Abstract
A rearranged T cell receptor (TCR) Vα and Jα gene from a cytochrome c-specific T cell hybridoma was introduced into the genomic Jα region. The introduced TCR α chain gene is expressed in a majority of CD3 positive and CD4 CD8 double-negative immature thymocytes. However, only a few percent of the double-positive and single-positive thymocytes express this TCR α chain. This decrease is caused by a rearrangement of TCR α chain locus, which deletes the introduced TCR gene. Analysis of the mice carrying the introduced TCR α chain and the transgenic TCR β chain from the original cytochrome c-specific T cell hybridoma revealed that positive selection efficiently rescues double-positive thymocytes from the loss of the introduced TCR α chain gene. In the mice with negatively selecting conditions, T cells expressing the introduced TCR αβ chains were deleted at the double-positive stage. However, a large number of thymocytes escape negative selection by using an endogenous TCR α chain created by secondary rearrangement maintaining normal thymocyte development. These results suggest that secondary rearrangements of the TCR α chain gene play an important role in the formation of the T cell repertoire.
A productive VDJ rearrangement of T cell antigen receptor (TCR) β chain inhibits a secondary TCR β gene rearrangement (1), a process called “allelic exclusion.” This process ensures that each T cell expresses only one TCR β chain. In contrast, VJ rearrangement of the TCR α chain is not subject to allelic exclusion, and the expression of a functional TCR α chain does not inhibit another TCR α gene rearrangement (2–4). A pre-existing, functionally rearranged Vα-Jα segment can be deleted by a secondary rearrangement on the same chromosome (5). These results clearly demonstrate that TCR α and β chain genes are regulated differently for the initiation of rearrangement and for allelic exclusion. The mechanisms controlling these differences are not known, and the biological significance of this differential regulation is not understood.
To study the regulation of TCR α chain rearrangement and expression during T cell maturation in the thymus, we established a genetically modified mouse line. In this knock-in (KI) mouse, the rearranged TCR VαJα gene from a cytochrome c (Cyt c)-specific T cell hybridoma, 2B4 (6), is inserted in the 5′ region of the germ-line Jα locus by using homologous recombination in an embryonic stem (ES) cell. Analysis of the TCR α chain KI mice revealed a rapid deletion of the introduced TCR α gene in the CD4 CD8 double-positive (DP) stage by rearrangement of endogenous TCR α chains. This finding indicates that the TCR α chain locus continuously rearranges and deletes pre-existing functionally rearranged VJ segments in vivo. By crossing the KI mouse to the 2B4 TCR β chain transgenic mouse, we demonstrate that the positive selection of T cells efficiently terminated secondary rearrangements and prevented deletion of the introduced TCR α chain gene. Furthermore, we demonstrate that T cells expressing the KI TCR/2B4 TCR β receptor are negatively selected by the expression of transgenic Cyt c. However, overall thymocyte development was not significantly altered in mice carrying the KI TCR/2B4 TCR β and Cyt c transgene. These data suggest that continuous rearrangement of the TCR α chain locus plays an important role in the formation of the TCR repertoire.
MATERIALS AND METHODS
Construction of TCR α Chain Gene Segment for the Generation of KI Mice.
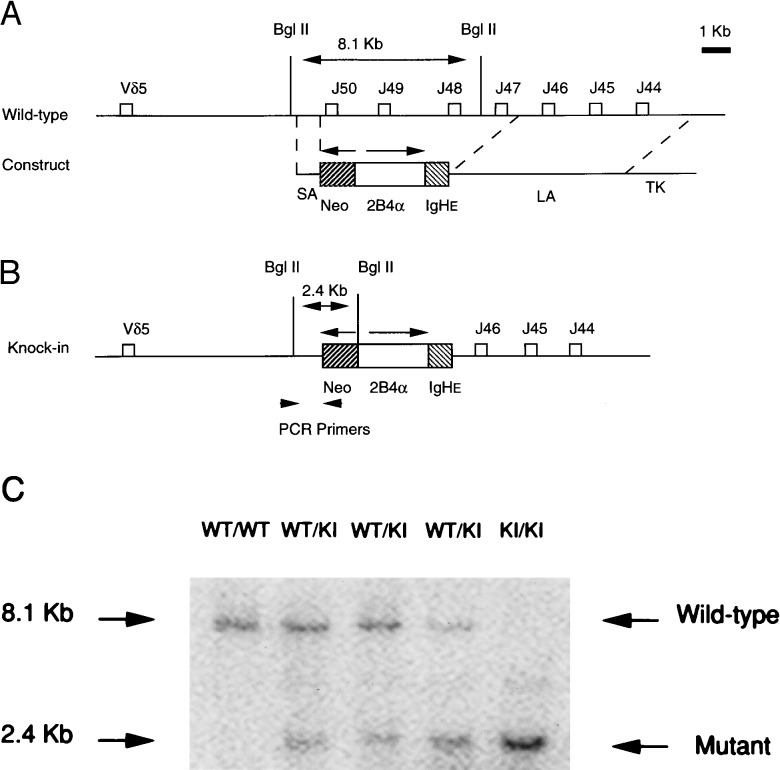
A positive-negative selection strategy was used to design the 2B4-TCR α KI vector for homologous recombination in ES cells derived from mouse 129. This vector used a cassette containing a PGK-NEO and 2B4 α-IgHɛ, a ClaI–SpeI fragment from 2B4 α transgenic construct (7) to replace the J47-J50 region of TCR α gene. At the end of the long arm, a cassette containing PGK-TK was inserted. The targeting vector was electroporated into 129 derived E14 ES cells, and the ES cells were selected in G418 and gancyclovir as described previously (8). Two hundred double-resistant clones were picked and screened by PCR [5′ primer: genomic sequence outside the short arm; 3′ primer: the PGK-poly(A) region of PGK-NEO] and Southern blotting (BglII digestion of DNA, and probed by a 110-bp fragment upstream of the short arm) (Fig. 1 A–C). Three ES cell clones with homologous recombination were injected into blastocysts of B6 mice to generate chimeras. The resulting male chimeras were bred to B6 female mice. The germ-line transmission of injected ES cells was scored by PCR and Southern analysis of the tail DNA of offspring.
Figure 1.
Generation of 2B4-TCR α KI mice. (A) The genomic structure of the Jα locus and the targeting vector. (B) The predicted structure of mutated alleles after homologous recombination. The expected sizes of the BglII fragments from hybridization are indicated. PCR primers to screen KI gene also are indicated. Arrows indicate transcriptional orientation of neo and TCR genes. (C) Southern blot analysis of tail DNA from 2B4-TCR α KI mice.
Generation of Experimental Mouse Lines.
TCR α−/− mice (H-2b) (9) were kindly provided by P. Mombearts and S. Tonegawa (Massachusetts Institute of Technology, Cambridge). 2B4 TCR α chain transgenic mice (H-2b) (7) and 2B4 TCR β chain transgenic mice (H-2k) (10) were kindly provided by L. Berg (Harvard University, Boston). Cyt C transgenic mice (H-2b) (11) were kindly provided by S. M. Hedreck (University of California, San Diego). These mice were used to generate KI TCR/TCR α− (H-2b), KI TCR/2B4 TCR β mice (kxb and bxb), and TCR αβ transgenic (kxb and bxb) mice. For negative selection experiments, KI TCR/2B4 TCR β mice (kxb) and αβ transgenic mice (kxb) were mated to Cyt c transgenic mice (bxb). Mice were screened for the appropriated genotype by using PCR analysis for KI gene and Cyt c transgene, FACS analysis of peripheral blood lymphocyte for Vβ3 expression and H-2 haplotype, and Southern blot analysis for TCR transgenes.
Immunofluorescence Analysis of Thymocytes.
Surface immunofluorescence analysis of thymocytes and T cells was performed as described previously (12). In brief, thymocytes were incubated with phycoerythrin (PE)-anti-CD4, fluorescein isothiocyanate (FITC)-anti-CD8, and biotin-labeled anti-TCR or CD3 antibodies at 4° for 30 min, washed and further incubated with RED613-streptavidin (GIBCO/BRL). Stained cells were analyzed by FACScan analyzer by using the cellquest program (Beckton-Dickinson). PE-labeled anti-CD4 and FITC-labeled anti-CD8 antibodies were purchased from PharMingen. The mAb specific for an idiotypic determinant of 2B4 TCR α chain (A2B4) (13) was a gift from L. Samelson (National Institutes of Health, Bethesda, MD) and biotinylated for three-color analysis. CD3 ɛ-specific antibody 500A2 was a gift from J. Allison (University of California, Berkeley) and was biotinylated for three-color analysis.
Generation of T Cell Hybridomas from 2B4-TCR α KI Mice.
Spleen cells (1 × 105 cells/ml) from the KI mice were stimulated in vitro with Con A (5 μg/ml). Three days after incubation, T cells were fused with BW5147 thymoma. Resulting hybridomas were cloned and tested for the expression of KI TCR by using A2B4 mAb staining (6% and 7% of hybridoma clones were positive in two separate experiments). Clones were tested for the presence of KI TCR by using PCR methods as described above.
RESULTS
Generation of 2B4-TCR α Chain KI Mice.
The TCR α chain construct contains a rearranged 2B4 TCR VαJα and Ig heavy chain enhancer (IgHɛ) from 2B4 TCR transgene construct (7). It has been shown that the inclusion of the IgHɛ is necessary for the efficient expression of the transgene TCR (7). We included this IgHɛ in our construct to ensure the expression of introduced 2B4 TCR α gene. This TCR segment was used to construct targeted KI genes to replace the genomic Jα50-Jα47 segment (Fig. 1 A and B and Materials and Methods). ES cells containing targeted replacement was used to establish the mouse line as described (8). Southern blot analysis of germ-line transmitted mice tail DNA showed that the 2B4 TCR α segment and IgHɛ were properly introduced into the 5′ region of Jα locus (Fig. 1C). In this study, we used KI heterozygous mice (KI TCR/α+) for the experiments unless otherwise stated.
The Expression of the 2B4 TCR α Gene in the KI Mice.
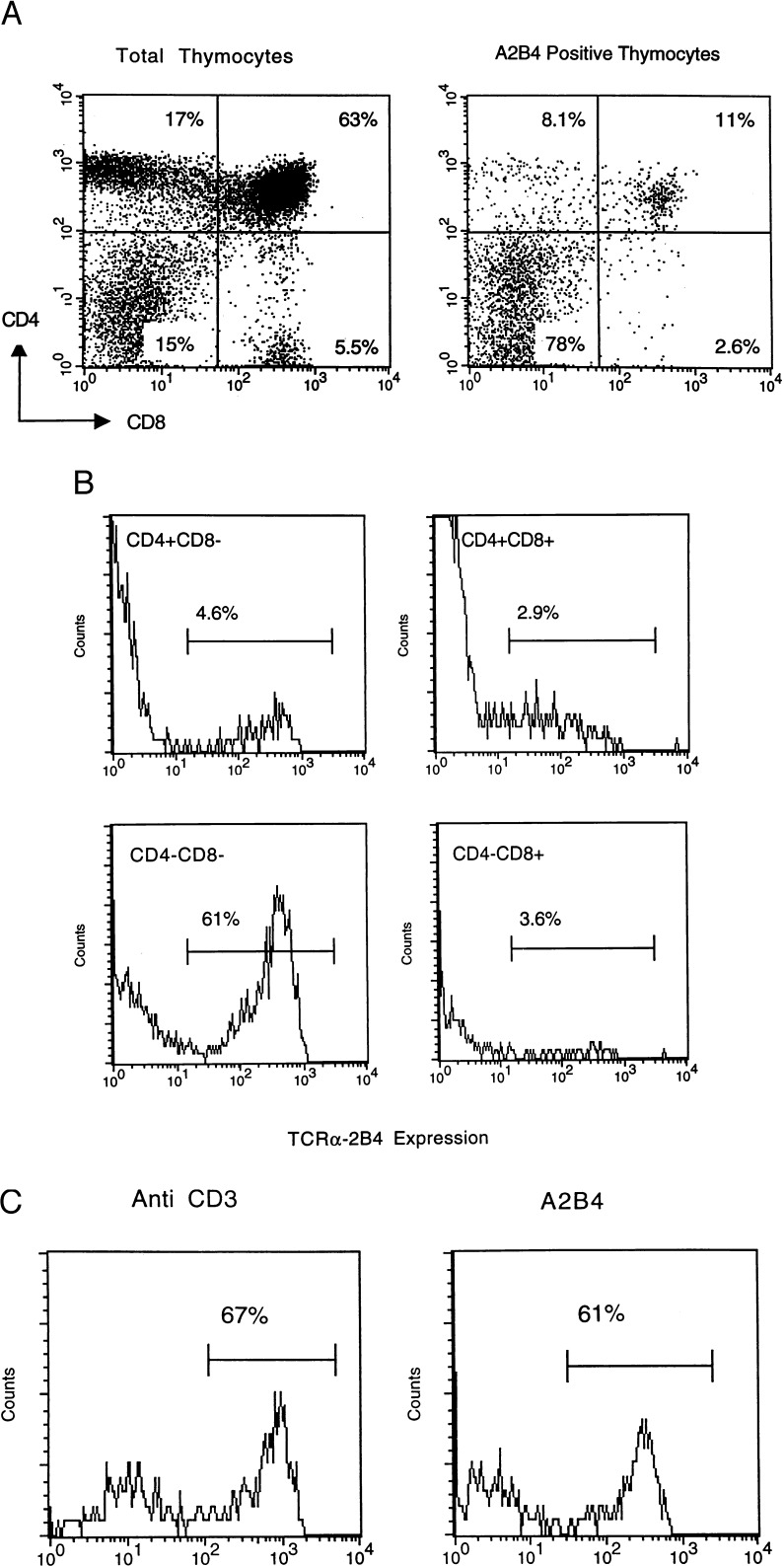
Expression of the introduced 2B4 α chain (KI TCR) on thymocytes was determined by using three-color surface fluorescence with anti-CD4, anti-CD8, and anti-2B4 α chain idiotypic, A2B4(13), antibodies. The KI TCR was expressed in a majority of CD3+ double-negative (DN) cells, but only 3–5% of the DP and single-positive (SP) T cells were stained with A2B4 antibody (Fig. 2 A–C).
Figure 2.
The expression of introduced 2B4TCR α gene in thymus. Thymocytes from KI mice were stained with phycoerythrin-anti-CD4 antibody, and fluorescein isothiocyanate-anti-CD8 antibody, and biotin-A2B4 antibody or biotin-anti-CD3 antibody as described in Materials and Methods. (A) CD4 and CD8 expression pattern of total thymocytes and A2B4 positive thymocytes. (B) Four different subpopulations (CD4+CD8−, CD4+CD8+, CD4−CD8−, and CD4−CD8+) of thymocytes of KI mouse were gated and analyzed for the expression of KI TCR gene. (C) The comparison of CD3 and KI TCR α chain expression in CD4−CD8− DN thymocyte population.
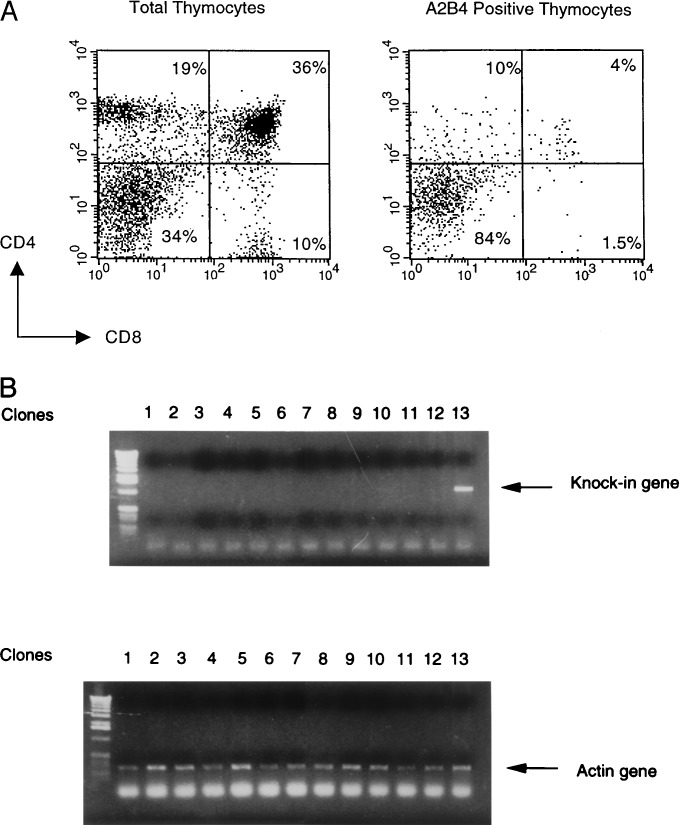
To assess whether the inefficient expression of KI TCR is caused by preferential expression of the TCR α from wild-type allele, the mice were mated to the TCR α−/− mice (9) to generate KI TCR/α− mice. These mice showed low KI TCR expression in the DP and SP thymocytes comparable to the KI TCR/α+ mice (Fig. 3A). Because thymocytes in KI TCR/α− mice can only use the KI TCR allele for generation of functional TCR α chain and KI TCR is located at the 5′ most of Jα region, surface KI TCR negative thymocyte in this mouse must have generated a functional TCR by deleting the KI gene with secondary rearrangement.
Figure 3.
Deletion of the KI TCR in the thymus. (A) Thymocytes from KI/TCR α− mice were stained as described in Fig. 2. (B) PCR analysis was performed on DNA extracted from 13 hybridoma clones from 2B4 KI mice to test the presence of the KI TCR gene. One hybridoma was stained positive with A2B4 antibody, and others were negative. Primers used were the same as the ones for the screening of homologous recombination. PCR analysis was performed on the same DNA by using actin primers.
Deletion of the KI TCR gene also was demonstrated by an analysis of T cell hybridomas derived from KI TCR mice. The splenic T cells from KI mice were activated by Con A in vitro and fused with BW5147 thymoma cells to generate T cell hybridomas. Surface immunofluorescence analysis of the hybridomas showed that 5–10% of the TCR αβ positive hybridomas expressed the KI TCR chain (data not shown). Genomic DNA from 13 individual hybridoma clones (one positive and 12 negative for the surface expression of KI TCR) were analyzed for the presence of the KI TCR gene by PCR. As shown in Fig. 3B, the KI TCR gene was absent from all 12 surface expression negative clones, whereas it was present in the hybridoma clone stained with A2B4 antibody. Further analysis of an additional 12 hybridomas and four T cell clones also demonstrated correlation between the lack of the KI TCR gene and the lack of surface expression.
These results suggest that the inefficient expression of the KI TCR gene is a result of the deletion of the gene caused by the rearrangement of the endogenous TCR Vα genes to downstream Jα segments. The competition with endogenous TCR α chains from non-KI allele for TCR β chain association seems to play a minor, if any, role in the decreased expression of KI TCR.
The Transgenic 2B4 TCR β Gene Rescues the KI TCR Gene from Deletion.
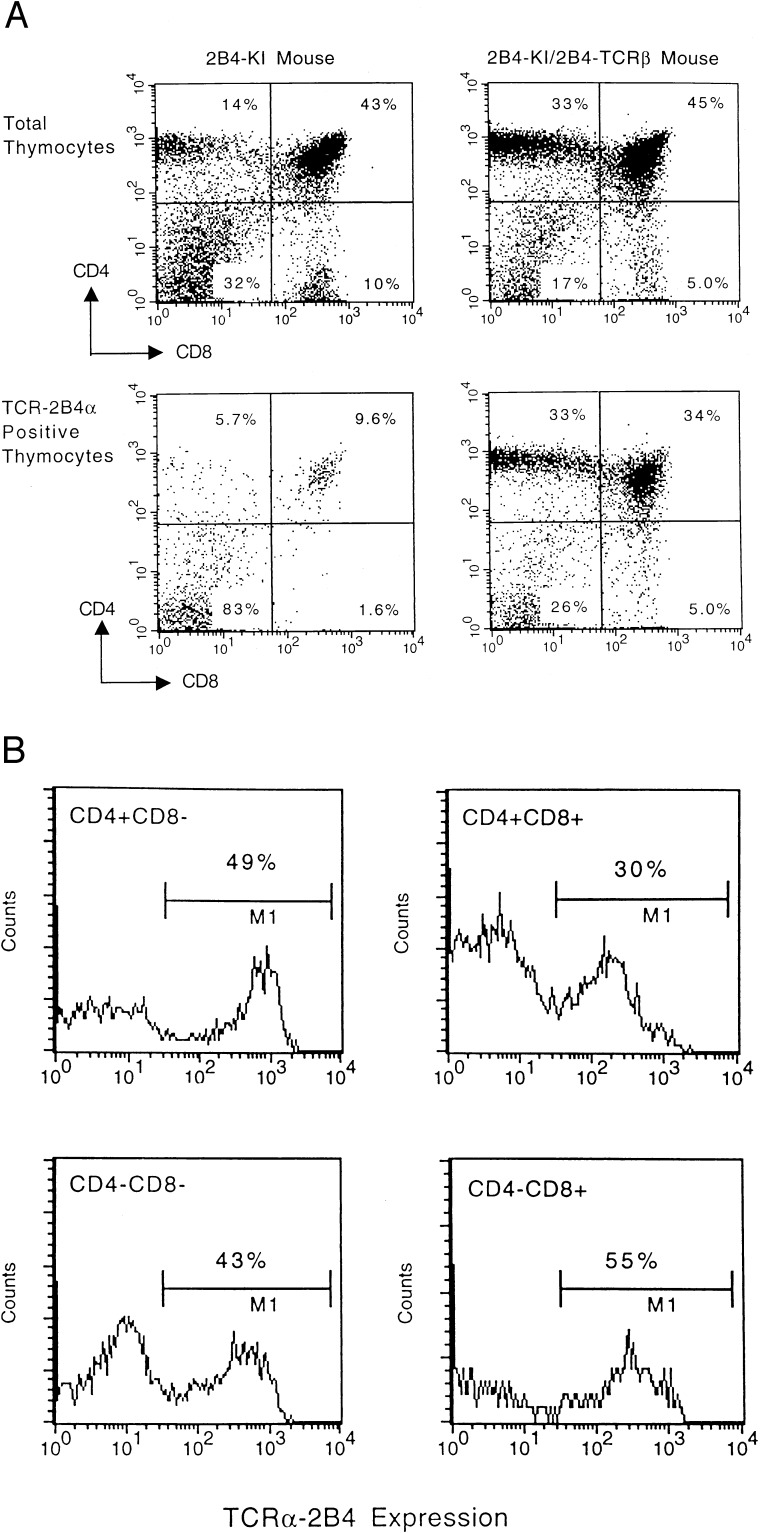
To examine the factors that affect the rearrangement of TCR α gene, we mated 2B4 TCR β chain transgenic mice to KI mice to generate KI TCR/2B4 TCR β mice (H-2 kxb). The analysis of these mice showed that a large number of DP and SP thymocytes are positive for the expression of the KI TCR together with the transgenic β chain (Fig. 4). The percentage of KI TCR positive cells in the DP cells rose from 3% in KI mice to 30% in KI/2B4 TCR β mice, and from 5% to 50% in SP T cells (compare Figs. 2B and 4B). This result showed that in this mouse deletion of the KI TCR by secondary rearrangement is greatly reduced and T cells expressing 2B4 TCR αβ chains (KI TCR α and transgene β chain) are effectively selected to mature into SP T cells. However this rescue is imperfect, because about half of the DP and SP T cells lack surface expression of the KI TCR α chain.
Figure 4.
Expression of introduced TCR α gene in the thymus of KI TCR/2B4 TCR β mice. Thymocytes from KI and KI/2B4 TCR β mice (H-2 kxb) were stained as described in Fig. 2. (A) 2B4+ thymocytes from KI TCR and KI TCR/2B4 TCR β mice were gated to illustrate their CD4 and CD8 expression patterns. (B) Four different subpopulations (CD4+CD8−, CD4+CD8+, CD4−CD8−, and CD4−CD8+) cells from KI TCR/2B4 TCR β thymus were gated and analyzed for the expression of the KI TCR gene in these populations.
The Requirement of Positive Selection of T Cells for the Rescue of KI TCR Gene Expression.
There are two apparent possibilities for the rescue of KI TCR gene expression in the KI TCR/2B4 TCR β transgenic mice (H-2 kxb). The “good” pairing of the KI TCR α and transgenic TCR β chain would allow T cells to maintain the expression of this TCR at the DP T cell stage and these T cells can be positively selected by I-Ek class II major histocompatibility complex (MHC) molecules (rescue by better pairing). The second possibility is that the interaction between 2B4 αβ TCR and the positively selecting MHC effectively down-regulates the rearrangement of TCR α loci and rescues the KI TCR from deletion (rescue by positive selection).
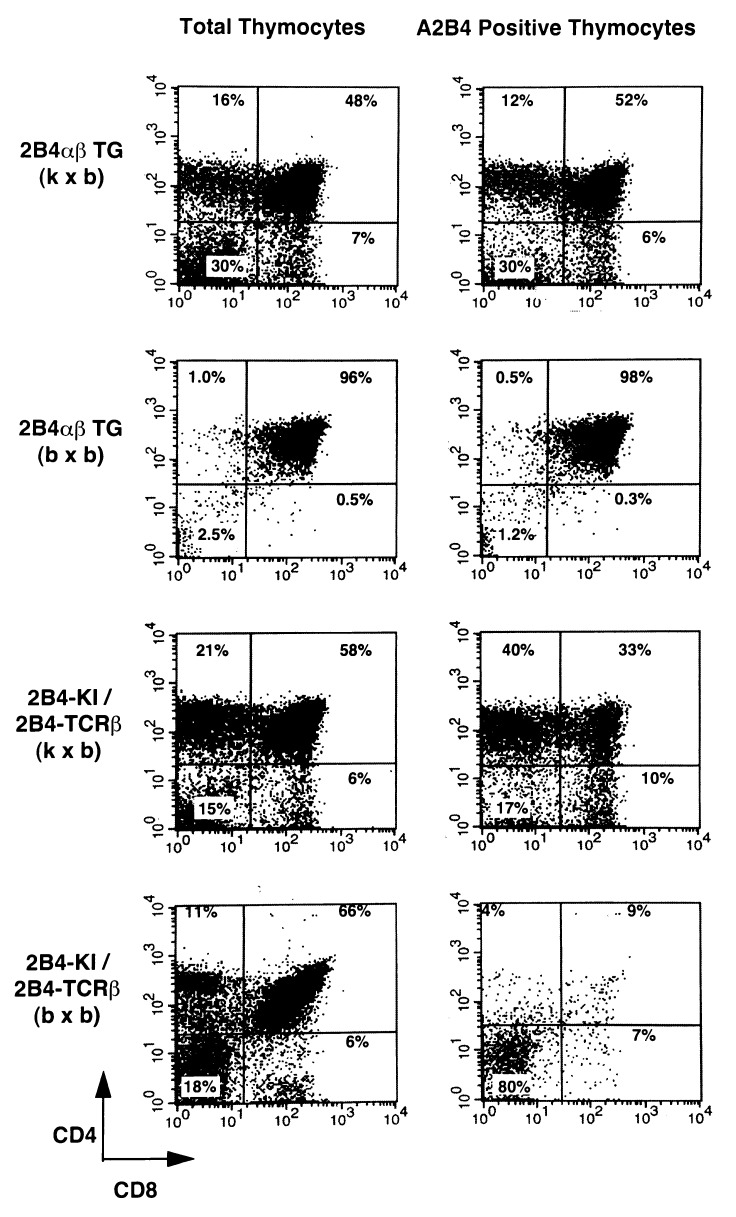
To distinguish these two possibilities, we compared the development of T cells expressing KI TCR in KI TCR/2B4 TCR β mice with kxb (positively selecting) and bxb (neutral for selection) MHC. We also examined mice with 2B4 αβ TCR transgenic mice with the same MHC backgrounds. In the mice with positively selecting MHC background (kxb), T cells expressing 2B4 TCR mature efficiently into the SP stage in both αβ transgenic and KI TCR/2B4 TCR β mice, except for a slightly lower number of 2B4 α chain positive DP and SP thymocytes in the KI TCR mice (Fig. 5).
Figure 5.
Expression of 2B4 TCR α gene in the thymus of 2B4 αβ TCR transgenic and KI TCR/2B4 TCR β mice with a nonpositive selecting (neutral) background. Thymocytes from 2B4 αβ TCR transgenic mice and KI TCR/2B4 TCR β with kxb and bxb MHC backgrounds were stained as described in Fig. 2. Thymocyte subsets distribution in total thymocyte and A2B4-positive thymocytes are shown.
In KI TCR/2B4 TCR β mice with a nonselecting MHC background, the number of KI TCR-expressing cells in the DP snd SP stage decreases to the level similar to that in KI TCR mice without 2B4 TCR β chain (compare Figs. 2 and 5). However, the overall development of thymocytes is not affected with normal number of DP and SP T cells formed. These results indicate that mere “good” pairing between the KI TCR and 2B4 TCR β chains could not inhibit the secondary rearrangement of TCR α gene in nonselecting H-2 bxb mice. Thus the rescue of the KI TCR gene in the KI TCR/2B4 TCR β mice depends on positive selection of T cells by I-Ek and self-peptide(s) complex.
In 2B4 αβ transgenic mice with nonselecting MHC background, a majority of the thymocytes accumulated in the DP stage. These thymocytes in the DP stage express transgene-derived TCR αβ chains and failed to give rise to SP thymocytes (Fig. 5). Thus, the development of thymocytes in a nonselecting condition differs significantly between KI TCR/2B4 TCR β and TCR αβ transgenic mice. This difference is likely caused by the different regulation of the KI and transgenic TCR α chain gene.
Impact of Negative Selection on Thymocytes Expressing Self-Reactive TCR.
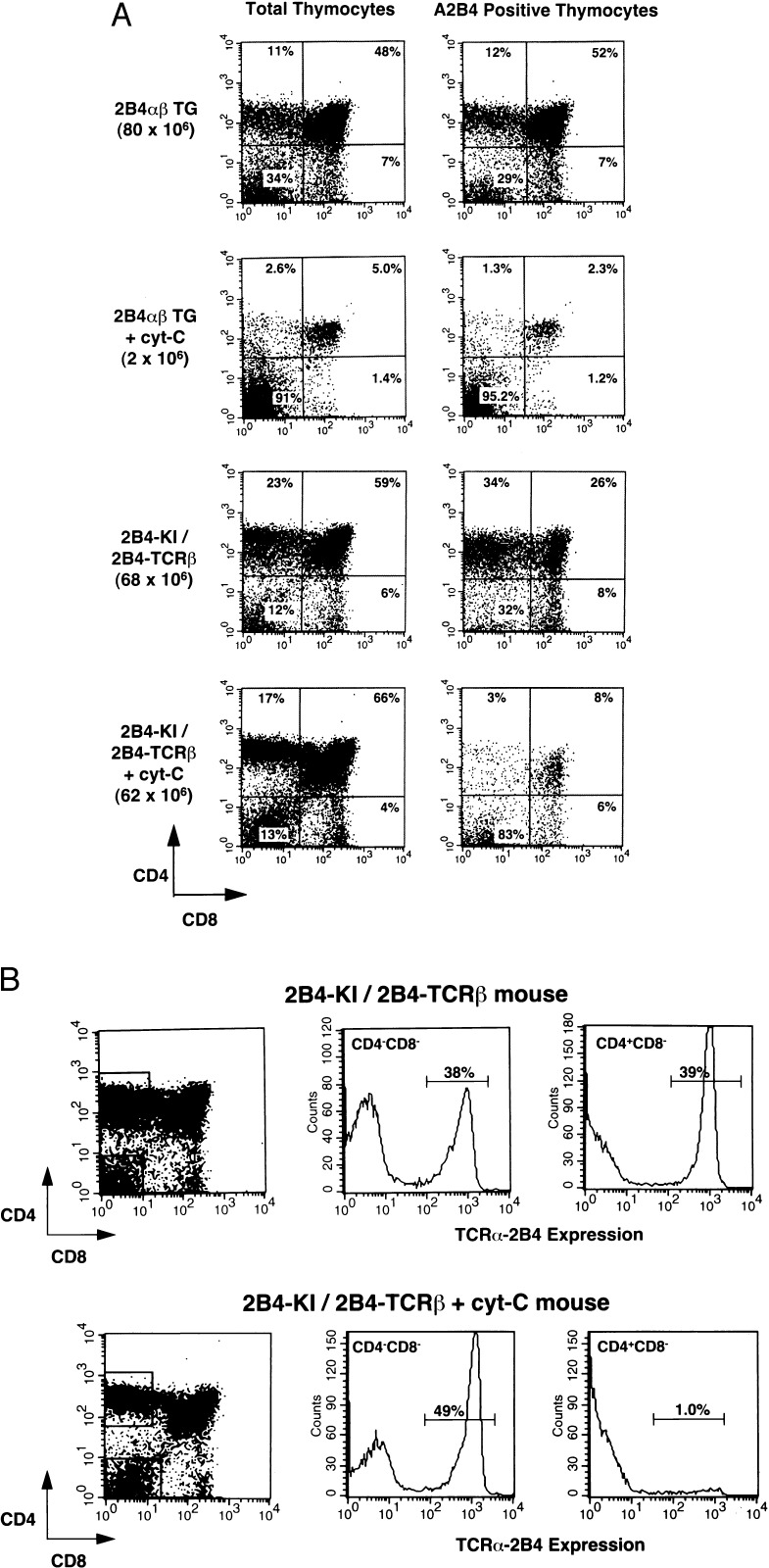
To study the impact of negative selection on self-reactive T cells, KI TCR/2B4 TCR β mice as well as 2B4 αβ transgenic mice were mated to the mice expressing the Cyt c transgene, the ligand to the 2B4 TCR (11). Thymocytes from mice with the appropriate genotype were analyzed by surface immunofluorescence as described. In the mice carrying 2B4 αβ TCR transgenes and Cyt c transgene, the number of thymocytes is greatly reduced (2–4 × 106 thymocytes in tolerant mice compared with 70–90 × 106 thymocytes in normal TCR transgenic mice). This reduction is caused by the decreased number of DP and SP thymocytes (Fig. 6A). In contrast, the presence of the Cyt c transgene has no effect on the total number of thymocytes in the KI TCR/2B4 TCR β mice (60–80 × 106 in both tolerant and nontolerant mice) (Fig. 6A). Furthermore, these mice have normal distribution of cells in all four thymocyte subsets.
Figure 6.
Effect of negative selection on thymocytes of 2B4 αβ TCR transgenic and KI TCR/2B4 TCR β mice. (A) Thymocytes from 2B4 αβ TCR transgenic mice and KI TCR/2B4 TCR β with or without transgenic Cyt c gene were stained as described in Fig. 2. Thymocyte subsets distribution in total thymocyte and A2B4-positive thymocytes are shown. All mice have MHC background kxb, and the number of total thymocytes is shown in parenthesis. (B) Thymocytes from KI TCR/2B4 TCR β mouse with or without Cyt c transgene were stained with anti-CD4, anti-CD8, and A2B4. Thymocytes subsets distribution is shown at left. CD4 CD8 DN and CD4 SP thymocytes were gated and analyzed for the expression of 2B4 TCR α chain.
Analysis of the expression of TCR α 2B4 in these mice revealed that the Cyt c transgene expression drastically reduces the number of Cyt c-reactive 2B4 TCR α positive T cells in DP and SP stage, whereas there was no change in the expression of the TCR α 2B4 in DN thymocytes (Fig. 6 A and B). Functional analysis revealed mice carrying Cyt c have no in vitro response to Cyt c, whereas spleen cells from KI TCR/2B4 TCR β mice mounted vigorous proliferative response (O.K., unpublished observation). These results clearly demonstrate that the presence of the Cyt c transgene deletes T cells expressing 2B4 αβ TCR in the thymus. However, the deletion of self-reactive T cells has no significant effect on the overall thymocyte maturation in KI TCR/2B4 TCR β mice. In this mouse T cells expressing endogenous TCR α chain created by secondary rearrangement undergo normal maturation steps. In contrast, in TCR αβ transgenic mice, negative selection of T cells affects overall thymocyte development by deleting a majority of DP and SP thymocyte. These results suggest that negative selection by self-antigen may not affect overall T cell development under physiological conditions, because potentially self-reactive T cells can change its specificity by replacing TCR α chain by secondary rearrangement.
DISCUSSION
We have generated a TCR α chain KI mouse line that has a rearranged TCR VαJα gene from a Cyt c-specific I-Ek restricted hybridoma, 2B4 (6), inserted in the Jα locus. The study of thymocyte development in the TCR KI mice revealed that the TCR α gene was expressed on virtually all TCR positive DN immature thymocytes. At this stage of development in a normal thymus, cells express the TCR β chain associating with the pTα chain but not with the TCR α chain (14). There are two apparent possibilities for the early expression of the KI TCR gene: (i) rearrangement itself is sufficient for the TCR α chain gene activation in the early stage of the development, or (ii) presence of IgHɛ and neo gene cassette in the KI construct promotes early expression. Nevertheless, having both a rearranged V-J segment and heterologous regulatory elements in the construct, we expected to have expression of introduced TCR in the DN stage and to use this TCR to monitor the deletion of the functionally rearranged and expressed TCR α chain by secondary rearrangement of TCR α chain genes.
The expression of the KI TCR gene decreased dramatically in the DP stage or in the transition stage from DN to DP cells. With the analysis of KI TCR/α− mice and hybridomas established from KI TCR mice, we demonstrate that this decreased expression is a result of rapid deletion of the introduced TCR gene by secondary rearrangement of the TCR α chain gene. These results, in agreement with a previous study with a T lymphoma line (5), demonstrate that an existing functionally rearranged TCR Vα-Jα segment can be deleted by a secondary rearrangement of upstream Vα chain to 3′ Jα segments in vivo. Because of the presence of an additional regulatory element (IgHɛ and neo gene cassette) in the KI gene, it is possible that the Jα locus may have an increased accessibility for gene rearrangement. However, we found this scenario unlikely because the KI TCR gene was expressed in the majority of the DN TCR positive cells (Fig. 2C). At this stage, there is extensive TCR β gene rearrangement (15, 16), but this recombination activity failed to induce TCR α chain rearrangement in the KI TCR allele to delete the KI TCR gene. Thus it seems that the introduced gene is only susceptible for deletion at the appropriate stage of the TCR α chain gene rearrangement. These results suggest that secondary rearrangement at the KI TCR loci is regulated similarly to the wild-type TCR α chain.
In the previous study of a transgenic mice carrying the TCR gene (the same VαJα from 2B4 hybridoma connected to the genomic Cα gene) (7), the transgene-derived TCR α chain was expressed on virtually all of the mature T cells. In contrast, in the TCR KI mouse, only a fraction of T cells expressing the 2B4 TCR α chain can be positively selected to mature. The drastic difference between the TCR KI and the TCR transgenic mouse is likely because of the persistent expression of the transgene-derived TCR α chain, because the transgenic TCR α chain is not subject to deletion by secondary rearrangement. This finding raises an intriguing possibility that the persistent presence of transgene TCR α chain may alter the normal positive selection process. Further comparison of the TCR transgenic and KI mice would be necessary to dissociate the role of a pre-existing functional TCR chain (both in KI and transgenic mice) and the impact of unregulated transgenic TCR α chain expression (only in transgenic mice) on T cell selection and the function of peripheral T cells.
In the TCR KI mouse, coexpression of the 2B4 TCR β chain in a positively selecting MHC background (H-2 kxb) can efficiently rescue the KI gene from deletion by secondary rearrangement. It has been shown that the positive selection signal down-regulates RAG-1 and RAG-2 gene expression in DP thymocytes (17, 18). The presence of the large number of KI TCR expressing T cells at the DP and SP stage is very likely because of this down-regulation, which effectively terminates secondary rearrangement. This result, in agreement with previous studies of TCR transgenic mouse models (19–21), demonstrates the role of positive selection for the maturation of antigen-specific T cells. These results further indicate that the KI TCR gene loci is under physiological regulation for the termination of the rearrangement machinery. However, the rescue by positive selection is not complete, because only half of the DP and SP T cells express the KI TCR/2B4 TCR β combination. This incomplete rescue may be because of the inefficient positive selection of the 2B4 TCR by bxk F1 MHC compared with the H-2k homozygous mouse (21). Alternatively, rapid rearrangement may delete the TCR α chain in T cells expressing positively selectable TCR αβ chains.
In the αβ TCR transgenic mouse with a neutral MHC background, transgene-derived αβ TCR positive T cells accumulated in the DP stage, and very few mature SP T cells were generated in the thymus (Fig. 5) (19–21). It was postulated that T cells expressing transgenic TCR αβ chains are arrested in DP stage because of a lack of positive selection. However, this accumulation of T cells expressing 2B4 αβ TCR was not observed in KI TCR/2B4 TCR β mice with a neutral MHC. This result clearly shows that the simple expression of a selectable TCR (2B4 αβ TCR) does not inhibit the ongoing secondary rearrangement in the absence of positive selection, allowing for the nonselectable TCR αβ combination (KI TCR α/transgene β chain) to be replaced by endogenous TCR α chains. This mechanism allows for the majority of the thymocytes to undergo normal positive selection by H-2b MHC to mature into SP T cells. Thus, thymocytes expressing a nonselectable TCR αβ chain can create new specificity by secondary rearrangement of the TCR α chain loci, and this mechanism plays an important role for the maturation and selection of thymocytes. In the transgenic mice, the lack of replacement of the transgene-derived TCR α chain seems to interfere with this process and thymocytes accumulate at the DP stage.
It has been demonstrated that T cells expressing self-reactive TCR can be deleted by negative selection in the thymus (19, 20). By using TCR transgenic mice, it has been shown that the interaction between thymocytes and antigen presented by self-MHC resulted in an apoptotic death of the DP thymocyte subpopulation both in vivo and in vitro (22, 23). However, analysis of the thymus using a sensitive staining method showed little evidence for an apoptotic death of immature thymocytes by negative selection (24). In KI TCR/2B4 TCR β mice with Cyt c transgene, almost all CD3 positive DN thymocytes express self-reactive TCR, but very few DP and no mature CD4 SP T cells express this self-reactive TCR. Thus mice are tolerant to the self-antigen, Cyt c, and self-reactive T cells are deleted in the DP stage. The deletion of self-reactive T cells also occurs in 2B4 αβ TCR transgenic mice. However, the overall T cell development differs drastically between KI TCR/2B4 TCR β mice and 2B4 αβ TCR transgenic mice. In TCR transgenic mice, the presence of the Cyt c transgene decreased the total number of thymocytes 30- to 40-fold, and very few thymocytes undergo natural development by using endogenous TCR α/transgenic β chain. In contrast, in KI TCR/2B4 TCR β mice, the presence of Cyt c transgene does not affect overall thymocyte development. Thus, our results with KI TCR support the findings of Surh and Sprent (24) and demonstrate that T cells, which initially express self-reactive T cells, can use new TCR αβ combination created by secondary rearrangement to mature normally from DP to SP in the thymus.
Our results indicate that the secondary rearrangement of the TCR α chain gene plays an important role in the formation of the mature T cell repertoire. We believe that selection process of T cells in the thymus is dynamic, involving continuous secondary rearrangement in TCR α chain locus and an interaction with self-MHC/peptide complex with different affinities for positive and negative selection. The KI TCR model, in which the TCR is under physiological regulation, is a useful model system to further investigate these interactions in detail.
Acknowledgments
We thank Drs. M. M. Davis and L. J. Berg for the 2B4 TCR α chain gene construct and 2B4 TCR transgenic mice, Dr. S. M. Hedrick for the cytochrome c transgenic mice, M. White for blastcysts injection, and J. Katz, R. Lorenz, R. Hayashi, and J. Russell for critically reading the manuscript. This work was supported by grants from the National Institutes of Health and the Human Frontier Science Program.
ABBREVIATIONS
- TCR
T cell antigen receptor
- KI
knock-in
- Cyt c
cytochrome c
- ES
embryonic stem
- IgHɛ
Ig heavy chain enhancer
- DN
double negative
- DP
double positive
- SP
single positive
- MHC
major histocompatibility complex
References
- 1. Uematsu Y, Ryser S, Dembic Z, Borgulya P, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Cell. 1988;52:831–841. doi: 10.1016/0092-8674(88)90425-4. [DOI] [PubMed] [Google Scholar]
- 2.Petrie H T, Livak F, Schatz D G, Strasser A, Crispe I N, Shortman K. J Exp Med. 1993;178:615–622. doi: 10.1084/jem.178.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malissen M, Trucy J, Letourneur F, Rebai N, Dunn D E, Fitch F W, Hood L, Malissen B. Cell. 1988;55:49–59. doi: 10.1016/0092-8674(88)90008-6. [DOI] [PubMed] [Google Scholar]
- 4.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 5.Marolleau J P, Fondell J D, Malissen M, Trucy J, Barbier E, Marcu K B, Cazenave P A, Primi D. Cell. 1988;55:291–300. doi: 10.1016/0092-8674(88)90052-9. [DOI] [PubMed] [Google Scholar]
- 6.Hedrick S M, Matis L A, Hecht T T, Samelson L E, Longo D L, Heber K E, Schwartz R H. Cell. 1982;30:141–152. doi: 10.1016/0092-8674(82)90020-4. [DOI] [PubMed] [Google Scholar]
- 7.Berg L J, Fazekas de St. Groth B, Ivars F, Goodnow C C, Gilfillan S, Garchon H J, Davis M M. Mol Cell Biol. 1988;8:5459–5469. doi: 10.1128/mcb.8.12.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motoyama N, Wang F, Roth K A, Sawa H, Nakayama K, Nakayama K, Negishi I, Senju S, Zhang Q, Fujii S, et al. Science. 1995;267:1506–1510. doi: 10.1126/science.7878471. [DOI] [PubMed] [Google Scholar]
- 9.Mombaerts P, Arnoldi J, Russ F, Tonegawa S, Kaufmann S H. Nature (London) 1993;365:53–56. doi: 10.1038/365053a0. [DOI] [PubMed] [Google Scholar]
- 10.Berg L J, Fazekas de St. Groth B, Pullen A M, Davis M M. Nature (London) 1989;340:559–562. doi: 10.1038/340559a0. [DOI] [PubMed] [Google Scholar]
- 11.Oehen S, Feng L, Xia Y, Surh C D, Hedrick S M. J Exp Med. 1996;183:2617–2626. doi: 10.1084/jem.183.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama K, Nakayama K, Negishi I, Kuida K, Louie M C, Kanagawa O, Nakauchi H, Loh D Y. Science. 1994;263:1131–1133. doi: 10.1126/science.8108731. [DOI] [PubMed] [Google Scholar]
- 13.Samelson L E, Schwartz R H. Immunol Rev. 1983;76:59–78. doi: 10.1111/j.1600-065x.1983.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 14.Fehling H J, Krotkova A, Saint R C, von Boehmer H. Nature (London) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 15.Raulet D H, Garman R D, Saito H, Tonegawa S. Nature (London) 1985;314:103–107. doi: 10.1038/314103a0. [DOI] [PubMed] [Google Scholar]
- 16.Snodgrass H R, Kisielow P, Kiefer M, Steinmetz M, von Boehmer H. Nature (London) 1985;313:592–595. doi: 10.1038/313592a0. [DOI] [PubMed] [Google Scholar]
- 17.Turka L A, Schatz D G, Oettinger M A, Chun J J, Gorka C, Lee K, McCormack W T, Thompson C B. Science. 1991;253:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 18.Kouskoff V, Vonesch J L, Benoist C, Mathis D. Eur J Immunol. 1995;25:54–58. doi: 10.1002/eji.1830250111. [DOI] [PubMed] [Google Scholar]
- 19.Loh D Y, Sha W C, Nelson C A, Newberry R D, Kranz D M, Russell J H. Cold Spring Harbor Symp Quant Biol. 1989;1:147–151. doi: 10.1101/sqb.1989.054.01.018. [DOI] [PubMed] [Google Scholar]
- 20.von Boehmer H. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 21.Berg L J, Pullen A M, Fazekas d S, Groth B, Mathis D, Benoist C, Davis M M. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 22.Murphy K M, Heimberger A B, Loh D Y. Science. 1990;250:1720–1723. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama K, Loh D Y. Science. 1992;257:94–96. doi: 10.1126/science.1621101. [DOI] [PubMed] [Google Scholar]
- 24.Surh C D, Sprent J. Nature (London) 1994;372:100–103. [Google Scholar]