Abstract
We purified native Myo2p/Cdc4p/Rlc1p (Myo2), the myosin-II motor required for cytokinesis by Schizosaccharomyces pombe. The Myo2p heavy chain associates with two light chains, Cdc4p and Rlc1p. Although crude Myo2 supported gliding motility of actin filaments in vitro, purified Myo2 lacked this activity in spite of retaining full Ca-ATPase activity and partial actin-activated Mg-ATPase activity. Unc45-/Cro1p-/She4p-related (UCS) protein Rng3p restored the full motility and actin-activated Mg-ATPase activity of purified Myo2. The COOH-terminal UCS domain of Rng3p alone restored motility to pure Myo2. Thus, Rng3p contributes directly to the motility activity of native Myo2. Consistent with a role in Myo2 activation, Rng3p colocalizes with Myo2p in the cytokinetic contractile ring. The absence of Rlc1p or mutations in the Myo2p head or Rng3p compromise the in vitro motility of Myo2 and explain the defects in cytokinesis associated with some of these mutations. In contrast, Myo2 with certain temperature-sensitive forms of Cdc4p has normal motility, so these mutations compromise other functions of Cdc4p required for cytokinesis.
Introduction
Life depends on cytokinesis but its mechanism is far from understood. Genetic studies in fission yeast and other systems have identified at least 50 proteins that contribute to cytokinesis (Guertin et al., 2002). Myosin-II is the motor protein responsible for constricting the cleavage furrow that separates daughter cells. Fission yeast have two myosin-II heavy chains encoded by myo2 and myp2. Myo2p, but not Myp2p (Bezanilla et al., 1997, Motegi et al., 1997), is required for cytokinesis under most conditions (Kitayama et al., 1997; Balasubramanian et al., 1998; May et al., 1998). Myo2p is one of the first proteins to join the contractile ring, whereas Myp2p joins much later (Bezanilla et al., 2000; Wu et al., 2003). Fission yeast have two established myosin light chains encoded by cdc4 and rlc1. Cdc4p is an “essential” light chain (ELC) that is required for cytokinesis (McCollum et al., 1995; Naqvi et al., 1999). Rlc1p is a “regulatory” light chain (RLC; Le Goff et al., 2000; Naqvi et al., 2000). rlc1Δ cells are viable, in spite of obvious cytokinesis defects (Le Goff et al., 2000; Naqvi et al., 2000). Immunoprecipitation experiments indicate that Myo2p and Myp2p heavy chains associate with both Cdc4p (Naqvi et al., 1999; Motegi et al., 2000) and Rlc1p (Le Goff et al., 2000; Naqvi et al., 2000). Although Myo2p is required for cytokinesis, little is known about its regulation. Cdc4p can be phosphorylated, but this modification is not required for cytokinesis (McCollum et al., 1999).
The protein comprised of the Myo2p/Cdc4p/Rlc1p (Myo2) heavy chain has a “conventional” structure in the sense that two heavy chains form a rod-shaped coiled-coil tail that is insoluble at low ionic strength, presumably owing to the formation of filaments (Bezanilla and Pollard, 2000). Given a dimeric tail and two IQ motifs in each heavy chain, the Myo2 protein is inferred to have two heads, each with one Cdc4p light chain and one Rlc1p light chain (Bezanilla and Pollard, 2000).
Myp2p forms an “unconventional” myosin-II because the isolated tail folds back on itself to form an antiparallel coiled-coil (Bezanilla and Pollard, 2000). Thus, the Myp2 protein is presumed to have just one head with Cdc4p and Rlc1p bound to each of two IQ motifs. Like Myo2p tails, Myp2p tails are insoluble at physiological salt concentrations (Bezanilla and Pollard, 2000).
Aside from its light chains, Myo2p has been inferred to interact with Rng3p, a member of the conserved Unc45-/Cro1p-/She4p-related (UCS) domain protein family (Wong et al., 2000). Like myo2, rng3 is essential for cytokinesis and is required for the assembly of tight Myo2p contractile rings that arise from medial accumulations of Myo2p (Wong et al., 2000). Unlike Myo2p and Cdc4p (Naqvi et al., 1999), Rng3p requires actin filaments for its localization to the division site (Wong et al., 2000). Furthermore, the Caenorhabditis elegans UCS domain protein, Unc45, binds myosin-II in vitro (Barral et al., 2002).
We purified native Myo2 from Schizosaccharomyces pombe to learn how it functions and is regulated. During purification, Myo2 lost the ability to support gliding motility of actin filaments in vitro, in spite of retaining full actin binding and Ca-ATPase activity. We ruled out denaturation and loss of light chains and discovered that addition of either native or recombinant Rng3p to pure Myo2 restores full motility activity and actin-activated Mg-ATPase activity. More specifically, the Rng3p UCS domain alone restored Myo2 gliding activity. Thus, Rng3p is essential for the function of the active motor. Our in vitro motility data demonstrate that Rng3p promotes the efficient interaction of Myo2 with actin filaments, which is essential for formation of a competent actomyosin ring. Rng3p concentrates in the contractile ring after Myo2p, which is consistent with a role as an activator of ring constriction. In vitro motility assays show that Myo2 function is compromised by mutations in the Myo2p head and Rng3p, and by the absence of Rlc1p. In contrast, Myo2 with some temperature-sensitive forms of Cdc4p (Cdc4-8p, Cdc4-31p, and Cdc4-A2p) has normal motility, indicating that these mutations compromise other functions of the ELC required for cytokinesis.
Results
Subunit composition of fission yeast conventional myosin-II, Myo2
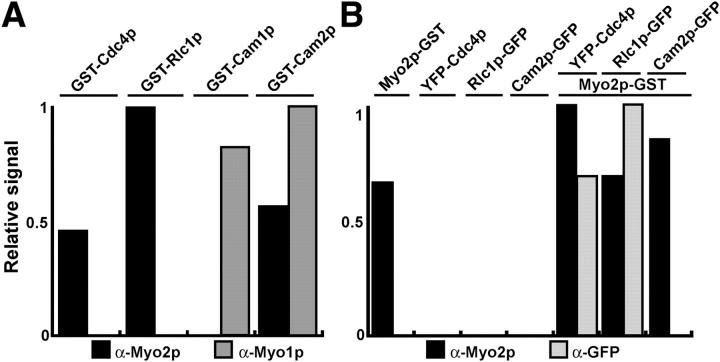
Before designing strategies to purify Myo2, we wished to confirm that Cdc4p and Rlc1p are the only light chains required for Myo2 function. The S. pombe genome encodes five proteins with sequences similar to Cdc4p: Cam1p (calmodulin; 38% identity/62% similarity to Cdc4p); SPAC29A4.05p (35%/58%); Cdc31p (27%/51%); Rlc1p (23%/49%); and a Cnb1p orthologue (21%/49%). We ruled out Cdc31p and Cnb1p as myosin light chains, because Cdc31p (a centrin) has a role in spindle pole body duplication (Paoletti et al., 2003), and Cnb1p is a regulatory subunit of calcineurin B phosphatase (Cyert and Thorner, 1992). Cam1p localizes at polarized growth sites and the contractile ring (Moser et al., 1997; Eng et al., 1998), whereas SPAC29A4.05p had not been characterized. We named SPAC29A4.05p Cam2p because its amino acid sequence is 41% identical/65% similar to Cam1p. Like GST-Cdc4p and GST-Rlc1p, overexpressed GST-Cam2p binds the Myo2 heavy chain to glutathione-Sepharose beads, whereas GST-Cam1p does not (Fig. 1 A). In contrast, only native Cdc4p and Rlc1p copurified with native Myo2p-GST (Fig. 1 B), suggesting that the Myo2p heavy chain binds these two light chains in preference to Cam2p. Cam2p localizes as patches at sites of polarized growth (unpublished data), like type-I myosin, Myo1p (Lee et al., 2000; Toya et al., 2001). In addition, like Cam1p (Toya et al., 2001), Cam2p associates with Myo1p (Fig. 1 A). Thus, both Cam1p and Cam2p are likely to be light chains for Myo1p. Given these results, we chose to purify Myo2p in combination with Cdc4p and Rlc1p.
Figure 1.
Cdc4p and Rlc1p are the light chains for Myo2p. Quantitative analysis of GST-pull down experiments with GST-tagged myosin light chains, GST-tagged Myo2p heavy chain, or GFP-tagged candidate myosin light chains. Proteins bound to glutathione-Sepharose beads were separated by SDS-PAGE, immunoblotted for Myo2p, Myo1p, or GFP, detected by ECL, and quantitated by densitometry of the bands. (A) TP 150 cells overexpressed GST-tagged candidate light chains cdc4, rlc1, cam1, or cam2. The GST-fusion proteins in cell extracts were affinity purified on glutathione-Sepharose and copurifying Myo2p (black bars) and Myo1p (gray bars) were quantitated by densitometry. Amounts were estimated relative to the maximal signal, which was given a value of 1. (B) Glutathione-Sepharose was used to purify GST-tagged proteins from cells expressing myo2-GST alone, YFP-cdc4 alone, rlc1-GFP alone, cam2-GFP alone, myo2-GST plus YFP-cdc4, myo2-GST plus rlc1-GFP, or myo2-GST plus cam2-GFP. Relative amounts of Myo2p (black bars) and copurifying light chains (gray bars) were quantitated by densitometry of bands detected after immunoblotting.
Purification of Myo2
We purified Myo2 in three steps starting with cells overexpressing GST-Cdc4p and GST-Rlc1p. Western blots showed that essentially all of the native Myo2p in extracts of these cells bound to glutathione-Sepharose (unpublished data). Gel filtration and hydroxylapatite chromatography yielded highly purified Myo2, but the yield from 45 g of cells (from 6 liters of culture medium) was exceedingly low and insufficient for biochemical analysis (unpublished data). The poor yield was likely the consequence of the low cellular concentration of Myo2.
To improve the yield of pure Myo2, we replaced the myo2 promoter with a thiamine repressible nmt1 promoter. Three-step purification from cells overexpressing Myo2p, Cdc4p, and Rlc1p in the absence of thiamine yielded sufficient quantities (25 μg per gram of cells) of pure Myo2 (Fig. 2 A) for further characterization. GST was removed from the light chains by thrombin-cleavage before gel filtration and hydroxylapatite chromatography.
Figure 2.
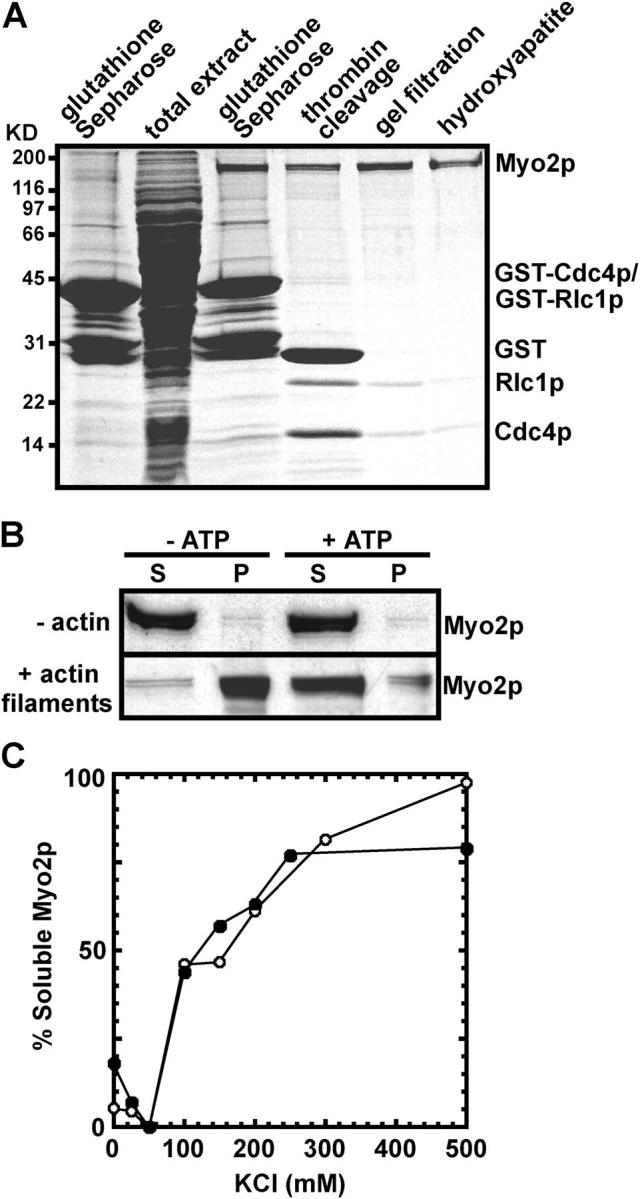
Purification and characterization of Myo2. (A) SDS-PAGE of proteins stained with Coomassie blue. The leftmost lane shows proteins affinity purified from a wild-type strain (MLP 479) overexpressing GST-tagged light chains from the plasmids pGST-cdc4 and pGST-rlc1. The right five lanes show samples from steps in the purification of Myo2 from a strain (MLP 374) overexpressing Myo2p from the 3nmt1 promoter and GST-tagged light chains from pGST-cdc4 and pGST-rlc1. The steps are the total cell extract after centrifuging lysed cells at 100,000 g, proteins eluted from glutathione-Sepharose, the products of thrombin cleavage, the pooled peak from gel filtration, and the pooled peak of purified Myo2 from hydroxyapatite chromatography. Polypeptides are named on the right. (B) ATP-sensitive binding of purified Myo2 to actin filaments evaluated by SDS-PAGE and staining with Coomassie blue. Samples containing 0.75 μM Myo2 in 0.5 M NaCl, 10 mM imidazole, pH 7.0, 1 mM EGTA, 2 mM MgCl2 with 0 or 10 μM actin filaments, and 0 or 2 mM ATP were centrifuged at 120,000 g for 45 min. (top) Myo2 remains in the supernatant in absence of actin filaments. (bottom) Myo2 pellets with actin filaments in the absence but not the presence of 2 mM ATP. S, supernatant; P, pellet. (C) Dependence of the solubility of Myo2 on KCl concentration. Samples of 0.5 μM Myo2 in 10 mM imidazole, 1 mM DTT, and 0 to 500 mM KCl were centrifuged at 120,000 g for 10 min. Soluble Myo2 in the supernatant was determined by both the Bradford protein assay and densitometry of samples stained on protein gels. Two independent experiments are shown based on densitometry data.
Enzyme activity and physical properties of Myo2
Like myosin-II from other sources, the Myo2 ATPase activity is low in Mg2+ and high in either Ca2+ or EDTA and KCl (Table I). Potassium is required for Myo2 EDTA ATPase activity because the EDTA-ATPase activity is negligible in the presence of NaCl (Table I). Purified Myo2 binds actin filaments and dissociates in ATP (Fig. 2 B). Actin filaments stimulated the Mg-ATPase activity of Myo2, but to a lesser extent than rabbit skeletal muscle myosin (Table I).
Table I. ATPase activities and actin filament gliding velocities.
| ATPase activity (sec−1)a ± SD
|
Gliding velocity (μm/sec−1) ± SD
|
||||
|---|---|---|---|---|---|
| Assay | Myo2 | RSMM | Sample | 24°C | 36°C |
| K+Mg2+ | 0.14 ± 0.02 | 0.06 ± 0.02 | Crude Myo2 samples | ||
| Na+ Mg2+ | 0.05 ± 0.01 | ND | wild-type | 0.47 ± 0.05 | 0.48 ± 0.05 |
| K+ Ca2+ | 3.8 ± 0.4 | 1.5 ± 0.1 | myo2-E1 (+/− nGST-Rng3pc) | NFA | |
| + rGST-Rng3c | 5.7 ± 0.6 | ND | myo2-IQ1Δ | 0.06 ± 0.03 | |
| + nGST-Rng3c | 6.3 ± 0.6 | ND | myo2-IQ2Δ | 0.03 ± 0.01 | |
| Na+ Ca2+ | 1.5 ± 0.1 | ND | rlc1Δ | 0.09 ± 0.01 | |
| K+ EDTA | 1.8 ± 0.1 | 2.4 ± 0.1 | rlc1Δ + nRlc1pd | 0.37 ± 0.06 | |
| Na+ EDTA | 0.02 ± 0.01 | ND | rlc1-N1Δ | 0.46 ± 0.04 | |
| Actin-activated Mg2+ ATPaseb | 0.75 ± 0.1 | 1.9 ± 0.2 | rng3-65 | 0.35 ± 0.03 | NFA |
| + rGST-Rng3c | 1.4 ± 0.2 | ND | cdc4-8 | 0.49 ± 0.03 | 0.53 ± 0.08 |
| + nGST-Rng3 c | 1.5 ± 0.2 | ND | cdc4-31 | 0.34 ± 0.04 | 0.46 ± 0.04 |
| cdc4-A2 | 0.43 ± 0.05 | 0.47 ± 0.08 | |||
| cdc4-C2 | 0.01 ± 0.01 | 0.01 ± 0.01 | |||
| Pure samples | |||||
| RSMM | 2.8 ± 0.5 | ||||
| Myo2 | NFA | ||||
| Myo2 + rGST-Rng3pc | 0.41 ± 0.04 | ||||
| Myo2 + nGST-Rng3pc | 0.39 ± 0.02 | ||||
| Myo2 + nGST-Rng3p-UCSc | 0.42 ± 0.07 | ||||
Activities: molecules of ATP hydrolyzed per head per second.
Actin filaments were included at a final concentration of 10 μm.
rGST-Rng3 and nGST-Rng3 denote recombinant and native protein, respectively.
nRlc1 was overexpressed and purified from S. pombe extracts.
NFA, no filaments attached; RSMM, rabbit skeletal muscle myosin.
Myo2 is soluble in high salt and insoluble at physiological salt concentrations (Fig. 2 C) like recombinant Myo2p tails (Bezanilla and Pollard, 2000) and other myosin-IIs. On gel filtration in high salt the Stokes' radius of Myo2 is 13.4 nm, similar to the Stokes' radius of 13.7 nm of Acanthamoeba myosin-II (Pollard et al., 1978), which has a 175-kD heavy chain like Myo2.
Myo2 supports actin filament gliding
Crude Myo2, purified on glutathione-Sepharose but not chromatographed further, bound to a glass coverslip, and supported the gliding motility of actin filaments at a velocity of ∼0.5 μm/s, somewhat slower than skeletal muscle myosin (Fig. 3 E, Table I, and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). Myo2 purified through three steps failed to bind or move actin filaments in this assay (Fig. 3 H). Given the good ATPase activity of this purified Myo2, we hypothesized that purification separates Myo2 from an activator required for motility activity. We ruled out Cdc4p and Rlc1p as the missing activators because both copurified with Myo2 through three purification steps, and because addition of excess purified Cdc4p or Rlc1p failed to activate purified Myo2 (unpublished data).
Figure 3.
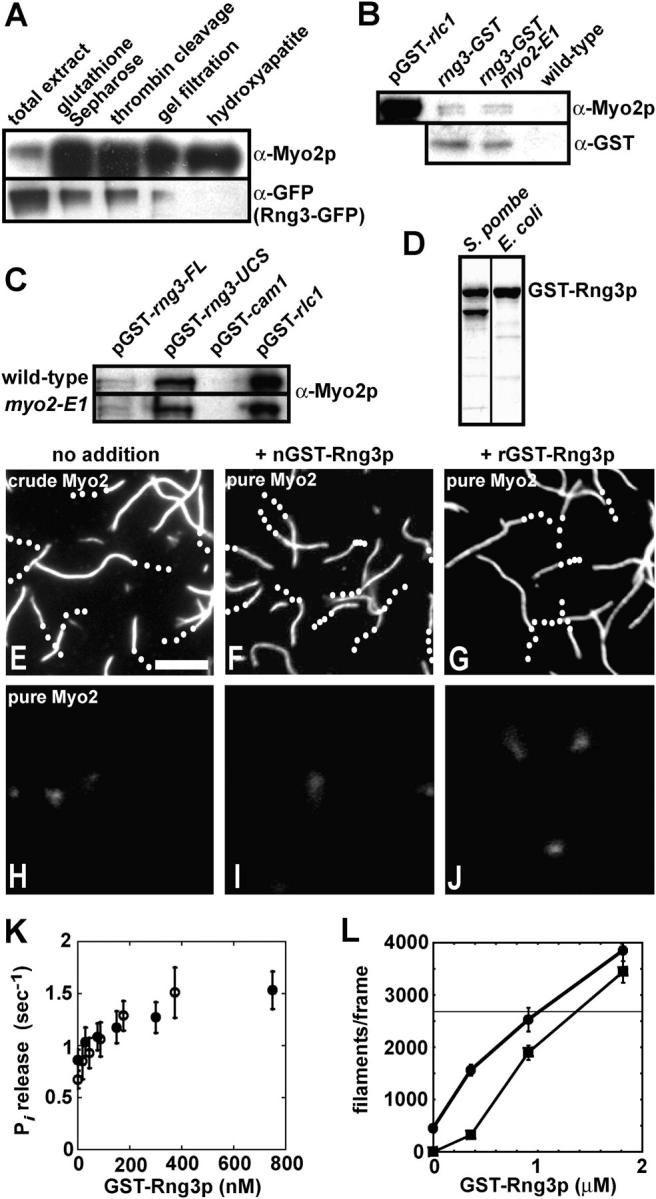
Rng3p stimulates Myo2 activity. (A) Lanes 1–5 show samples from steps in the purification of Myo2 from a strain (MLP 676) containing a chromosomal rng3-GFP 3 fusion overexpressing Myo2p from the 41nmt1 promoter and GST-tagged light chains from pGST-cdc4 and pGST-rlc1. Samples were run on an SDS-PAGE gel, immunoblotted and probed with Myo2 heavy chain antibodies (top) and GFP antibodies (bottom). (B) Bead binding assay for interaction of chromosomal Rng3p-GST with Myo2. Strains: FY 435 carrying pGST-rlc1, MLP 694 (rng3-GST), MLP 693 (rng3-GST, myo2-E1), and MLP 695 (wild-type). GST proteins from extracts were affinity purified on glutathione-Sepharose. Bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with antibodies to Myo2p heavy chain (top) and Rng3p-GST (α-GST; bottom). pGST-rlc1, positive control with GST-Rlc1p. wild type, negative control lacking a GST fusion. (C) Bead binding assay for interaction of overexpressed GST-Rng3p (pGST-rng3-FL) and GST-Rng3-UCS domain (pGST-rng3-UCS) with Myo2. Strains: FY 435 (wild-type) and TP 73 (myo2-E1) carrying plasmids pGST-rng3, pGST-rng3-UCS, pGST-cam1, and pGST-rlc1. GST proteins from extracts were affinity purified on glutathione-Sepharose. Bound proteins were separated by SDS-PAGE and analyzed by immunoblotting with antibodies to Myo2p heavy chain. pGST-cam1, negative control with GST-Cam1p. pGST-rlc1, positive control with GST-Rlc1p. (D) GST-Rng3p purified from S. pombe and recombinant GST-Rng3p purified from Escherichia coli. SDS-PAGE gel stained with Coomassie blue. Lower band in the S. pombe lane represents a breakdown product. (E–J) Actin filament gliding assays. Time-lapse fluorescence micrographs of filaments labeled with rhodamine-phalloidin (also, see Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). Trajectories are indicated with white dots marking the trailing end of filaments at 2-s intervals. Bar, 5 μm. Conditions: indicated concentrations of Myo2 and GST-Rng3p were applied to flow cells in 25 mM imidazole, pH 7.4, 25 mM KCl, 4 mM MgCl2, 1 mM ATP, 100 mM DTT, and 10 nM labeled actin filaments. (E) Crude one-step purified Myo2 (0.25 mg/ml impure protein). (F) Three-step–purified 75 nM Myo2 preincubated with 250 nM native S. pombe GST-Rng3p. (G) Three-step–purified 75 nM Myo2 preincubated with 250 nM recombinant GST-Rng3p. (H) Three-step–purified 75 nM Myo2 alone. (I) S. pombe GST-Rng3p (250 nM) alone. (J) recombinant GST-Rng3p (250 nM) alone. (K) Dependence of the actin-activated ATPase activity of three-step–purified Myo2 as a function of the concentrations of native (open circles) and recombinant (closed circles) GST-Rng3p. Conditions: 30 nM Myo2 and 10 μM actin filaments in 2 mM ATP, 3 mM MgCl2, 0.1 mM CaCl2, and 75 mM KCl. Error bars show SD. (L) Dependence of the number of actin filaments captured by two-step–purified Myo2 on the concentration of recombinant GST-Rng3p. All filaments in a 130 μm2 frame of a fluorescence micrograph were counted. (squares) 20 nM two-step–purified Myo2. No gliding was observed in the absence of GST-Rng3p. (circles) 200 nM two-step–purified Myo2. These samples supported gliding in the absence of GST-Rng3p. (horizontal line) 150 nM crude Myo2 purified after overexpression from MLP 374 (3nmt1 promoter-myo2 plus pGST-cdc4 and pGST-rlc1). These samples supported robust gliding.
Rng3p interacts with and stimulates Myo2 activity
We took a candidate protein approach to identify the activating factor for Myo2, beginning with the UCS protein Rng3p. Fission yeast Rng3p is required to localize Myo2 to the contractile ring and for cytokinesis (Wong et al., 2000). Rng3p is related to C. elegans Unc45 (Barral et al., 1998), Podospora anserina Cro1p (Berteaux-Lecellier et al., 1998), and budding yeast She4p (Jansen et al., 1996). The UCS domains of these proteins are conserved in yeast and animals and participate in the function of certain myosins from classes I, II, and V, possibly as chaperones that fold the myosin head (Yu and Bernstein, 2003).
Consistent with Rng3p being the missing activating factor for Myo2 motility activity, Rng3p copurified with crude Myo2 but was depleted upon further purification of Myo2 (Fig. 3 A). Likewise, Myo2 associated with native or overexpressed Rng3p, although in smaller amounts than with overexpressed Rlc1p (Fig. 3, B and C). Genetic studies demonstrated a synthetic interaction between rng3 mutations and a myo2-E1 mutation (Wong et al., 2000), but GST-Rng3p pulled down the same amount of Myo2-E1p as Myo2p (Fig. 3, B and C).
Highly purified recombinant Rng3p or GST-Rng3p partially purified from S. pombe (Fig. 3 D) restored in vitro motility of three-step purified Myo2 (Fig. 3, F and G; and Video 2 A, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1) with rates similar to crude Myo2 (Table I). Native or recombinant Rng3p also doubled the actin-activated Mg-ATPase (Fig. 3 K and Table I). Note that recombinant Rng3p came from bacteria lacking fungal Hsp70, Hsp90, or kinases. Neither three-step purified Myo2 nor GST-Rng3p alone bound actin filaments to coverslips in ATP buffer (Fig. 3, H–J; and Video 2, B and C, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1).
Rng3p also enhanced actin filament binding and motility powered by two-step purified Myo2. High concentrations but not low concentrations of this preparation supported gliding of a few attached filaments (Fig. 3 L). Recombinant Rng3p dramatically increased the number of filaments captured on the surface in a concentration-dependent fashion (Fig. 3 L).
Repression of Rng3p expression compromises concentration of Myo2 in the contractile ring and cytokinesis (Fig. 4 A), similar to a rng3 mutant (Wong et al., 2000). This experiment used a strain with two chromosomal replacements: a thiamine repressible 41nmt1 promoter replaced the rng3 promoter; and a GFP-myo2 fusion replaced the Myo2p coding sequence. Control cells expressing Rng3p in the absence of thiamine had no defects in cytokinesis or Myo2p/actin localization. Repression of rng3 expression with thiamine resulted in cells with Myo2 and actin mislocalized together in disorganized structures that failed to condense into contractile rings. Cytokinesis and septation failed, so these cells became multinucleate.
Figure 4.
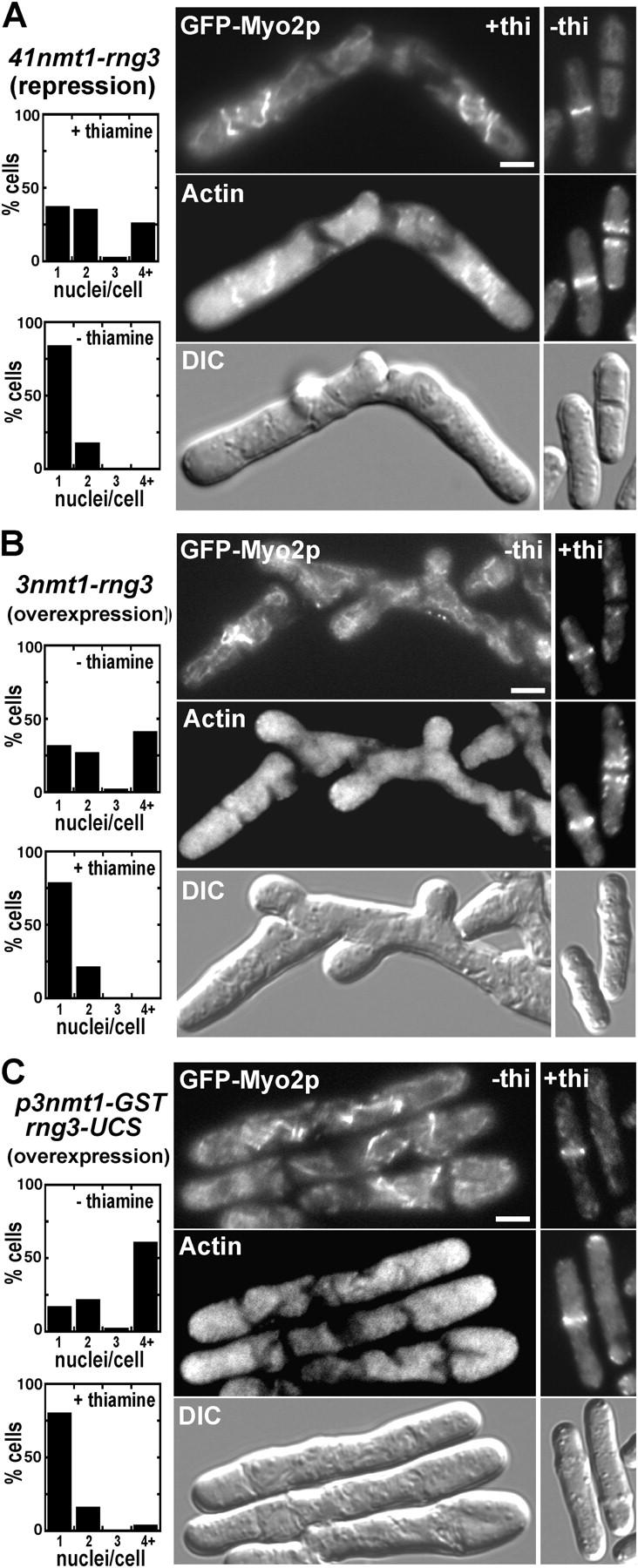
Effects of Rng3p repression or overexpression on Myo2p and actin localization and cytokinesis. Fluorescence and DIC micrographs of yeast cells expressing GFP-Myo2p and stained for actin with rhodamine phalloidin. Bars, 5 μm. Cells were mounted on 25% gelatin pads in the appropriate EMM medium. In a separate experiment, cells were treated with Hoescht stain to mark nuclei. (A) Repression of Rng3p expression: MLP 640 (41nmt1 promoter-rng3, GFP-myo2) was grown in liquid EMM medium with 5 μg/ml thiamine to repress Rng3p expression (left micrographs) or without thiamine (right micrographs). (top) GFP-Myo2p; (middle) actin; (bottom) DIC. Plots on the left summarize the phenotypes of cells grown in either nonrepressing (− thiamine) or repressing (+ thiamine) conditions as quantitated by scoring nuclei/cell using fluorescence microscopy. (B) Overexpression of Rng3p and (C) overexpression of the Rng3p UCS domain. MLP 639 (3nmt1 promoter-rng3, GFP-myo2) and FY 435 carrying pGST-rng3-UCS were grown in liquid EMM medium supplemented with 5 μg/ml thiamine (to repress Rng3p and Rng3p-UCS expression to a modest level) or without thiamine (to overexpress Rng3p and Rng3p-UCS). (top) GFP-Myo2p; (middle) actin; (bottom) DIC. Plots on the left summarize the phenotypes of cells grown in either inducing (− thiamine) or uninducing (+ thiamine) conditions.
Overexpression of rng3 or the rng3-UCS domain by induction of an integrated (strong) 3nmt1 promoter (in the absence of thiamine) also caused defects in Myo2 localization and cytokinesis (Fig. 4, B and C). Myo2p mislocalized as disorganized structures, whereas actin was diffuse, often accumulating at the tips of elongated cells. Control cells expressing low levels of Rng3p in the presence of thiamine lacked these defects (Fig. 4, B and C). Thus, cells require an optimal concentration of Rng3p for Myo2 localization and cytokinesis.
Rng3p UCS domain alone restores the motility activity of pure Myo2
The COOH-terminal UCS domain of Rng3 has all of the activities of full-length Rng3p that we have detected. (a) Overexpressed GST-Rng3p UCS domain pulled down Myo2 somewhat more effectively than full-length Rng3p (Fig. 3 C). (b) Overexpression of Rng3 UCS compromised the formation of contractile rings resulting in cytokinesis defects (Fig. 4 C). (c) The purified UCS domain of Rng3p fully restored the in vitro motility of pure Myo2 (Table I and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). The UCS domain is the only region of Rng3p having significant homology with other UCS proteins (Wong et al., 2000; Hutagalung et al., 2002).
Rng3p localizes to the contractile ring
Wong et al. (2000) observed Rng3p-GFP in contractile rings in temperature-sensitive myo2-E1 cells but not wild-type cells. One explanation is that Rng3p has a chaperone function and only concentrates in contractile rings with defective Myo2-E1p. Alternatively, if Rng3p activates Myo2, it may be recruited in greater amounts to the contractile ring of myo2-E1 cells to compensate for reduced Myo2 activity. To distinguish these hypotheses, we integrated triple GFP or YFP tags at the COOH terminus of a fully functional rng3 locus under the control of the native promoter.
Rng3p-GFP3 and Rng3p-YFP3 concentrated in contractile rings from anaphase B through constriction (Fig. 5). CFP-Myo2p joins the contractile ring earlier, beginning with a broad band of small dots before condensing into the contractile ring (Wu et al., 2003). Thus, 21% of full-diameter CFP-Myo2p contractile rings lacked detectable Rng3p-YFP3, whereas both CFP-Myo2p and Rng3p-YFP3 concentrated in all constricting contractile rings (Fig. 5, C–E; note the positions of the nuclei in Fig. 5 D; Video 4, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). The Rng3p-YFP3 signal was weak, so we cannot rule out low concentrations of Rng3-YFP3 in immature contractile rings, but Rng3p lags behind Myo2p incorporation into the ring.
Figure 5.
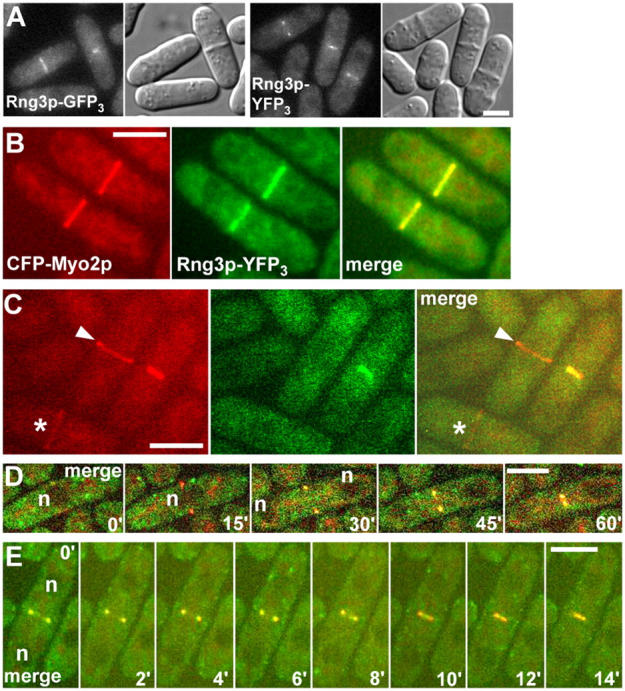
Rng3p colocalizes with Myo2p at the contractile ring. (A) Epi-fluorescence and DIC micrographs of live yeast cells expressing Rng3p-GFP3 (MLP 662) or Rng3p-YFP3 (MLP 660). (B–E) Spinning disk confocal fluorescence micrographs of live yeast cells expressing CFP-Myo2p and Rng3p-YFP3 (MLP 665). Bars, 5 μm. Cells were mounted on 25% gelatin pads in EMM medium and photographed with GFP, YFP, or CFP filters. (A) Images of Rng3p-GFP3 (left panels) and Rng3p-YFP3 (right panels) in dividing cells. (B) Images of Myo2p (left), Rng3p (middle), and their merge (right) in two cells from a stack of 12 confocal z sections of 0.6 μm. (C) Images of Myo2p, Rng3p, and their merge are shown (as in B). Asterisk denotes Myo2p broad band; arrowhead indicates Myo2p ring lacking Rng3p. (D and E) Time-lapse series of a cell at intervals of 15 min (D) and 2 min (E). To avoid bleaching a single z section was collected at each time point. Myo2p and Rng3p are shown in merged format. Myo2p is detected at the contractile ring (D, 15' panel) before Rng3p. Both constrict together (D and E; Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). n, position of nuclei.
Genetic requirements for Myo2 activity
The ability of crude Myo2 (purified by affinity chromatography) to support actin filament motility provided an assay for the consequences of mutations on Myo2 activity. Despite the fact that cells with the temperature-sensitive myo2-E1 mutation grow at the permissive temperature, Myo2-E1 from these cells was unable to capture actin filaments, let alone power gliding even at the permissive temperature (Table I). Preincubation of Myo2-E1 with purified Rng3p did not rescue gliding (Table I).
Rlc1p is not required for viability (Le Goff et al., 2000; Naqvi et al., 2000), but cells lacking Rlc1p die under stressful conditions (Fig. 6 B). Myo2 from rlc1Δ cells moved actin filaments at only 20% the rate of wild-type Myo2 (Fig. 6 E, Table I, and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). Pre-incubation of one step purified Myo2 from rlc1Δ cells with purified GST-Rlc1p restored in vitro motility to rates close to those of wild type (Table I).
Figure 6.
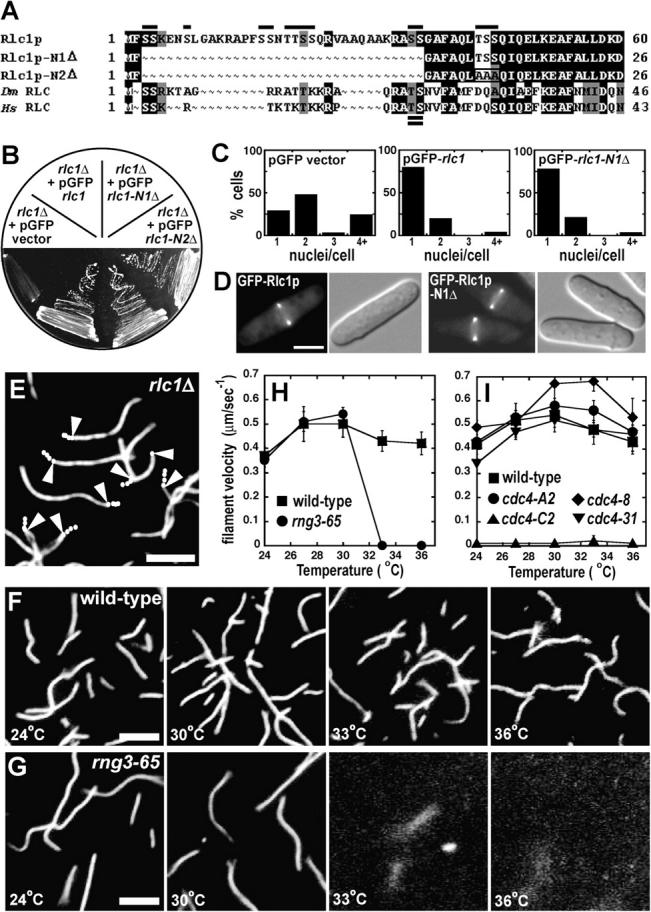
Effects of mutations in Myo2p light chains and Rng3p on motility activity. (A) Amino acid sequence alignment of the NH2-terminal regions of Rlc1p, Rlc1p-N1Δ, Rlc1p-N2Δ, D. melanogaster RLC, and Homo sapien RLC homologues. Alignment was generated with MacVector 7.1.1 and Boxshade software. Single (above) and double (below) lines on the alignment denote potential Rlc1p Ser/Thr phosphorylation sites and the phospho-regulatory Thr-Ser characteristic of higher eukaryotic RLCs, respectively. The box in Rlc1p-N2Δ indicates the additional amino acid substitutions. Black, amino acid identities; gray, amino acid similarities. (B) Viability of an rlc1Δ strain (MLP 7) carrying empty vector (negative control), pGFP-rlc1 (positive control), pGFP-rlc1-N1Δ, or pGFP-rlc1-N2Δ. Transformants were streaked on an EMM Ura- plate containing 1M KCl. (C) Phenotypic quantitation of MLP 7 carrying empty vector (negative control), pGFP-rlc1 (positive control), or pGFP-rlc1-N1Δ. Transformants were grown in liquid EMM Ura− media and their nuclei stained. Nuclei/cell were visualized and scored by fluorescence microscopy. (D) Localization of GFP-Rlc1p in MLP 7 containing either pGFP-rlc1 (left panels) or pGFP-rlc1-N1Δ (right panels). Cells were grown in liquid EMM Ura− media. Fluorescence and DIC micrographs are shown. Bar, 5 μm. (E–I) Actin filament gliding assays using crude Myo2 (0.25 mg/ml impure protein) isolated in one step on glutathione-Sepharose from strains with mutations in Myo2p light chains and Rng3p. (E–G) Fluorescence micrographs of filaments labeled with rhodamine-phalloidin. Bars, 5 μm. (H and I) Quantitation of gliding rates. Conditions: samples were applied to flow cells in 25 mM imidazole, pH 7.4, 25 mM KCl, 4 mM MgCl2, 1 mM ATP, 100 mM DTT, and 10 nM labeled actin filaments. (E) Myo2 from a strain lacking Rlc1p (MLP 534 rlc1Δ 41nmt1 promoter-myo2 plus pGST-cdc4). Trajectories are indicated with white dots marking the trailing end of filaments at 2-s intervals. Arrowheads mark the 6-s time point in filaments that moved. (F) Temperature dependence of actin filament attachment to crude, wild-type Myo2 (0.25 mg/ml) purified from MLP 509 (41nmt1 promoter-myo2 plus pGST-cdc4 and pGST-rlc1). (G) Temperature dependence of actin filament attachment to crude Myo2 (0.25 mg/ml) purified from a strain with a temperature-sensitive mutation, rng3-65 (MLP 586 rng3-65 41nmt1 promoter-myo2 plus pGST-cdc4 and pGST-rlc1). (H) Temperature dependence of in vitro motility rates of crude Myo2 purified from wild-type (MLP 509) and rng3-65 (MLP 586) temperature-sensitive backgrounds. (I) Temperature dependence of in vitro motility rates of crude Myo2 purified from wild-type (MLP 509) and cdc4 temperature-sensitive backgrounds. Strains: MLP 539 cdc4-8 41nmt1 promoter-myo2, plus pGST-cdc4-8 and pGST-rlc1; MLP 641 cdc4-31 41nmt1 promoter-myo2 plus pGST-cdc4-31 and pGST-rlc1; MLP 647 cdc4-C2 41nmt1 promoter-myo2 plus pGST-cdc4-C2 and pGST-rlc1; MLP 648 cdc4-A2 41nmt1 promoter-myo2 plus pGST-cdc4-A2 and pGST-rlc1.
None of the 14 potential regulatory phosphorylation sites at the NH2 terminus of Rlc1p is required for biological function. Truncation of 36 NH2-terminal residues including 11 potential phosphorylation sites did not compromise in vitro motility (Table I), Rlc1p function in vivo, or localization at the contractile ring (Fig. 6, A–D). Like wild-type Rlc1p, Rlc1p-N1Δ grew on plates containing 1M KCl (Fig. 6 B) and displayed none of the cytokinesis defects associated with loss of Rlc1p function (Fig. 6 C). Further, a truncated form of Rlc1p (Rlc1p-N2Δ) with alanines replacing three additional potential phosphorylation sites (Thr43-Ser44-Ser45; Fig. 6 A) was also fully functional and localized to the contractile ring (Fig. 6 B and not depicted).
In vitro motility is temperature sensitive for crude Myo2 purified from the rng3-65 ts mutant. Motility of crude Myo2 from rng3-65 cells was normal up to 30°C (Table I), but when the chamber was warmed to 33°C, the filaments stopped moving and dissociated from the surface of the coverslip (Fig. 6, F–H). Wild-type Myo2 is active up to 36°C. This result is strong evidence that Rng3p is the factor that supports the motility activity of crude Myo2 (Fig. 3 A).
Although temperature-sensitive mutations in the cdc4 ELC gene are lethal at the restrictive temperature owing to cytokinesis defects (Balasubramanian et al., 1998; Desautels et al., 2001), the motility activity of crude Myo2 from cdc4 conditional alleles was defective only in strain cdc4-C2 but not strains cdc4-8, cdc4-31, and cdc4-A2 (Table I and Fig. 6 I). Overexpression of wild-type GST-Cdc4p but not GST-Cdc4-8p, GST-Cdc4-31p, or GST-Cdc4-A2p corrected the cytokinesis defects of these cdc4 mutants at the restrictive temperature (unpublished data). Thus, the normal motility of crude Myo2 from cells overexpressing mutant forms of Cdc4p was not caused by simply overexpressing defective Cdc4p. The motility activity of crude Myo2 from cdc4-C2 temperature-sensitive cells was severely compromised, even at permissive temperatures (Table I and Fig. 6 I). Crude Myo2 from cdc4-C2 cells bound to the slide only 4% the number of actin filaments as wild-type Myo2, and bound filaments moved little or not at all. These results suggest that cytokinesis defects in the cdc4-8, cdc4-31, and cdc4-A2 mutants are not due to lack of Myo2 function, but instead reveal another role of Cdc4p in cytokinesis.
Deletion of Myo2 ELC or RLC binding sites compromises activity
Strains with deletions of either the IQ1 motif (Cdc4p binding site) or the IQ2 motif (Rlc1p binding site) from the myo2 gene grow normally, even in the absence of myp2 (Naqvi et al., 2000; D'souza et al., 2001). Crude preparations of Myo2-IQ1Δp/Rlc1p and Myo2-IQ2Δp/Cdc4p both supported motility, but the velocity with Myo2-IQ1Δp/Rlc1p was 12% that of wild-type Myo2 and that of Myo2-IQ2Δp/Cdc4p was only 6% of wild type (Table I and Video 6, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1). Myo2 with just a single light chain and low motility activity suffices for cytokinesis and growth.
Discussion
We report the first purification and characterization of a native fungal myosin. The native protein comes from an environment with all of the factors required for optimal posttranslational modifications. Further, purified native myosin provides a means to test whether or not mutations that compromise cytokinesis have a direct effect on Myo2. In addition to contributing to understanding the role of myosin-II in cytokinesis, our work establishes fission yeast as a genetically tractable model system to relate biochemical activities to biological function.
We confirmed that Cdc4p and Rlc1p associate with Myo2p and showed that none of the other candidate light chains are subunits of Myo2. When overexpressed, the calmodulin-like protein Cam2p can bind Myo2p but does not purify with native Myo2. Instead, Cam2p is likely a light chain for Myo1p, the single type-I myosin of fission yeast. Cam1p and Cam2p may bind the two Myo1p IQ motifs, one complete and the other incomplete (Toya et al., 2001).
Purified Myo2 has most of the characteristics of a functional myosin-II: large Stokes' radius (13.7 nm); an 87-nm tail (Bezanilla and Pollard, 2000); insolubility at physiological ionic strength, likely reflecting its ability to form filaments; and ATP-sensitive binding to actin filaments. The ATPase profile is characteristic of myosin-II, activated by EDTA in high K+, and activated by actin filaments in Mg2+. In spite of these characteristics of a native myosin-II, Myo2 lost its ability to support actin filament gliding in in vitro motility assays as it was purified.
Our central finding is that the UCS domain of Rng3p restores the ability of purified Myo2 to move actin filaments. Previous work established a connection between UCS proteins and myosins. The C. elegans UCS-domain protein, Unc45, is required for the organization of myosin-II in muscle (Barral et al., 1998). S. pombe Rng3p is required for incorporation of Myo2p into the contractile ring, as well as the constriction of the ring (Wong et al., 2000). Budding yeast She4p, although not required for function of the essential type V myosin (Myo2p) and the type-II myosin (Myo1p), is required by both type-I myosins (Myo3p and Myo5p) and Myo4p, a second type-V myosin (Toi et al., 2003; Wesche et al., 2003). Work on Unc45 suggested that UCS proteins are molecular chaperones or cochaperones required for folding the heads of myosin-II and possibly other myosins (Barral et al., 2002; Yu and Bernstein, 2003).
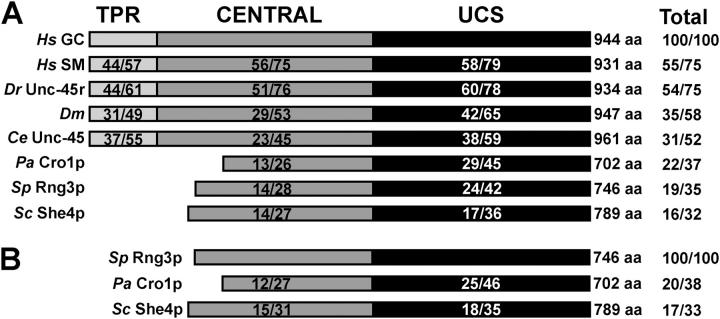
Domain organization of UCS proteins and association with known chaperones
Like Unc45, most UCS-domain proteins consist of three domains: an NH2-terminal tetratricopeptide repeat (TPR); a central region; and a COOH-terminal UCS domain (Hutagalung et al., 2002). TPR domains can bind either Hsp70 or Hsp90 chaperones (Blatch and Lassle, 1999). The TPR domain of Unc45 is specific for Hsp90 in vitro (Barral et al., 2002). The central domain is well conserved only in animals, whereas the UCS domain is well conserved in all UCS proteins (Hutagalung et al., 2002; Fig. 7 A). When overexpressed in Sf9 cells, a truncated form of Unc45 lacking its TPR domain copurified with endogenous insect Hsp70, but not Hsp90. (Barral et al., 2002). This truncated form of Unc45 also bound C. elegans myosin-II in vitro. Because Unc45 inhibited thermal aggregation of scallop myosin subfragment 1, Barral et al. (2002) concluded that Unc45 acts both as a molecular chaperone and a Hsp90 cochaperone supporting proper folding of myosin heads and subsequent assembly of muscle thick filaments.
Figure 7.
Domain organization and sequence comparisons of UCS proteins. Amino acid sequence comparisons (identities/similarities) were generated from sequence alignments (MacVector 7.1.1). Conservation amongst NH2-terminal tetratricopeptide repeat (TPR) domains (light gray), central regions (gray), and COOH-terminal UCS domains (black) are compared individually. Amino acid sequence length and total conservation are shown on the right for each protein. (A) Comparison of animal and fungal UCS proteins with H. sapiens general cell form (Hs GC; Price et al., 2002), H. sapiens striated muscle form (Hs SM; Price et al., 2002), Danio rerio (Dr Unc45r; zebrafish; Etheridge et al., 2002), D. melanogaster (Dm; GenBank/EMBL/DDBJ accession no. AAK93568), C. elegans (Ce Unc-45; Venolia et al., 1999); P. anserina (Pa Cro1p; Berteaux-Lecellier et al., 1998); S. pombe (Sp Rng3p; Wong et al., 2000); and Saccharomyces cerevisiae (Sc She4p; Jansen et al., 1996). (B) Comparison of fungal UCS proteins with Rng3p.
Animal and fungal UCS proteins diverged significantly from a common ancestor. All three known fungal UCS proteins lack the TPR domain and lack, even amongst themselves, any obvious sequence similarity in their central domains (Fig. 7, A and B). Budding yeast She4p is the most divergent, with relatively low sequence similarity to animal and other fungal UCS proteins in both its central and UCS domains (Fig. 7, A and B).
Evidence that Rng3p activates the Myo2 motor and is not simply a chaperone
Our evidence shows that Rng3p is an accessory subunit required for the motor activity of purified Myo2. Rng3p doubles the actin-activated ATPase activity of Myo2 and in the presence of ATP promotes binding and movement of actin filaments in the in vitro motility assay. Thus, we propose that Rng3p promotes the interaction of the “weak-binding” Myo2-ATP and –ADP-Pi intermediates with actin filaments. Biophysical analysis of single molecules will be required to learn how Rng3p participates in force production by Myo2. Two-hybrid and coprecipitation experiments are also consistent with the budding yeast UCS protein She4p promoting interaction of Myo5p with actin (Toi et al., 2003; Wesche et al., 2003).
Activation of Myo2 does not appear to reflect an ability of Rng3p to promote the folding of the Myo2 head. First, Rng3p purified from bacteria, which lack the requisite Hsp90, activates Myo2. Second, purified Myo2 binds reversibly to actin filaments and has robust ATPase activity, so it must be folded without associated Rng3p. Third, cells with excess Rng3p fail to coalesce Myo2 into a contractile ring. This finding is easier to explain by over-activation than by excess chaperone activity. Fourth, Rng3p is required to maintain contractile rings. Fully formed contractile rings disintegrate when rng3-65 cells are shifted to the restrictive temperature (Wong et al., 2000). If Rng3p were simply required to fold Myo2p before its incorporation in a contractile ring, one would not expect it to be required later.
Our finding that the UCS domain of Rng3p interacts with Myo2 and stimulates its activity is consistent with previous work. Both rng3 temperature-sensitive alleles known to compromise Myo2 function have point mutations in the UCS domain (Wong et al., 2000). The UCS domain of budding yeast She4p associates with Myo5p (Toi et al., 2003; Wesche et al., 2003). The activities and functions of the NH2-terminal regions of Rng3p are not known.
Evidence that other UCS proteins activate myosins
Budding yeast UCS proteins may also activate myosin motors. Each of four independent MYO5 point mutations that suppress the defects of a she4Δ strain is located inside the cleft between the upper and lower 50-kD domains of this myosin-I head (Toi et al., 2003). Conformational changes involving this cleft are thought to modulate affinity of the myosin head for actin filaments (Rayment et al., 1993). We find it unlikely that each of these single amino acid substitutions would allow an unfolded myosin to fold in the absence of a required chaperone. Considering their position, these MYO5 mutations likely act as enhancers that bypass the requirement for She4p activation, possibly increasing affinity for actin filaments in ATP.
The dependence of myofibrillogenesis on UCS proteins has been attributed to chaperone or chaperone-binding activity, but is also consistent with myosin activation. For example, inhibiting myosin-II activity in Xenopus laevis myocytes with N-benzyl-p-toluenesulphonamide compromises myofibril formation (Ramachandran et al., 2003). Similarly, fission yeast with reduced Rng3p fail to coalesce Myo2 into a contractile ring. Thus, myosin-II motor activity may contribute to assembly of sarcomeres and contractile rings. Furthermore, Unc45 colocalizes with myosin-II at A-bands of sarcomeres (Ao and Pilgrim, 2000), whereas Hsc70 and Hsp90 chaperones only colocalize with myosin-II before its incorporation into sarcomeres (Srikakulam and Winkelmann, 2004). The presence of Unc45 in sarcomeres suggests a role in the function of folded myosin-II.
Roles of Rng3p and Myo2 subunits in cytokinesis
Formation of a functional contractile ring depends on Rng3p. Myo2p appears to concentrate around the equator independent of Rng3p, but maturation and constriction of the contractile ring depend on Rng3p (Wong et al., 2000). Consistent with the idea that the function of Rng3p is to activate Myo2, Rng3p colocalizes with Myo2p in mature and constricting contractile rings.
Biochemical assays of crude Myo2 from different genetic backgrounds provide an assay for the roles of the three Myo2 subunits. Myo2-E1 with a temperature-sensitive point mutation in the catalytic domain failed to support filament gliding even at permissive temperatures. Residual activity not detected here may allow myo2-E1 cells to divide at permissive temperatures. Alternatively, because deletion of Myp2p accentuates the cytokinesis defects of myo2-E1 cells (Motegi et al., 1997), Myp2p may suffice for effective cytokinesis, providing that enzymatically inactive Myo2 contributes some other essential function.
RLC Rlc1p is not essential for viability, although deletion strains die when stressed. The ability of cells to function without Rlc1p is explained by the fact that Myo2 from a strain lacking Rlc1p still moved actin filaments at 20% the wild-type rate.
None of the potential phosphorylation sites near the NH2 terminus of Rlc1p is required for motor activity or cytokinesis. Thus, other factors, such as Rng3p must regulate Myo2. Phosphorylation of RLCs controls the motor activity of many myosin-IIs, either activating or inhibiting motility depending on the phosphorylated residue (Bresnick, 1999).
Biochemical analysis of Myo2 from strains with temperature sensitive alleles of cdc4 show that Cdc4p contributes at least one function beyond its structural role in the lever arm of Myo2. Three (cdc4-8, cdc4-31, and cdc4-A2) of four cdc4 mutations had no effect on the biochemical activity of isolated Myo2 in spite of severe defects in cytokinesis at the restrictive temperature. Only the cdc4-C2 mutation strongly affected Myo2 activity. The cdc4-C2 mutation substitutes lysine for strictly conserved arginine-33, a residue contacting the IQ helix in a crystal structure of an ELC-IQ complex (Terrak et al., 2003). The cdc4-C2 mutation may compromise Cdc4p binding to Myo2p or its activity.
Unlike deletion of cdc4, loss of the ELC binding site (IQ1) from myo2 has no effect on cell growth (D'souza et al., 2001) despite Myo2-IQ1Δ exhibiting relatively low in vitro motility activity (0.06 μm/s). Consistent with our biochemical findings with mutant Cdc4p forms, the myo2-IQ1Δ mutation partially suppressed the cdc4-C2 allele but not the cdc4-8, cdc4-31, and cdc4-A2 alleles (D'souza et al., 2001).
Genetics first suggested ELC bifunctionality because some cdc4 mutant alleles exhibit interallelic complementation (Nurse et al., 1976; Desautels et al., 2001). Coprecipitation experiments demonstrated that Cdc4p also interacts with a phosphatidylinositol 4 (PI 4)-kinase and IQGAP/Rng2p (Desautels et al., 2001; D'souza et al., 2001). Of the reported mutant forms of Cdc4, Cdc4-8p fails to interact with PI 4-kinase (Desautels et al., 2001). Although PI 4-kinase has no established role in fission yeast cytokinesis, PI 4-kinase is required for cytokinesis in Drosophila melanogaster spermatids (Brill et al., 2000). Similar to Cdc4-8p, Cdc4-31p and Cdc4-A2p may lack an important protein interaction critical for cytokinesis.
The ability of a myo2-IQ2Δ mutation (deletion of the RLC binding site) to suppress cytokinesis defects associated with an rlc1Δ mutation led to the idea that Rlc1p overcomes auto-inhibition of Myo2 imposed by the unoccupied IQ2 region (Naqvi et al., 2000). Our biochemical results suggest another view. The motility activity of Myo2 from an rlc1Δ strain (0.09 μm/s) is actually greater than Myo2-IQ2Δ (0.03 μm/s). Because neither the myo2-IQ1Δ and myo2-IQ2Δ strains exhibit cytokinesis defects (Naqvi et al., 2000; D'souza et al., 2001) in spite of low motility activity, the defects in rlc1Δ cells may stem from rogue light chains invading the open IQ2 domain and compromising cellular functions.
Reconciliation of cellular and biochemical phenotypes
Our results emphasize how difficult it is to interpret the effects of mutations on cellular function without parallel biochemical analysis. We now see that cells can complete cytokinesis and survive with Myo2 having little (myo2-IQ1Δ, myo2-IQ2Δ, rlc1Δ, cdc4-C2) or no (myo2-E1) detectable motility. In contrast, several cdc4 conditional alleles die at the restrictive temperature in spite of Myo2 with full activity, presumably due to loss of other Cdc4p functions.
Very little Myo2 function is required to complete cytokinesis. This finding makes sense because the contractile ring constricts slowly, at a maximum rate of 20 nm/s (Wu et al., 2003). Because native Myo2 moves actin filaments at 470 nm/s, a bipolar filament of Myo2 would pull together oppositely polarized actin filaments at 940 nm/s. Around a ring with a circumference of 15 μm, several Myo2 bipolar filaments are likely to function in series, adding their sliding velocities together. Just five such units in series would constrict an unloaded actin ring at 4700 nm/s, over two orders of magnitude faster than constriction of the contractile ring. Neither the resistance in the cell nor the force velocity relationship for Myo2 is known, but the fact that Myo2 with only 2% of wild-type motility activity suffices for cytokinesis suggests that the system has vast excess capacity, perhaps as a fail-safe system for this vital function.
Materials and methods
Myo2 purification
Myo2p/Rlc1p/Cdc4p (Myo2): MLP 509 was cotransformed with pGST-rlc1 and pGST-cdc4, and transformants were isolated on EMM Leu− Ura− plates containing 5 μg/ml of thiamine. Liquid cultures were grown to saturation in EMM Leu− Ura− with 5 μg/ml of thiamine. Cells were harvested and washed three times in EMM Leu− Ura− medium and diluted to an optical density at 595 nm (OD595) of ∼0.05 in 4 liters of the same medium. Overexpression of Myo2p, GST-Rlc1p, and GST-Cdc4p was achieved after 24–28 h of incubation at 32°C, by which time the OD had reached 3. When purifying Myo2p from temperature-sensitive mutants, cells were grown at 25°C with overexpression without thiamine for 30–36 h. Cells were harvested and washed once in water and once in ice-cold lysis buffer (750 mM KCl, 25 mM Tris-HCl, pH 7.4, 4 mM MgCl2, 20 mM Na4P2O7, 2 mM EGTA, and 0.1% Triton X-100). Pellets were resuspended in an equal volume of ice-cold lysis buffer with additives consisting of 1 mM DTT, 4 mM ATP, 2 mM PMSF, complete EDTA-free protease inhibitors (Roche), and diisopropyl fluorophosphate at a final concentration of 0.5 mM. From this point forward all work was performed at 4°C and samples were stored on ice. Cells were lysed by glass bead beating with a Fastprep (BIO101) bead beater. The lysate was centrifuged at 500 g for 5 min to remove unlysed cells and beads and further centrifuged at 100,000 g for 45 min to remove insoluble matter. The supernatant was batch incubated with 2 ml of glutathione-Sepharose (Amersham Biosciences) for 90 min, and then transferred to a 20-ml disposable column. The bound sample was washed with lysis buffer with additives (4 × 15 ml) and eluted in 5 ml of lysis buffer plus additives and 10 mM glutathione. Eluate contained affinity-purified GST-Rlc1p and GST-Cdc4p enriched with copurifying Myo2p. The sample was filtered (0.22 μm filter; Millipore) and dialyzed into A15 buffer (0.5 M KCl, 10 mM imidazole, pH 7.0, 10 mM EDTA, 1 mM DTT, and 0.3 mM NaN3) overnight in the presence of thrombin (Amersham Biosciences; 10 units/mg Myo2p/GST-Cdc4p/GST-Rlc1p) to detach GST from the light chains. This one-step purified Myo2 was either stored at −20°C in A15 buffer containing 50% glycerol or further purified by gel filtration on a 2.5 × 50 cm column of Bio-Gel A15m, 200–400 mesh (Bio-Rad Laboratories) equilibrated and eluted with A15 buffer. Gel filtration fractions enriched in Myo2 were applied to a 1 ml hydroxyapatite (Bio-Gel HT; Bio-Rad Laboratories) column equilibrated with A15 buffer lacking EDTA. A 10–200-mM potassium phosphate pH 7.0 gradient (in 75 ml of A15 buffer lacking EDTA) was used to elute Myo2 (peak elution at 100 mM phosphate). Myo2 fractions were pooled and dialyzed extensively into A15 buffer. Samples were stored at −20°C in A15 buffer/50% glycerol. Protein concentrations were determined by Bradford assay with rabbit skeletal muscle myosin as the standard. This three-step purification typically yielded 0.5 mg Myo2 per 4 liters of culture. Myo2 was purified in one step by affinity chromatography on glutathione-Sepharose purified from a variety of genetic backgrounds expressing appropriate GST-light chains. Where appropriate, the Myo2 concentration in one-step samples was estimated by densitometry from stained protein gels carrying known concentrations of pure Myo2.
Image acquisition
Fluorescent proteins/stains were imaged with a microscope (model IX71; Olympus) at ambient temperature by epi-fluorescence illumination with a PlanApo 60× (1.4) objective and recorded with a cooled CCD camera (model Orca-ER; Hamamatsu). For temperatures above ambient, the objective and specimen were heated with a Flexible Heater (model Omegalux KHLV0504/2-P; Kapton). For Spinning Disk confocal microscopy, the microscope was connected to a confocal scanner (model Ultraview RS; PerkinElmer) and the images were captured with a PlanApo 100× (1.4) objective. Images were acquired and processed using Metamorph, Ultraview RS, and ImageJ software.
Online supplemental material
Standard fission yeast methods and lists of strains (Table SI) and plasmids (Table SII) used in this study (and details of their construction) are provided online. Details of additional cell biological and biochemical methods are available online. Video 1 shows filament gliding by crude Myo2. Videos 2 and 3 are stimulations of Myo2 actin filament gliding by Rng3p and Rng3p-UCS domain, respectively. Video 4 shows colocalization of Rng3p with Myo2p during cytokinesis. Videos 5 and 6 show the effects of rlc1Δ and myo2-IQ1Δ and myo2-IQ2Δ, respectively, on filament gliding by Myo2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200404045/DC1.
Acknowledgments
We are grateful to Rachel Mahaffy for assistance with fluorescence methods and Volodia Sirotkin and Jian-Qiu Wu for help with cell imaging. We thank Jian-Qiu Wu for GFP/CFP strains; Mohan Balasubramanian for myo2 and rng3 mutants; Fred Chang and Sean Hemmingsen for cdc4 mutants; Takashi Toda for the protease-deficient strain; Susan Forsburg for numerous strains and pREP plasmids; Dave Kovar for the triple FP plasmids; Magdalena Bezanilla for the Myo2 antibody; and Jeremy Hyams for the Myo1p antibody. We acknowledge Zina Gorelik and Yuling Jiao for technical assistance and members of our lab for helpful discussions.
This work was supported by National Institutes of Health grant GM-26132 to T.D. Pollard.
Abbreviations used in this paper: ELC, essential light chain; Myo2, Myo2p/Cdc4p/Rlc1p (myosin-II); PI 4, phosphatidylinositol 4; RLC, regulatory light chain; TPR, tetratricopeptide repeat; UCS, Unc45-/Cro1p-/She4p-related.
References
- Ao, W., and D. Pilgrim. 2000. UNC-45 is a component of muscle thick filaments and colocalizes with myosin heavy chain B, but not myosin heavy chain A. J. Cell Biol. 148:375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian, M.K., D. McCollum, L. Chang, K.C. Wong, N.I. Naqvi, X. He, S. Sazer, and K.L. Gould. 1998. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 149:1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, J.M., C.C. Bauer, I. Ortiz, and H.F. Epstein. 1998. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J. Cell Biol. 143:1215–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral, J.M., A.H. Hutagalung, A. Brinker, F.U. Hartle, and H.F. Epstein. 2002. Role of myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 295:669–671. [DOI] [PubMed] [Google Scholar]
- Berteaux-Lecellier, V., D. Zickler, R. Debuchy, A. Panvier-Adoutte, C. Thompson-Coffe, and M. Picard. 1998. A homologue of the yeast SHE4 gene is essential for the transition between the syncytial and cellular stages during sexual reproduction of the fungus Podospora anserina. EMBO J. 17:1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla, M., and T.D. Pollard. 2000. Myosin-II tails confer unique functions in Schizosaccharomyces pombe: characterization of a novel myosin-II tail. Mol. Biol. Cell. 11:79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla, M., S.L. Forsburg, and T.D. Pollard. 1997. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol. Biol. Cell. 8:2693–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla, M., J.M. Wilson, and T.D. Pollard. 2000. Fission yeast myosin-II isoforms assemble into contractile rings at distinct times during mitosis. Curr. Biol. 10:397–400. [DOI] [PubMed] [Google Scholar]
- Blatch, G.L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 21:932–939. [DOI] [PubMed] [Google Scholar]
- Bresnick, A.R. 1999. Molecular mechanisms of nonmuscle myosin-II regulation. Curr. Opin. Cell Biol. 11:26–33. [DOI] [PubMed] [Google Scholar]
- Brill, J.A., G.R. Hime, M. Scharer-Schuksz, and M.T. Fuller. 2000. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 127:3855–3864. [DOI] [PubMed] [Google Scholar]
- Cyert, M.S., and J. Thorner. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'souza, V.M., N.I. Naqvi, H. Wang, and M. Balasubramanian. 2001. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct. Funct. 26:555-565. [DOI] [PubMed] [Google Scholar]
- Desautels, M., J.P. Den Haese, C.M. Slupsky, L.P. McIntosh, and S.M. Hemmingsen. 2001. Cdc4p, a contractile ring protein essential for cytokinesis in Schizosaccharomyces pombe, interacts with a phosphatidylinositol 4-kinase. J. Biol. Chem. 276:5932–5942. [DOI] [PubMed] [Google Scholar]
- Eng, K., N.I. Naqvi, K.C. Wong, and M.K. Balasubramanian. 1998. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr. Biol. 8:611–621. [DOI] [PubMed] [Google Scholar]
- Etheridge, L., P. Diiorio, and C.G. Sagerstrom. 2002. A zebrafish unc-45-related gene expressed during muscle development. Dev. Dyn. 224:457–460. [DOI] [PubMed] [Google Scholar]
- Guertin, D.A., S. Trautmann, and D. McCollum. 2002. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66:155–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung, A.H., M.L. Landsverk, M.G. Price, and H.F. Epstein. 2002. The UCS family of myosin chaperones. J. Cell Sci. 115:3983–3990. [DOI] [PubMed] [Google Scholar]
- Jansen, R.P., C. Dowzer, C. Michaelis, M. Galova, and K. Nasmyth. 1996. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 84:687–697. [DOI] [PubMed] [Google Scholar]
- Kitayama, C., A. Sugimoto, and M. Yamamoto. 1997. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 137:1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff, X., F. Motegi, E. Salimova, I. Mabuchi, and V. Simanis. 2000. The S. pombe rlc1 gene encodes a putative myosin regulatory light chain that binds the type II myosins myo3p and myo2p. J. Cell Sci. 113:4157–4163. [DOI] [PubMed] [Google Scholar]
- Lee, W.L., M. Bezanilla, and T.D. Pollard. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, K.M., S.P. Wheatley, V. Amin, and J.S. Hyams. 1998. The myosin ATPase inhibitor 2,3-butanedione-2-monoxime (BDM) inhibits tip growth and cytokinesis in the fission yeast, Schizosaccharomyces pombe. Cell Motil. Cytoskeleton. 41:117–125. [DOI] [PubMed] [Google Scholar]
- McCollum, D., M.K. Balasubramanian, L.E. Pelcher, S.M. Hemmingsen, and K.L. Gould. 1995. Schizosaccharomyces pombe cdc4 + gene encodes a novel EF-hand protein essential for cytokinesis. J. Cell Biol. 130:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., A. Feoktistova, and K.L. Gould. 1999. Phosphorylation of the myosin-II light chain does not regulate the timing of cytokinesis in fission yeast. J. Biol. Chem. 274:17691–17695. [DOI] [PubMed] [Google Scholar]
- Moser, M.J., M.R. Flory, and T.N. Davis. 1997. Calmodulin localizes to the spindle pole body of Schizosaccharomyces pombe and performs an essential function in chromosome segregation. J. Cell Sci. 110:1805–1812. [DOI] [PubMed] [Google Scholar]
- Motegi, F., K. Nakano, C. Kitayama, M. Yamamoto, and I. Mabuchi. 1997. Identification of Myo3, a second type-II myosin heavy chain in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 420:161–166. [DOI] [PubMed] [Google Scholar]
- Motegi, F., K. Nakano, and I. Mabuchi. 2000. Molecular mechanism of myosin-II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 113:1813–1825. [DOI] [PubMed] [Google Scholar]
- Naqvi, N.I., K. Eng, K.L. Gould, and M.K. Balasubramanian. 1999. Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J. 18:854–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi, N.I., K.C.Y. Wong, X. Tang, and M.K. Balasubramanian. 2000. Type II myosin regulatory light chain relieves auto-inhibition of myosin-heavy-chain function. Nat. Cell Biol. 2:855–858. [DOI] [PubMed] [Google Scholar]
- Nurse, P., P. Thuriaux, and K. Nasmyth. 1976. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 146:167–178. [DOI] [PubMed] [Google Scholar]
- Paoletti, A., N. Bordes, R. Haddad, C.L. Schwartz, F. Chang, and M. Bornens. 2003. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Mol. Biol. Cell. 14:2793–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., W.F. Stafford, and M.E. Porter. 1978. Characterization of a second myosin from Acanthamoeba castellanii. J. Biol. Chem. 253:4798–4808. [PubMed] [Google Scholar]
- Price, M.G., M.L. Landsverk, J.M. Barral, and H.F. Epstein. 2002. Two mammalian UNC-45 isoforms are related to distinct cytoskeletal and muscle-specific functions. J. Cell Sci. 115:4013–4023. [DOI] [PubMed] [Google Scholar]
- Ramachandran, I., M. Terry, and M.B. Ferrari. 2003. Skeletal muscle myosin cross-bridge cycling is necessary for myofibrillogenesis. Cell Motil. Cytoskeleton. 55:61–72. [DOI] [PubMed] [Google Scholar]
- Rayment, I., H.M. Holden, M. Whittaker, C.B. Yohn, M. Lorenz, K.C. Holmes, and R.A. Milligan. 1993. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 261:58–65. [DOI] [PubMed] [Google Scholar]
- Srikakulam, R., and D.A. Winkelmann. 2004. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117:641–652. [DOI] [PubMed] [Google Scholar]
- Terrak, M., G. Wu, W.F. Stafford, R.C. Lu, and R. Dominguez. 2003. Two distinct myosin light chain structures are induced by specific variations within the bound IQ motifs-functional implications. EMBO J. 22:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toi, H., K. Fujimura-Kamada, K. Irie, Y. Takai, S. Todo, and K. Tanaka. 2003. She4p/Dim1p interacts with the motor domain of unconventional myosins in budding yeast, Saccharomyces cerevisiae. Mol. Biol. Cell. 14:2237–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya, M., F. Motegi, K. Nakano, I. Mabuchi, and M. Yamamoto. 2001. Identification and functional analysis of the gene for type I myosin in fission yeast. Genes Cells. 6:187–199. [DOI] [PubMed] [Google Scholar]
- Venolia, L., W. Ao, S. Kim, C. Kim, and D. Pilgrim. 1999. Unc-45 gene of Caenorhabditis elegans encodes a muscle-specific tetratricopeptide repeat-containing protein. Cell Motil. Cytoskeleton. 42:163–177. [DOI] [PubMed] [Google Scholar]
- Wesche, S., M. Arnold, and R.P. Jansen. 2003. The UCS-domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr. Biol. 13:715–724. [DOI] [PubMed] [Google Scholar]
- Wong, K.C.Y., N.I. Naqvi, Y. Lino, M. Yamamoto, and M.K. Balasubramanian. 2000. Fission yeast rng3p: an UCS-domain protein that mediates myosin II assembly during cytokinesis. J. Cell Sci. 113:2421–2432. [DOI] [PubMed] [Google Scholar]
- Wu, J.-Q., J.R. Kuhn, D.R. Kovar, and T.D. Pollard. 2003. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 5:1–20. [DOI] [PubMed] [Google Scholar]
- Yu, Q., and S.I. Bernstein. 2003. UCS proteins: managing the myosin motor. Curr. Biol. 13:R525–R527. [DOI] [PubMed] [Google Scholar]