10 μCi/μl; Perkin-Elmer) according to the supplier's protocol. An aliquot of 35S-Elk-1 was incubated with the beads in 100 μl B buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween 20) for 30 min at RT (Sampson et al., 2001). After three washes in B buffer, the beads were resuspended in SDS-PAGE sample mix, boiled, and separated by SDS-PAGE. 35S-Elk-1 was visualized by autoradiography.
In vitro SUMOylation reactions contained 1 μl 35S-Elk-1, 2 μl HeLa cell SUMO E1 activation fraction II.4 (provided by M. Rodriguez, Institut Jaques Monod, Paris; Rodriguez et al., 1999), 10 μl ATP regenerating system (50 mM Tris-HCl, pH 7.6, 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate, 3.5 U/ml creatine kinase and 0.6 U/ml inorganic pyrophosphatase), 10 μg purified GST-SUMO and 1 μg purified, recombinant Ubc9. The reactions were incubated 3 h at 37°C, then stopped by adding 1/5 vol 10% SDS-5% β-mercaptoethanol, boiled and separated by SDS-PAGE. 35S-Elk-1 was visualized by autoradiography.
Denaturing purification and analysis of SUMO-Elk conjugates
HeLa cells were transfected with the appropriate Elk-1 expression vector, along with pCDNA3(His)SUMO-1, -2, or -3. 36 h later, cells were lysed in 1 ml 6 M Guanidinium-HCl, 0.1 M NaH2PO4, 10 mM Tris-HCl, pH 8.0 (Rodriguez et al., 1999). 50 μl lysate was precipitated with 1 ml 5% TCA for total protein. Lysates were incubated 3 h at RT with 75 μl NTA agarose (QIAGEN) prewashed with lysis buffer. The slurry was washed with lysis buffer; 8 M urea, 0.1 M Na2HPO4, 10 mM Tris-HCl, pH 8.0 (buffer A); buffer A at pH 6.3; then buffer A, 6.3 containing 0.2% Triton X-100. After denaturation, proteins were separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes and Elk visualized by immunoblotting as described previously (Nissen et al., 2001).
SUMOylation of endogenous Elk
Total protein was extracted from 106 PC12 cells using the denaturing buffer above or 1% Triton lysis buffer (Nissen et al., 2001), precipitated with 5% TCA, separated on SDS-PAGE and Elk visualized by immunoblotting. For immunoprecipitation, 108 PC12 cells were lysed in IP buffer (0.15 M NaCl, 20 mM Tris-HCl, pH 7.5, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS, 20 mM N-ethyl-maleimide). After preclearing with protein A–Sepharose, proteins were precipitated using 40 μg anti-Elk (sc-355; Santa Cruz Biotechnology, Inc.) or anti-p38 (sc-728) and protein A–Sepharose. After three washes with IP buffer, proteins were separated on SDS-PAGE and immunoblotted for SUMO-1 (sc-9060).
Acknowledgments
We thank R. Hay and M. Rodriguez for materials, and M. Rodriguez, H. Enslen, and M. Le Clech for their insight.
This work was supported by the Association pour la Recherche sur le Cancer (A. Debant, I. Jariel-Encontre, and R.A. Hipskind), the Ligue Nationale contre le Cancer (A. Debant), and the Centre National de la Recherche Scientifique. S. Salinas and A. Briancon-Marjollet are doctoral fellows of the French Research and Education Ministry. S. Salinas has a 6-mo doctoral fellowship from the Ligue.
Abbreviations used in this paper: CPITC, coumarin phenyl isothiocyanate; DN, dominant negative; SRF, serum response factor; WT, wild-type.
References
- Caceres, J.F., G.R. Screaton, and A.R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach, S., S. Schmidt, S. Diriong, A. Penna, A. Blangy, P. Fort, and A. Debant. 2002. The Human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr. Biol. 12:307–312. [DOI] [PubMed] [Google Scholar]
- Janknecht, R., W.H. Ernst, V. Pingoud, and A. Nordheim. 1993. Activation of ternary complex factor Elk-1 by MAP kinases. EMBO J. 12:5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht, R., R. Zinck, W.H. Ernst, and A. Nordheim. 1994. Functional dissection of the transcription factor Elk-1. Oncogene. 9:1273–1278. [PubMed] [Google Scholar]
- Kim, K.I., S.H. Baek, and C.H. Chung. 2002. Versatile protein tag, SUMO: its enzymology and biological function. J. Cell. Physiol. 191:257–268. [DOI] [PubMed] [Google Scholar]
- Kirsh, O., J.S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., M. Liang, Y.Y. Liang, F.C. Brunicardi, and X.H. Feng. 2003. SUMO-1/Ubc9 promotes nuclear accumulation and metabolic stability of tumor suppressor Smad4. J. Biol. Chem. 278:31043–31048. [DOI] [PubMed] [Google Scholar]
- Miyauchi, Y., S. Yogosawa, R. Honda, T. Nishida, and H. Yasuda. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277:50131–50136. [DOI] [PubMed] [Google Scholar]
- Nissen, L.J., J.C. Gelly, and R.A. Hipskind. 2001. Induction-independent recruitment of CREB-binding protein to the c-fos serum response element through interactions between the bromodomain and Elk-1. J. Biol. Chem. 276:5213–5221. [DOI] [PubMed] [Google Scholar]
- Rodriguez, M.S., J.M. Desterro, S. Lain, C.A. Midgley, D.P. Lane, and R.T. Hay. 1999. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 18:6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, M.S., C. Dargemont, and R.T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654–12659. [DOI] [PubMed] [Google Scholar]
- Sampson, D.A., M. Wang, and M.J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664–21669. [DOI] [PubMed] [Google Scholar]
- Seeler, J.S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690–699. [DOI] [PubMed] [Google Scholar]
- Sgambato, V., P. Vanhoutte, C. Pages, M. Rogard, R. Hipskind, M.J. Besson, and J. Caboche. 1998. In vivo expression and regulation of Elk-1, a target of the extracellular-regulated kinase signaling pathway, in the adult rat brain. J. Neurosci. 18:214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf, T., K. Jensen, B. Reich, and H. Will. 1999. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274:12555–12566. [DOI] [PubMed] [Google Scholar]
- Tatham, M.H., E. Jaffray, O.A. Vaughan, J.M. Desterro, C.H. Botting, J.H. Naismith, and R.T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368–35374. [DOI] [PubMed] [Google Scholar]
- Vanhoutte, P., J.L. Nissen, B. Brugg, B.D. Gaspera, M.J. Besson, R.A. Hipskind, and J. Caboche. 2001. Opposing roles of Elk-1 and its brain-specific isoform, short Elk-1, in nerve growth factor-induced PC12 differentiation. J. Biol. Chem. 276:5189–5196. [DOI] [PubMed] [Google Scholar]
- Wasylyk, B., J. Hagman, and A. Gutierrez-Hartmann. 1998. Ets transcription factors: nuclear effectors of the Ras-MAP-kinase signaling pathway. Trends Biochem. Sci. 23:213–216. [DOI] [PubMed] [Google Scholar]
- Yang, S.H., D.C. Bumpass, N.D. Perkins, and A.D. Sharrocks. 2002. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol. Cell. Biol. 22:5036–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S.H., E. Jaffray, R.T. Hay, and A.D. Sharrocks. 2003. Dynamic interplay of the SUMO and ERK pathways in regulating Elk-1 transcriptional activity. Mol. Cell. 12:63–74. [DOI] [PubMed] [Google Scholar]


