Abstract
Here, we describe a new muscle LIM domain protein, UNC-95, and identify it as a novel target for the RING finger protein RNF-5 in the Caenorhabditis elegans body wall muscle. unc-95(su33) animals have disorganized muscle actin and myosin-containing filaments as a result of a failure to assemble normal muscle adhesion structures. UNC-95 is active downstream of PAT-3/β-integrin in the assembly pathways of the muscle dense body and M-line attachments, and upstream of DEB-1/vinculin in the dense body assembly pathway. The translational UNC-95::GFP fusion construct is expressed in dense bodies, M-lines, and muscle–muscle cell boundaries as well as in muscle cell bodies. UNC-95 is partially colocalized with RNF-5 in muscle dense bodies and its expression and localization are regulated by RNF-5. rnf-5(RNAi) or a RING domain deleted mutant, rnf-5(tm794), exhibit structural defects of the muscle attachment sites. Together, our data demonstrate that UNC-95 constitutes an essential component of muscle adhesion sites that is regulated by RNF-5.
Keywords: UNC-95; RNF-5; LIM; RING; E3 ligase
Introduction
Focal adhesions are the sites in which the actin cytoskeleton is linked via integral membrane proteins to the ECM in cultured cells (for review see Geiger and Bershadsky, 2001). Structures analogous to vertebrate focal adhesions, both in composition and function, are the dense bodies and M-lines in the body wall muscle of the nematode Caenorhabditis elegans (Waterston, 1988; Moerman and Fire, 1997). Muscle thick (myosin) and thin (actin) filaments are anchored to the plasma membrane by the M-lines and dense bodies, respectively, to form the A bands and I bands of the contractile unit.
The dense bodies and M-lines establish the adhesion sites of the muscle cells to the basal lamina (ECM), and are essential both for the organization of the actin–myosin contractile structure into sarcomeres and the maintenance of the muscles in their correct position. The assembly process of the myofilament lattice during embryogenesis initiates at the basal lamina, proceeds through the cell membrane (sarcolemma) into the cell to form the dense bodies and M-lines, and ends with the formation of thin and thick filaments of the myofilament lattice (Francis and Waterston, 1985, 1991; Hresko et al., 1994; Williams and Waterston, 1994). On the basis of mutant analysis, genetic pathways have been described for dense bodies and M-line formation (Lin et al., 2003). UNC-52/perlecan, a basement membrane heparan sulfate proteoglycan, is initially deposited in the basal lamina (Francis and Waterston, 1991; Rogalski et al., 1993; Williams and Waterston, 1994). Later on, integrin is polarized to the basal sarcolemma to initiate myofilament assembly (Francis and Waterston, 1985, 1991; Hresko et al., 1994). After this step, cytoplasmic dense bodies and M-line proteins such as talin, vinculin, and α-actinin are recruited from the cytosol and enter into the nascent attachment sites. Finally, actin- and myosin-containing filaments are recruited into I and A bands, respectively. By hatching, the mature body wall muscle cells are polarized such that all the contractile filaments are located on the basal surface of the cell associated with the hypodermis (Barstead and Waterston, 1991). Proteins that have been shown to be required for the proper assembly of muscle attachments are DEB-1/vinculin, which has a critical role in thin filament organization (Francis and Waterston, 1985; Barstead and Waterston, 1991), α-actinin (Francis and Waterston, 1985), the UNC-112/Mig-2 FERM domain protein (Rogalski et al., 2000), PAT-4/ILK (Mackinnon et al., 2002), PAT-6/actopaxin(Lin et al., 2003), talin (Moulder et al., 1996), UNC-89 (Benian et al., 1996), the zinc finger protein UNC-98 (Mercer et al., 2003), and the LIM domain protein UNC-97/PINCH (Hobert et al., 1999).
Studies of vertebrate focal adhesion plaques and C. elegans muscle adhesion structures have recently demonstrated the important role of LIM domain proteins in the assembly of these structures (Labouesse and Georges-Labouesse, 2003). LIM domain proteins are double zinc finger–like structures that mediate protein–protein interactions (Schmeichel and Beckerle, 1994). Interactions of LIM domains with specific protein partners influence subcellular localization and mediate the assembly of multimeric protein complexes (for review see Dawid et al., 1998; Bach, 2000). In C. elegans, UNC-97, a LIM domain protein of the PINCH family, has been shown to have a central role in the assembly of muscle attachment structures (Hobert et al., 1999).
Here, we describe a new LIM domain protein, UNC-95, that is required for the proper assembly of muscle attachment sites. As first described by Zengel and Epstein (1980), adult animals homozygous for the unc-95(su33) and unc-95(su106) mutation are very slow to paralyzed (UNCoordinated) and have altered body wall muscle cell structure. Lack of striations and birefringent structures of varying size were observed in these animals by polarized light microscopy. The altered structures correspond to disorganized thick and thin filaments as observed by EM of the su106 allele (Zengel and Epstein, 1980). We originally isolated UNC-95 (Y105E8A.6) as a positive interactor of the C. elegans RNF-5 in a yeast two-hybrid screen (Didier et al., 2003) and here, by searching neighboring candidates on the genetic map, we identified it as the unc-95 gene. RNF-5 is a RING finger protein with high homology to the human gene Rnf5 (Kyushiki et al., 1997; 36% identity). RING finger proteins have been demonstrated to function as ubiquitin protein ligases (E3s) in the ubiquitin modification system (for review see Fang et al., 2003). RNF-5 contains a RING finger domain (C3HC4 type) followed by a proline/serine-rich domain. We demonstrated previously that C. elegans RNF-5 exhibits E3 ligase activity (Didier et al., 2003; unpublished data). Through its E3 ligase activity, RNF-5 affects the cellular localization and abundance of its associated proteins, as was demonstrated for human paxillin. Overexpression of human Rnf5 causes exclusion of paxillin from focal adhesions, resulting in inhibition of cell motility (Didier et al., 2003). Here, we characterize the role of UNC-95 in the assembly of muscle attachment structures and identify RNF-5 as a regulator of the localization and expression of UNC-95 within the C. elegans body wall muscle cells.
Results
unc-95 is required for the integrity of muscle dense bodies
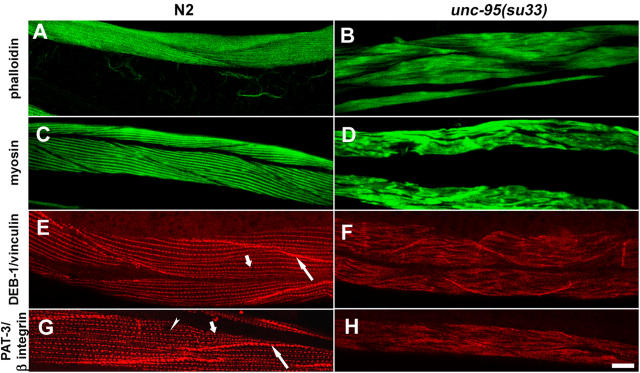
As first described by Zengel and Epstein (1980), adult animals homozygous for the unc-95(su33) mutation have aberrant muscle structure. To further characterize the structural defects in the unc-95 mutant muscle, we stained the body wall muscle actin-containing filaments with phalloidin and found that the muscle I bands are disorganized as compared with wild-type muscles (Fig. 1, A and B). Staining with myosin antibody demonstrated the disorganization of the muscle A bands (Fig. 1, C and D). Thus, the unc-95 gene affects both the thin and thick filaments. In the C. elegans body wall muscles, the anchorage of the sarcomeres to the plasma membrane is mediated by the dense bodies and M-lines. To analyze the integrity of the muscle dense bodies, immunofluorescence was performed on adult animals with the DEB-1/vinculin mAb. The staining revealed that the unc-95(su33) mutant animals have disorganized dense bodies (Fig. 1, E and F). In wild-type muscles, the vinculin staining is in rows of discrete spots and also in the cell–cell boundaries, whereas in the mutant animals the staining forms a disorganized pattern of stripes that are mostly not parallel to one another. Staining of adult animals with PAT-3/β-integrin antibody, which is localized both to dense bodies and M-lines, shows a similar disorganized pattern (Fig. 1, G and H). To further characterize the structural aberrations of the su33 allele, we performed EM on the mutant adult animals (Fig. 2). The EM images demonstrate that the entire sarcomeric structure is aberrant. The dense bodies are impaired with variable, irregular shapes. The spaces between the dense body structures appear to be random and vary between the different sarcomeres. The M-line structures could not be recognized. As shown in Fig. 1 by phalloidin and anti-myosin staining, the EM images reveal that both the thin and thick filaments are not normally organized. In addition, the I band region of thin filaments surrounding the dense bodies (Fig. 2 A, asterisk) is not recognized in the unc-95(su33) mutant. Schematic model of the aberrant muscle structure in unc-95(su33) mutant compared with the wild type is shown in Fig. 2 C.
Figure 1.
Analysis of muscle structure in unc-95(su33) mutant adult animals. Left column shows wild-type staining; right column shows unc-95(su33) staining. (A and B) Actin filament organization in body wall muscles visualized by staining with phalloidin. (C and D) Anti-myosin staining of body wall muscles. (E and F) Anti-DEB-1/vinculin staining of muscle dense bodies and muscle–muscle cell boundaries. Short arrow indicates the muscle dense bodies. Long arrow indicates the muscle–muscle cell boundaries. (G and H) Anti PAT-3/β-integrin staining of dense bodies, M-lines, and muscle–muscle cell boundaries. Short and long arrows are as in E. Arrowhead indicates the muscle M-lines. It is not possible to distinguish between the dense bodies and M-line structures in the unc-95(su33) mutant. Bar, 10 μm.
Figure 2.
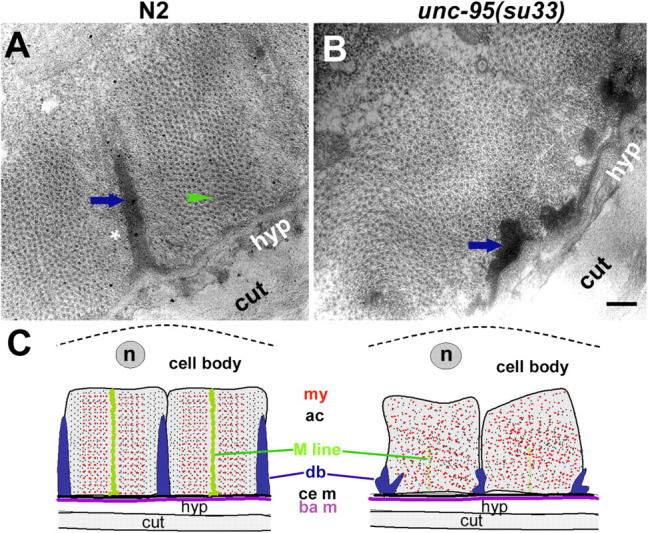
EM of unc-95(su33) adult animals. EM of body wall muscles from wild-type (A) and unc-95(su33) mutant (B). Shown are cross sections of the myofilament lattice; thin and thick filaments appear as dots. Arrows indicate the dense body, arrowheads indicate the M-line, and the asterisk indicates the I band region of thin filaments. Bar, 200 nm. (C) Schematic diagram of cross section of the body wall muscle of wild type and the unc-95(su33) mutant based on the EM data. The body wall muscle cells are associated through the basement membrane (ba m) to the hypodermis (hyp) and cuticle (cut). Dense bodies (db) and M-lines project from the cell membrane (ce m) into the cell and attach actin-containing thin filaments (ac) and myosin-containing thick filaments (my), respectively, to the cell membrane (ce m). The muscle cell body is shown above the sarcomeres including the nucleus (n). The disrupted dense bodies and M-lines of the mutant cause the disorganization of the sarcomeres in the unc-95(su33) mutant.
unc-95 encodes a LIM domain protein
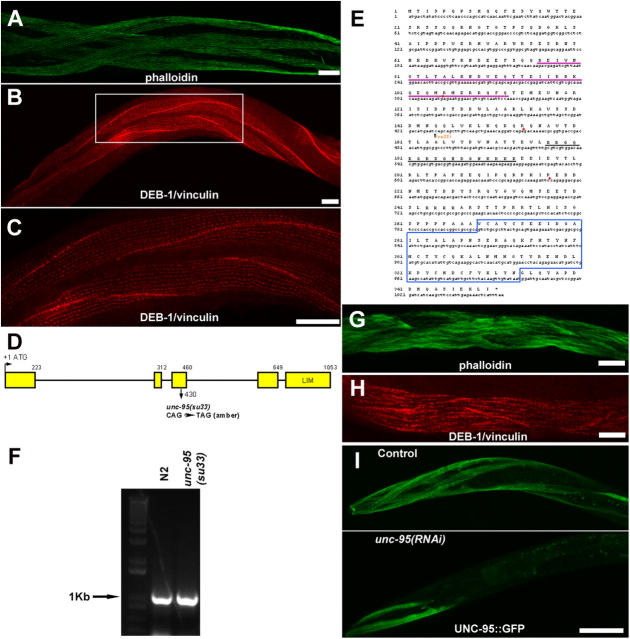
We originally identified the putative protein Y105E8A.6 as an interactor of RNF-5 in the yeast two-hybrid system. On the basis of this result, we hypothesized that this LIM domain protein may be a specific substrate for the E3 ligase activity of RNF-5. A candidate gene encoding for this protein was unc-95 (see Materials and methods). We sequenced Y105E8A.6 in unc-95(su33) mutant background and revealed an amber mutation (CAG→TAG; see Zengel and Epstein, 1980 regarding the type of mutation) in aa 144 (Glutamine→Stop), predicting the translation of a shorter protein without the LIM domain (Fig. 3, D and E). A GFP translational fusion construct including 2.5 Kb upstream sequences and the genomic sequence of Y105E8A.6 including all exons and introns fully rescued the unc-95(su33) muscle phenotype. Rescued animals exhibited normal motility. Muscle structure was assayed by phalloidin and DEB-1/vinculin staining of transgenically rescued strains (n = 4) that are homozygous for the unc-95(su33) allele and carry a rol reporter (i.e., animals carrying this dominant reporter are right-handed rollers; Fig. 3, A–C). To check if a transcript of unc-95 is detected in the unc-95(su33) mutant animals, we performed RT-PCR analysis. A single product of 1 Kb corresponding to the full-length unc-95 cDNA was observed in both wild type (N2) and unc-95(su33) mutant (Fig. 1 F), suggesting that the unc-95 mRNA in the mutant background is not completely degraded by the system of nonsense-mediated mRNA decay (Pulak and Anderson, 1993). It also suggests that a truncated inactive protein is translated in these animals.
Figure 3.
unc-95 encodes a LIM domain protein. (A and B) Rescue of unc-95(su33) mutant animals with the UNC-95::GFP translation fusion construct. Phalloidin staining (A), anti DEB-1/vinculin staining (B), and enlargement of inset in B (C). Use of the rol reporter in the rescue experiments enables the identification of the rescued animals. Bars, 10 μm. (D) Structure of the unc-95 gene (Y105E8A.6). Exons are in yellow. (E) The sequence of unc-95 cDNA and protein. Note the mutation in the su33 allele, the LIM domain (marked in blue) at the COOH terminus, the coiled-coil domain (underlined in purple), and the putative NLS (underlined in black). Introns are labeled in red dots. The sequence data are available from GenBank/EMBL/DDBJ under accession no. NM_170938. Comparison of the LIM domain of UNC-95 to other LIM domain proteins in C. elegans revealed 35% homology to the fourth LIM domain of C28H8.6 (additional C. elegans homologue of vertebrate paxillin, which includes four LIM domains, but not the NH2-terminal extension characteristic of the paxillin family members), 30% homology to the UNC-97 LIM domains, and 30% homology to the predicted protein MLP-1 (T04C9.4b), which includes only one LIM domain and belongs to the MLP/CRP family of muscle LIM domain proteins. (F) RT-PCR analysis of mixed stage RNA purified from N2 and unc-95(su33) cultures. The full-length unc-95 cDNA is 1053 bp. (G and H) unc-95(RNAi) analysis performed on wild-type animals. (G) Phalloidin staining. (H) Anti DEB-1/vinculin staining. Note the phenotype similarity to the su33 allele in Fig. 1 (B and F). Bars, 10 μm. (I) unc-95(RNAi) analysis performed on Ex[unc-95::GFP; rol-6] transgenic animals. Residual GFP expression in body wall muscle cells in the anterior region is shown. In >50% of the RNAi animals analyzed (n = 100), no GFP expression was detected. In all images anterior is to the left. Bars, 50 μm.
To confirm the molecular identity of unc-95 and to assess the nature of the su33 allele, we performed RNA interference (RNAi) experiments. This enabled us to characterize the loss-of-function phenotype of unc-95 obtained by decreased expression after injection of dsRNA. Wild-type hermaphrodites injected with Y105E8A.6 double-stranded RNA (dsRNA) produced viable progeny with reduced motility. No paralyzed arrest at embryonic two-fold stage (Pat) phenotype or other embryonic or larval lethality was observed. Staining of the unc-95(RNAi) progeny with phalloidin and DEB-1/vinculin antibody revealed a phenotype similar to the unc-95(su33) allele (Fig. 3, G and H; n > 100). As a control for the efficiency of the RNAi method, transgenic worms harboring the UNC-95::GFP translational fusion (see below) were injected with the same dsRNA. The level of GFP expression decreased dramatically in the progeny of the injected animals (Fig. 3 I). Thus, we confirmed that unc-95 encodes the Y105E8A.6 LIM domain protein. In addition, the high similarity between the RNAi phenotype and the unc-95(su33) mutant suggests that the su33 allele represents a strong loss-of-function phenotype.
The UNC-95 protein is a 350-aa LIM-only protein. It includes a single LIM domain at the COOH terminus that is highly homologous to the LIM domain of Drosophila and vertebrate focal adhesion protein, paxillin (for review see Turner, 2000). High homology was detected specifically to LIM1 (30%) and LIM4 (43%) of human paxillin. The LIM domain of UNC-95 is located at the COOH terminus, as in the paxillin family proteins. The UNC-95 sequence also contains a predicted coiled-coil motif characteristic of muscle proteins and NLS (Fig. 3 E). Comparison with the predicted UNC-95 protein sequence from the closely related species Caenorhabditis briggsae revealed complete conservation of the LIM domain and 97% homology along the entire protein sequence. Blast search of the genome did not identify clear UNC-95 relatives in C. elegans.
UNC-95 is required for the initial assembly of muscle attachment sites
To determine whether UNC-95 is required for the assembly of muscle attachment sites (dense bodies and M-lines) during embryogenesis, we stained homozygous unc-95(su33) embryos with mAbs to UNC-52/perlecan, PAT-3/β-integrin, and DEB-1/vinculin. Staining of unc-95(su33) embryos with UNC-52/perlecan antibody demonstrated that perlecan is concentrated in the muscle basal lamina in a pattern similar to that of wild-type embryos (Fig. 4, A–F). The MH27 antibody for the hypodermal cell junction protein, AJM-1 (Francis and Waterston, 1991) was used to follow the body wall muscles in the context of the embryo shape. Staining with the PAT-3/β-integrin antibody revealed that UNC-95 is not required for the deposition of integrin in the muscle attachment sites as well, and the pattern is very similar to the wild-type pattern. At the 1.5-fold stage, staining is detected as a band along the anterior–posterior axis of the embryo, whereas at the two- and threefold embryos the structural elements of the muscle (dense bodies and M-lines) can be detected in both wild-type and unc-95(su33) embryos (Fig. 4, G–L). Only after hatching does the organization of β-integrin begin to deteriorate, as shown by the disorganized pattern of staining in the already mature but defective dense bodies (Fig. 4, M and N). In contrast, the dense body protein DEB-1/vinculin is not recruited to the nascent muscle attachments. In 1.5-fold and twofold embryos, staining is much more diffuse than in the wild-type embryos. At the threefold stage, DEB-1/vinculin staining remains diffuse and does not form the regular pattern of the mature dense bodies as shown in the wild-type threefold embryos (Fig. 4, O–T; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200401133/DC1). Therefore, during embryonic muscle development UNC-95 is neither required for the normal localization of UNC-52/perlecan in the muscle basement membrane nor for the recruitment of β-integrin to the basal sarcolemma and the formation of nascent muscle attachments. However, UNC-95 is necessary for the subsequent recruitment of vinculin to these focal adhesion-like sites.
Figure 4.
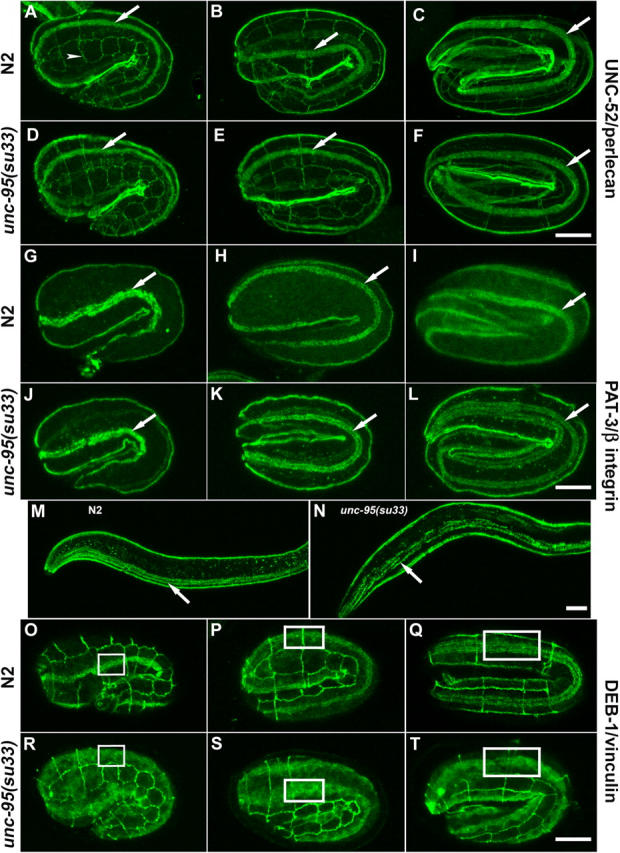
Muscle assembly during embryogenesis of unc-95(su33) animals. (A–F) Anti-UNC-52/perlecan and MH27 (arrowhead indicates hypodermal cells junctions) staining of wild-type (A–C) and unc-95(su33) (D–F) in 1.5-, 2-, and 3-fold embryos (left, middle, and right panels, respectively). (G–L) Anti-PAT-3/β-integrin staining of wild type (G–I) and unc-95(su33) (J–L). Arrows indicate the pattern of staining in one body wall muscle quadrant. The images show a lateral or dorso-lateral view of the embryo. (M) Wild-type and unc-95(su33) (N) L1 larvae stained with anti-PAT-3/β-integrin. Note the random distribution of dots and lines associated with the muscle dense bodies and M-lines at the L1 stage. (O–T) Anti-DEB-1/vinculin and MH27 staining of wild-type (O–Q) and unc-95(su33) embryos (R–T). Note the disorganized staining and the cytoplasmic accumulation of vinculin in the mutant embryos. Insets are used to indicate the pattern of staining in one muscle quadrant and to highlight the difference in organization between the wild-type and unc-95(su33) mutant embryo. All panels show lateral view of the embryo. Bars, 10 μm.
UNC-95 is expressed in the developing muscle during embryogenesis and in body wall, vulval, and anal muscle cells in larvae
To determine the expression pattern of unc-95, we first made a transcriptional GFP fusion, including 2.5 Kb of the Y105E8A.6 upstream sequences. Transgenic lines (n = 4) showed strong GFP expression in body wall muscles, vulval muscles, and the anal muscles (Fig. 5 A). Next, we analyzed developmental expression and cellular localization using GFP translation fusion (n = 6 independent lines analyzed showed a similar pattern of expression). The translation fusion construct was the same construct used to rescue unc-95(su33). We observed strong GFP expression at the embryonic stage ∼350 min after the first cell division (Fig. 5 B, comma). The staining is dispersed in the muscle cell cytoplasm as was also shown for vinculin and integrin at this stage of muscle development, when muscle cell precursors are positioned in lateral rows adjacent to the seam cells (Hresko et al., 1994). The staining gradually becomes more localized at the membrane until the muscles are fully positioned into quadrants and the expression is shown in two separate parallel stripes (Fig. 5, C and D; 1.5- and 2-fold). Later on the expression becomes localized to the discrete spots of the newly formed muscle dense bodies, but through all stages GFP is also present in the cytoplasm (Fig. 5 E, threefold). Nuclear localization of UNC-95 could be clearly observed from the threefold stage (Fig. 5 E). In adult animals, UNC-95 is expressed in body wall muscles in the dense bodies, M-lines, and muscle–muscle cell boundaries (Fig. 5 F). Strong expression is also detectable in muscle cell cytoplasm and nucleus (Fig. 5 G). From the head muscle cells we could detect the muscle arms (extensions that form the neuromuscular junctions) that extend near the back of the pharynx to reach the nerve ring (Fig. 5 G, arrow). In the vulval muscles the protein is strongly localized at sites of muscle attachment to the hypodermis (Fig. 5 H). Expression was also detected in the anal depressor muscle (Fig. 5 I). A bright ring at the tip of the buccal cavity (shown in Fig. 7 and Fig. 8) seems to be the attachment between the arcade cells to the hypodermal anterior hyp1 cell, the pharyngeal epithelium, and the associated cuticles. Thus, in different expressing cells UNC-95 is mainly localized to the cellular attachment sites between the cell membrane and the ECM.
Figure 5.
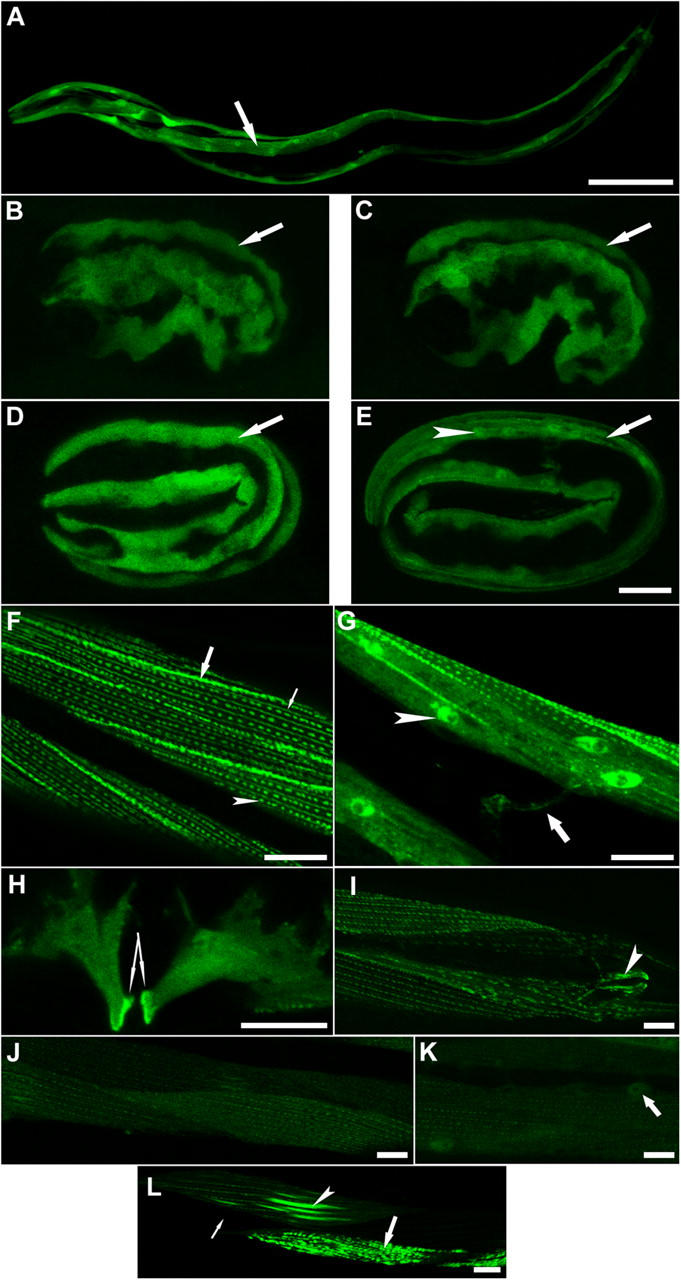
UNC-95 is expressed in muscle attachment sites. (A) Expression of the transcriptional fusion reporter including 2.5 Kb of unc-95 upstream sequences. Arrow points at one of the four body wall muscle quadrants. Bar, 50 μm (B–E). Expression of the translational fusion UNC-95::GFP in developing embryos. The cytoplasmic expression is maintained through embryogenesis. Localization to the muscle attachment sites is shown in E. Arrow indicates one muscle quadrant. Arrowhead indicates the nucleus. Lateral view of the embryo is shown. Bars, 10 μm. (F and G) Expression of the translational fusion UNC-95::GFP in the body wall muscles of adults. (F) UNC-95 is expressed in muscle dense bodies (arrowhead), in M-lines (thin arrow), and in muscle–muscle cell boundaries (thick arrow). (G) UNC-95 localization in the nucleus and cytoplasm of body wall muscles. Arrowhead indicates the nucleus. Expression is also detected in muscle arms (arrow). (H) UNC-95 is localized in the vulval muscles and is highly concentrated in the sites of muscle attachments to the hypodermis (two arrows indicate the strong GFP labeling in the region of attachment). (I) Localization to the anal depressor muscle (arrowhead). (J–L) Expression of the mutant protein unc-95(su33)::GFP translational fusion. (J) Weak staining could be detected in dense bodies and M-lines, but there is no staining in muscle–muscle cell boundaries. (K) Localization of the unc-95(su33)::GFP is maintained in nuclei (arrow). (L) Irregular pattern of staining of the unc-95(su33)::GFP translational fusion. Thin arrow indicates cell in which the transgene was silenced; arrowhead indicates cell in which the M-lines are much thicker; thick arrow indicates cell in which the dense bodies aggregate to abnormal structures. Bars, 10 μm.
Figure 7.
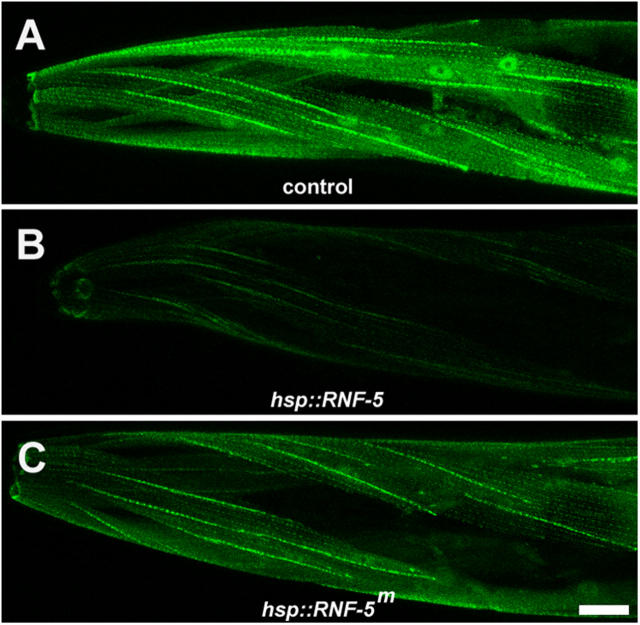
Ectopic expression of RNF-5 in the UNC-95::GFP transgenic animals affects GFP expression levels. (A) Control level of expression of the translational fusion UNC-95::GFP. (B) Overexpression of wild-type RNF-5 under the heat shock promoter. UNC-95::GFP levels are lower compared with control. (t test; P < 0.005, n = 10; based on measurement of GFP fluorescence). (C) Overexpression of a RING mutant form of RNF-5 (RNF-5m) under the heat shock promoter. Note weaker effect of the mutant form on GFP levels (t test; P < 0.05, n = 10; based on measurement of GFP fluorescence). The representative images shown were captured using the same confocal parameters; all panels were enhanced in order to enable the detection of the expression pattern in B. Bar, 10 μm.
Figure 8.
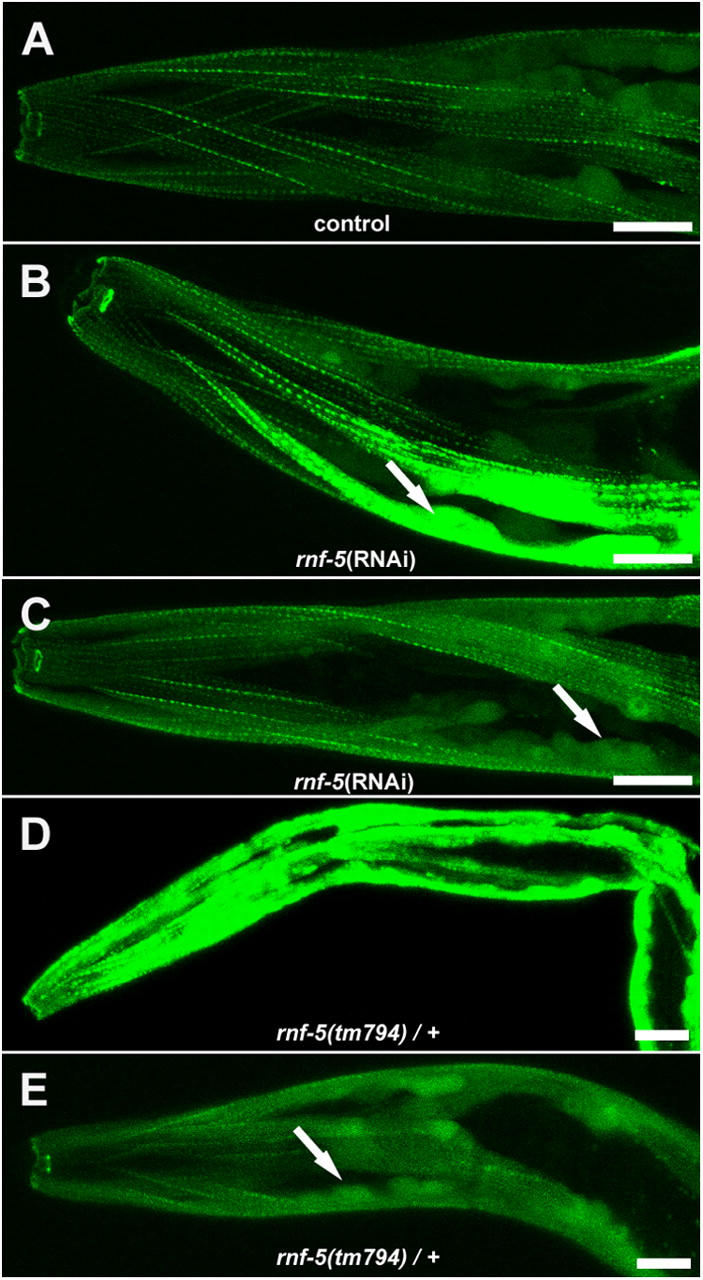
rnf-5(RNAi) and rnf-5(tm794) heterozygous caused accumulation of the UNC-95::GFP transgene in muscle cell bodies. (A) Control level of expression of the translational fusion UNC-95::GFP. (B and C) Two representative images of rnf-5(RNAi) animals. The muscle cell body is indicated by an arrow. Note increased GFP expression, especially in muscle cell body. (D and E) Two representative images of rnf-5(tm794) heterozygous animals. Arrow as in C. All images were captured using the same confocal parameters. Bars, 10 μm.
Based on sequence analysis supported by RT-PCR experiments (Fig. 3, E and F), a shorter protein is predicted to be expressed in the unc-95(su33) mutant. To better characterize the strong loss-of-function phenotype of this allele, we next analyzed the pattern of expression of this putative truncated protein. We made transgenic lines (n = 4) in the wild-type N2 background, harboring a shorter GFP fusion protein including only the first 144 amino acids of UNC-95, which corresponds to the predicted su33 allele, Ex[unc-95(su33)::GFP; rol-6]. Low GFP expression was detected in the dense bodies and M-lines of body wall muscles; however, there was no expression in the muscle–muscle cell boundaries (Fig. 5, J and K). Weak expression was also detected in muscle nuclei (Fig. 5 K). In addition, the expression was not equal along the body wall muscle cells and appeared in a mosaic pattern. In several cells (observed in all lines), the protein accumulated to very high levels that were not observed in the wild-type UNC-95 construct. In those cells, the organization of the M-lines and dense bodies was disrupted, and these structures appeared as intense stripes or aggregates (Fig. 5 L). On the other hand, additional surrounding cells had no GFP expression at all (Fig. 5 L).
RNF-5 colocalizes with UNC-95 in muscle dense bodies
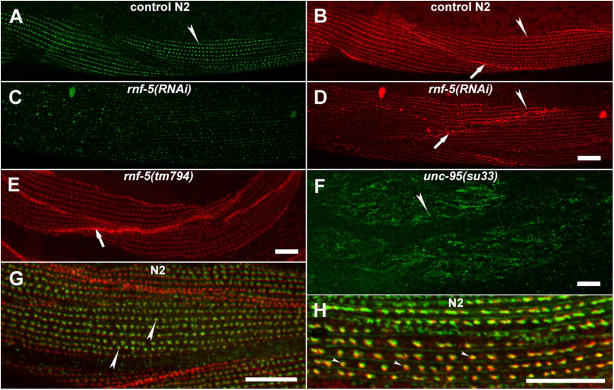
Immunostaining with a pAb that we raised against RNF-5 revealed its expression in the larval muscle dense bodies (Fig. 6 A), the apical cell junctions of the spermatheca (septate junctions), and the junctions between the gonadal sheath cells and the gonadal basal lamina (unpublished data). During embryogenesis, RNF-5 is not expressed specifically in the developing muscle and is localized to the nucleus of most of the embryo cells (unpublished data). The interaction between RNF-5 and UNC-95 identified by the yeast two-hybrid system (Didier et al., 2003), together with their similar pattern of expression in muscle dense bodies suggest that RNF-5 may have a regulatory role in the muscle and that UNC-95 is likely to be one of its substrates. To test this hypothesis, we first performed rnf-5(RNAi) analysis on N2 animals. Motility and viability of the RNAi animals were not affected. However, when the dense body structure was analyzed in adult animals using the DEB-1/vinculin antibody, we found that the dense bodies are not organized in linear stripes as in control animals; in addition, the cell–cell boundaries were abnormal (Fig. 6, A–D; n > 100). Next, we analyzed the motility and muscle structure of the RNF-5 RING-deleted mutant rnf-5(tm794) that is expected to express the RNF-5 form that lacks the E3 ligase activity (see Materials and methods). Although the motility of the mutant is normal, an irregular pattern of staining of the DEB-1/vinculin antibody was detected, especially in the muscle–muscle cell contacts. The contacts are thicker than normal, suggesting higher levels of vinculin accumulation at these sites (Fig. 6 E; n > 100).
Figure 6.
RNF-5 has a role in the maintenance of muscle dense bodies. (A–D) rnf-5(RNAi) analysis. (A and B) Control animals double stained with the anti-RNF-5 (A; green) and anti-DEB-1/vinculin (B; red) antibodies. (C and D) rnf-5(RNAi) animals. Note weak staining with the RNF-5 antibody in C, demonstrating that the RNAi treatment indeed depleted the endogenous RNF-5. Note the aberrant structure of dense bodies and muscle–muscle cell boundaries in (D). (E) Staining of the rnf-5(tm794) homozygous animals with anti-DEB-1/vinculin antibody. (F) Staining of unc-95(su33) adult muscle with anti-RNF-5 antibody. Note the aberrant dense body structure as compared with A. (G) RNF-5 (green) and DEB-1/vinculin (red) are localized in close proximity in the muscle dense bodies. (H) Partial colocalization (yellow) of RNF-5 (red) with GFP (green) of the UNC-95::GFP expression construct in muscle dense bodies. Arrowheads indicate the dense bodies. Arrows indicate muscle–muscle cell boundaries. Bars, 10 μm.
Immunostaining of the unc-95(su33) mutant animals with the RNF-5 antibody demonstrates that RNF-5 is recruited to the aberrant dense body structures of the adult mutant (Fig. 6 F). Thus, loss of function of UNC-95 does not interfere with the cellular localization and recruitment of RNF-5 to the larval muscle attachment sites, even if those structures are not properly assembled.
Next, we analyzed the relative localization of RNF-5, UNC-95, and DEB-1/vinculin in the muscle dense bodies. RNF-5 does not colocalize with DEB-1/vinculin at the muscle dense bodies (Fig. 6 G). However, staining of the transgenic lines harboring the extrachromosomal array of the UNC-95::GFP translation fusion construct with both anti-GFP and anti-RNF-5 antibodies revealed that RNF-5 and UNC-95 are partially colocalized (Fig. 6 H, yellow region between the red and green area), and thus may be part of the same protein complexes within the muscle attachment sites.
RNF-5 regulates UNC-95 levels and subcellular localization
To determine whether UNC-95 level of expression is affected by RNF-5 E3 ligase activity, we analyzed the pattern of UNC-95::GFP (translational fusion construct) expression when RNF-5 was overexpressed under a heat shock promoter. When the wild-type form of RNF-5 was overexpressed in adult animals, we detected a marked decrease in the intensity of GFP labeling after heat shock, especially in the muscle cell body, as compared with control animals (Fig. 7, A and B; n > 100; see Materials and methods). However, upon overexpression of RNF-5, which harbors a mutation within the RING finger domain (see Materials and methods), there was a less pronounced effect on the GFP levels (Fig. 7 C; n > 40). These findings suggest that RNF-5 affects the stability of UNC-95 and requires an intact RING domain for this activity. These observations are in line with our finding that the RING finger domain is necessary for the E3 ligase activity of RNF-5 (Didier et al., 2003; unpublished data).
Next, we determined the expression pattern of the translational fusion UNC-95::GFP upon reduced RNF-5 expression using the RNAi method. We detected a major increase in GFP levels, especially in the muscle cells bodies (that include the nuclei) of the RNAi animals (Fig. 8, A–C; n = 55/85). Such a pattern of expression could not be detected in control animals (n > 100). In parallel, crossing the RING finger–deleted mutant rnf-5(tm794) to the strain carrying the UNC-95::GFP extrachromosomal array resulted in a very high level of GFP expression in the heterozygous animals (see Materials and methods), which is mainly localized to the muscle cell body (Fig. 8, D and E; n = 60/60). Moreover, the homozygous rnf-5(tm794) mutant harboring the UNC-95::GFP transgene is zygotic or early embryonic lethal. In addition, when analyzing the strain carrying the UNC-95::GFP extrachromosomal array in the wild-type background, we observed low levels of early larval arrest (6.2 ± 1.3%; the progeny of n = 4 hermaphrodites were scored), suggesting that high levels of UNC-95 may cause lethality. Collectively, these data suggest that RNF-5 regulates UNC-95 levels and localization in the muscle. When RNF-5 is overexpressed using the hsp::RNF-5 construct, it causes a reduction in UNC-95::GFP levels. However, either depletion of RNF-5 or a mutant lacking the RNF-5 RING finger domain result in the accumulation of UNC-95, especially in the muscle cell body. These data also suggest that the high levels of UNC-95::GFP transgene expression lead to lethality in the rnf-5(tm794) homozygous background.
Discussion
Role of UNC-95 in sarcomere organization
The initial deposition of UNC-52/perlecan in the basal lamina and PAT-3/β-integrin accumulation in the basal sarcolemma occurs normally in unc-95(su33) mutant embryos. The pattern of integrin staining confirms the primary formation of the structural elements of the muscle. However, the recruitment of DEB-1/vinculin to these nascent attachments sites requires UNC-95. Thus, UNC-95 is not necessary for the initiation of the assembly complex, but for downstream processes of sarcomere organization. We assume that in addition to DEB-1/vinculin, UNC-95 is required for the recruitment of additional cytoplasmic factors during embryonic muscle formation because not only the dense bodies, but also the M-lines, which do not contain vinculin, are disorganized. The finding that both the I and A bands are disrupted/disorganized in unc-95(su33) mutant and unc-95(RNAi) animals suggests that UNC-95 has a role in both the dense bodies and M-line assembly pathways (Lin et al., 2003).
It is interesting that unc-95(su33) and unc-95(RNAi) animals (likely null phenotype) are not embryonic lethal, exhibiting the Pat phenotype, but develop into adults with reduced motility and some degree of paralysis. The Pat phenotype is characteristic of mutants in dense bodies and M-line components (Williams and Waterston, 1994; Moerman and Fire, 1997). We suggest that the reason is the capability of the actin- and myosin-containing filaments to be recruited to the aberrant dense bodies and M-lines of the mutant. Although the A and I bands are aberrant, they do assemble into the myofilament lattice, which contains linkages to the basement membrane that are stable enough to proceed past the twofold stage, complete elongation, and allow larval development. It is possible that other muscle LIM domain proteins (for example UNC-97) contribute either redundantly or additively and function as molecular adaptors to enable the recruitment of essential structural components as vinculin to the muscle adhesion sites, and in this way substitute for the lack of UNC-95 activity.
UNC-95 belongs to the LIM-only protein group
A central role in the assembly of multiprotein complexes at mammalian focal adhesions in cultured cells was shown for LIM-only proteins such as paxillin, zyxin, enigma, and others. These proteins are primarily cytoplasmic and are associated with the actin cytoskeleton (Dawid et al., 1998; for review see Turner, 2000). UNC-95 is a LIM-only protein and may similarly function as an adaptor protein, allowing the recruitment of various structural and signaling molecules to the nematode muscle focal adhesion–like structures. This possibility is supported by the analysis of transgenic animals harboring the UNC-95::GFP translational fusion construct, which is expressed not only in the dense bodies and M-lines, but also in the muscle cell body. The cytoplasmic and nuclear localization of UNC-95 suggests that it shuttles between the cytoplasm, nucleus, and attachment sites, and by its association with specific proteins may affect both structural and signaling processes. Shuttling between cytoplasmic and nuclear compartments was shown for mammalian focal adhesion LIM domain proteins such as paxillin, zyxin, and Hic5, and is thought to facilitate signaling between adhesion sites and the nucleus (for review see Wang and Gilmore, 2003). In parallel to the focal adhesion proteins, a common feature of a variety of LIM domain proteins that have a role in muscle differentiation or assembly is their bimodal subcellular distribution. Examples are the muscle LIM-proteins of the CRP/MLP/TLP family (for review see Weiskirchen and Gunther, 2003), and the four-and-a-half LIM (FHL) proteins (Li et al., 2001). UNC-95 may also have a dual role in the nematode body wall muscles. In C. elegans muscle, UNC-97 (PINCH) and UNC-98 (zinc finger) are required for the assembly of muscle attachment sites and, like UNC-95, are localized to the nucleus in addition to muscle attachment sites (Hobert et al., 1999; Mercer et al., 2003). Because UNC-95, -97, and -98 exhibit a similar pattern of expression, it is possible that they interact to form a multidomain adaptor complex.
Using the COOH-terminal truncated form of UNC-95 in the translation fusion unc-95(su33)::GFP, which lacks the LIM domain, we demonstrated that the LIM domain of UNC-95 is not required for the primary localization of the truncated protein to the dense bodies and M-lines, but for its accumulation in these sites. This is in contrast to paxillin localization in focal adhesions, which requires the LIM domain (for review see Turner, 2000). The truncated unc-95(su33)::GFP construct enabled nuclear localization, although the putative NLS (aa 177–193) is not included. Most important, the truncated transgene was not expressed in muscle cell boundaries, which are the regions of contact between adjacent muscle cells. Thus, the COOH-terminal region of UNC-95 (from aa 144), which includes the LIM domain, is essential for the localization of UNC-95 to the muscle–muscle cell boundaries. This suggests that additional signaling pathways and protein interactions are involved in the recruitment of UNC-95 to the cell–cell contacts. The “mosaic-like” pattern of expression of the su33::GFP allele could be a dominant-negative effect caused by overexpression of the mutant form of UNC-95 in the wild-type background expressing the wild-type UNC-95 protein.
It was shown previously that paxillin binds to vinculin through its LD motifs (Turner, 2000). It is possible that UNC-95 also binds vinculin directly to recruit it to the dense bodies and muscle–muscle cell boundaries and that the vinculin-binding domain lies in the COOH-terminal part of the UNC-95 protein, which is missing in the unc-95(su33) mutant embryo. Along these lines, aa 234–265 of UNC-95 show high similarity to the LD4 region of vertebrate paxillin (see alignment in Didier et al., 2003).
RNF-5 and UNC-95
RNF-5 is detected in muscle dense bodies at the L1 larval stage. This observation suggests that RNF-5 is not required for the assembly of the I bands in the body wall muscle cells, but probably for subsequent regulatory events that maintain and stabilize the thin filaments within the I bands during contraction of the mature muscle.
Overexpression of wild-type RNF-5 through the heat shock promoter caused a marked decrease in the UNC-95::GFP expression levels. This decrease was detected as soon as 6–8 h following induction of RNF-5, which is not likely to be a result of transcriptional repression of the transgene. Therefore, we suggest that RNF-5 plays a key role in the regulation of UNC-95 stability in the mature muscle. Accumulation of UNC-95 (as shown when RNF-5 was depleted) may induce inadvertent signaling pathways and cause damage to the muscle. This is prevented by the specific E3 ligase activity of RNF-5 that limits the levels of UNC-95 in the mature muscle. In addition, an RNF-5 RING mutant exhibits a substantially weaker effect on the UNC-95::GFP expression. In accordance with this observation, we demonstrated earlier that mutation within the conserved cysteines in the RING finger domain or RNF-5 that is deleted of its RING domain were no longer capable of exhibiting E3 ligase activity (Didier et al., 2003), further indicating that the RING finger domain is required for RNF-5 ability to elicit its E3 ligase activity. Along these lines, rnf-5(RNAi) or the RING-deleted mutant rnf-5(tm794) caused a major increase in the level of UNC-95::GFP predominantly in the muscle cell body, and lethality in the rnf-5(tm794) homozygous background. Of interest to note is that considerable efforts to detect association between RNF-5 and UNC-95 did not succeed, either due to the localization of these proteins within insoluble membrane fractions, which results in their dissociation by methods used for extraction, or because RNF-5 mediates its effect on UNC-95 via an intermediate protein/ligase, as commonly seen for RING finger proteins. Consistent with this observation is the weak association seen for RNF-5 with paxillin in vitro (Didier et al., 2003).
The COOH-terminal tail of the Arabidopsis RNF-5 has been implicated in membrane association (Matsuda and Nakano, 1998). In addition, the COOH-terminal tail of human RNF-5 was shown to be required for paxillin ubiquitination (Didier et al., 2003). Together with the confocal analysis presented in this paper, it is likely that RNF-5 is localized through its COOH terminus to the muscle cell membrane at the base of the dense bodies where it regulates UNC-95 localization and stability. Although UNC-95 is expressed both in the muscle attachment sites and the muscle cell body, we suggest that RNF-5 regulates the levels and distribution of UNC-95 between these muscle cell compartments.
Finally, because the homology of UNC-95 to paxillin is limited to the LIM domain and the LD4 region, it is possible that RNF-5 regulates the stability and localization of additional LIM/LD domain proteins that participate in the assembly of muscle attachment sites in C. elegans.
Materials and methods
Strains and genetics
The following strains were used: N2 wild-type strain, unc-95(su33) (Zengel and Epstein, 1980), and rnf-5(tm794) (deletion mutant isolated by the Mitani laboratory, Tokyo Women's Medical University, Tokyo, Japan). The deleted region includes exon 2 of rnf-5, which encodes the RING finger domain and transgenic strains Ex[Punc-95::GFP; rol-6], Ex[unc-95::GFP; rol-6], Ex[unc-95(su33)::GFP; rol-6], Ex[hsp::RNF-5; rol-6], Ex[hsp::RNF-5C26C29R; rol-6], and rnf-5(tm794); Ex[unc-95::GFP; rol-6] (this paper). Heterozygosity to the deletion in the rnf-5(tm794); Ex[unc-95::GFP; rol-6] strain was determined by PCR performed after fluorescence or confocal microscopy analysis.
unc-95(su33) was mapped by Zengel and Epstein (1980) to linkage group I and appeared to be near and to the left of the unc-54 gene. The cosmid Y105E8A maps to this area and contains a predicted LIM domain protein, Y105E8A.6, identified by the C. elegans genome sequencing consortium. The complete gene structure (exon–intron) was determined by RT-PCR. The unc-95 gene mRNA includes 1,053 nucleotides and is encoded by five exons.
The unc-95(su33) allele is affected by temperature. The decrease in motility and paralysis is less severe in animals raised in 15°C compared with animals raised at 20°C.
Rescue of the unc-95(su33) mutant phenotype was performed by mating males carrying the extrachromosomal array Ex[unc-95::GFP; rol-6] with the unc-95(su33) hermaphrodite. F2 fluorescent rol animals homozygous to the unc-95(su33) allele were analyzed. Homozygosity to unc-95(su33) was confirmed by the non-rol progeny, which all displayed the slow/paralyzed phenotype.
Electron microscopy
Transmission EM was performed as described by Hall (1995).
Sequence analysis
The unc-95(su33) amber mutation was identified by PCR amplification of the Y105E8A.6 sequence from DNA purified from unc-95(su33) mutant animals. Subsequent DNA sequencing of the amplified region was performed on the two strands. In addition, single worm PCR and DNA sequencing of five independent animals was performed to verify that this was indeed a molecular lesion and not an error introduced randomly by the PCR.
RT-PCR
RNA from N2 and unc-95(su33) mutant animals was purified from mixed stage cultures using the RNeasy kit (QIAGEN). Nonquantitative RT-PCR was performed using the OneStep RT-PCR kit with primers designed for the amplification of the full-length unc-95 cDNA.
Transcriptional and translational unc-95::GFP fusion constructs
The transcriptional GFP construct, Ex[Punc-95::GFP; rol-6], included a 2.5-Kb fragment upstream of the predicted initiator methionine of Y105E8A.6 fused to the GFP reporter that includes the unc-54 3′ untranslated region at the 3′ end (a gift from A. Fire, Stanford University, Stanford, CA). The translational GFP construct, Ex[unc-95::GFP; rol-6], which rescues the unc-95(su33) mutant phenotype, includes the same 2.5-Kb upstream sequences fused to the entire genomic sequence of Y105E8A.6 (unc-95), including all exons and introns and fused in the COOH terminus to the GFP reporter that includes the unc-54 3′ untranslated region at the 3′ end. The mutated construct that mimics the su33 allele, Ex[unc-95(su33)::GFP; rol-6], included the first three exons and two introns of Y105E8A.6 until position 429 and fused at the COOH terminus to the same GFP reporter. An irregular pattern of staining (as shown in Fig. 5 L) could be observed in few cells of the body wall muscle in most of the transgenic animals of this fusion construct in all lines generated (n = 4 independent lines).
The GFP reporter constructs were generated by PCR fusion according to Hobert (2002). The template was N2 (wild-type) genomic DNA. Transgenic arrays were constructed through DNA microinjection into the N2 background with the dominant marker pRF4/rol-6. The expression of the above constructs was monitored in live animals.
RNF-5 overexpression constructs
Full-length rnf-5 cDNA and the rnf-5 RING mutant were cloned into vectors containing the heat shock promoters hsp16-2 and hsp16-41 (Stringham et al., 1992). In the RING-mutated form of RNF-5, both cysteine 26 and cysteine 29, which are part of the RING finger domain (C3HC4 type), were mutated to arginine. This mutagenesis disrupts the structure of the RING. Each form was cloned into both vectors and the two vectors were mixed before injection. Transgenic lines were crossed to the Ex[unc-95::GFP; rol-6] strains. Two independent lines were constructed for each RNF-5 construct. Heat shock was performed for 1 h at 32°C. Five independent experiments were performed for the RNF-5 wild-type construct and in each experiment ∼20 animals were scored (n > 100). Three independent experiments were performed for the RNF-5 mutant construct (n > 40). Confocal analysis of live animals was done 6–8 h after heat shock. Images shown were chosen as representative examples. Two control experiments were performed: first, identical heat shock treatment and analysis of the Ex[unc-95::GFP; rol-6] parental strain (n > 50), and second, no heat shock of the examined strain Ex[unc-95::GFP; rol-6]; Ex[hsp::rnf-5; rol-6] (n > 100; served as the control group for t test). Measurement of GFP fluorescence (mean pixel values) was performed using the Laser Sharp processing program (Bio-Rad Laboratories) and the Confocal Assistant program on n = 10 animals of each group. t tests were performed on the GFP fluorescence values.
RNAi
For RNAi experiments (Fire et al., 1998) of the unc-95 gene, we used the first 600 bp of the unc-95 cDNA, which does not include the conserved LIM domain. The gene-specific primers included T7 and Sp6 sites at their 5′ end and were used for PCR from the unc-95 cDNA. RNA was synthesized from the amplified fragment with either T7 or Sp6 RNA polymerase (Riboprobe®; Promega). Equal amounts of sense and antisense RNA were annealed to create dsRNA. dsRNA was injected into the gonad of wild-type hermaphrodites (n = 40) or to the Ex[unc-95::GFP; rol-6] transgenic animals (n = 10) at a concentration of 0.5–1.0 mg/ml. Scoring of the phenotype was done by allowing single injected animals to lay eggs at 20°C starting at 24 h after injection and analysis of the progeny (n > 100). For RNAi experiments of the rnf-5 gene, we used the spliced form that does not contain the RING domain. dsRNA was synthesized as above and injected to wild-type animals or to the Ex[unc-95::GFP; rol-6] transgenic animals. At least five independent experiments (n = 20 injected animals per experiment) were performed for each strain. GFP pattern of expression was analyzed in the progeny (n = 85). Images shown are representative examples.
Antibody staining and confocal microscopy
Fixation of larvae and adult animals for antibody staining was performed as described by Finney and Ruvkun (1990). Permeabilization of embryos was done by freeze-cracking and fixation by methanol acetone. Incubation times were overnight at 4°C with the primary antibodies, and 2 h at RT with the secondary antibodies. Antibodies used were: monoclonal mouse anti-vinculin antibody MH24 (used at 1:150; Francis and Waterston, 1985), monoclonal mouse anti-β-integrin antibody MH25 (1:100; Francis and Waterston, 1985), monoclonal mouse anti-perlecan antibody MH2 (1:100; Francis and Waterston, 1985), monoclonal mouse anti-myosin myoA (1:100; Miller et al., 1983), monoclonal mouse anti-GFP antibody (1:100; MBL International Corporation), and rabbit polyclonal anti-RNF-5 antibody (1:150; this paper).
Staining of the rnf-5(RNAi) animals and rnf-5(tm794) was repeated three independent times in parallel to staining of wild-type animals in order to verify that the moderate phenotype observed was not a technical artifact.
Confocal microscopy images were captured as a stacked series using a confocal scanning microscope (MRC 1024; Bio-Rad Laboratories) as described in Kolotuev and Podbilewicz (2004), and were processed using the Confocal Assistant (CAS) program and Adobe Photoshop®.
Online supplemental material
Online supplemental material contains enlarged insets from Fig. 4 (Q and T), and is available at http://www.jcb.org/cgi/content/full/jcb.200401133/DC1.
Acknowledgments
We thank the Caenorhabditis Genetic Center (University of Minnesota, Minneapolis, MN) for providing strains, the Mitani laboratory for isolation of the rnf-5(tm794) deletion mutant, Henry Epstein (Baylor College of Medicine, Houston, TX) for the anti-myoA antibody and discussion, Pamela Hoppe (Washington University School of Medicine, St. Louis, MO) for the MH24 and MH25 antibodies, and David Hall for helpful discussions. We thank Yehuda (Jojo) Yosef and Sagit Argaman for their professional help in EM. We also thank the members of the Ronai and Podbilewicz labs for discussions, and Gidi Shemer for critical reading of this manuscript.
Support by National Institutes of Health grant (CA97105 to Z. Ronai) and the Fund for Promotion of Research at the Technion (to B. Podbilewicz) is gratefully acknowledged.
The online version of this article includes supplemental material.
Abbreviations used in this paper: dsRNA, double-stranded RNA; Pat, paralyzed arrest at embryonic two-fold stage; RNAi, RNA interference.
References
- Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5–17. [DOI] [PubMed] [Google Scholar]
- Barstead, R.J., and R.H. Waterston. 1991. Vinculin is essential for muscle function in the nematode. J. Cell Biol. 114:715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian, G.M., T.L. Tinley, X. Tang, and M. Borodovsky. 1996. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 132:835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawid, I.B., J.J. Breen, and R. Toyama. 1998. LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet. 14:156–162. [DOI] [PubMed] [Google Scholar]
- Didier, C., L. Broday, A. Bhoumik, S. Israeli, S. Takahashi, K. Nakayama, S.M. Thomas, C.E. Turner, S. Henderson, H. Sabe, and Z. Ronai. 2003. RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 23:5331–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, S., K.L. Lorick, J.P. Jensen, and A.M. Weissman. 2003. RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin. Cancer Biol. 13:5–14. [DOI] [PubMed] [Google Scholar]
- Finney, M., and G. Ruvkun. 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 63:895–905. [DOI] [PubMed] [Google Scholar]
- Fire, A., S. Xu, M.K. Montgomery, S.A. Kostas, S.E. Driver, and C.C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391:806–811. [DOI] [PubMed] [Google Scholar]
- Francis, G.R., and R.H. Waterston. 1985. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101:1532–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis, G.R., and R.H. Waterston. 1991. Muscle cell attachment in Caenorhabditis elegans. J. Cell Biol. 114:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, B., and A. Bershadsky. 2001. Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol. 13:584–592. [DOI] [PubMed] [Google Scholar]
- Hall, D.H. 1995. Electron microscopy and three-dimensional image reconstruction. Methods Cell Biol. 48:395–436. [DOI] [PubMed] [Google Scholar]
- Hobert, O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 32:728–730. [DOI] [PubMed] [Google Scholar]
- Hobert, O., D.G. Moerman, K.A. Clark, M.C. Beckerle, and G. Ruvkun. 1999. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J. Cell Biol. 144:45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko, M.C., B.D. Williams, and R.H. Waterston. 1994. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotuev, I., and B. Podbilewicz. 2004. Pristionchus pacificus vulva formation: Polarized division, cell migration, cell fusion and evolution of invagination. Dev. Biol. 266:322–333. [DOI] [PubMed] [Google Scholar]
- Kyushiki, H., Y. Kuga, M. Suzuki, E. Takahashi, and M. Horie. 1997. Cloning, expression and mapping of a novel RING-finger gene (RNF5), a human homologue of a putative zinc-finger gene from Caenorhabditis elegans. Cytogenet. Cell Genet. 79:114–117. [DOI] [PubMed] [Google Scholar]
- Labouesse, M., and E. Georges-Labouesse. 2003. Cell adhesion: parallels between vertebrate and invertebrate focal adhesions. Curr. Biol. 13:R528–R530. [DOI] [PubMed] [Google Scholar]
- Li, H.Y., E.K. Ng, S.M. Lee, M. Kotaka, S.K. Tsui, C.Y. Lee, K.P. Fung, and M.M. Waye. 2001. Protein-protein interaction of FHL3 with FHL2 and visualization of their interaction by green fluorescent proteins (GFP) two-fusion fluorescence resonance energy transfer (FRET). J. Cell. Biochem. 80:293–303. [PubMed] [Google Scholar]
- Lin, X., H. Qadota, D.G. Moerman, and B.D. Williams. 2003. C. elegans PAT-6/actopaxin plays a critical role in the assembly of integrin adhesion complexes in vivo. Curr. Biol. 13:922–932. [DOI] [PubMed] [Google Scholar]
- Mackinnon, A.C., H. Qadota, K.R. Norman, D.G. Moerman, and B.D. Williams. 2002. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr. Biol. 12:787–797. [DOI] [PubMed] [Google Scholar]
- Matsuda, N., and A. Nakano. 1998. RMA1, an Arabidopsis thaliana gene whose cDNA suppresses the yeast sec15 mutation, encodes a novel protein with a RING finger motif and a membrane anchor. Plant Cell Physiol. 39:545–554. [DOI] [PubMed] [Google Scholar]
- Mercer, K.B., D.B. Flaherty, R.K. Miller, H. Qadota, T.L. Tinley, D.G. Moerman, and G.M. Benian. 2003. Caenorhabditis elegans UNC-98, a C2H2 Zn finger protein, is a novel partner of UNC-97/PINCH in muscle adhesion complexes. Mol. Biol. Cell. 14:2492–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.M., III, I. Ortiz, G.C. Berliner, and H.F. Epstein. 1983. Differential localization of two myosins within nematode thick filaments. Cell. 34:477–490. [DOI] [PubMed] [Google Scholar]
- Moerman, D.G., and A. Fire. 1997. Muscle: structure, function and development. C. Elegans II. D.L. Riddle, T. Blumenthal, B.J. Meyer and J.R. Priess, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 417–470. [PubMed]
- Moulder, G.L., M.M. Huang, R.H. Waterston, and R.J. Barstead. 1996. Talin requires β-integrin, but not vinculin, for its assembly into focal adhesion-like structures in the nematode Caenorhabditis elegans. Mol. Biol. Cell. 7:1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7:1885–1897. [DOI] [PubMed] [Google Scholar]
- Rogalski, T.M., B.D. Williams, G.P. Mullen, and D.G. Moerman. 1993. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 7:1471–1484. [DOI] [PubMed] [Google Scholar]
- Rogalski, T.M., G.P. Mullen, M.M. Gilbert, B.D. Williams, and D.G. Moerman. 2000. The UNC-112 gene in Caenorhabditis elegans encodes a novel component of cell-matrix adhesion structures required for integrin localization in the muscle cell membrane. J. Cell Biol. 150:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel, K.L., and M.C. Beckerle. 1994. The LIM domain is a modular protein-binding interface. Cell. 79:211–219. [DOI] [PubMed] [Google Scholar]
- Stringham, E.G., D.K. Dixon, D. Jones, and E.P. Candido. 1992. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol. Biol. Cell. 3:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, C.E. 2000. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2:E231–E236. [DOI] [PubMed] [Google Scholar]
- Wang, Y., and T.D. Gilmore. 2003. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 1593:115–120. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H. 1988. Muscle. The Nematode Caenorhabditis elegans. W.B. Wood, editor. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 281–335.
- Weiskirchen, R., and K. Gunther. 2003. The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. Bioessays. 25:152–162. [DOI] [PubMed] [Google Scholar]
- Williams, B.D., and R.H. Waterston. 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124:475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel, J.M., and H.F. Epstein. 1980. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1:73–97. [DOI] [PubMed] [Google Scholar]