Abstract
When the protein folding capacity of the endoplasmic reticulum (ER) is challenged, the unfolded protein response (UPR) maintains ER homeostasis by regulating protein synthesis and enhancing expression of resident ER proteins that facilitate protein maturation and degradation. Here, we report that enforced expression of XBP1(S), the active form of the XBP1 transcription factor generated by UPR-mediated splicing of XBP1 mRNA, is sufficient to induce synthesis of phosphatidylcholine, the primary phospholipid of the ER membrane. Cells overexpressing XBP1(S) exhibit elevated levels of membrane phospholipids, increased surface area and volume of rough ER, and enhanced activity of the cytidine diphosphocholine pathway of phosphatidylcholine biosynthesis. These data suggest that XBP1(S) links the mammalian UPR to phospholipid biosynthesis and ER biogenesis.
Introduction
The ER is a dynamic protein-folding compartment that can be expanded according to the demands placed upon the exocytic pathway. This is exemplified by the highly developed ER network present in specialized secretory cells such as insulin-producing β cells of the pancreas and antibody-secreting plasma cells. Yet, the molecular mechanisms that coordinate the synthesis of protein and lipid components necessary for ER biogenesis remain poorly understood.
Homeostasis of the ER is regulated in large part by the unfolded protein response (UPR), a complex signaling system emanating from the ER membrane that regulates translation and transcription in response to increased demands on the protein folding capacity of the ER (Rutkowski and Kaufman, 2004). One branch of the UPR is directed by the ER membrane-bound activating transcription factor 6α and 6β (ATF6α/β). Regulated intramembrane proteolysis releases the soluble cytosolic domains of ATF6α/β to function as basic leucine zipper transcription factors that up-regulate expression of various ER resident chaperones and folding enzymes (Haze et al., 1999, 2001).
A second branch of the UPR that induces transcription involves X-box binding protein 1 (XBP1). The ER transmembrane kinase/endoribonucleases IRE1α (expressed ubiquitously; Tirasophon et al., 1998) and β (expressed in gut epithelium; Wang et al., 1998) initiate a novel UPR-mediated mRNA splicing mechanism that modifies XBP1 transcripts to encode a potent basic leucine zipper transcription factor, XBP1(S). In contrast, unspliced XBP1 mRNA encodes a basic leucine zipper protein, XBP1(U), that lacks transactivation activity and is more labile than XBP1(S) (Yoshida et al., 2001; Calfon et al., 2002). Genes identified as targets of XBP1(S) encode proteins that function in the ER (Lee et al., 2003; Yoshida et al., 2003). Thus, like ATF6α/β, the IRE1-XBP1 pathway appears to enhance the ability of the ER to properly handle an increased load of client proteins.
The differentiation of B-lymphocytes into antibody-secreting cells requires XBP1 and its modulation by UPR-mediated splicing (Iwakoshi et al., 2003), and pancreatic tissue exhibits robust splicing of XBP1 mRNA (Iwawaki et al., 2004). Furthermore, synthesis of phosphatidylcholine (PtdCho), the predominant phospholipid in mammalian ER membranes (Lykidis and Jackowski, 2001), elevates in B-cells differentiating in response to lipopolysaccharide (Rush et al., 1991). PtdCho biosynthesis also increases in macrophages subjected to free cholesterol loading (Shiratori et al., 1994), another example of physiologic UPR activation (Feng et al., 2003). Therefore, UPR activation correlates with the presence of an elaborate ER in specialized secretory cells and with induction of phospholipid biosynthesis, but a molecular mechanism linking the UPR to ER biogenesis has not been elucidated.
Given the roles of the UPR in maintenance of ER homeostasis, we hypothesized that the UPR might regulate overall ER abundance. Here, we report that enforced expression of a single transcription factor, XBP1(S), is sufficient to induce PtdCho biosynthesis, elevate the level of membrane phospholipids, and trigger expansion of the ER in fibroblasts. These data suggest that XBP1(S) regulates ER biogenesis, revealing another means by which cells use the UPR to cope with increased demands on the exocytic pathway.
Results and discussion
Effect of XBP1(S) on phospholipid biosynthesis and ER expansion
To determine whether mammalian UPR transcriptional activators regulate phospholipid biosynthesis, we transduced NIH-3T3 fibroblasts with retroviral vectors encoding either XBP1(U), XBP1(S), or a constitutively active form of ATF6α (ATF6α(373); Yoshida et al., 2000). Flow cytometric assessment of GFP, expressed via the bicistronic mRNA encoded by the retroviral vectors, revealed ≥95% transduction efficiency for all retroviral constructs tested in NIH-3T3 cells (see online supplemental material, available at http://www.jcb.org/cgi/content/full/jcb.200406136/DC1). As expected, ATF6α(373) induced expression of immunoglobulin heavy chain binding protein (BiP) and the disulfide-isomerase-like protein ERp72, two ER resident chaperones. In addition, ATF6α(373) up-regulated ERdj3 (also known as HEDJ), an ER chaperone cofactor that has been identified as an XBP1 target (Lee et al., 2003) (Fig. 1 A). The XBP1(U) retroviral vector yielded the ∼30-kD pXBP1(U) and no detectable pXBP1(S), whereas the XBP1(S) virus exclusively yielded the ∼54-kD pXBP1(S). Cells transduced with XBP1(S), but not XBP1(U), exhibited increased levels of ERdj3 and BiP (Fig. 1 A). These data demonstrate that ATF6α(373), XBP1(U), and XBP1(S) all functioned as predicted upon enforced expression in NIH-3T3 cells.
Figure 1.
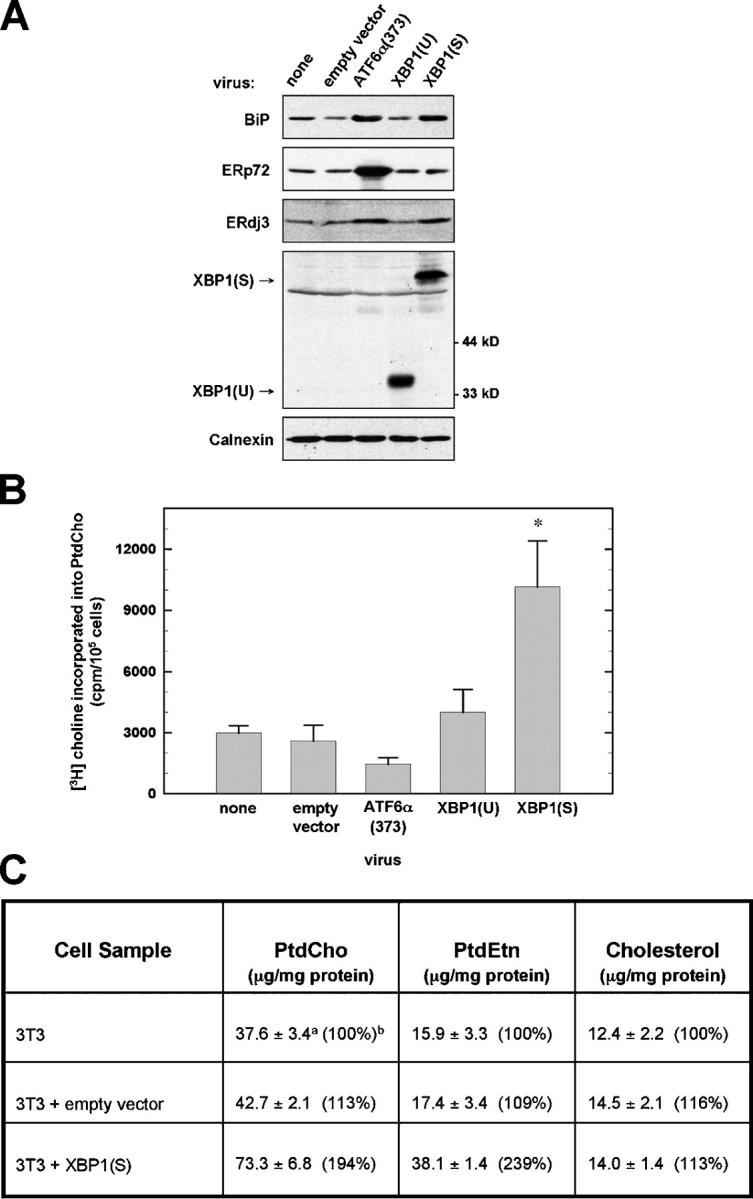
Phospholipid synthesis in NIH-3T3 fibroblasts retrovirally transduced with UPR transcriptional activators. NIH-3T3 cells were transduced with the indicated pBMN-I-GFP retroviral vectors. (A) At 48 h after transduction, cells were harvested and equivalent amounts of cell lysate protein were resolved by standard SDS-PAGE under reducing conditions. Chemiluminescent immunoblotting was performed. A nonspecific band just below the position of pXBP1(S) was detected by the anti-XBP1 antibody. Calnexin served as a loading control. (B) Cells were metabolically labeled with [3H]choline for 2 h, a labeling interval in which [3H]choline is incorporated exclusively into PtdCho. The amount of radiolabel present in cellular lipid extracts was normalized as cpm/105 cells and was plotted as the mean ± SD (n = 3; asterisk denotes P = 0.002 for comparison of XBP1(S) to empty vector). (C) Cells were harvested and total cellular mass of PtdCho, PtdEtn, and cholesterol was determined by flame ionization. Superscript a: average of triplicate determinations from two independent experiments (n = 6) ± SD. Superscript b: control NIH-3T3 values set at 100% for comparison.
In contrast to ATF6α(373) and XBP1(U), enforced expression of XBP1(S) resulted in a large (approximately fourfold) increase in PtdCho synthesis (Fig. 1 B). Furthermore, the total amount of both PtdCho and phosphatidylethanolamine (PtdEtn), also a major lipid in intracellular membranes (Lykidis and Jackowski, 2001), was increased approximately twofold in XBP1(S)-transduced cells. However, the mass of cholesterol, most prevalent in the plasma membrane, was unchanged (Fig. 1 C). Therefore, XBP1(S) has the capacity to mediate substantial increases in phospholipids that constitute the membranes of intracellular organelles like the ER. It is important to note that the yeast UPR has also been implicated in lipid metabolism (Cox et al., 1997).
Electron microscopy revealed that XBP1(S)-transduced cells contained an increased number of intracellular membrane–bound structures that were frequently studded with ribosomes, indicating their identity as RER (Fig. 2 A). Indeed, the surface area and volume of RER were significantly increased in XBP1(S)-transduced cells, 2.5- and 3.4-fold, respectively (Fig. 2 B). Immunofluorescence microscopy demonstrated that KDEL-containing proteins were distributed throughout the cytoplasm of XBP1(S)-transduced cells (Fig. 2 C), consistent with an overall increase in the amount of ER. In addition, XBP1(S) induced a 44% increase in cell size (empty vector cells, 103.9 ± 1.8 μm2; XBP1(S) cells, 149.8 ± 2.5 μm2). Cells overexpressing XBP1(S) exhibited a slower rate of proliferation 24 h after transduction, but their viability was not affected through 48 h. Importantly, while this manuscript was under review, Shaffer et al. (2004) reported that XBP1(S) can mediate increased cell size and expansion of intracellular organelles including the ER, thereby corroborating our findings.
Figure 2.
Microscopy analysis of the ER. (A) At 48 h after transduction with the indicated retroviral vectors, thin sections were prepared from NIH-3T3 cells and examined by transmission EM. Representative micrographs of increasing magnification (from top to bottom) are shown with scale bar (1 μm) in top right corner: middle panels, arrow indicates representative ER; bottom panels, arrow indicates membrane-bound ribosomes. N, nucleus. (B) Stereological analysis of RER volume (top) and RER surface area (bottom) was performed on electron micrographs and the mean ± SEM are plotted (n = 4 stereological sets with 2–16 micrographs per set; *, P < 0.01). (C) Immunofluorescence analysis (using anti-KDEL mAb) and confocal microscopy was performed on NIH-3T3 cells grown on coverslips and transduced with the indicated retroviral vectors for 48 h.
Synthesis of XBP1(S) can also be elicited by ER stress-inducing agents such as the glycosylation inhibitor tunicamycin. In a series of experiments, we observed a trend toward a small increase (∼20%) in PtdCho biosynthesis within 3 to 4 h of tunicamycin treatment, but this was not sustained in a statistically significant fashion for longer intervals (unpublished data). Perhaps gross inhibition of protein folding in the ER and/or complete UPR activation in cells subjected to pharmacologic agents that poison the ER is not compatible with increased phospholipid biosynthesis.
Effect of XBP1(S) on the cytidine diphosphocholine pathway of PtdCho biosynthesis
The predominant pathway for PtdCho biosynthesis in mammalian cells is the cytidine diphosphocholine (CDP-choline) or Kennedy pathway (Lykidis and Jackowski, 2001). First, choline kinase (CK) phosphorylates choline in the presence of ATP to yield phosphocholine. Second, choline cytidylyltransferase (CCT) converts phosphocholine to CDP-choline in the presence of CTP, and this is considered to be the rate-limiting step in the CDP-choline pathway (Lykidis and Jackowski, 2001). Third, cholinephosphotransferase 1 (CPT1) transfers the phosphocholine moiety of CDP-choline to DAG, producing PtdCho. This final step can also be catalyzed by choline/ethanolaminephosphotransferase (CEPT1), a bifunctional enzyme that can synthesize both choline- and ethanolamine-containing phospholipids. Here, we refer to the activities of CPT1 (Henneberry et al., 2000) and CEPT1 (Henneberry and McMaster, 1999) collectively as CPT activity. In XBP1(S)-transduced cells, CK activity was unchanged (Fig. 3 A), CCT activity increased ∼30% (Fig. 3 B), and CPT activity elevated approximately fivefold (Fig. 3 C). Remarkably similar to these data, increased PtdCho biosynthesis in lipopolysaccharide-stimulated splenic B cells was previously shown to correlate with unchanged CK activity, a small increase in CCT activity, and an approximately sixfold increase in CPT activity (Rush et al., 1991). Therefore, it follows that XBP1(S) might mediate the increased synthesis of phospholipids necessary for ER biogenesis as B cells transition into high-rate antibody secretion.
Figure 3.

Enzymatic activities in the CDP-choline pathway of PtdCho synthesis. The relative activities of CK, CCT, and CPT, which constitute the CDP-choline pathway of PtdCho biosynthesis (right side of figure), were determined using lysates or microsomes prepared from NIH-3T3 cells (▪), or NIH-3T3 cells transduced with XBP1(S) (•) or empty vector (○). The rates of production of phosphocholine (P-Choline), CDP-Choline, and PtdCho were compared as a function of total protein in each assay. (A) Data for CK are averaged from triplicate determinations and are representative of two independent experiments. (B) Data for CCT are averaged from five determinations obtained in two independent experiments. (C) Data for CPT are averaged from duplicate determinations and are representative of three independent experiments.
There are three isoforms of CCT: α, β2, and β3. CCTα is ubiquitously expressed, whereas the β isoforms exhibit tissue-specific expression that is ∼10-fold lower compared with CCTα (Jackowski et al., 2004). Quantitative real-time RT-PCR revealed no change in transcript levels for any of the CCT isoforms in XBP1(S)-transduced cells (Fig. 4 A). Likewise, Northern blotting revealed no change in the steady-state levels of CPT1 and CEPT1 transcripts (Fig. 4 B). These data suggest that enforced expression of XBP1(S) leads to post-transcriptional and/or post-translational regulation of the CCTα and CPT1/CEPT1 enzymes, thereby augmenting their activities and increasing synthesis of PtdCho (Fig. 5).
Figure 4.
Expression of enzymes that function in the CDP-choline pathway of PtdCho synthesis. Total RNA was prepared from NIH-3T3 fibroblasts harvested at 24 and 48 h after transduction with the indicated retroviral vectors. (A) The relative levels of expression of the CCTα, β2, and β3 isoform cDNAs at the 48-h interval were measured by quantitative real-time PCR. The amount of target RNA was normalized to the endogenous GAPDH reference and related to the amount of target CCTα in NIH-3T3, which was set as the calibrator at 1.0 (superscript a). The mean ± SD (superscript b) of triplicate determinations is shown. (B) Northern blot analysis using 32P-labeled cDNA probes specific for CEPT1 and CPT1, the two genes encoding enzymes possessing CPT activity; ERdj3, a known XBP1 target as a positive control; and γ-actin as a loading control.
Figure 5.
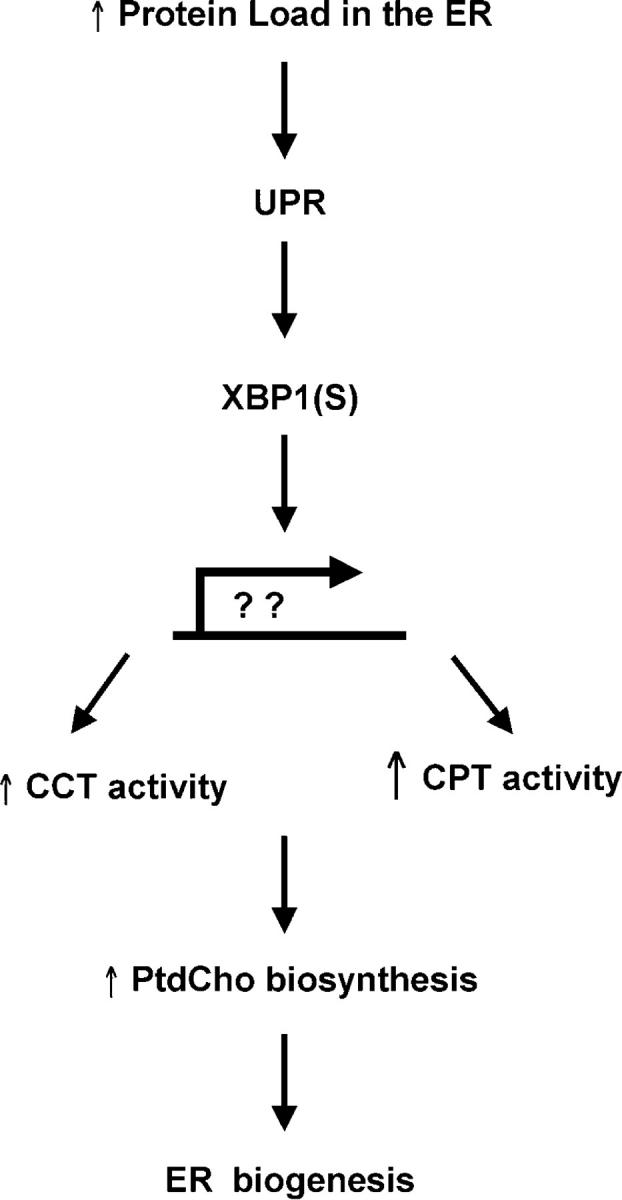
A model for how XBP1 links the UPR to ER biogenesis. In response to increased demand on the protein folding capacity of the ER, UPR-mediated splicing of XBP1 mRNA yields the XBP1(S) transcriptional activator. We propose that XBP1(S) induces expression of unknown gene products that lead to an increase in CCT activity and a large increase in CPT activity, thereby augmenting synthesis of PtdCho. Increased production of PtdCho, the major phospholipid of the ER membrane, is critical for expansion of the ER compartment.
CCTα is an amphitropic enzyme as it transitions from a soluble form, largely sequestered in the nucleus, into a more active form associated with ER membranes (Cornell and Northwood, 2000). Conversion of CCTα into the membrane-associated form correlates with dephosphorylation of serine residues near its carboxy terminus, and this influences its affinity for membranes (Lykidis and Jackowski, 2001). Thus, there are a number of steps at which an XBP1(S)-regulated gene product might modulate CCTα activity. Both CPT1 and CEPT1 contain an estimated seven membrane-spanning helices and are predicted to reside in the ER membrane (Henneberry and McMaster, 1999; Henneberry et al., 2000), but very little is known concerning control of their respective activities. Perhaps XBP1(S) regulates expression of an allosteric modulator or a cofactor that enhances the stability of CPT1 and/or CEPT1.
Summary
Our data regarding XBP1(S) provide the first example of a transcription factor being sufficient to induce phospholipid biosynthesis and ER expansion in mammalian cells. Whether the expanded ER in XBP1(S)-transduced cells is properly organized and fully functional is under investigation. We propose that in a physiologic UPR, the IRE1-XBP1 pathway regulates synthesis of phospholipids according to cellular needs for ER membrane components (Fig. 5). Interestingly, inhibition of PtdCho biosynthesis via a thermosensitive mutation in the CCT enzyme has been correlated with UPR activation (van der Sanden et al., 2003), providing further evidence that the UPR might provide a means to rapidly respond to an increased need for phospholipids. Coordinating induction of phospholipid biosynthesis with up-regulated expression of ER resident proteins would allow the mammalian UPR to both build and equip the ER.
Materials and methods
Plasmids
pBMN-I-GFP (Dr. G. Nolan, Stanford University, Palo Alto, CA) encodes a bicistronic mRNA with a GFP cassette 3′ of the internal ribosomal entry site (I). pBMN-hATF6(373)-I-GFP encodes aa 1–373 of human ATF6α (Yoshida et al., 2000). pBMN-hXBP1(U)-I-GFP encodes full-length human XBP1 not modified by UPR-mediated splicing. pBMN-hXBP1(S)-I-GFP encodes full-length human XBP1 generated by UPR-mediated splicing.
Cell culture and retroviral transduction
NIH-3T3 fibroblasts and Phoenix-Eco cells (Dr. G. Nolan) were maintained in DME (Gass et al., 2002). Ecotropic retroviruses were produced using Phoenix-Eco packaging cells (Gunn et al., 2004) and pBMN plasmids. Cells were transduced with retroviruses (Brewer and Diehl, 2000) with ≥95% efficiency, as measured by GFP fluorescence using a FACSCalibur flow cytometer (BD Biosciences). Tunicamycin (Sigma-Aldrich) was used at 0.5 μg/ml.
Immunoblotting and Northern blotting
Chemiluminescent immunoblotting of BiP, XBP1, and calnexin was performed as described previously (Gass et al., 2002). The ERdj3 antisera was provided by Dr. Linda Hendershot (St. Jude Children's Research Hospital, Memphis, TN). Rabbit anti–mouse ERp72 antibody (SPA-720; Stressgen Biotechnologies) and mouse anti-β actin mAb (clone AC-15; Sigma-Aldrich) were purchased. Northern blotting was performed as described previously (Gass et al., 2002). Probes used in this work are described in the online supplemental material.
Detection of CCT isoform mRNAs by real-time PCR
Quantitative real-time PCR of the CCTα, β2, and β3 isoform cDNAs was performed as described previously (Karim et al., 2003). For details, see online supplemental material.
Analysis of PtdCho synthesis
Cells were cultured for 2 h at 37°C in DME containing 2 μCi/ml methyl[3H]choline chloride (81 Ci/mmol; Amersham Biosciences). In tunicamycin experiments, labeling was performed during the final 2 h of treatment. After labeling, cells were washed with cold PBS and scraped in methanol/water (2:0.8, vol/vol). Lipids were extracted as described previously (Bligh and Dyer, 1959). Radiolabel incorporated into chloroform-soluble metabolites was determined by scintillation spectroscopy. The organic phase was subjected to TLC in chloroform/methanol/water (65:35:4, vol/vol/vol) to verify that detectable radioactivity was present exclusively in PtdCho.
Phospholipid mass determinations
Frozen cell pellets (2 × 107 cells) were thawed on ice and resuspended in 1 ml of water. The total volume was measured and a 100-μl aliquot was removed for protein determination. The remainder of the lysate was extracted as described previously (Bligh and Dyer, 1959). Lipids were detected by flame ionization using an Iatroscan instrument (Iatron Laboratories) with PEAK SIMPLE software (SRI Instruments). Peaks were identified by comigration with authentic standards. PtdCho, PtdEtn, and cholesterol mass were calculated using standard curves for each prepared with either the polar lipid (no. 1127) or the neutral lipid (no. 1129) mixture (Matreya, Inc.). For details, see online supplemental material.
CK, CCT, and CPT assays
Pelleted frozen cells (2 × 107 cells) were used for assays of CK and CCT enzymatic activity as described previously (Lykidis et al., 1999, 2001). CPT activity was assessed as described previously (Henneberry and McMaster, 1999) using microsomes prepared from frozen cell pellets (2 × 107 cells). The assays were linear through the time of incubation at all protein concentrations. For details, see online supplemental material.
Immunofluorescence microscopy
NIH-3T3 cells were seeded on glass coverslips in 60-mm dishes. At 48 h after transduction, cells were fixed on coverslips with 95% ethanol/5% acetic acid for 10 min at −20°C, blocked with PBS containing 5% FCS at RT, and then stained with mouse anti-KDEL mAb (SPA-827; StressGen Biotechnologies) or mouse IgG (Southern Biotechnology Associates) as an isotype control, followed by biotinylated goat anti–mouse IgG (Southern Biotechnology Associates) and R-phycoerythrin–conjugated streptavidin (Molecular Probes, Inc.). Coverslips were mounted in ProLong Antifade (Molecular Probes, Inc.) and were examined on a confocal microscope (LSM-510; Carl Zeiss MicroImaging, Inc.). Images were assessed using the LSM Browser (Carl Zeiss MicroImaging, Inc.) and were processed using Adobe Photoshop software.
Electron microscopy
Retrovirally transduced NIH-3T3 cells were trypsinized, pelleted by centrifugation, fixed in cacodylate buffer containing 4% glutaraldehyde, and washed in cacodylate buffer. Cells were then postfixed with 1% osmium tetroxide prepared in cacodylate buffer, dehydrated by sequential extraction with graded concentrations of ethanol from 25 to 100%, and embedded in Embed 812 resin. Thin sections were cut on an ultramicrotome (Reichert), mounted on nickel grids, stained with saturated uranyl acetate and Reynolds lead citrate, and examined at 2,500, 8,000, and 20,000× at RT using a transmission electron microscope (model H-600; Hitachi). Images were captured on 35-mm film and were processed using Adobe Photoshop software.
Stereological measurements of RER and cell size determinations
Four independent stereological sets containing electron micrographs of empty vector– and XBP1(S)-transduced cells at magnifications of 8,000 and 20,000 were analyzed. RER volume was measured by determining the volume fraction (Vv) with a grid containing test points in the linear array (1-cm spacing). Vv is equal to the number of test points falling on RER divided by the number of test points falling on the containing space. RER surface area was calculated by determining the surface density (Sv). Sv is equal to two times the number of line intersections with RER membrane, divided by the total length of line on the containing space (Weibel and Bolender, 1973). Average cell size was determined from light microscopy analysis of thick sections using the Scion image program. The mean cell area (μm2) was determined in a minimum of 38 fields, and SEM was calculated.
Statistical analysis
Both one-tailed and two-tailed independent t tests were used to compare test samples to controls. In all cases, both tests were in agreement. P values in figure legends correspond to results of one-tailed t tests.
Online supplemental material
Fig. S1 shows flow cytometric analysis of GFP expression in NIH-3T3 cells transduced with pBMN-I-GFP retroviral vectors encoding either ATF6α(373), XBP1(U) or XBP1(S). Details are provided for: detection of CCT isoform mRNAs by real-time PCR; northern hybridization probes; assays for enzymatic activities of CK, CCT, and CPT; phospholipid mass determinations. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200406136/DC1.
Acknowledgments
We thank Jennifer N. Gass and Kathryn E. Gunn for critical reading of the manuscript; Dr. John McNulty, Linda Fox (Loyola University Imaging Facility), and Patricia Simms (Loyola University Flow Cytometry Facility) for expert technical assistance; Dr. David Keating for assistance with lipid extraction and TLC; Hemamalini Bommiasamy for assistance with immunofluorescence; and LeeTerry Moore for technical assistance. We also thank Christopher Gunter for real-time PCR and enzymatic analysis, and Daren Hemingway for enzymatic analysis and lipid mass determinations.
S. Jackowski is supported by National Institutes of Health (NIH) grant GM45737, Cancer Center (CORE) support grant CA21765, and the American Lebanese Syrian Associated Charities. J.W. Brewer. is funded by NIH grant GM61970.
Abbreviations used in this paper: ATF, activating transcription factor; BiP, binding protein; CCT, choline cytidylyltransferase; CDP-choline, cytidine diphosphocholine; CEPT, choline/ethanolaminephosphotransferase; CK, choline kinase; CPT, cholinephosphotransferase; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; UPR, unfolded protein response; XBP1, X-box binding protein 1; XBP1(S), XBP1 (spliced); XBP1(U), XBP1(unspliced).
References
- Bligh, E.G., and W.J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Med. Sci. 37:911–917. [DOI] [PubMed] [Google Scholar]
- Brewer, J.W., and J.A. Diehl. 2000. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc. Natl. Acad. Sci. USA. 97:12625–12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon, M., H. Zeng, F. Urano, J.H. Till, S.R. Hubbard, H.P. Harding, S.G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. [DOI] [PubMed] [Google Scholar]
- Cornell, R.B., and I.C. Northwood. 2000. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem. Sci. 25:441–447. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., R.E. Chapman, and P. Walter. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell. 8:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, B., P.M. Yao, Y. Li, C.M. Devlin, D. Zhang, H.P. Harding, M. Sweeney, J.X. Rong, G. Kuriakose, E.A. Fisher, et al. 2003. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat. Cell Biol. 5:781–792. [DOI] [PubMed] [Google Scholar]
- Gass, J.N., N.M. Gifford, and J.W. Brewer. 2002. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277:49047–49054. [DOI] [PubMed] [Google Scholar]
- Gunn, K.E., N.M. Gifford, K. Mori, and J.W. Brewer. 2004. A role for the unfolded protein response in optimizing antibody secretion. Mol. Immunol. 41:919–927. [DOI] [PubMed] [Google Scholar]
- Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 10:3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze, K., T. Okada, H. Yoshida, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2001. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 355:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberry, A.L., and C.R. McMaster. 1999. Cloning and expression of a human choline/ethanolaminephosphotransferase: synthesis of phosphatidylcholine and phosphatidylethanolamine. Biochem. J. 339:291–298. [PMC free article] [PubMed] [Google Scholar]
- Henneberry, A.L., G. Wistow, and C.R. McMaster. 2000. Cloning, genomic organization, and characterization of a human cholinephosphotransferase. J. Biol. Chem. 275:29808–29815. [DOI] [PubMed] [Google Scholar]
- Iwakoshi, N.N., A.H. Lee, P. Vallabhajosyula, K.L. Otipoby, K. Rajewsky, and L.H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329. [DOI] [PubMed] [Google Scholar]
- Iwawaki, T., R. Akai, K. Kohno, and M. Miura. 2004. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10:98–102. [DOI] [PubMed] [Google Scholar]
- Jackowski, S., J.E. Rehg, Y.-M. Zhang, J. Wang, K. Miller, P. Jackson, and M.A. Karim. 2004. Disruption of CCTβ2 expression leads to gonadal dysfunction. Mol. Cell. Biol. 24:4720–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, M., P. Jackson, and S. Jackowski. 2003. Gene structure, expression and identification of a new CTP:phosphocholine cytidylyltransferase β isoform. Biochim. Biophys. Acta. 1633:1–12. [DOI] [PubMed] [Google Scholar]
- Lee, A.H., N.N. Iwakoshi, and L.H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykidis, A., and S. Jackowski. 2001. Regulation of mammalian cell membrane biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 65:361–393. [DOI] [PubMed] [Google Scholar]
- Lykidis, A., I. Baburina, and S. Jackowski. 1999. Distribution of CTP:phosphocholine cytidylyltransferase (CCT) isoforms. Identification of a new CCTβ splice variant. J. Biol. Chem. 274:26992–27001. [DOI] [PubMed] [Google Scholar]
- Lykidis, A., J. Wang, M.A. Karim, and S. Jackowski. 2001. Overexpression of a mammalian ethanolamine-specific kinase accelerates the CDP-ethanolamine pathway. J. Biol. Chem. 276:2174–2179. [DOI] [PubMed] [Google Scholar]
- Rush, J.S., T. Sweitzer, C. Kent, G.L. Decker, and C.J. Waechter. 1991. Biogenesis of the endoplasmic reticulum in activated B lymphocytes: temporal relationships between the induction of protein N-glycosylation activity and the biosynthesis of membrane protein and phospholipid. Arch. Biochem. Biophys. 284:63–70. [DOI] [PubMed] [Google Scholar]
- Rutkowski, D.T., and R.J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20–28. [DOI] [PubMed] [Google Scholar]
- Shaffer, A.L., M. Shapiro-Shelef, N.N. Iwakoshi, A.H. Lee, S.B. Quian, H. Zhao, X. Yu, L. Yang, B.K. Tan, A. Rosenwald, et al. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 21:81–93. [DOI] [PubMed] [Google Scholar]
- Shiratori, Y., A.K. Okwu, and I. Tabas. 1994. Free cholesterol loading of macrophages stimulates phosphatidylcholine biosynthesis and up-regulation of CTP: phosphocholine cytidylyltransferase. J. Biol. Chem. 269:11337–11348. [PubMed] [Google Scholar]
- Tirasophon, W., A.A. Welihinda, and R.J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sanden, M.H., M. Houweling, L.M. van Golde, and A.B. Vaandrager. 2003. Inhibition of phosphatidylcholine synthesis induces expression of the endoplasmic reticulum stress and apoptosis-related protein CCAAT/enhancer-binding protein-homologous protein (CHOP/GADD153). Biochem. J. 369:643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.Z., H.P. Harding, Y. Zhang, E.M. Jolicoeur, M. Kuroda, and D. Ron. 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17:5708–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel, E.R., and R.P. Bolender. 1973. Stereological techniques for electron microscopic morphometry. Principles and Techniques of Electron Microscopy. Vol. 3. M.A. Hayat, editor. Van Nostrand-Reinhold Company, New York. 237–296.
- Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755–6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., T. Matsui, N. Hosokawa, R.J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell. 4:265–271. [DOI] [PubMed] [Google Scholar]




